Abstract
In the study of gene regulation, it is often necessary to employ functional assays that investigate the action or mechanism of specific promoters or enhancer binding factors and their role in transcription by RNA Polymerase II. There are many assays that measure the transcription of a gene under the control of an endogenous or model activator in vivo. However, to study specific regulatory mechanisms it can be useful to recreate transcription in vitro. In this unit, we will present protocols and techniques that will allow the investigator to perform transcription in vitro using preinitiation complexes (PICs) assembled from cellular extracts on either naked DNA or chromatin templates.
Keywords: In vitro transcription, Chromatin reconstitution, Immobilized template
Unit introduction/overview
The development of a HeLa cell nuclear extract protocol optimized for transcription of an adenovirus Major Late Promoter naked DNA template represented a major milestone in the analysis of regulatory mechanisms (Dignam et al. 1983). Subsequently, investigators have prepared transcriptionally active extracts from yeast (Lue and Kornberg 1987; Verdier et al. 1990), Drosophila embryos (Kamakaka et al. 1991), and other mammalian cell lines (Snoek et al. 1996) to study the biochemical mechanisms of both sequence-specific factors and the general transcription machinery. For a more detailed description of various approaches to preparing extracts, the investigators may wish to consult Chapter 12 of Carey, Peterson, and Smale, 2009. These assays are largely dependent on the formation of functional preinitiation complexes (PICs) from factors within the crude nuclear extract. However, when combined with immunodepletion studies and immobilized template approaches, extracts are extremely useful in dissecting regulatory mechanisms that govern transcription initiation.
The immobilized template technique employs end-biotinylated promoter DNA fragments immobilized on streptavidin-coated Dynabeads™ (Ranish et al. 1999; Johnson et al. 2004; Black et al. 2006). These beads can be isolated from suspension using a magnetic particle concentrator (MPC). These templates can be easily prepared by PCR using 5’ end-labeled biotinylated primers from a commercial source (e.g. Eurofins MWG Operon). The low dissociation constant of the biotin-streptavidin interaction (~10−15 mol/L) facilitates manipulation of the immobilized template DNA. These immobilized templates can be used to purify PICs from extract for transcription assays and to study, in parallel, the composition of these PICs by western blotting or other protein identification technologies (Ranish et al. 2003). This approach also allows the investigator to characterize the effects of specific proteins/reagents on both PIC assembly and transcription.
Strategic Planning
One of the most important considerations before starting a time- and resource-intensive biochemical study such as in vitro transcription is the practicality of the chosen system.
Do the questions being addressed warrant the use of biochemical analysis? While a biochemical study can provide in depth analyses of precise mechanism, in vivo assays often reflect conditions that are more physiologically relevant. Additionally, biochemical assays are complicated and involve special training and a long-term commitment.
Will the study focus on the activity of a specific activator or promoter? If the study focuses on a specific activator, is that protein available in sufficient quantity from recombinant sources to launch the investigation? Moreover, transcription from endogenous promoters and activators in vitro is not as robust as that of model templates and activators like those constituting the GAL4-VP16 model system (Carey et al. 1990). If biochemical studies using the endogenous promoter or activator are required, it would be useful to first compare transcription signals to that of the GAL4-VP16 model system. Should the investigators decide to proceed, it may be necessary or beneficial to make certain modifications to the template such as multimerization of the activator binding site, the addition of an upstream enhancer element from then endogenous activator-responsive gene, or the incorporation of strong inducible core promoter elements such as the adenoviral Major Late Promoter (MLP) or E4 TATA boxes.
Another point to consider is whether to assemble the template into a chromatin array. Although transcription in vitro of naked DNA is much more efficient than chromatin, it does not address the regulatory steps that govern the assembly of a transcription complex on or passage of Pol II through chromatin. If the investigator decides to proceed, be aware that effective transcription on chromatin from nuclear extract requires the addition of acetyl-CoA, a key reagent for histone acetylation. It is crucial to carefully prepare the chromatin to minimize any non-acetyl-CoA-dependent signal (from residual naked DNA templates). It will be up to the investigator to determine which template will be more appropriate for their study because the use of chromatin templates adds another layer of complexity.
In vitro transcription using nuclear extract is best suited for assaying functional transcription initiation. However, transcription initiation can also be recreated in a fully purified system with the basal transcription machinery on naked DNA templates (Sawadogo and Roeder 1985; Malik et al. 1998). This type of study, however, requires extensive protein purification from recombinantly produced factors and from cell lines bearing tagged subunits of major factors like TFIID and Mediator (Zhou et al. 1992; Malik et al. 2000; Sato et al. 2003). The use of these pure systems is advisable when a specific function of one of the aforementioned complexes is being investigated. However, we and others have identified numerous other protein complexes recruited by the model activator GAL4-VP16 to DNA and chromatin templates. Although the assembly of a basally functional PIC on naked DNA has been extensively characterized (Orphanides et al. 1996; Roeder 1996), the composition of the proteins recruited to the PIC is highly more complex than simply the GTFs, Pol II, TFIID and Mediator. The specific roles of many of these other recruited proteins, such as the super elongation complex (Lin et al. 2010; Takahashi et al. 2011), have yet to be fully dissected in vitro in the context of a PIC. Hence, the use of crude systems may be a more logical starting point when the precise targets and mechanism of a new activator are unknown.
Will the study require the characterization of specific protein complexes to investigate their effect on transcription? In this scenario, the goals of the study will be best addressed using immobilized DNA templates. The immobilized template approach permits the use of immunoblotting or mass spectrometry to identify the specific composition of the complexes formed on a DNA template using either extracts or pure complexes like Mediator and TFIID. An additional advantage of immobilizing the DNA templates is the ability to manipulate the protein-bound templates before or after the addition of NTPs to initiate transcription (Johnson et al. 2004; Black et al. 2006). Biotinylated DNA templates immobilized on magnetic Dynabeads™ can be isolated using the MPC, and subjected to buffer changes, wash steps and other manipulations. Additionally, by combining immobilized template with extracts depleted of important proteins and add-back experiments using purified proteins, one can begin to address the specific mechanisms of gene activation including chromatin remodeling and modification and numerous other important aspects of the transcription process.
Contents
In this unit, we will describe two basic protocols: in vitro transcription from nuclear extract to assay functional transcription initiation (Basic Protocol 1), and the immobilized template assay in nuclear extract to assay PIC assembly (Basic Protocol 2). Support Protocol 1 describes primer extension analysis to measure the RNA product from Basic Protocol 1. If in vitro transcription and PIC assembly are to be assayed on chromatin templates, the investigators will also want to consult Support Protocol 2, which describes a straightforward method for the preparation of reconstituted chromatin templates. Support Protocol 3 describes the preparation of nuclear extract as adapted from the Roeder lab protocol (Dignam et al. 1983).
Basic Protocol 1: In vitro transcription from nuclear extract
Recreating transcription from nuclear extracts relies on the recruitment and formation of a functional preinitiation complex on the template. In the following protocol, DNA template will be incubated with a sequence-specific model activator, GAL4-VP16, to recruit the transcription machinery from crude extract. After pre-incubation with Acetyl-CoA (may be skipped for naked DNA templates) to ensure histone acetylation, nucleoside triphosphates (NTPs) are added to initiate transcription by RNA Pol II. The reaction is terminated with buffer containing EDTA and SDS, and mRNA transcripts are isolated. The RNA may then be analyzed directly if 32P-labeled NTPs were used for runoff transcription, or may be subjected to a primer extension reaction with a labeled probe (Support Protocol 1: Primer Extension).
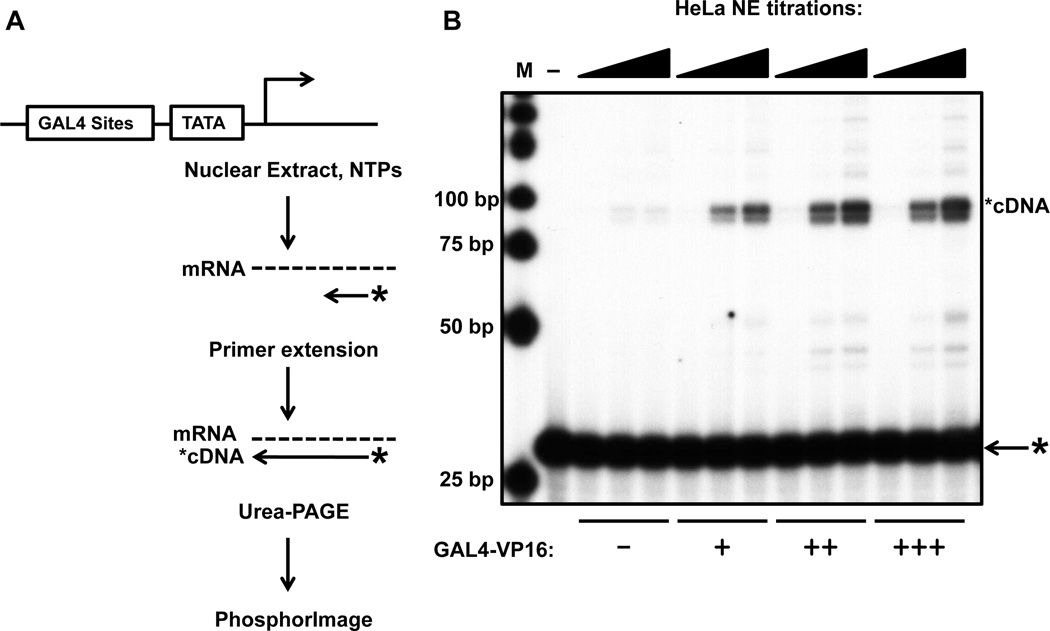
The amount of nuclear extract and templates in this protocol has been optimized for transcription using the GAL4-VP16 model activator and G5E4T model promoter bearing 5 GAL4 sites immediately upstream of the adenovirus E4 gene TATA box, which exhibits strong activator-stimulated transcription (Carey et al. 1990). Concentrations of extract, activator, and template will need to be titrated and optimized for the investigator’s own systems. An example of such a titration is shown in Figure 1.
Figure 1. In vitro transcription from nuclear extract.
A schematic of in vitro transcription assayed by a primer extension assay is shown on the left (A). To the right, is an example of an in vitro transcription experiment determining the optimal levels of both nuclear extract and GAL4-VP16 activator on the model promoter, G5E4T (B).
Reagents
Acetyl-Coenzyme A (1 mM)
Activator protein (200 ng/µL in 0.1 M Buffer D)
Buffer D (0.1 M KCl; See Recipes)
Chloroform
DNA template (in this protocol, the authors use a 602-bp linear chromatin template containing the G5E4T promoter at a starting concentration of 5 ng/µL)
Dry Ice
Ethanol (100%)
Formamide Loading Dye (See Recipes)
MgCl2 (0.1 M)
Nonspecific Competitor DNA (in this protocol the authors use the plasmid vector used in cloning the G5E4T template promoter: pGEM3 at a starting concentration of 100 µg/mL)
NTPs (25 mM each, ATP, UTP, CTP and GTP)
HeLa Nuclear Extract (Support Protocol 3)
Pasteur Pipettes
Novagen™ Pellet Paint Coprecipitant
Phenol
Proteinase K
Transcription Stop Buffer (See Recipes)
Transcription Buffer Preparation/Activator binding
NOTE: If immobilized templates are to be used, beads containing the immobilized promoter should be resuspended in the appropriate amount of 0.1 M Buffer D and incubated with the desired proteins (e.g., activator, co-activator complexes) as described in Basic Protocol 2 (Immobilized DNA template assay) before resuspending in up to 10 µL of 0.1 M Buffer D. Skip to Step 3.
-
1)
In a microcentrifuge tube, mix 20 ng of chromatin or naked DNA template with 1µL of Activator protein and 1 µL of 10 mg/mL BSA. Bring the final volume up to 10 µL with 0.1 M Buffer D.
-
2)
Incubate 5 min on ice.
-
3)During this incubation, in a separate tube, prepare the transcription reaction mixture by adding in this order on ice, mixing by tapping the tube after addition of each reagent:
Buffer D (0.1 M) 5 µL pGEM3 Carrier DNA (100 µg/mL) 200 ng MgCl2 (0.1 M) 3 µL Acetyl-Coenzyme A (1 mM) 2 µL HeLa Nuclear Extract 15 µL ddH2O to 30 µL - When optimizing the amount of HeLa extract in the reaction, the appropriate volume adjustments should be compensated by increasing or decreasing the volume of Buffer D. This total should not exceed 20 µL.
In vitro transcription reaction
-
4)Add the reaction mixture from step 3 to the prepared templates, and incubate at 30 degrees C for 15 minutes (rotating, if necessary, to prevent bead settling when using immobilized templates) to allow chromatin hyperacetylation (when applicable) and the subsequent formation of full PICs.
- If transcription is to be assayed on the fully-formed, pre-captured PICs, the immobilized templates can be quickly washed with the 0.1 M Wash Buffer from Basic Protocol 2 (Immobilized DNA template assay) to remove unbound proteins, and resuspended in Step 3’s transcription reaction mix, with 0.1 M Buffer D in place of the nuclear extract.
-
5)
Add 0.8 µL of 25 mM NTPs (A, U, G and CTP) to the reaction, mix, and incubate at 30 degrees for an additional 60 minutes.
-
6)Warm the Transcription Stop Buffer to 37 degrees C to ensure that the SDS is fully dissolved, and add Proteinase K (10 mg/ mL) at a 1:100 dilution, and carrier yeast tRNA (50 mg/mL) at a 1:200 dilution.
- Commercial nucleic acid co-precipitant may be used in place of yeast tRNAs.
-
7)
Add 100 µL of the Transcription Stop Buffer to terminate the transcription reaction and incubate at 55 degrees C for fifteen minutes to allow the proteinase K to degrade the proteins. Proteinase K treatment facilitates RNA extraction with phenol and chloroform.
mRNA isolation
-
8)
Extract the nucleic acids from the transcription reaction by adding an equal volume (140 µL) of phenol. Vortex for 10 full seconds and centrifuge at max speed (~12–14,000 × g) in a microfuge for 2 minutes to separate the phases. Take appropriate care when using phenol and consult the materials safety data sheet (MSDS) before using.
-
9)
Transfer the (upper) aqueous layer to a new tube and repeat step 8 with a 1:1 phenol:chloroform mix. Take appropriate care when using phenol: chloroform and consult the MSDS before using.
-
10)
Carefully transfer ~90% or 125 µL of the aqueous layer to a new tube. It is important not to take the entire supernatant in order to avoid contamination with the organic phase and to ensure uniform sample recovery. Add 2× volume (250 µL) of 100% ethanol to the aqueous layer. Mix well and incubate on dry ice for 10–15 minutes.
-
11)Centrifuge at max speed (~12–14,000 × g) for 15 minutes.
- The pellet should be visible, particularly if using a co-precipitant dye like Novagen Pellet Paint™ obtained from EMD/Millipore. Pellet paint co-precipitant should be added according to manufacturer specifications (the authors find that 0.5 µL of Novagen Pellet Paint™ per reaction is generally sufficient).
-
12)Using a drawn-out glass pasteur pipette (>0.5 mm diameter), remove the supernatant and allow the pellet to air-dry. The use of drawn out Pasteur pipettes permits complete removal of the ethanol and buffers and prevents carryover of contaminants into subsequent steps.
- At this point, the investigators can proceed directly to Support Protocol 1 (Primer extension analysis), or the mRNA pellets can be stored at −80 degrees C overnight. Do not store pellets in ethanol. Only the dried pellets may be stored overnight. If 32P-labeled NTPs were used for runoff transcription, the mRNA transcripts can be resuspended in 1× Formamide Dye and analyzed directly by polyacrylamide/urea gel electrophoresis.
Basic Protocol 2: Immobilized DNA template PIC capture assay
Immobilizing the DNA template offers a unique advantage to the investigator studying transcription in vitro. Because transcription involves the assembly of a functional PIC on the template of interest, the most obvious advantage to its immobilization is the ability to analyze PIC composition by western blot or other protein detection methods, and to correlate that with transcriptional activity (Ranish et al. 1999; Ranish et al. 2003). Furthermore, multi-step manipulations can be made to the template to compare transcription across a variety of conditions, such as the use of extracts depleted of key proteins or pre-binding and saturation of the template with certain co-activators or chromatin modifying/remodeling enzymes (Johnson et al. 2002; Johnson and Carey 2003; Black et al. 2006). This protocol describes a PIC-capture experiment, where immobilized templates, pre-bound with activator, are used to capture and analyze fully-formed PICs from HeLa nuclear extract.
Reagents
Acetyl-Coenzyme A (1 mM)
Activator protein (200 ng/µL)
Buffer D (0.1 M; See Recipes)
BW Buffer (2×; See Recipe)
BSA (10 mg/mL)
DNA template (in this protocol, the authors use a 602-bp linear chromatin template containing the G5E4T promoter at a starting concentration of 5 ng/uL)
MgCl2 (0.1 M)
- Magnetic Particle Concentrator (MPC)
- (Invitrogen 120.26, 120.27, 120.28D)
Nonidet-P-40 (NP-40) or NP-40 alternative (1%)
Nonspecific Competitor DNA (in this protocol the authors use the plasmid vector used in cloning the G5E4T template promoter: pGEM3 at a starting concentration of 100 µg/mL)
HeLa Nuclear Extract (Support Protocol 3)
- Streptavidin M-280 Dynabeads™
- (Invitrogen 112-05D)
Template immobilization
-
1)Resuspend Dynabeads from stock and aliquot 50 µL into a micro-centrifuge tube.
- This volume has been empirically determined to completely bind minimally 1 µg of biotinylated G5E4T template DNA. The investigators will need to determine the appropriate quantity of beads to fully bind their template of interest. Generally, the authors find that the bead-binding capacity information provided by the manufacturer is a good starting point (50 uL of M-280 Dynabeads™ for 1ug of G5E4T template DNA).
-
2)Concentrate the Dynabeads into a pellet onto the side of the tube using the magnetic particle concentrator (MPC) and remove the supernatant buffer with a pipet.
- Be sure to work quickly when manipulating the magnetic beads, being careful not to let the beads dry out.
-
3)
Wash the beads by adding 200 µL of 1× BW Buffer, mixing well, and replacing the tube in the MPC.
-
4)
Remove the supernatant and wash with an additional 200 µL of 1× BW Buffer.
-
5)
Add 1 µg of template DNA to an equal volume of 2× BW Buffer. Remove the wash solution from the beads, add the DNA solution to the Dynabeads and incubate at 4 degrees C for 3 hours on an end-over-end rotator.
-
6)
Remove the supernatant and wash the beads with 200 µL of 1× BW Buffer, followed by 200 µL of Buffer D.
-
7)
Remove the Buffer D and resuspend the immobilized template beads in 20 µL of Buffer D, or the appropriate volume for the desired concentration.
Activator binding
-
8)Set up the immobilized template activator binding reaction by mixing over ice:
ddH2O 8.5 µL pGEM3 Carrier DNA (100 µg/mL) 2 µL NP-40 (1%) 0.5 µL MgCl2 (0.1 M) 3 µL BSA (10 µg/uL) 1 µL Buffer D (0.1 M) 24 µL DNA-Bead slurry (50ng/uL) 1 µL - This recipe results in a final KCl concentration of 62.5 mM. Should the investigators wish the change the salt concentration, a higher concentration of Buffer D KCl may be used (e.g. 24 µL 0.2 M Buffer D for a final KCl concentration of 125 mM).
-
9)
Add 1 µL activator protein, mix well, and incubate at room temperature for 30 minutes, rotating end-over-end to ensure uniform bead suspension.
Bead wash and Nuclear extract Binding
-
10)During the incubation in step 9, prepare 50 µL of additional buffer from the recipe in step 8 and set up the immobilized template PIC assembly binding reaction by mixing on ice:
ddH2O 6.5 µL Acetyl-CoA (1 mM) 2 µL pGEM3 Carrier DNA (100 µg/mL) 2 µL NP-40 (1%) 0.5 µL MgCl2 (0.1 M) 3 µL BSA (10 µg/uL) 1 µL Buffer D (0.1 M) 14 µL HeLa nuclear extract 10 µL - This protocol uses 10 µL of nuclear extract per reaction. Should the investigators wish to change the volume of nuclear extract, the appropriate amount of Buffer D can be used to adjust the reaction. However, the total extract+Buffer D volume should total 25 µL.
-
11)
Pulse the reaction mix from step 9 quickly in the microcentrifuge to consolidate the beads, then use the MPC and remove the supernatant. Wash the beads with 50 µL of the step 8 buffer, remove the supernatant and resuspend the beads with the PIC assembly binding reaction buffer (with nuclear extract) from step 10.
-
12)
Mix well and rotate end-over-end for the appropriate amount of time (60 mins for full PIC assembly).
Bead wash and resuspension
-
13)
Pulse the reaction quickly in the microcentrifuge and remove the supernatant using the MPC. Wash the beads with 50 µL of the step 8 buffer.
-
14)
Repeat step 13 with an additional 50 µL of the step 8 buffer.
-
15)
If in vitro transcription is to be performed, resuspend the beads in 10 µL of 0.1 M Buffer D and proceed to step 3 of Basic Protocol 1 (In vitro transcription from nuclear extract). Otherwise PICs can be eluted using SDS-gel loading dye mix and analyzed by SDS-PAGE followed by western blotting. Typically, 1 µL of the nuclear extract (or the appropriate volume to visualize 10% of the input protein) should be loaded in a separate lane to compare the efficiency of PIC recruitment to the template. Typically, the blot is cut into cross slices to probe for different PIC components (see Figure 2).
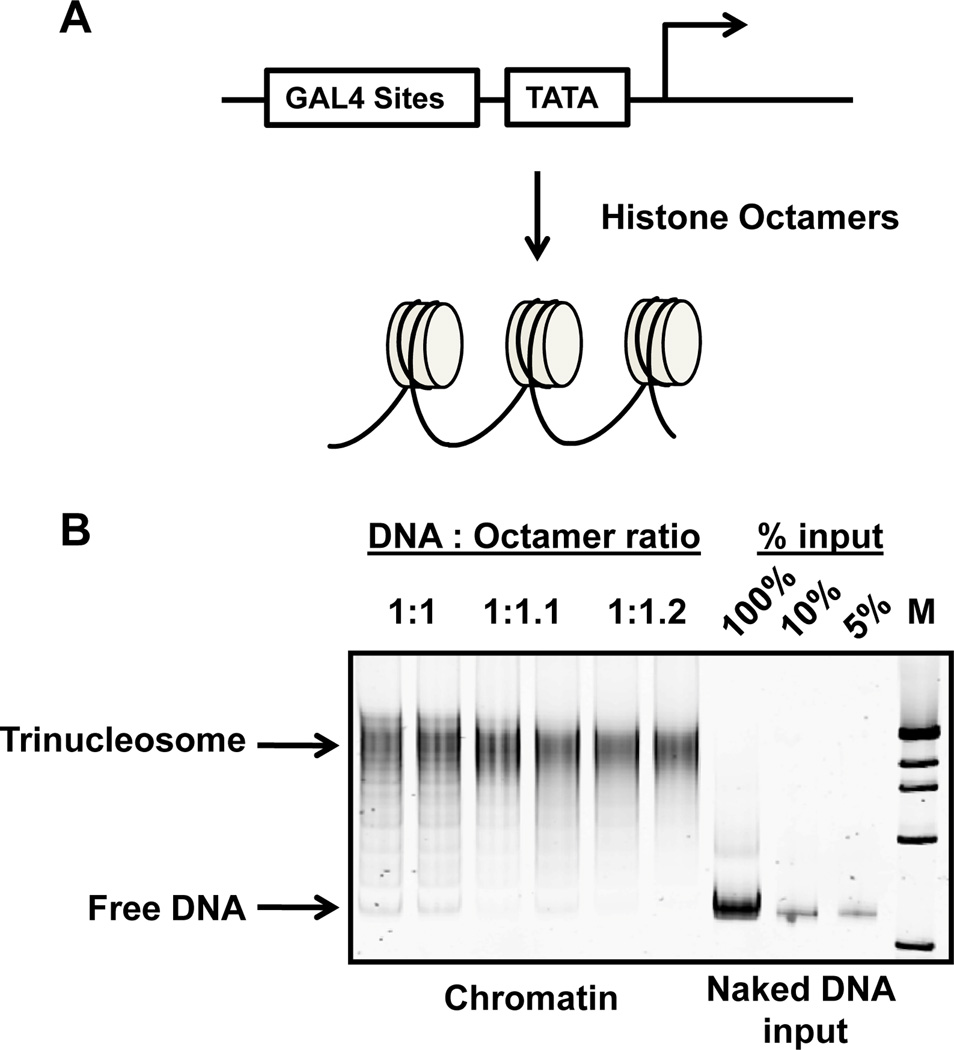
Figure 2. Chromatin assembly by rapid salt dilution.
A schematic of the chromatin assembly reaction (A), followed by a native PAGE EMSA of chromatin assembly reactions with an octamer:DNA ratio titration (B). Two separate reactions for each titration point are shown here. Free DNA inputs are run alongside, on the right side of the gel, for comparison. The DNA is visualized by SYBR-green staining.
Support Protocol 1: Primer extension analysis
The primer extension reaction is a method of converting the RNA products of the in vitro transcription reaction into a uniform, labeled population of complementary DNA. This is achieved by the use of a 32P-labeled oligonucleotide primer that has been designed to hybridize to a specific sequence downstream of the RNA start site (usually 60 to 90 nucleotides downstream of the transcription start site). The isolated RNA from the in vitro transcription reaction is incubated with a saturating amount of this primer before subsequent purification of the DNA-RNA hybrid. Avian reverse transcriptase is added to fully extend the primer and create a 32P-complementary DNA strand. This method of RNA analysis allows the investigators to easily quantify the amount of transcription that has reached the position of the labeled primer by polyacrylamide gel electrophoresis and also permits verification that transcription initiated at the proper location on the promoter. Alternatively, if the investigators wish to visualize the total population of RNA transcripts generated by the reaction, 32P-labeled NTPs may be substituted for their cold counterparts during the in vitro transcription reaction (Usually at a final concentration of 200 µM unlabeled ATP, GTP and UTP, 10 µM unlabeled CTP, and 5 µCi of α32P-CTP). These products can be analyzed by PAGE directly after RNA isolation, skipping the primer extension step. The reaction products using 32P-labeled NTPs are considerably more complex as random transcription occurs frequently in crude extracts.
Reagents
32P-gamma-ATP (Perkin Elmer BLU002Z500UC)
Ammonium Acetate 1 M pH 7 (Ammonium Acetate is a neutral salt)
dNTPs (10 mM each)
Geiger counter
GE Biosciences™ G50 de-salting column
Hybridization Buffer (2×)
Isopropyl Alcohol
- 10× PNK buffer
- (Supplied with T4 PNK)
- Reverse Transcriptase
- (Promega M5108)
- Reverse Transcriptase Buffer (5×)
- (Promega M515A1)
- Reverse transcription primer
- In this protocol, the primer begins +92 nucleotides downstream of the mRNA transcription start site. It is important to design the primer to be G-C rich, with a melting temperature of ~70 degrees C, and without any hairpin-forming dyads.
- RNAsin
- (Promega N2511)
- T4 Polynucleotide Kinase (PNK)
- (NEB M0201)
Primer end-labeling
-
1)In a tube, combine the following reagents:
Primer for primer extension (10 µM) 10 pmol T4 PNK (10 units/µL) 1 µL PNK reaction buffer (10×) 5 µL gamma ATP(P32) (>5000 Ci/mmol) 10 µL DTT (100 mM) 0.5 µL ddH2O to 50 µL -
2)
Mix well and incubate at 37 degrees C for 30 minutes.
-
3)
Heat-inactivate enzyme at 65 degrees C for 10 minutes.
-
4)
Purify 32P-end-labeled primer using GE Biosciences™ G50 de-salting column.
Primer hybridization
-
5)Resuspend the appropriate mRNA-containing pellet in 9 uL ddH2O
- It is imperative to fully resuspend the mRNA pellet. Mix by flicking or vortexing until pellet is completely dissolved.
-
6)
Add 1 µL 32P-labeled primer (~0.2 pmols) (from step 4) and 10 µL 2× Hybridization Buffer to the resuspended RNA.
-
7)Incubate for 2 hr at 37 degrees C
- This temperature should be determined empirically, and will differ from primer to primer.
-
8)
Mix together ammonium acetate (1 M) and isopropyl alcohol 1:1, and add 400 µL to the hybridization reaction. Mix well and let stand at room temperature for 10 minutes to allow nucleic acid precipitation.
-
9)Centrifuge at max speed (≥13000 × g) for 15 minutes and aspirate the supernatant from the visible pellet with a drawn-out Pasteur pipette.
- This pellet will be loose and small. We recommend tracking the pellet with a Geiger counter and/or using a co-precipitant dye like Pellet Paint™. Pellet paint co-precipitant should be added according to manufacturer specifications (the authors find that 0.5 µL per reaction is generally sufficient). If Pellet Paint™ has been used previously, it does not need to be added again.
-
10)
Prepare 75% ice cold Ethanol and add 400 µL to the pellet. Wash and centrifuge again for 5 minutes.
-
11)
Remove the supernatant and air-dry the pellet thoroughly. It is very important that all liquid be removed as excess ammonium acetate carryover from the previous precipitation can inhibit the reverse transcriptase.
Primer extension reaction
-
12)
Resuspend the DNA-RNA duplex pellet in 10 µL 1× Reverse Transcriptase Buffer, making sure to completely resuspend the pellet.
-
13)Chill the resuspended DNA-RNA solution on ice. Meanwhile, prepare the primer extension buffer:
5× RT buffer 2 µL dNTP mix (10 mM each) 1.5 µL RNAsin 0.4 µL ddH2O 4.9 µL DTT (100 mM) 0.2 µL AMV Reverse Transcriptase 1 µL -
14)
Chill the primer extension buffer and add to the resuspended pellet over ice. Mix well and quickly transfer to 45 degrees C and incubate for 30 minutes.
-
15)End the reaction by the addition of 20 µL 2× Formamide Dye. Mix well and heat to 90 degrees for 3 minutes and chill immediately on ice.
- The primer extension reaction should be analyzed by polyacrylamide/urea gel electrophoresis. Imaging can be performed using either X-ray film or using a phosphorimager.
Support Protocol 2: Rapid Chromatin Assembly by Salt Dilution
Depending on the aims of the study, the investigators may choose to assay in vitro transcription on chromatinized templates. This requires a source of purified core histone octamers in equimolar concentration, generally obtained either from recombinant sources (Luger et al. 1997), or purified from nuclear chromatin fractions (Workman et al. 1991). This protocol describes a method of chromatin assembly by rapid salt dilution beginning with 2M NaCl and stepwise dilution down to 0.2M NaCl. This procedure takes roughly three hours from setup to completion, assuming the templates and histones are in hand, and typically yields chromatin at concentrations of 5 ng/uL. It is important for investigators using this method to determine empirically the optimal ratio of DNA to total octamer used in each assembly, as this will vary depending on the octamer source/type. As a general rule of thumb, a 1:1 octamer to template ratio generally results in chromatinization of 90% of the DNA template while avoiding non-specific DNA-histone interactions that cause chromatin aggregation. However, it will be up to the investigators to carefully titrate the octamer to DNA template ratio in order to achieve optimal chromatin assembly. The authors suggest using a 1:1 ratio as a midpoint, and increase or decrease the octamers in 0.1:1 steps (e.g. 0.8:1, 0.9:1, 1:1, 1.1:1 octamer:DNA ratio, respectively). Note that in the absence of nucleosome positioning sequences, it is very difficult to predict the location of nucleosomes. Some protocols utilize enzymes like ISWI, which space the nucleosomes evenly. We avoid the introduction of exogenous enzymes into the mix because their effect on the reaction is unknown.
Reagents
Bovine Serum Albumin (1mg/mL)
Assembly Buffer (See Recipes)
DNA template (in H2O or standard Tris-EDTA pH 7.5–8.0)
Histone Octamers (recombinant source, see Luger et al. 1997 for preparation protocols)
NaCl (2 M and 5 M)
Assembly reaction preparation
-
1)In a tube, mix the following reagents for each point of the desired octamer titration as described above in the protocol summary:
NaCl (5 M) 5.6 µL BSA (1mg/mL) 2 µL DNA template 1 µg ddH2O to 14 µL -
2)Add the appropriate amount of histone octamer, using a 1:1 DNA:octamer ratio as the midpoint, and bring the reaction volume up to 20 µL with 2 M NaCl
- It will be up to the investigators to carefully titrate the octamer to achieve an optimal degree of chromatinization. Remember to prepare a mock reaction without the addition of octamers as a control.
-
3)
Mix well and incubate at 30 degrees C for 15 minutes.
Rapid Salt Dilution
-
4)Add the appropriate volume of 1× Assembly Buffer as indicated sequentially in the table below, incubating for 15 minutes at 30 degrees C after each dilution step.
Add Vol Total Vol Salt Conc. Start 20.00 2.0 M 6.67 µL 26.67 1.5 M 13.33 µL 40.00 1.0 M 10.00 µL 50.00 0.8 M 7.14 µL 57.14 0.7 M 9.52 µL 66.67 0.6 M 13.33 µL 80.00 0.5 M 20.00 µL 100.00 0.4 M 33.33 µL 133.33 0.3 M 66.67 µL 200.00 0.2 M -
5)
Assay the degree of chromatin assembly by native PAGE electrophoretic mobility assay, and use candidate points in the in vitro transcription reaction. These are generally between 90–95% conversion of the naked DNA template to chromatin. A loading titration of the mock reaction should allow the investigators to determine accurately the quantity of free DNA in the assembly reaction (see Figure 3).
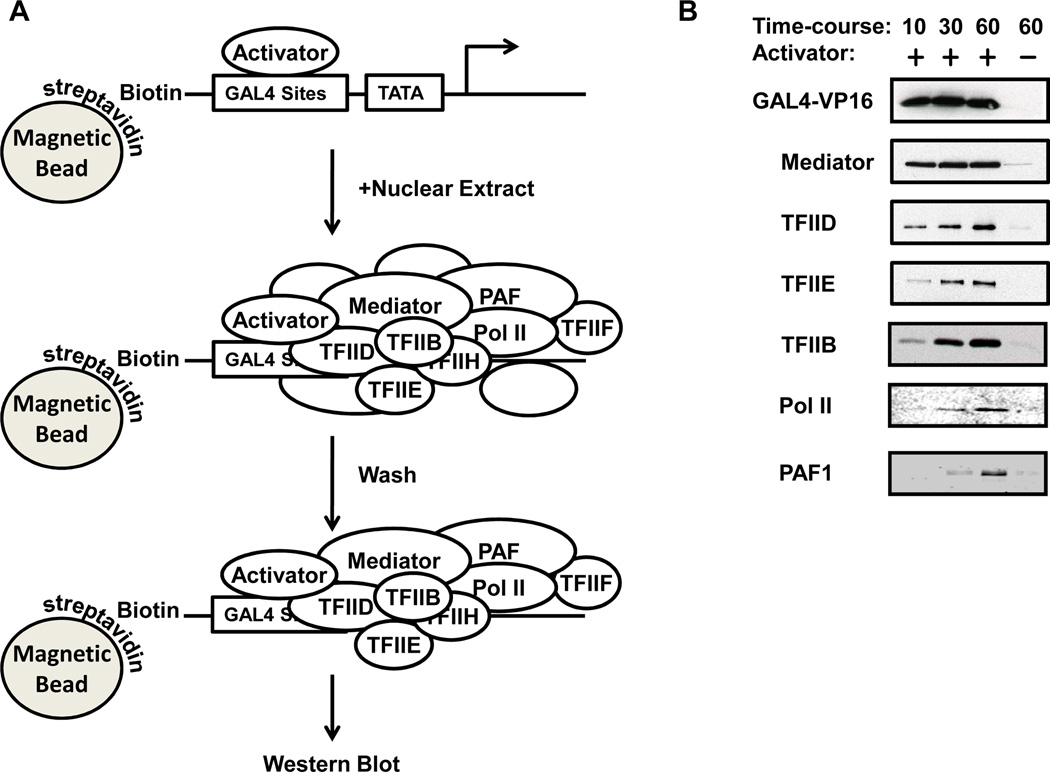
Figure 3. Immobilized template PIC capture assay.
A schematic of the PIC capture and purification process from nuclear extract using the immobilized model promoter, G5E4T, is shown on the left (A). To the right, western blots of a time-course assay of stepwise PIC assembly demonstrates the kinetics of co-activator and GTF recruitment from nuclear extract (B).
Support Protocol 3: HeLa Nuclear Extract preparation
Groundbreaking work in the field of eukaryotic transcription was accomplished in 1983 by Dignam and Roeder, who experimentally optimized a method of obtaining a transcriptionally active fraction from HeLa nuclei by salt-extraction (Dignam et al. 1983). This protocol remains a universal starting point for any investigator attempting to recreate transcription in vitro using extracts prepared from tissue-culture cells. The basic principles outlined within this protocol are applicable to a wide spectrum of cell types, from HeLa to cancer cell lines, including even embryonic stem cells. However, the salt concentration used for extraction described in this protocol was determined empirically as the optimal concentration for HeLa cell nuclei. This may vary for other cell types, and should be established by the investigator using the method described by Dignam et al in 1983. In addition, it is important to mention here, that this protocol describes the isolation of nuclei from roughly 1.6 × 1010 HeLa cells (32 liters of spinner culture), which will yield 30–40 mL of nuclear extract at a concentration of 5 to 8 mg/ml. This protocol can be effectively scaled down to a lower limit of roughly 4.0 × 109 cells. However, for smaller preparations (as few as 1.0 × 107 cells), it is advisable to consult mini-extract protocols that have been optimized for lower cell numbers (for details, see Chapter 12, Carey et al. 2009)
NOTE: The authors recommend the harvest of fresh cells for this procedure. These are lysed by hypotonic swelling and hand-dounce homogenization to release the nuclei, which are collected by high-speed centrifugation. Once isolated, nuclei may be rapidly frozen for long-term storage. However, it is highly advised to proceed quickly through the subsequent steps of nuclear salt-extraction and extract dialysis, as the transcriptional activity has a half-life of roughly 12 hours at 4 degrees C.
Reagents
Bradford Dye
Bovine Serum Albumin
Buffer A (See Recipes)
Buffer C (See Recipes)
Buffer D (See Recipes)
Centrifuge Bottles
Conductivity Meter
Dialysis Tubing
Dounce Homogenizer
Dry Ice
HeLa cells in culture
Hemocytometer
Microscope
PBS (1×)
Rotisserie rotator
Sorvall Refrigerated Centrifuge (All centrifugation steps should be performed at 4 degrees C.)
Trypan Blue Dye
Cell harvest and hypotonic swelling
-
1)
Harvest HeLa cells at a density of 4.0 × 105 cell/mL by centrifugation at 3000 × g for 10 minutes at 4 degrees C.
-
2)
Wash the cell pellet with 10 mL of 1× PBS per liter of cell culture. Centrifuge for 3000 × g for 10 minutes.
-
3)
Remove supernatant and Resuspend cell pellet with 3 mL of 1× PBS per liter of original cell culture and transfer to graduated centrifuge tubes. Centrifuge for 3000 × g for 10 minutes.
-
4)
Using the graduations, determine the volume of the cell pellet and resuspend the pellet in 5 packed cell volumes of Buffer A. Centrifuge this wash, remove the supernatant and resuspend the cell pellet in 3 packed cell volumes of Buffer A.
-
5)
Allow the cells to swell on ice for 5 minutes and transfer the resuspension to a Dounce Homogenizer.
Dounce-homogenizer lysis and nuclei isolation
-
6)
Homogenize the cells using a tight-fitting dounce pestle the cells to roughly 95% total cell population lysis, checking by Hemacytometer-aided counting of Trypan Blue stained cells.
-
7)Separate the nuclei from the cytoplasm by centrifugation at 10,000 × g for 30 minutes.
- Do not exceed 10,000 × g to avoid lysis by centrifugal shear forces.
-
8)
Carefully remove the lipid layer at the top of the supernatant with a kimwipe or by pipetting. Aspirate the cytoplasmic supernatant and discard unless the lab has other uses for it.
Nuclear extraction and dialysis
-
9)Resuspend the crude nuclei in Buffer C by gentle dounce homogenization using 2 or 3 strokes of a loose dounce.
- Be careful not to lyse any of the fragile nuclei and contaminate your extract with the chromatin fraction of the nuclei at this stage. This is usually indicated by an increase in viscosity.
-
10)
Agitate the suspended nuclei by either magnetic stirring or by end-over-end rotation at 4 degrees C for 30 minutes.
-
11)Centrifuge the suspension at 10,000 × g for 30 minutes to separate the nuclear extract from the nuclei and transfer the supernatant to the dialysis bag. Dialyze against at least 40 volumes of 0.1 M Buffer D, changing the external buffer every 2 hours while monitoring the conductivity of the extract to match that of the 0.1 M Buffer D.
- If the nuclear extract is still cloudy, clarification can be performed by centrifugation for an additional 30 minutes.
-
12)
During the dialysis, prepare a BSA standard curve and the Bradford Assay reagents. As soon as dialysis is complete, centrifuge the nuclear extract at 10,000 × g to clear the lysate of any protein precipitate and then measure protein concentration of the cleared extract by Bradford Assay.
-
13)
Aliquot and freeze the nuclear extract in working volume aliquots (0.05–1.0 mL) on dry ice, and may be stored for years at −80 degrees C.
Recipes
<!> Add fresh, immediately before use
Assembly Buffer (3×)
150 mM Hepes pH 7.5
3 mM EDTA
300 µg/uL BSA
30% Glycerol
2% NP-40
0.5 mM DTT <!>
1 mM PMSF <!>
Buffer A
10 mM HEPES pH 7.9
1.5 mM MgCl2
10 mM KCl
0.5 mM DTT <!>
5 mM PMSF <!>
Buffer C
20 mM HEPES pH 7.9
25% Glycerol
0.42 M NaCl
1.5 mM MgCl2
0.2 mM EDTA
0.5 mM DTT <!>
5 mM PMSF <!>
1 uM Leupeptin <!>
1 uM Pepstatin <!>
Buffer D
20 mM HEPES pH 7.9
20% Glycerol
100 mM KCl
0.2 mM EDTA
0.5 mM DTT <!>
1 mM PMSF <!>
Transcription Stop Buffer
10 mM EDTA
0.2% SDS <!>
0.3 M Sodium Acetate
50 µg/mL yeast tRNA
Hybridization Buffer (2×)
2 mM EDTA
600 mM NaCl
20 mM Tris-HCl pH 7.6
0.2 % SDS <!>
BW Buffer (2×)
20 mM Hepes pH 7.9
600 mM KCl
25 µg/mL BSA
1 mM EDTA
COMMENTARY
Background information
DNA transcription in vitro is most commonly and easily recapitulated using protein complexes and factors extracted from isolated intact nuclei. The widely-used and adapted Dignam and Roeder method (Dignam et al. 1983), which resuspends the isolated nuclei in hypertonic buffer, extracts these proteins using high salt concentrations. Many of these proteins will be extracted at varying salt concentrations, and nuclear extract prepared using different salt concentrations will have varying nuclear protein compositions. Typically, preparation of transcriptionally active nuclear extract seeks to strike a balance between extracting sufficient concentrations of transcription machinery and minimizing the extraction of transcription repressors such as histones and non-specific DNA-binding proteins, which are extracted at higher salt concentrations (Dignam et al. 1983; Croston et al. 1991). While the Dignam-derived protocol described in this unit uses a salt concentration of (0.42 M NaCl) that was empirically determined to best activate transcription on the adenovirus Major Late Promoter using HeLa cells as a source of nuclei, protocols using other sources of nuclei, such as Drosophila embryos have been optimized at low salt concentrations (0.12 M NaCl) (Kamakaka et al. 1991).
In vitro transcription from these extracts relies on the efficient recruitment of functional PICs to the template of interest. Biochemical studies of activator-stimulated PIC assembly from these transcriptionally active extracts on immobilized templates have demonstrated the step-wise assembly of co-activators and GTFs (Black et al. 2006). In time-course assays, the activator-specific recruitment of the Mediator co-activator was shown to coincide with histone acetyltransferase recruitment and activity (Black et al. 2006), which is required for efficient transcription from chromatinized templates (Kundu et al. 2000). It is important, therefore, to include acetyl-CoA in the reaction mix when transcribing chromatin, and to ensure that any signal from these templates is dependent on this substrate. Any residual acetyl-CoA-independent transcription observed is most likely signal from free DNA templates resulting from incomplete chromatin assembly. Histone acetyltransferase recruitment and activity precedes the recruitment of TFIID, GTFs, and Pol II (Black et al. 2006).
The ATP-dependent transition between closed complex polymerase, to open-complex polymerase, where the DNA surrounding the start-site is melted, can be bypassed if an accessible region of (pre-melted) single-stranded DNA upstream of the start site already exists. (Pan and Greenblatt 1994; Tantin and Carey 1994; Holstege et al. 1995; Dvir et al. 1996). ATP-dependent promoter melting is mediated by TFIIH, which is recruited in the later stages of PIC assembly (as reviewed in (Dvir et al. 2001)). Permanganate probing of assembled PICs incubated with ATP alone reveals the functional transition to the transcriptionally-primed open-complex state (Wang et al. 1992).
Although the immobilized template assay has been used by many labs previously to study activated RNA Polymerase II transcription (for example, see Lin and Green 1991), it came into broader use in the late 1990s after many of the components of the transcriptional machinery had been identified and cloned. One of the first uses that identified key steps in activator function was performed in yeast extracts to investigate the effect of mutations in GTFs, Pol II, and Mediator subunits on PIC assembly (Ranish et al. 1999). Among the subsequent studies, the immobilized template was used to compare the composition of pre- and post-initiation complexes at the promoter in vitro (Yudkovsky et al. 2000). The immobilized template approach can be used either in parallel with, or directly incorporated into, in vitro transcription assays to link regulatory steps in PIC formation with functional transcription. Our lab has used this assay to demonstrate that purified Mediator and TFIID assemble cooperatively on an immobilized template in vitro, and that this complex can stimulate the formation of a PIC and subsequent transcription initiation from Med- and TFIID-depleted extract (Johnson et al. 2002; Johnson and Carey 2003). Time-course studies of PIC formation from nuclear extracts revealed that full assembly of a VP16-stimulated PIC on chromatin was limited by a Mediator-dependent histone acetyltransferase (p300) recruitment step, whose activity was required for full PIC assembly and functional transcription initiation (Black et al. 2006). These and other studies have demonstrated that functional transcription initiation closely coincides with the assembly of a completed PIC. Indeed, fully-formed, closed-complex PICs, assembled in the absence of ATP and captured on immobilized templates, have demonstrated transcriptional activity upon the addition of NTPs (Baek et al. 2006).
The limitations imposed by antibody availability and variable sensitivity prevent investigators from the comprehensive and quantitative detection of all PIC components. Typically, detection of select subunits from a number of various PIC components is used as an indication of a properly formed PIC. The presence of the co-activators Mediator and TFIID, GTFs, and RNA Pol II are generally deemed the hallmark of proper PIC formation. However, more recent studies of the PIC using more sophisticated proteomic analyses reveal that a large number of additional proteins and protein complexes are recruited by activators to a PIC, although their specific roles in transcription have not yet been fully dissected (for example, see Ranish et al. 2003; Kim et al. 2007; Takahashi et al. 2011).
Critical Parameters and Troubleshooting
Due to the relative instability of RNA in comparison to DNA samples and its susceptibility to ubiquitous rampant RNA nucleases, it is imperative to use clean and sterile high-grade reagents and lab ware to prevent nuclease contamination during both extract and template preparation, and subsequent steps in the in vitro transcription reactions.
Temperature changes during steps outlined by the protocols above should be performed rapidly, especially during transcription and primer-extension reactions. This ensures efficient transcription and reverse-transcription, and reduces non-specific products such as snap-back reverse transcription, and other aberrant polymerase activity which may complicate the data and make signal quantitation difficult.
Time-courses and specific time-points during immobilized template recruitment assays may need to be optimized to observe the maximal recruitment of certain cofactors. The concentration of nuclear extracts used in these experiments may also need to be carefully titrated.
Additionally, when using nuclear extracts for transcription, it is crucial to maintain relatively high protein concentrations for the efficient assembly of functional PICs. Dilute extract preparations (< 5mg/ml) will often result in incomplete PIC assembly and marginally active transcription. Protocols for extract concentration using ammonium sulfate (0.3–0.4 g/ml) have been successfully used to precipitate transcription proteins for resuspension in lesser volumes of Buffer D (Scopes 1994). These samples will need to be re-dialyzed to reduce the salt concentration from residual ammonium sulfate. It is important to note that since HeLa nuclear extract has a transcriptional half-life of 12 hours at 4 degrees C, preparation should be carried out expediently. Extract dialysis should be monitored frequently with a conductivity meter to reduce dialysis time to a minimum. Extracts should be aliquoted into working volumes and kept frozen at −80 degrees C, and should not be freeze-thawed more than twice.
If weak to no transcription signal is detected, the investigator may wish to purchase a commercially available control extract and template to test their experimental conditions. Typically, natural promoters and activators are relatively weak in comparison to viral promoters and activators such as VP16. In addition, the investigators may want to run a positive control for the reverse transcription reaction during primer extension. These kits are available commercially through Promega (A1260). If overall signals are weak, it may be prudent to verify that the primer itself is labeled efficiently with fresh 32P. Typically, a band of 10 fmol of labeled primer alone should give a robust signal within a 3 hour exposure to x-ray film.
Time Considerations
The protocols described in this unit are all relatively time-intensive, between 3–4 hours for the actual in vitro transcription reaction, an additional 3–4 hours for primer extension, up to 8 hours for nuclear extract preparation, and 4–5 hours for chromatin assembly. Purification of specific templates and proteins will vary depending on the needs of the investigators.
Anticipated Results
Transcription from HeLa nuclear extract prepared to a concentration of roughly 5–10 mg/ml will yield relatively strong activator-stimulated signals from a promoter utilizing the GAL4-VP16 model activator. This artificial trans-activator has already demonstrated robust activity in in vitro transcription assays using both nuclear extract, and highly purified factors. Analysis of these transcription products by primer extension should ideally yield a product of uniform size. 32P-labeled DNA markers can be added to the gel to measure the size of the primer extension products to ensure that transcription initiated at the proper location. Figure 1 shows a 60-minute in vitro transcription reaction from HeLa nuclear extract using a titration of both GAL4-VP16 activator (2.0, 6.7, and 20 ng per reaction) as well as a titration of HeLa nuclear extract (5, 15, and 25 µL per reaction; ~8 mglml extract) on naked DNA template. Note that both the medium and high points of GAL4-VP16 give equal signal in these reaction conditions, indicating that 6.7 ng/reaction is a saturating concentration. Weaker or natural activators rarely display the strong trans-activation capabilities of GAL4-VP16. However, this does not mean that in vitro transcription is not feasible. Carefully titrating template, activator and extract concentrations can optimize levels of in vitro transcription.
In vitro chromatin assembly using the rapid salt dilution method described above can be scaled up or down in DNA template concentration, up to 1ug DNA template per assembly reaction. Octamer quantities should be carefully titrated, but a 1:1 DNA: histone octamer ratio on a 600 bp template without nucleosome positioning sequences should yield approximately 95% conversion of the naked template into chromatin (5% free DNA remaining). This is typically indicated in an electrophoretic mobility shift assay (EMSA) in which the free DNA band shifts to a higher-migrating chromatin band. Figure 2 shows an EMSA gel of a DNA/histone octamer titration. A titration of naked DNA template (to the right of the chromatin) allows careful quantitation of the free DNA remaining in the chromatin assembly reactions. Note that the ladder of intermediate products between the free DNA and the trinucleosome chromatin templates is typical of templates that lack specific nucleosome-positioning sequences such as the 601-positioning sequence (Lowary and Widom 1998).
If immobilized templates are to be used for either in vitro transcription or PIC capture assays, a titration of streptavidin-coated Dynabeads™ should be performed in the template immobilization reaction to ensure complete binding of the DNA template. This way, the concentration of DNA in the bead slurry can be calculated for subsequent experiments. Figure 3 shows a time-course of PIC assembly at 10, 30, and 60 minute time-points from 10 µL HeLa nuclear extract using 50 ng of G5E4T template with a loading control of 10% input HeLa nuclear extract to the right. MED23 was probed as a representative subunit of the Mediator, TAF1 as a representative subunit of TFIID, subunit alpha for TFIIE, and ASH2L to represent a core SET/MLL subunit. PIC capture assays using the immobilized template using GAL4-VP16 should pull down approximately 10% of the input of the major PIC co-activators, Mediator and TFIID, depending on the length of nuclear extract incubation assayed in the PIC assembly assay. Recruitment of other PIC components may not be as robust, sometimes below 1% of the input, depending on the stringency of the washes and various other binding conditions, and the original abundance of the complex within the nuclear extract preparation. It is important to remember that only a fraction of the pre-assembled PICs will be transcriptionally active. Increasing the concentration of extract or purified components will increase the population of complete PICs and the overall transcription signal.
Acknowledgement
This work was supported by grants to Michael Carey from the National Institutes of Health 5R01GM074701.
References
- Baek HJ, Kang YK, Roeder RG. Human Mediator Enhances Basal Transcription by Facilitating Recruitment of Transcription Factor IIB during Preinitiation Complex Assembly. Journal of Biological Chemistry. 2006;281:15172–15181. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- Black JC, Choi JE, Lombardo SR, Carey M. A Mechanism for Coordinating Chromatin Modification and Preinitiation Complex Assembly. Molecular Cell. 2006;23:809. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Carey M, Leatherwood J, Ptashne M. A potent GAL4 derivative activates transcription at a distance in vitro. Science (New York, NY) 1990;247:710–712. doi: 10.1126/science.2405489. [DOI] [PubMed] [Google Scholar]
- Carey M, Peterson C, Smale S. Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques. Cold Spring Harbor: Cold Springs Harbor Laboratory Press; 2009. [Google Scholar]
- Croston GE, Lira LM, Kadonaga JT. A general method for purification of H1 histones that are active for repression of basal RNA polymerase II transcription. Protein Expression and Purification. 1991;2:162–169. doi: 10.1016/1046-5928(91)90066-r. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Research. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir A, Conaway JW, Conaway RC. Mechanism of transcription initiation and promoter escape by RNA polymerase II. Curr Opin Genet Dev. 2001;11:209–214. doi: 10.1016/s0959-437x(00)00181-7. [DOI] [PubMed] [Google Scholar]
- Dvir A, Garrett KP, Chalut C, Egly J-M, Conaway JW, Conaway RC. A Role for ATP and TFIIH in Activation of the RNA Polymerase II Preinitiation Complex Prior to Transcription Initiation. Journal of Biological Chemistry. 1996;271:7245–7248. doi: 10.1074/jbc.271.13.7245. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Tantin D, Carey M, van der Vliet PC, Timmers HT. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, Carey M. Assembly of a Mediator/TFIID/TFIIA Complex Bypasses the Need for an Activator. Current Biology. 2003;13:772–777. doi: 10.1016/s0960-9822(03)00283-5. [DOI] [PubMed] [Google Scholar]
- Johnson KM, Wang J, Smallwood A, Arayata C, Carey M. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes & Development. 2002;16:1852–1863. doi: 10.1101/gad.995702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, Wang J, Smallwood A, Carey M. In: Methods in Enzymology. Jo MLJ, Holt M, Gary KA, editors. Academic Press; 2004. pp. 207–219. [DOI] [PubMed] [Google Scholar]
- Kamakaka RT, Tyree CM, Kadonaga JT. Accurate and efficient RNA polymerase II transcription with a soluble nuclear fraction derived from Drosophila embryos. Proceedings of the National Academy of Sciences. 1991;88:1024–1028. doi: 10.1073/pnas.88.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proceedings of the National Academy of Sciences. 2007;104:16068–16073. doi: 10.1073/pnas.0704573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu TK, Palhan VB, Wang Z, An W, Cole PA, Roeder RG. Activator-Dependent Transcription from Chromatin In Vitro Involving Targeted Histone Acetylation by p300. Molecular Cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a Component of the ELL/P-TEFb Elongation Complex and a Shared Subunit of MLL Chimeras, Can Link Transcription Elongation to Leukemia. Molecular Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-S, Green MR. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of Molecular Biology. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Lue NF, Kornberg RD. Accurate initiation at RNA polymerase II promoters in extracts from Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences. 1987;84:8839–8843. doi: 10.1073/pnas.84.24.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Malik S, Gu W, Wu W, Qin J, Roeder RG. The USA-Derived Transcriptional Coactivator PC2 Is a Submodule of TRAP/SMCC and Acts Synergistically with Other PCs. Molecular Cell. 2000;5:753–760. doi: 10.1016/s1097-2765(00)80254-3. [DOI] [PubMed] [Google Scholar]
- Malik S, Guermah M, Roeder RG. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proceedings of the National Academy of Sciences. 1998;95:2192–2197. doi: 10.1073/pnas.95.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Development. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Pan G, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. Journal of Biological Chemistry. 1994;269:30101–30104. [PubMed] [Google Scholar]
- Ranish JA, Yi EC, Leslie DM, Purvine SO, Goodlett DR, Eng J, Aebersold R. The study of macromolecular complexes by quantitative proteomics. Nat Genet. 2003;33:349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes & Development. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends in Biochemical Sciences. 1996;21:327–335. [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, Banks CAS, Parmely TJ, Sorokina I, Brower CS, Conaway RC, Conaway JW. A Mammalian Homolog of Drosophila melanogaster Transcriptional Coactivator Intersex Is a Subunit of the Mammalian Mediator Complex. Journal of Biological Chemistry. 2003;278:49671–49674. doi: 10.1074/jbc.C300444200. [DOI] [PubMed] [Google Scholar]
- Sawadogo M, Roeder RG. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proceedings of the National Academy of Sciences. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes RK. Protein Purification: Principles and Practice. Springer Advanced Texts in Chemistry; 1994. [Google Scholar]
- Snoek R, Rennie PS, Kasper S, Matusik RJ, Bruchovsky N. Induction of cell-free, in vitro transcription by recombinant androgen receptor peptides. The Journal of Steroid Biochemistry and Molecular Biology. 1996;59:243–250. doi: 10.1016/s0960-0760(96)00116-1. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Parmely Tari J, Sato S, Tomomori-Sato C, Banks Charles AS, Kong Stephanie E, Szutorisz H, Swanson Selene K, Martin-Brown S, Washburn Michael P, et al. Human Mediator Subunit MED26 Functions as a Docking Site for Transcription Elongation Factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D, Carey M. A heteroduplex template circumvents the energetic requirement for ATP during activated transcription by RNA polymerase II. Journal of Biological Chemistry. 1994;269:17397–17400. [PubMed] [Google Scholar]
- Verdier J-M, Stalder R, Roberge M, Amati B, Sentenac A, Gasser SM. Preparation and characterization of yeast nuclear extracts for efficient RNA polymerase B (II)-dependent transcription in vitro. Nucleic Acids Research. 1990;18:7033–7039. doi: 10.1093/nar/18.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Carey M, Gralla JD. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- Workman JL, Taylor ICA, Kingston RE. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell. 1991;64:533–544. doi: 10.1016/0092-8674(91)90237-s. [DOI] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Lieberman PM, Boyer TG, Berk AJ. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes & Development. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]