SUMMARY
There is considerable heterogeneity in immunological parameters between individuals, but its sources are largely unknown. To assess the relative contribution of heritable versus non-heritable factors, we have performed a systems-level analysis of 210 healthy twins between 8–82 years of age. We measured 204 different parameters, including cell population frequencies, cytokine responses, and serum proteins, and found that 77% of these are dominated (> 50% of variance) and 58% almost completely determined (> 80% of variance) by non-heritable influences. In addition, some of these parameters become more variable with age, suggesting the cumulative influence of environmental exposure. Similarly, the serological responses to seasonal influenza vaccination are also determined largely by non-heritable factors, likely due to repeated exposure to different strains. Lastly, in MZ twins discordant for cytomegalovirus infection, over half of all parameters are affected. These results highlight the largely reactive and adaptive nature of the immune system in healthy individuals.
INTRODUCTION
The study of monozygotic (MZ) and dizygotic (DZ) twin pairs, has provided a powerful means for separating heritable from non-heritable influences on measured traits for almost 100 years (Jablonski, 1922). Such studies have been used to study autoimmune diseases, vaccine responses (Jacobson et al., 2007), serum cytokines (de Craen et al., 2005) or the frequencies of major immune cell populations (Clementi et al., 1999; Evans et al., 2004). Most of these studies have found that both heritable and non-heritable factors contribute to the resulting phenotype. Recent advances in technology now allow much more comprehensive surveys to be conducted across the many different components of the immune system and thus we performed a very broad, “systems level” study in which we measured 51 serum cytokines, chemokines and growth factors, the frequencies of 95 different immune cell subsets, and cellular responses to cytokine stimulation (Figure 1A). Our results show that these functional units of immunity vary across individuals primarily as a consequence of non-heritable factors with a generally limited influence of heritable ones. This indicates that the immune system of healthy individuals is very much shaped by the environment and most likely by the many different microbes that an individual encounters in their lifetime.
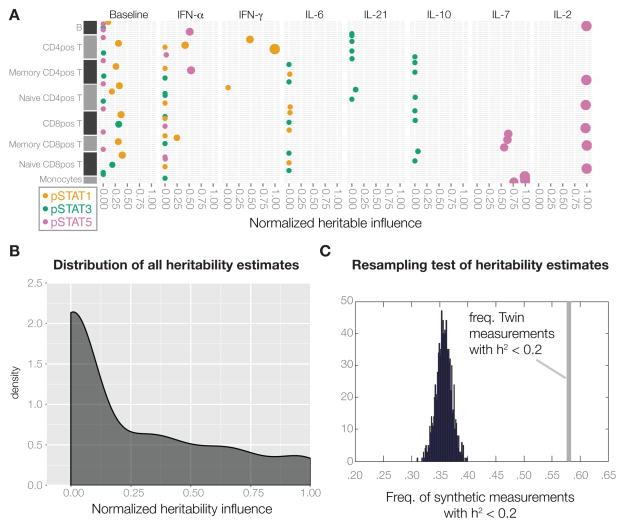
Figure 1. Systems-level analysis of healthy human twins.
(A) Overview of data collected covering the functional units of the immune system, the cells and proteins in circulation. (B) Summary of all heritability estimates for 72 immune cell population frequencies as determined by Flow (2009) and Mass cytometry (2010–2011)(Experimental Procedures, section 3), See also Table S3. (C) Heritability estimates of 43 serum proteins as determined by a fluorescent bead assay, see also Table S4. Error bars represent 95% confidence intervals for the heritability estimate. Grey area is heritability < 0.2, our detection limit.
RESULTS
A systems-level analysis of the immune system in healthy twins
Our study cohort was recruited from the Twin Research Registry at SRI International (Krasnow et al., 2013) in the years 2009–2011 with demographic data detailed in Table S1. The subjects were all apparently healthy, without any symptoms of disease (Experimental procedures, section 1). To minimize biological variability, the time between blood sampling of each twin in a pair was kept to a minimum (Experimental procedures, section 2). Immunological assays were performed by the Human Immune Monitoring Center where assays are continually benchmarked to minimize technical variability (http://iti.stanford.edu/himc/) (Maecker et al., 2005). However, some technical variability is inevitable and thus we corrected for this in all of our models. We did this by analyzing aliquots of the same control sample many (>17) times to estimate the technical variance and subtract this from our estimates of heritability (Experimental procedures, section 8). We also analyzed longitudinal samples in an unrelated cohort over 2–5 consecutive, yearly samplings, and found the variation was largely due to technical variability (Table S2). A total of 204 different immune measurements were included in our analyses.
Estimating heritable and non-heritable influences
Heritability for each parameter was estimated by comparing observed MZ and DZ covariance matrices to the expected values based on a structural equation model that partitioned the observed variance into three components, heritable (A), shared (C) and unique (E) non-heritable factors. This model is based on the assumptions that, I) heritable factors correlate perfectly between MZ twins (rMZ=1) but only to 50% between DZ twins (rDZ=0.5), and II) that shared non-heritable influences are equally similar (rMZ = rDZ) between MZ and DZ twin pairs (Experimental procedures, section 7). For each measurement, we subtracted the technical variance estimate from the E-component prior to normalization to correct for noise (Experimental procedures, section 8). We also corrected all measurements for the effects of age (Dorshkind et al., 2009) and gender (Furman et al., 2014), by regressing out such effects and using only residual variance for estimating heritability. Finally, we performed jack-knife bootstrapping tests to obtain 95% confidence intervals (Experimental procedures, section 7). Importantly, as our model estimates heritability by comparing MZ and DZ twins, heritable influences include genomic and shared epigenetic traits (Bell and Spector, 2011), and non-heritable influences include environmental factors and stochastic epigenetic changes (Fraga et al., 2005).
We first ran a simulation experiment to verify that our cohort size of 210 twins (78 MZ and 27 DZ pairs) would be enough to test our hypothesis that most immunological traits are explained more by non-heritable than heritable influences. We found this to be the case, and we estimate 20% heritability to be our detection limit, under which we cannot distinguish small heritable influences from zero (Figure S1).
Most cell population frequencies and serum proteins are dominated by non-heritable influences
While it is well known that the frequencies of different types of immune cells in blood often vary widely between individuals, in most cases it is not known how much of this can be attributed to heritable or non-heritable factors respectively. To address this question, we used antibodies against cell surface markers to quantify 95 different cell subset frequencies, but used the 72 most non-redundant ones and estimated the influence of heritable and non-heritable factors on their variation (Experimental Procedures, section 3). Among these a few had very strong influences from heritable factors, especially naïve, CD27+ and central memory CD4+ T-cells (Figure 1B and Table S3), but for most, non-heritable influences were clearly dominant. In fact, for 61% of all cell populations, the influence of heritable factors was undetectable (<20% of the total variation)(Figure 1B and Table S3). This was true of both adaptive (T- and B-cells) and innate cell types (granulocytes, monocytes and NK-cells).
Serum cytokines and chemokines also have important functions as immune mediators and biomarkers of disease (Villeda and Wyss-Coray, 2013) and thus we measured 51 serum proteins, but eliminated eight that were often at or below the limits of detection (Experimental Procedures, section 5). This left 24 cytokines, 10 chemokines, 6 growth factors and 3 other serum proteins for which we estimated the influences of heritable and non-heritable factors (Figure 1C, Table S4). Some cytokines were particularly heritable, such as IL-12p40 (Figure 1C, Table S4). Interestingly, variants in the IL12B gene that contribute to the IL-12p40 protein have been associated with immune-mediated diseases such as psoriasis (Nair et al., 2009) and asthma (Morahan et al., 2002). In the latter condition, the susceptibility locus was also associated with a reduced serum concentration of IL-12p40 (Morahan et al., 2002). For many other measurements, such as IL-10 and a group of chemokines, the heritable influence was low (Figure 1C, Table S4).
Homeostatic cytokine responses are largely heritable, while most other cell responses are highly non-heritable
Since these serum proteins often regulate immune cells, we assessed the responses of eight different cell populations stimulated in vitro with seven different cytokines for the phosphorylation of three important and used transcription factors STAT1, 3 and 5 using phospho-specific antibodies in flow cytometry (Krutzik and Nolan, 2006). We performed a total of 192 different measurements, but focused on the 24 baseline measurements and the 65 strongest induced responses (Experimental Procedures, section 4). Baseline measurements were generally driven by non-heritable factors, with possible minor contributions from heritable factors (Figure 2A). The important homeostatic cytokines IL-2 and IL-7, known to stimulate the proliferation and differentiation of T-cells, were found to induce STAT5 phosphorylation in both CD4+ and CD8+ T-cell populations and these responses were highly heritable (Figure 2A, Table S5). In contrast, most signaling responses such as interferon-induced STAT1 phosphorylation, and in particular the IL-6, IL-21 and regulatory IL-10 induced phosphorylation of STAT3, were dominated by non-heritable influences (Figure 2A, Table S5). In total, 69% of all signaling responses had no detectable heritable influence (e.g. < 20%)(Figure 2A, Table S5). This lack of heritability was not related to the strength of responses and explained by a bias towards weak and variable responses (Figure S2).
Figure 2. Heritable factors explain only a fraction of the variation for most immune measurements.
(A) Heritability estimates for immune cell signaling states, upon stimulation with the indicated cytokines. Only unstimulated controls and induced responses > 1.5-fold are shown, see also Table S5. (B) The overall distribution of heritability for all 204 measurements. (C) The maximum number of measurements with heritability < 0.2 across 1000 synthetic datasets with the same MZ/DZ-ratio as in our twin cohort is < 40%, significantly less than our results of 58% of measurements with heritability < 20% (grey bar), p-value < 0.001.
Non-heritable influences are major factors determining immune variation
Taken together, these results show that variation in blood cell frequencies and functions and soluble factors is largely driven by non-heritable factors, with 58% of all measurements having less than 20% of their total variance explained by heritable influences (Figure 2B). There was no relationship between the absolute degree of measurement variability in the cohort and estimated heritability (Figure S3), and we could also rule out any underestimation of heritability due to the skewed ratio of MZ/DZ twin pairs in our cohort by a resampling test. Briefly, by creating 1000 synthetic datasets with uniform heritability and the same MZ/DZ ratio as in our cohort, we estimated heritability and found that none of the 1000 datasets ever had more than 40% of measurements with an estimated heritability < 0.2 (p-value < 0.001) (Figure 2C), thus suggesting that the low heritability estimates obtained are not a result of study design or overall measurement variation in the cohort.
Heritable and non-heritable measurements are interrelated in the immune network
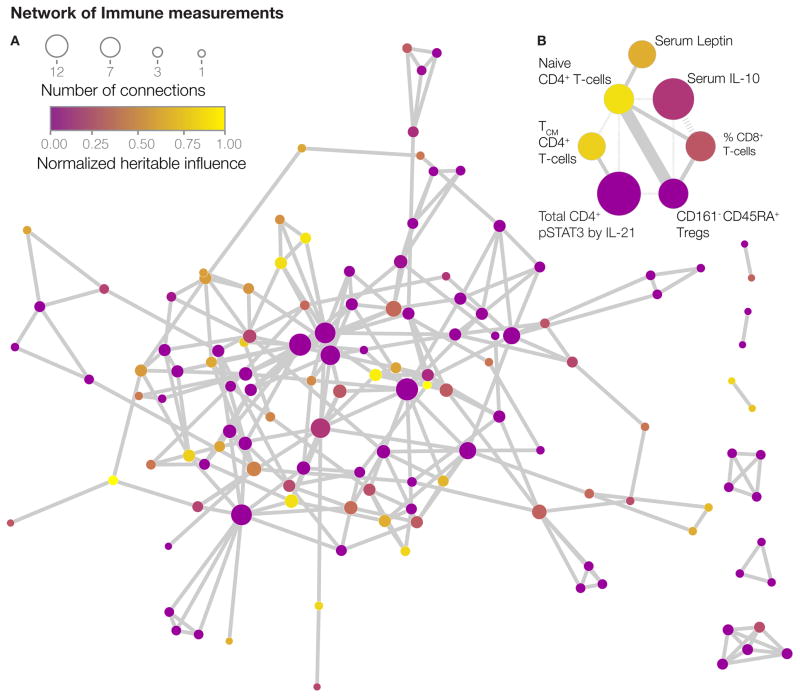
Our analysis also allowed us to analyze the interrelationships between the different components of the immune system. To construct such a network model, we calculated a precision matrix derived from a Spearman correlation matrix (Liu et al., 2012). A matrix of this type captures partial correlations between variables and avoids spurious, indirect interactions that might occur in standard correlation analyses. By penalizing non-zero entries in this matrix (Friedman et al., 2008), we could pursue what is referred to as a sparse (rather than dense) network model, making it more interpretable. After removing unconnected nodes, and validating the edges by a permutation test (Experimental procedures, section 9), this model consists of 126 nodes and 142 edges (Figure 3A and Table S6). An interactive version is available online (http://www.brodinlab.com/twins.html). We found that heritable nodes (yellow) were generally connected to non-heritable nodes (purple) throughout the network (Figure 3A). One example shows how the weakly heritable cytokine IL-10 and CD161− CD45RA+ regulatory T-cells are connected to the strongly heritable frequency of naïve CD4+ T-cells (Figure 3B). We found that all hubs in the network were dominated by non-heritable influences, like the network as a whole, showing that heritable factors are not isolated by themselves, but are buffered by connected non-heritable ones (Figure 3A–B). This may explain why the many gene polymorphisms found (for example CTLA4 (Gregersen et al., 2009)), outside of the HLA locus that are associated with immune mediated disease, only explain a small fraction of the total disease risk (Todd, 2010).
Figure 3. Network of dependencies between immune measurements.
(A) Undirected network model of the healthy human immune system showing 126 nodes (measurements), connected by 142 undirected edges illustrating conditional measurement dependencies. Nodes are colored by their estimated heritability and sized by their number of edges.(B) Sub-network exemplifying direct relationships between heritable and non-heritable nodes. Solid edges represent positive relationships and dashed edges represent negative relationships. The edge weight represents the strength of relationships. See also Table S6.
With age, genetically identical twins diverge as a consequence of non-heritable influences
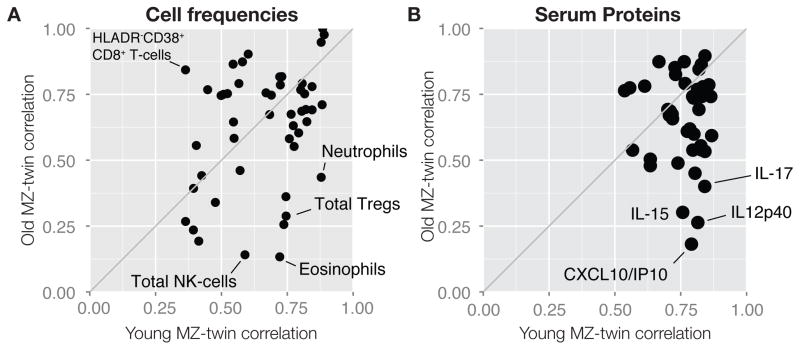
As a major source of non-heritable influence is likely to be environmental, particularly microbial exposure, we reasoned that such influences would increase with time. To this end, we compared twin-twin correlations for all immune measurements between the oldest (> 60 years, median: 72 years) and the youngest (< 20 years, median: 13 years) MZ pairs in our cohort. Here we also note that twins in the younger (<20 years) cohort are in most cases living together, while the older (>60 years) twins, have lived apart for decades, so concordance can also be a result of either shared environment and/or shared exposure, in addition to genetic similarity. For several cell population frequencies, we found much reduced correlations with age (Figure 4A). In the most striking example, the frequency of eosinophils between the youngest MZ twins correlated very strongly at 0.72, but was highly uncorrelated at 0.13 between the oldest MZ twin pairs (Figure 4A). Similarly, several serum proteins showed remarkably reduced correlations between older as compared to younger MZ twins (Figure 4B). In particular the chemokine CXCL10/IP10 showed a strong correlation (0.79) between the youngest MZ twins but was greatly reduced (0.18) in the older MZ twins (Figure 4B). Similar patterns were found for many cell signaling responses (data not shown), suggesting that this immune divergence between genetically identical twins with age is a common phenomenon, consistent with a major role for environmental exposure in driving variation, although some epigenetic changes could also contribute (Fraga et al., 2005).
Figure 4. Increased variability in the immune system with age.
(A) Twin-twin correlations (Spearman’s rank) for all cell frequencies within the youngest MZ twin pairs (≤ 20yrs, median: 13.5, n=25 pairs), and the oldest MZ twin pairs (≥ 60yrs, median: 72yrs, n=16). (B) Twin-twin correlations (Spearman’s rank) for all serum protein concentrations within the youngest MZ twin pairs (≤ 20yrs, median: 13yrs, n=26), and the oldest MZ twin pairs (≥ 60yrs, median: 73yrs, n=13).
Cytomegalovirus infection has a broad influence on immune variation
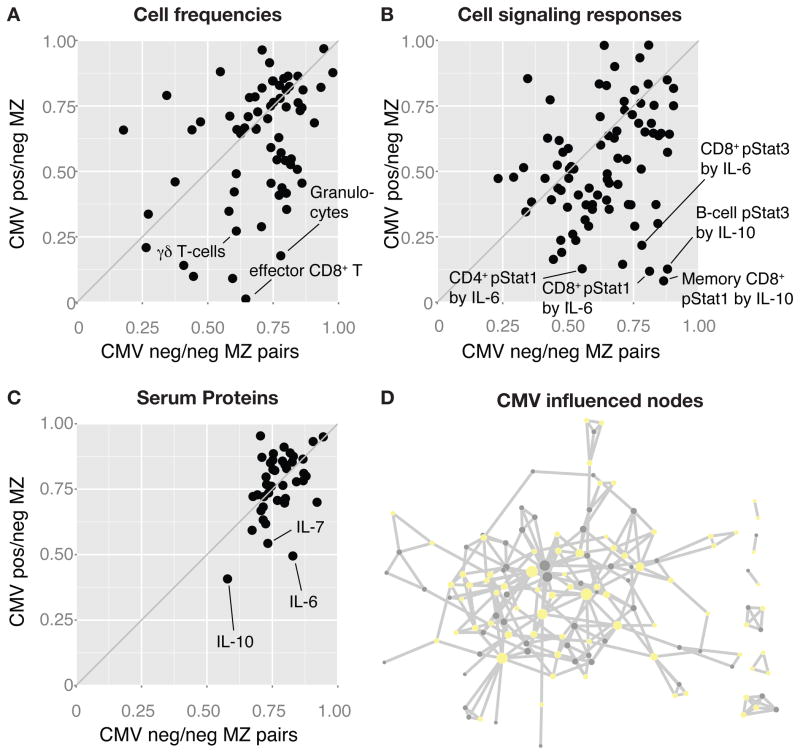
As we postulate that microbial exposure is a likely driver of immune variation with age, a particularly interesting example is cytomegalovirus (CMV), a lifelong viral infection which has striking effects on the immune phenotypes of both humans and rhesus macaques (Sylwester et al., 2005). In our twin cohort, 16 MZ pairs were discordant for CMV seropositivity, and we compared their twin-twin correlations for all measurements to those of 26 CMV concordant (negative) MZ-pairs. Here we found that the CMV discordant MZ twins showed greatly reduced correlations for many immune cell frequencies such as effector CD8+ and gamma-delta T-cells (Figure 5A), as compared to CMV negative MZ-twins. The same was true for cell signaling responses, especially in response to IL-10 and IL-6 stimulation (Figure 5B), as well as the concentrations of these same cytokines in serum (Figure 5C). In general, the influence of CMV was very broad, affecting 119 of all 204 measurements (58%), dispersed throughout the immune network (Figure 5D), and illustrating how at least one type of microbial exposure can dramatically modulate the overall immune profile of healthy individuals.
Figure 5. Broad non-heritable influences in the healthy immune system.
(A) Twin-twin correlations (Spearman’s rank) for all cell frequency measurements made in CMV concordant negative (neg./neg.) MZ twin pairs (n=26 pairs), and CMV discordant (pos./neg.) MZ twin pairs (n=16 pairs). (B) Twin-twin correlations (Spearman’s rank) for cell signaling responses to cytokine stimulation, and (C) serum protein measurements between CMV neg/neg and CMV pos/neg MZ twin pairs. (D) 58% of all 126 nodes in the immune network model with reduced correlations in CMV pos/neg as compared to CMV neg/neg MZ-twin pairs.
Antibody responses to seasonal Flu-vaccines in adults have no detectable heritable component
Finally, we immunized all of the subjects with seasonal flu vaccines in the year of participation (2009, 2010 or 2011), and assessed antibody responses using a standard hemagglutination-inhibition (HAI) assay (Experimental procedures, section 6). We were surprised to find no detectable contribution from heritable factors on any of these vaccine responses (Table 1). As pre-existing antibodies are known to influence flu vaccine responses (Sasaki et al., 2008), we excluded subjects with a pre-vaccination titer above 40, but were still unable to find any heritable influences (Table 1). While preliminary due to a small sample size, this result suggests that responses to seasonal flu vaccines in healthy adults (median age: 44 years) are dominantly influenced by non-heritable factors, likely due to multiple previous vaccinations and/or infections involving this pathogen (Table 1).
Table 1. Influenza vaccine responses in adults are determined mainly by non-heritable factors.
Table of published heritability estimates for vaccine responses to various vaccines as well as the seasonal influenza vaccine responses analyzed in this study with or without the removal of subjects with a pre-vaccine antibody titer ≥40.
| Vaccine response | Age at vaccination | Heritability | Comment |
|---|---|---|---|
| Measles | 1–6 years a | 89% | (Tan et al., 2001) |
| Polio (oral vaccine) | <1 year | 60% | (Newport et al., 2004) |
| H. influenzae b | <1 year | 51% | (Lee et al., 2006) |
| Diphtheria | <1 year | 49% | (Newport et al., 2004) |
| Rubella | 1–6 years a | 46% | (Tan et al., 2001) |
| Tetanus | <1 year | 44% | (Newport et al., 2004) |
| Mumps | 1–6 years a | 39% | (Tan et al., 2001) |
| Combined Hepatitis A/B | 18–65 years | 36% (HAV)/61% (HBsAg) | (Höhler et al., 2002) |
| Hepatitis B virus | <1 year | 77%/91% | (Newport et al., 2004; Yan et al., 2013) |
| B/Brisbane/60/2008 | 12–82 yrs (median: 45) | 0% (< 20%) | |
| A/Cal/07/2009 (H1N1) | 12–82 yrs (median: 44) | 0% (< 20%) | |
| A/Perth/16/2009 (H3N2) | 12–82 yrs (median: 44) | 0% (< 20%) | |
| B/Brisbane/60/2008 | 13–77 yrs (median: 49) | 0% (< 20%) | Pre-vaccine titer < 40 |
| A/Cal/07/2009 (H1N1) | 12–76 yrs (median: 45) | 0% (< 20%) | Pre-vaccine titer < 40 |
| A/Perth/16/2009 (H3N2) | 12–82 yrs (median: 44) | 0% (< 20%) | Pre-vaccine titer < 40 |
Vaccine responses analyzed between 2–18 years of age
In summary our findings strongly suggest that a healthy human immune system adapts to non-heritable influences such as pathogens, nutritional factors and more, and that this overshadows the influences of most, although not all, heritable factors.
DISCUSSION
The vertebrate immune system consist of thousands of different components and the application of systems biology (Davis, 2008; Duffy et al., 2014; Li et al., 2013; Querec et al., 2009; Sekaly, 2008; Tsang et al., 2014) holds great promise as a way to understand the interactions between these during immune health and disease. Here we combine a classical twin study approach with the most recent advances in immune monitoring technologies to assess the balance between heritable and non-heritable influences on the functional units of the immune system, namely serum proteins and cell populations. In every category, we find that non-heritable influences dominantly influence 77% of all measurements (>50% of variance), and almost exclusively drive 58% of the measurements (>80% of the variance). Since most of measurements made in this study focus on the adaptive immune system, partly due to the availability of reagents, one possibility is that these low levels of heritability are related to the stochastic nature of antigen receptor recombination. Indeed, previous work has shown that there are significant differences in the immunoglobulin-sequence repertoires of MZ-twins (Glanville et al., 2011). But this is unlikely, since we find low heritability estimates also for many innate immune cell frequencies (NK cells, Monocytes and Granulocytes, Figure 1B) and differences within, and between CD4+ and CD8+ subsets of T cells, which share the same antigen receptor apparatus. Although this study is not powered to completely rule out all heritable influences on any of the measurements made, the overall dominance of non-heritable factors is independent of this. The low estimates of heritable influences are also not explained by technical noise as this is rigorously corrected for in our models (Figure S6A). We are also able to rule out that measurement variability over time is an important source of bias (Figure S6B). Therefore, the low heritability estimates for the majority of measurements cannot to be explained by either technical noise or biological variability over time.
Given the concordance rates for common autoimmune diseases between 25–50% (Cooper et al., 1999), and the many studies finding associations between specific genetic variants and immunological traits and disorders, we were surprised to find such a dominance by non-heritable factors on these functional units of the immune system. Several large population studies have associated specific loci with white blood cell counts showing some heritable influence, although the amount of variation explained is typically low (Okada et al., 2011; Reiner et al., 2011). A recent study by Orrù, et al estimated the heritability of 272 immune cell traits from a non-twin cohort of healthy individuals on the island of Sardinia. They found that 220 of these (~81%) had an estimated heritability lower than 50% (Orrù et al., 2013), which is comparable to our results. Although all the most heritable subpopulations identified by Orrù, et al, expressed the marker CD39, which was not analyzed in our study, a number of the cell populations gave quite similar values, although others were different, such as NKT-cells and central memory CD8+ T-cells (Table S7), possibly reflecting different environments (Bell and Spector, 2011), or a more diverse Palo Alto cohort vs. a less diverse Sardinian one.
It is important to note that the twins in our cohort are healthy and without any known immunological deficiencies. Interestingly, two serious immunodeficiency syndromes are caused by defects in the genes IRAK-4 and MyD88 and are associated with invasive bacterial infections due to defects in TLR or IL-1R signaling (Bernuth et al., 2008; Ku et al., 2007). Despite being associated with severe, often lethal infections in young children, both of these conditions improve significantly with age, starting in late childhood (Bernuth et al., 2008; Ku et al., 2007). Although this could be explained by developmental immaturity, an alternative explanation for the improvement with age could be that these children’s immune systems become more capable with greater environmental exposure.
A striking example of how broad the influence from one non-heritable factor can be is shown here for CMV, influencing 58% of all parameters measured in discordant MZ twins. These striking differences illustrate how non-heritable factors, alone or in combination, can affect the immune system broadly. We suggest that repeated environmental influences like herpes viruses and other pathogens, vaccinations and nutritional factors, cause shifts in immune cell frequencies and other parameters and, with time, outweigh most heritable factors. As an example, the lifelong need to control CMV seems to cause a broad shift in the magnitude and complexity of many cell subsets (Chidrawar et al., 2009; Wills et al., 2011), and approximately 10% of all T cells in CMV+ individuals can be directed against this virus (Sylwester et al., 2005). The microbiome also clearly has a major influence on the immune system (Hooper et al., 2012; Mazmanian et al., 2005), and shifts in its composition might cause significant changes in the immune system. Also interesting in the context of how infectious disease exposure can shape subsequent immunity are the influenza vaccine results, where we could not detect any heritable influence on the antibody responses to influenza vaccination (Table 1). This recalls the “original antigenic sin” hypothesis (Francis, 1960), where it was postulated that previous encounters with influenza strains strongly impact the response to a novel strain. But our finding contrasts with other vaccine studies in twins, most often performed in very young children and involving vaccines against pathogens less frequently encountered in the population (Table 1) (Jacobson et al., 2007). Specifically vaccines against mumps, measles, rubella(Tan et al., 2001), oral polio, tetanus and diphtheria vaccines (Newport et al., 2004) have all been shown to be strongly heritable (Table 1) in studies with young children. The one study that we know of from adult twins is a study of Hepatitis A/B vaccine responses and was conducted across a similar age range (18–65 years) as ours and reported a heritable influence of 36% for Hepatitis A and 61% for Hepatitis B antibody responses (Table 1). However, two other studies of Hepatitis B vaccine responses performed in young children showed much higher estimates of heritability of 91% (Yan et al., 2013) and 77% (Newport et al., 2004), respectively. In addition, responses to vaccines given at birth (oral polio vaccine (OPV) and Bacillus Calmette–Guérin (BCG)) are more heritable than even those administered only two Months later (Diphtheria and Tetanus) (Newport et al., 2004; O’Connor and Pollard, 2013). In addition, Evans et al analyzed twelve year old twins in Australia (Evans et al., 1999). Although there are only six broad categories of immune cell subsets that can be compared with our study and those of Orrù, et al, it is interesting that in all cases, the estimates of heritability in the 12 year old twins were higher than either ours or Orrù, et al., where the mean age is about 38 and 40 years, respectively (Table S7). These observations are consistent with our data (Figure 4), showing an increasing non-heritable influence on many variables with age, and suggests quite strongly that many, if not most of the less heritable traits that we measure here in our mostly adult population, may be much more heritable if measured in young children.
Before the advent of childhood vaccines, antibiotics and improvements in human hygiene, almost half of all children under five years of age died of infectious diseases. Casanova and colleagues have proposed a genetic theory of infectious disease to account for the inter-individual differences in susceptibility (Alcaïs et al., 2009). Our data, and the vaccine studies cited here, suggests that such genetic predisposition would be most pronounced in young children, but that later on the adaptive nature of the immune system is able to overcome many defects. This is similar to the hypothesis proposed by Alcais and colleagues, to explain the discrepancies in genetic lesions underlying the susceptibility to primary and secondary infections respectively (Alcaïs et al., 2010). Adaptations of the immune system with time could be the result of well known immune mechanisms, such as specific antibodies and T-cells or cross-reactive immunity (Su et al., 2013), or some as yet to be defined maturational process. We would also speculate that the immune system may have feedback mechanisms that allow it to skew its mix of cell types and functional properties in order to compensate for a given individual’s particular mix of gene polymorphisms and microbial exposures.
In summary we find that in an examination of many of the component parts of the immune system, as well as some response metrics, that much of the considerable variation in human beings is driven by non-heritable influences. This variation increases with age and is likely due in large part to exposure to pathogens and other microbes, as we see for CMV discordant MZ twins and in the responses to influenza vaccination. Lastly, we expect that other complex systems in higher organisms, such as the nervous system, will also show this pronounced influence of non-heritable factors, as there is also a need (and ability) of such systems, to adapt to environmental stimuli.
EXPERIMENTAL PROCEDURES
1. Twin cohort
In collaboration with the Twin Research Registry at SRI International (Krasnow et al., 2013), 105 healthy twin pairs were recruited over the years 2009, 2010 and 2011. The study protocol was approved by the Stanford University Administrative Panels on Human Subjects in Medical Research and written informed consent was obtained from all participants. We excluded anyone having received the seasonal Influenza vaccine in the last 6 months, anyone with known or suspected impairment of immunologic function, with clinically significant liver disease, diabetes mellitus treated with insulin, moderate to severe renal disease or any other chronic disorder including autoimmune diseases or severe hypertension. We also excluded anyone who had received blood products in the last 6 months and pregnant or lactating women. The complete inclusion/exclusion criteria are available (Data S1).
2. Blood sampling, PBMC preparation and zygosity testing
Blood samples were collected in heparinized vacutainer tubes by venipuncture at day 0 (and day +28 for HAI responses after seasonal influenza vaccination) at the Clinical and Translational Research Unit, Stanford University Hospital. Whole blood collected in sodium heparin tubes was either analyzed immediately using our whole-blood flow cytometry protocol below or enriched for PBMCs using 15ml of Ficoll-Paque PLUS (GE Health care, Inc. Piscataway, NJ) and frozen at −80° C over night, transferred to liquid nitrogen and stored until further analysis. Zygosity was determined by comparing 384 SNP-loci using a discriminatory DNA-polymerase and ligase assay (GoldenGate genotyping assay, Illumina, Inc, San Diego, CA) and were performed by IGenix (Bainbridge Island, WA). Twins were considered fraternal if similarities in DNA markers were below 99.0%. Twins of the same pair was almost exclusively analyzed in the same experimental batch, irrespective of technology used in order to minimize technical variation.
3. Immune cell phenotyping by mass cytometry and flow cytometry
All experiments were performed by the Human Immune Monitoring Center at Stanford University. For years 2010 and 2011, two million thawed PBMCs were stained without prior resting or fixation using a panel of 26 different metal-tagged probes to surface antigens and DNA (Data S2A). After repeated washes, cells were analyzed by mass cytometry (CyTOF™, Fluidigm, South San Francisco, CA) with the following instrument settings: High resolution mode analysis with cell length set to 10–75 pushes, a lower de-convolution threshold of 10 and instrument run in dual-count detection mode and noise reduction turned off. FCS3.0 files were manually analyzed using FlowJo v9.3 (TreeStar, Inc. Ashland OR) as shown in Figure S4. Year 2009 PBMC samples were similarly processed, but instead analyzed using a set of custom-made Lyoplates (BD Biosciences, San Jose, CA) covering 7 different antibody panels of fluorescently labeled antibodies (Data S2B–C). These samples were acquired using a LSRII flow cytometer (BD Biosciences) and similarly analyzed manually using the same FlowJo v9.3 (TreeStar, Inc.).
4. Immune cell signaling experiments
PBMCs were thawed in warm media, washed twice and resuspended at 0.5×106 viable cells/microliter. 200 microliter of cells were plated per well in 96-well deep-well plates. After resting for 1 hour at 37° C, cells were, stimulated by addition of 50 microliter solutions of cytokines: IFNα, IFNγ, IL-6, IL-7, IL-10, IL-2, or IL-21 (Data S2D) respectively and incubated at 37°C for 15 minutes. Cells were then immediately fixed in 1.6% paraformaldehyde, permeabilized with 100% cold methanol, and kept at −80° C overnight. Each well was bar-coded by a unique combination of Pacific Orange and Alexa-750 dye concentrations (Invitrogen/Life Technologies, Inc.), the cells were washed with FACS buffer (PBS supplemented with 2% FBS and 0.1% sodium azide), and stained with our phospho-Flow antibody panel (Data S2E). Finally cells were washed and resuspended in FACS buffer and 100,000 cells per stimulation condition were collected using DIVA 6.0 software on an LSRII flow cytometer (BD Biosciences). Data analysis was performed using FlowJo v9.3 (TreeStar, Inc.) as shown in Figure S5 and the Mean Fluorescence Intensity (MFI) of the 90th percentile was used for downstream analysis. All stimulated samples were compared to unstimulated control (baseline) samples and fold-changes calculated. Responses above 1.5-fold change as well as baseline MFI values were used for heritability estimates.
5. Serum protein quantification
Blood samples were centrifuged and stored at −80° C awaiting analysis. Human 51-plex were purchased from Affymetrix (Santa Clara, CA) and used according to the manufacturer’s recommendations with modifications as described below. Briefly, samples were mixed with antibody-linked polystyrene beads on 96-well filter-bottom plates and incubated at room temperature for 2 h followed by overnight incubation at 4°C. Room temperature incubation steps were performed on an orbital shaker at 500–600 rpm. Plates were vacuum filtered and washed twice with wash buffer, then incubated with biotinylated detection antibody for 2 h at room temperature. Samples were then filtered and washed twice as above and resuspended in streptavidin-PE. After incubation for 40 minutes at room temperature, two additional vacuum washes were performed, and the samples resuspended in Reading Buffer. Each sample was measured in duplicate. Plates were read using a Luminex 200 instrument with a lower bound of 100 beads per sample per protein. Each sample was measured in duplicate. Plates were read using a Luminex LabMap200 instrument with a lower bound of 100 beads per sample per protein per well. The Luminex LabMap200 outputs the fluorescence intensity of a given protein in a sample. For each well, we considered the median fluorescence intensity (MFI) for a serum protein as its abundance and averaged the MFI of these replicates. To ignore low abundance proteins, only measurements with mean concentrations higher than a negative control serum were included in our analysis.
6. Hemagglutination Inhibition (HAI) Assays
The HAI assay was performed on sera from day 0 and day 28-post influenza vaccination. Fold-changes day28/day0 was used for analyses. In one analysis, subjects with a pre-vaccination titer of 40 or more were excluded (Table 1). Serially diluted 25 microliter aliquots of serum samples in PBS were mixed with 25 microliter aliquots of virus, corresponding to 4 HA units, in V-bottom 96-well plates (Nunc, Rochester, NY, USA). These were then incubated for 15 min at room temperature. At the end of the incubation, 50 microliters of 0.5% chicken red blood cells (cRBC) were added and the plate incubated for one hour at room temperature and HAI activity was read as follows: I) Postive result: hemagglutination is present, the well is hazy with no cRBC button, or II) Negative result: hemagglutination is absent, the well is relatively clear with cRBC button. The HAI titer is defined as the reciprocal of dilution of last well that inhibits hemagglutination.
7. Structural equation modeling to estimate heritable and non-heritable influences
For all the measurements made, a structural equation modeling approach was used to estimate heritability (Rijsdijk and Sham, 2002). This classical twin modeling approach is based on the assumption that MZ twins are genetically identical while DZ twins share approximately 50% of their polymorphic genes, and that MZ and DZ twin pairs are equally similar with respect to their shared environmental influence. The covariance matrix for each measurement made in MZ and DZ twin pairs can then be decomposed into three parameters: I) Additive genetic parameter II) Common environmental parameter and III) Environmental parameter unique to one twin. A linear ACE model then estimates the contribution of each of these parameters by maximum likelihood. After correcting the E-parameter for technical measurement errors, as described in detail below, all parameters were scaled as a proportion of the total variance (A+C+E). All data was corrected for age and gender prior to ACE modeling. We performed resampling tests on all heritability estimates, using a jackknife resampling approach leaving one twin pair out of the calculation in each iteration, and using the mean values of all such iterations as our final estimate with 95% confidence intervals. All calculations were performed both using our own implementation of ACE-fitting in MATLAB, version 8.3, and using the openMX library version 1.4 running in R version 3.0.3. Both platforms were used in order to prevent any bias due to the software used.
8. Correction of model estimates for technical variability
For all measurements, standard samples were analyzed repeatedly (>17 times). These were either PBMC aliquots frozen at the same time and thawed for every experiment or pooled serum used as standards for Luminex and HAI assays. By calculating pooled variance estimates for these technical replicates and subtracting this from the E-component in our ACE models prior to normalization, we prevented the underestimation of heritability due to such stochastic measurement errors. This procedure overestimates the technical noise level of the actual twin samples by being collected across multiple batches, while twin samples compared were always analyzed within the same batch and after correction no relationship between technical variability and heritability estimates are seen (Figure S6A). We also assessed the biological variability over time in an unrelated cohort, by calculating coefficients of variance (CVs) from longitudinal samples drawn once yearly for 2–5 years. No relationship between longitudinal CVs and estimated heritability was found (Figure S6B).
9. Identification of pairwise dependencies between measurements and the creation of an immune network model
To identify meaningful relationships between measurements, we used the recently developed non-paranormal SKEPTIC approach (Liu et al., 2012), with a transformed Spearman/rank correlation matrix as input. To make the network model interpretable, we pursued sparse precision matrices with a graphical lasso approach penalizing non-zero entries in the matrix (Friedman et al., 2008). A zero entry in the precision matrix encodes conditional independencies of pairs of measurements given the state of all other measurements and is less sensitive to spurious indirect connections as compared to simple correlation analyses. To validate the inferred structural relationships, two tests were performed, 1) a permutation of samples on per phenotype basis was done to obtain null distributions for entries of the precision matrix. By repeatedly producing permuted samples and running our procedure on those samples, we obtain a distribution of precision matrices that is mostly dominated by very sparse, diagonal, matrices but also has occurrences of precision matrices that have non-zero entries. The non-zero entries thus obtained were false positive and hence we can estimate which entries obtained on the actual data exceed the size of these false positives. 2) Given relative infrequent occurrences of non-zero entries when fitting sparse precision matrices, we opted to estimate confidence intervals for each entry in the precision matrix. We deemed entries whose confidence interval contained 0 insignificant. To obtain these confidence intervals we performed bootstrap analysis, by resampling the real samples.
10. CMV serology
CMV serology was determined using a commercially available ELISA kit (CMV IgG, Gold Standard Diagnostics, Davis, CA) as per manufacturer’s instructions. Briefly, sera stored at −80°C were thawed to room temperature (20–25°C) and diluted 1:51 in kit diluent. Diluted samples were added to wells coated with CMV antigen from strain AD169 and incubated at room temperature for 30 minutes. Wells were washed and drained, followed by the addition of goat anti-human IgG antibodies labeled with calf alkaline phosphatase, and incubated at room temperature for 30 minutes. Wells were again washed and drained, followed by addition of p-nitrophenyl phosphate substrate, and incubated at room temperature for 30 minutes. After the addition of 0.5M trisodium phosphate stop solution, the absorbance of each well at 405nm was read and results analyzed using the manufacturer’s instructions.
11. MZ twin-twin correlations
All measurements performed in the youngest set of MZ-twin pairs (<20 years) and oldest MZ-twin pairs (>60 years) was extracted and Spearman’s rank correlation coefficients were calculated and compared between these independent groups for every measurement made. Similarly, CMV serologically negative/negative MZ-twin pairs were compared to CMV positive/negative (discordant twins).
Supplementary Material
Acknowledgments
The authors thank all members of the Davis and Y. Chien labs for inspiring discussions and the staff at the Human Immune Monitoring Center and the Stanford-LPCH Vaccine Program for sample collection, and data generation. Thanks also to Rob Tibshirani and Chiara Sabatti for input on statistical analyses. Twins were recruited from Twin Research Registry at SRI International, Menlo Park, CA and the authors wish to thank the following individuals: Mary McElroy, Lisa Jack, Ruth Krasnow, Jill Rubin, Dina Basin, Lucia Panini, and Marty Ritchey. The Stanford-LPCH program thanks Project Manager Sally Mackey, Research Nurses Susan Swope, Tony Trela and Cynthia Walsh, phlebotomist Michele Ugur, and CRAs Ashima Goel, Thu Quan, Kyrsten Spann, Sushil Batra, Isaac Chang and Raquel Fleischmann. The authors also wish to thank the contributions and commitment to science provided by the twins through their ongoing participation in the Registry and various research studies. Funds to the Twin Registry were provided through SRI’s Center for Health Sciences and through grants from the NIH including: DA011170, DA023063, AI057229, AI090019, and ES022153. This work was also supported by the Wenner-Gren Foundation and Sweden-America Foundation to PB, and NIH grants U19 AI090019 and U19 AI057229 to MMD as well as an NIH/NCRR CTSA award number UL1 RR025744 to the Clinical and Translational Research Unit, Stanford University Hospital.
Footnotes
AUTHOR CONTRIBUTIONS
PB and VJ performed all analyses. TG helped with the network analysis. SB and SSO provided input on heritability estimates. All data was generated by the Human Immune Monitoring Center under the direction of HTM except for the 2009 HAI data generated by DF and CMV serology by CJLA. GES directs the twin registry and provided input on the twin analyses and CLD was responsible for regulatory approvals, protocol design, study conduct and clinical data management. PB and MMD wrote the paper with input from coauthors.
Supplemental information includes 6 figures, 7 tables, and 2 Supplementary data documents.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcaïs A, Abel L, Casanova JL. Human genetics of infectious diseases: between proof of principle and paradigm. J Clin Invest. 2009;119:2506–2514. doi: 10.1172/JCI38111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaïs A, Quintana-Murci L, Thaler DS, Schurr E, Abel L, Casanova JL. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann N Y Acad Sci. 2010;1214:18–33. doi: 10.1111/j.1749-6632.2010.05834.x. [DOI] [PubMed] [Google Scholar]
- Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends Genet. 2011;27:116–125. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bernuth H, Picard C, Jin Z, Pankla R, Xiao H, Ku CL, Chrabieh M, Mustapha IB, Ghandil P, Camcioglu Y, et al. Pyogenic Bacterial Infections in Humans with MyD88 Deficiency. Science. 2008;321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidrawar S, Khan N, Wei W, McLarnon A, Smith N, Nayak L, Moss P. Cytomegalovirus-seropositivity has a profound influence on the magnitude of major lymphoid subsets within healthy individuals. Clin Exp Immunol. 2009;155:423–432. doi: 10.1111/j.1365-2249.2008.03785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi M, Forabosco P, Amadori A, Zamarchi R, De Silvestro G, Di Gianantonio E, Chieco-Bianchi L, Tenconi R. CD4 and CD8 T lymphocyte inheritance. Evidence for major autosomal recessive genes. Hum Genet. 1999;105:337–342. doi: 10.1007/s004399900140. [DOI] [PubMed] [Google Scholar]
- Cooper GS, Miller FW, Pandey JP. The role of genetic factors in autoimmune disease: implications for environmental research. Environ Health Perspect. 1999; 107(Suppl 5):693–700. doi: 10.1289/ehp.99107s5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MM. A Prescription for Human Immunology. Immunity. 2008;29:835–838. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Craen AJM, Posthuma D, Remarque EJ, van den Biggelaar AHJ, Westendorp RGJ, Boomsma DI. Heritability estimates of innate immunity: an extended twin study. Genes Immun. 2005;6:167–170. doi: 10.1038/sj.gene.6364162. [DOI] [PubMed] [Google Scholar]
- Dorshkind K, Montecino-Rodriguez E, Signer RAJ. The ageing immune system: is it ever too old to become young again? Nat Rev Immunol. 2009;9:57–62. doi: 10.1038/nri2471. [DOI] [PubMed] [Google Scholar]
- Duffy D, Rouilly V, Libri V, Hasan M, Beitz B, David M, Urrutia A, Bisiaux A, Labrie ST, Dubois A, et al. Functional analysis via standardized whole-blood stimulation systems defines the boundaries of a healthy immune response to complex stimuli. Immunity. 2014;40:436–450. doi: 10.1016/j.immuni.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Evans DM, Frazer IH, Martin NG. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res. 1999;2:250–257. doi: 10.1375/136905299320565735. [DOI] [PubMed] [Google Scholar]
- Evans DM, Zhu G, Duffy DL, Frazer IH, Montgomery GW, Martin NG. A major quantitative trait locus for CD4-CD8 ratio is located on chromosome 11. Genes Immun. 2004;5:548–552. doi: 10.1038/sj.gene.6364126. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis TJ. On the doctrine of original antigenic sin. Proceedings of the American Philosophical Society. 1960;104:572–578. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9:432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman D, Hejblum BP, Simon N, Jojic V, Dekker CL, Thiébaut R, Tibshirani RJ, Davis MM. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:869–874. doi: 10.1073/pnas.1321060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville J, Kuo TC, von Büdingen H-C, Guey L, Berka J, Sundar PD, Huerta G, Mehta GR, Oksenberg JR, Hauser SL, et al. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20066–20071. doi: 10.1073/pnas.1107498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen PK, Amos CI, Lee AT, Lu Y, Remmers EF, Kastner DL, Seldin MF, Criswell LA, Plenge RM, Holers VM, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41:820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhler T, Reuss E, Evers N, Dietrich E, Rittner C, Freitag CM, Vollmar J, Schneider PM, Fimmers R. Differential genetic determination of immune responsiveness to hepatitis B surface antigen and to hepatitis A virus: a vaccination study in twins. Lancet. 2002;360:991–995. doi: 10.1016/S0140-6736(02)11083-X. [DOI] [PubMed] [Google Scholar]
- Jablonski W. A contribution to theheredity of refraction in human eyes. Arch Augenheilk. 1922;91:308–328. [Google Scholar]
- Jacobson RM, Ovsyannikova IG, Targonski PV, Poland GA. Studies of twins in vaccinology. Vaccine. 2007;25:3160–3164. doi: 10.1016/j.vaccine.2007.01.048. [DOI] [PubMed] [Google Scholar]
- Krasnow RE, Jack LM, Lessov-Schlaggar CN, Bergen AW, Swan GE. The Twin Research Registry at SRI International. Twin Res Hum Genet. 2013;16:463–470. doi: 10.1017/thg.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nature Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- Ku C-L, von Bernuth H, Picard C, Zhang S-Y, Chang H-H, Yang K, Chrabieh M, Issekutz AC, Cunningham CK, Gallin J, et al. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity. J Exp Med. 2007;204:2407–2422. doi: 10.1084/jem.20070628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Newport MJ, Goetghebuer T, Siegrist CA, Weiss HA, Pollard AJ, Marchant A MRC Twin Study Group. Influence of genetic and environmental factors on the immunogenicity of Hib vaccine in Gambian twins. Vaccine. 2006;24:5335–5340. doi: 10.1016/j.vaccine.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, et al. Molecular signatures of antibody responses deriv… [Nat Immunol. 2014] - PubMed - NCBI. Nature Immunology. 2013;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Han F, Yuan M, Lafferty J, Wasserman L. High-dimensional semiparametric Gaussian copula graphical models. The Annals of Statistics. 2012;40:2293–2326. [Google Scholar]
- Maecker HT, Rinfret A, D’Souza P, Darden J, Roig E, Landry C, Hayes P, Birungi J, Anzala O, Garcia M, et al. Standardization of cytokine flow cytometry assays. BMC Immunology. 2005;6:13. doi: 10.1186/1471-2172-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Morahan G, Huang D, Wu M, Holt BJ, White GP, Kendall GE, Sly PD, Holt PG. Association of IL12B promoter polymorphism with severity of atopic and non-atopic asthma in children. Lancet. 2002;360:455–459. doi: 10.1016/S0140-6736(02)09676-9. [DOI] [PubMed] [Google Scholar]
- Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport MJ, Goetghebuer T, Weiss HA, Whittle H, Siegrist CA, Marchant A MRC Gambia Twin Study Group. Genetic regulation of immune responses to vaccines in early life. Genes Immun. 2004;5:122–129. doi: 10.1038/sj.gene.6364051. [DOI] [PubMed] [Google Scholar]
- O’Connor D, Pollard AJ. Characterizing Vaccine Responses Using Host Genomic and Transcriptomic Analysis. Clinical Infectious Diseases. 2013;57:860–869. doi: 10.1093/cid/cit373. [DOI] [PubMed] [Google Scholar]
- Okada Y, Hirota T, Kamatani Y, Takahashi A, Ohmiya H, Kumasaka N, Higasa K, Yamaguchi-Kabata Y, Hosono N, Nalls MA, et al. Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population. PLoS Genet. 2011;7:e1002067. doi: 10.1371/journal.pgen.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrù V, Steri M, Sole G, Sidore C, Virdis F, Dei M, Lai S, Zoledziewska M, Busonero F, Mulas A, et al. Genetic Variants Regulating Immune Cell Levels in Health and Disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature Immunology. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner AP, Lettre G, Nalls MA, Ganesh SK, Mathias R, Austin MA, Dean E, Arepalli S, Britton A, Chen Z, et al. Genome-wide association study of white blood cell count in 16,388 African Americans: the continental origins and genetic epidemiology network (COGENT) PLoS Genet. 2011;7:e1002108. doi: 10.1371/journal.pgen.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijsdijk FV, Sham PC. Analytic approaches to twin data using structural equation models. Brief Bioinformatics. 2002;3:119–133. doi: 10.1093/bib/3.2.119. [DOI] [PubMed] [Google Scholar]
- Sasaki S, He XS, Holmes TH, Dekker CL, Kemble GW, Arvin AM, Greenberg HB. Influence of Prior Influenza Vaccination on Antibody and B-Cell Responses. PLoS ONE. 2008;3:e2975. doi: 10.1371/journal.pone.0002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekaly RP. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? Journal of Experimental Medicine. 2008;205:7–12. doi: 10.1084/jem.20072681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan PL, Jacobson RM, Poland GA, Jacobsen SJ, Pankratz VS. Twin studies of immunogenicity determining the genetic contribution to vaccine failure. Vaccine. 2001;19:2434–2439. doi: 10.1016/s0264-410x(00)00468-0. [DOI] [PubMed] [Google Scholar]
- Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, Wang E, Olnes MJ, Narayanan M, Golding H, et al. Global Analyses of Human Immune Variation Reveal Baseline Predictors of Postvaccination Responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeda SA, Wyss-Coray T. The circulatory systemic environment as a modulator of neurogenesis and brain aging. Autoimmun Rev. 2013;12:674–677. doi: 10.1016/j.autrev.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Wills M, Akbar A, Beswick M, Bosch JA, Caruso C, Colonna-Romano G, Dutta A, Franceschi C, Fülöp T, Gkrania-Klotsas E, et al. Report from the second cytomegalovirus and immunosenescence workshop. Immun Ageing. 2011;8:10. doi: 10.1186/1742-4933-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Cai W, Cao F, Sun H, Chen S, Xu R, Wei X, Shi X, Yan W. Genetic effects have a dominant role on poor responses to infant vaccination to hepatitis B virus. J Hum Genet. 2013;58:293–297. doi: 10.1038/jhg.2013.18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.