Abstract
Human primary neural tissue is a vital component for the quick and simple determination of chemical compound neurotoxicity in vitro. In particular, such tissue would be ideal for high-throughput screens that can be used to identify novel neurotoxic or neurotherapeutic compounds. We have previously established a high-throughput screening platform using human induced pluripotent stem cell (iPSC)-derived neural stem cells (NSCs) and neurons. In this study, we conducted a 2,000 compound screen with human NSCs and rat cortical cells to identify compounds that are selectively toxic to each group. Approximately 100 of the tested compounds showed specific toxicity to human NSCs. A secondary screen of a small subset of compounds from the primary screen on human iPSCs, NSC-derived neurons, and fetal astrocytes validated the results from >80% of these compounds with some showing cell specific toxicity. Amongst those compounds were several cardiac glycosides, all of which were selectively toxic to the human cells. As the screen was able to reliably identify neurotoxicants, many with species and cell-type specificity, this study demonstrates the feasibility of this NSC-driven platform for higher-throughput neurotoxicity screens.
1. Introduction
Pluripotent stem cells (PSCs) have the unique potential to be differentiated to any cell type. This capacity makes them attractive starting materials for the generation of large quantities of differentiated cells that can be utilized in cellular transplantation therapies and high-throughput compound screens. The last few decades of basic research have yielded enormous insights on the development of the central nervous system and the neurons and glia that are its primary components. This research has been leveraged in vitro to differentiate neural stem cells (NSCs), neurons, astrocytes, and oligodendrocytes from PSCs (Chambers et al., 2009; Hu et al., 2009; Swistowski et al, 2009; Shaltouki et al, 2013; Yan et al, 2013) thus making it possible to conduct research on cells of human origin to study fundamental processes in normal and pathological nervous system function. Although animal models have been and will continue to be important for such work there are critical differences in nervous system development not only between humans and rodents but also between human and non-human primates (Rice and Barone Jr, 2000). Because of this, the availability of human NSCs and their differentiated derivatives is critical for proper understanding of human nervous system biology.
A field in which NSCs and their neural derivatives could be particularly valuable is predicting the neurotoxicity of particular chemicals in the human nervous system. Most neurotoxicity assays are currently performed either in animal models or in vitro with immortalized tumor cell lines. The animal models, as mentioned above, may not truly replicate human physiology. Additionally, whole animal experiments are expensive, labor and time intensive and not amenable to high-throughput screens. In vitro models bypass these issues but require use of tumor cell lines of neural origin and thus do not reflect a tissue state that represents normal human physiology. Because of these limitations it is important to develop assay platforms so that future neurotoxicity studies can test large numbers of compounds at greater speed and lower cost in neural cells that are not of tumorigenic origin (National Research Council, 2007; Llorens et al., 2012). As such, human NSCs represent an excellent alternative that offers the capability for high-throughput toxicity testing on a wide array of neural cell types (Breier et al., 2010). The ability to screen NSCs and neural cell types provides an opportunity to not only predict neurotoxicity of compounds at high-throughput but also identify drugs that are selectively toxic to NSCs. With recent findings indicating that many glioblastoma tumors are seeded by NSC-like tumor stem cells that are resistant to currently used therapies compounds specifically killing NSCs could be tested for their clinical efficacy (Cho et al., 2013).
We have previously reported on the development of a screening platform that utilizes PSC-derived NSCs as the starting cells in a high-throughput assay (Efthymiou et al., 2014). This platform showed high reproducibility for viability assays on neurons differentiated from PSC-derived NSCs. We have also previously discovered small compounds that eliminate human NSCs but not dopaminergic neurons in a screen of a 720 compound library (Han et al., 2009). Based upon these earlier results we decided to assay a 2,000 compound library for toxicity against human NSCs and mixed cultures of rat cortical cells that we have previously studied (Efthymiou et al., 2014; Haughey et al., 2004; Nath et al., 2012).
Compounds that were toxic to NSCs but not mixed cultures of rat cortical neurons were validated and tested against human iPSCs, NSC-differentiated neurons, and fetal astrocytes to further determine the specificity of their toxicity. The screen identified ~100 compounds toxic to human NSCs but not mixed rat cortical neurons. One class of compounds that we identified as being particularly toxic to human but not rat neural cells was cardiac glycosides. While there is an extensive literature on the anti-tumorigenic effects of cardiac glycosides in a variety of cancers including glioblastoma, to our knowledge this is the first report demonstrating their toxicity to NSCs (Badr et al, 2011; Joshi et al., 2011; Slingerland et al., 2013; Lee et al., 2014). The findings described in this paper could be of particular relevance for both performing future neurotoxicology screens in an effort to predict more accurately the neurotoxicity profiles of select drugs and for the identification of selective NSC toxicants that could have potential therapeutic value in the treatment of glioblastoma.
2. Methods
2.1. Cell culture and maintenance
Targeted and parent line NSCs from the NCRM1 line were cultured and maintained as previously described (Efthymiou et al., 2014). Briefly, the cells were maintained in neural stem cell medium (NSCM) consisting of Neurobasal base medium supplemented with GlutaMAX, NEAA, 1× B27 (all from Life Technologies, Grand Island, NY, USA), and 10 ng/mL bFGF (Peprotech, Rocky Hill, NJ, USA). Media was changed every other day and cells were passaged using Accutase about every 4 days.
Neuronal differentiation was accomplished as previously described (Efthymiou et al., 2014) with some adjustments. Briefly, NSCs were passaged to about ~70% confluence in 6-well plates and cultured in neuronal differentiation medium (NDM). NDM consisted of DMEM/F12, GlutaMAX, 1% BSA, 1× hESC supplement, BDNF at 10ng/mL, and GDNF at 10 ng/mL (growth factors from Peprotech). NDM was changed every two days.
Primary rat neuronal cultures were prepared from Sprague-Dawley (SD) rat fetal brains (Charles River Laboratories, Wilmington, MA, USA) at 18 days of gestation. Brains were harvested from 10 to 15 pups, all meninges were removed and the cortex was dissected. Tissues were dissociated by gentle trituration with a fire polished glass pipette in calcium-free Hank’s balanced salt solution. The single cell suspension was centrifuged at 200×g and re-suspended in minimal essential medium containing 10% (v/v) heat-inactivated fetal bovine serum and 1% (v/v) antibiotic and antimycotic solution (Sigma, St Louis, MO, USA). Cells were allowed to attach for 3 hours before the media was replaced with serum-free neurobasal medium containing 5% fetal bovine serum, 2% (v/v) B-27 supplement (Gibco, Rockville, MD, USA) and 1% antibiotic and antimycotic solution. Rat cortical neuronal cultures were used between 7–12 days in vitro and were plated in 96-well plates at a density of 4× 105 cells per ml. This mixed rat cortical cultures consisted of 40–45% β–III tubulin expressing neurons, 50–55% GFAP expressing astrocytes and about 1% microglia (Haughey et al., 2004).
Fetal astrocytes and growth medium were purchased from Lonza (Walkersville, MD) and cultured according to the manufacturer’s instructions.
An episomally generated iPSC line (NCRM5) was cultured in Essential 8 Medium (Life Technologies) on a Matrigel (Corning, Corning, NY) coated surface (Chen et al, 2011). Medium was supplemented with ROCK-inhibitor after each passage.
Characterization of the mixed rat cortical cultures, human NSCs, NSC-derived neurons, and fetal astrocytes has been described previously and summary of microarray data for specific markers of these cells is summarized in Supplementary Table 1 (Efthymiou et al., 2014, Malik et al., 2014, Haughey et al., 2004).
2.2. Screening Cells
The set-up for the screen is schematically described in Figure 1. The Microsource Discovery Spectrum collection (Supplementary Table 2) contains 2,000 compounds, with about 50% known drugs (800 USP/USAN + 200 INN & BAN & JAN), 30% natural products (580 compounds) and about 20% other bioactive compounds, many with prior human exposure (420 compounds). The compound library consisted of 25 plates. Each plate was screened on the same day with each compound tested against the cell type of interest in triplicate on three different 96-well plates. The compound library is plated in 10 mM DMSO and was diluted 1,000-fold in the appropriate cell culture medium to achieve a concentration of 10 µM for the primary screen. For secondary screens including dose responses fresh compounds were purchased, dissolved to an appropriate concentration in DMSO, and diluted to the indicated concentrations with cell culture medium with a 24 hour treatment just like the primary screen. The positive controls for neurotoxicity included 3 mM 3-nitropropionic acid, a fungal toxin that inhibits mitochondrial respiration and is known to be toxic to neurons (Ludolhp et al., 1991), which showed moderate toxicity to the rat cultures (ranging from 25–40%), but less to the NSCs (5–15%). The 0.1% DMSO vehicle control was compared to media alone as a negative control for these assays. Percentage toxicity was calculated by comparing treated cells to the 0.1% DMSO vehicle control.
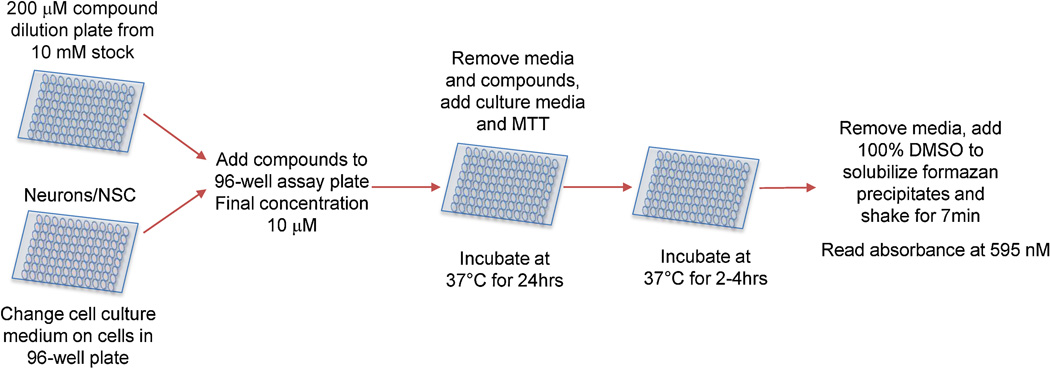
Figure 1.
Flow chart depicting how NSCs and mixed rat cortical cultures were assayed for toxicity against the 2,000 compound Spectrum collection.
NSCs, iPSCs, and fetal astrocytes were passaged 4–5 days before screening onto Geltrex-coated 96-well plates at a ratio of one million cells per plate. The initial plating density was ~10,000 cells per well grown in 100 µl of NSCM. NSCM was changed every other day, iPSC medium daily, and fetal astrocyte medium every third day.
Rat cultures were prepared as described above and plated onto 96 well plates for assay.
Neurons were first cultured in bulk as described above. On day 4–5 of neuronal differentiation, cells were passaged using Accutase and plated on Geltrex-coated 96-well plates at a ratio of ~3 million cells per plate (~30,000cells/well in 100 µl of NDM). NDM was then changed every two days for the remaining five days by removing 30% of the medium and replacing it with 50% of fresh NDM. Screens were performed on day 10 of neuronal differentiation. Day 10 was chosen because we have found that there is very strong beta-III-tubulin (TuJ1) staining (>90% cells TuJ1+) by day 10 and the intensity of staining and percentage of cells stained does not change by day 14 (data not shown). At this stage, there is expression of neuron-specific markers including GAD1, MYT1, Neurog2), and decreased expression of NSC markers (MSI1, LIN28 and LIN28B). There was continued expression of Nestin, Prom1 and Sox2, which although often used as markers of NSC status, have also been reported to be expressed in neurons and astrocytes (Hendrickson et al., 2011; Cavalloro et al., 2008). See Supplemental Table 1 for more details.
On screening days, cells received a fresh media change and were screened on anautomated system containing a Thermo CRS F3 robotic gripper arm, Cytomat, delidder, Perkin Elmer Janus Mini-liquid handler, BioTek EL-406 plate washer and Thermo Varioskan Flash multimode reader. The compound plates were diluted in NSC assay buffer prior to addition to the cell cultures at a final concentration of 10 µM. The cultures were incubated at 37 °C in humidified 5% CO2 atmosphere in the Cytomat for 24 hours. Afterward, the media was removed and replaced with fresh assay buffer. The cells were then incubated with 5 mg/mL MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) for 2 hours to assay for cell viability (Mosmann, 1983).
All compounds were screened in triplicate and a subset of the hits from the primary screen were validated on NSCs and tested against other cell types. Dose responses were then performed for selected compounds. All data are represented as mean ± SEM and analyzed by one-way analysis of variance (ANOVA). Group-wise post hoc comparisons were assessed by the Tukey method of multiple comparison tests, with statistical significance at p<0.05. For the dose response analyses, the differences between groups were evaluated by 2-way ANOVA, comparing the effect of dose and the effect of the 2 different cell types and the interaction of the dose effect on the cells. Statistical significance was set with an alpha of p<0.05.
2.3. Live imaging of treated cells
An NSC line ubiquitously expressing tdTomato was grown in 96-well plates at 10,000 cells per well for 48 hours prior to the addition of compounds of interest. The reporter line was generated by inserting a tdTomato cDNA driven by the chicken beta-actin promoter to the AAVS1 safe harbor locus by TALEN-mediated targeting into the NCRM1 NSC line (Luo et al., 2014). The plate was cultured in NSC media and the drug treatments were made at a concentration of 10 µM. Time lapse live cell imaging of the NSC was accomplished at 40× magnification on the GE INCell Analyzer 2000 imager, acquiring images of four fields per well every hour for up to 60 hours. GE Developer Toolbox was used to quantitate cell number, cellular area, fluorescence intensity and cellular elongation from NSC or neuronal cultures that contained tdTomato fluorescence throughout these cells. Cellular elongation was calculated as the ratio of the length of the cell’s minor axis in um compared to the target’s major axis (the longer of two perdicular axes of symmetery) in um. Four fields per well were acquired by the INCell Analyzer 2000 instrument every hour for the time lapse experiments. These fluorescent images were segmented, and analyzed at a medium sensitivity setting of 60. Visual inspection of 10–12 random images showed that we captured and measured more than 95% of the cells in these fields.
3. Results
3.1. Screening human NSCs and mixed rat cortical cultures with the Spectrum Collection Library
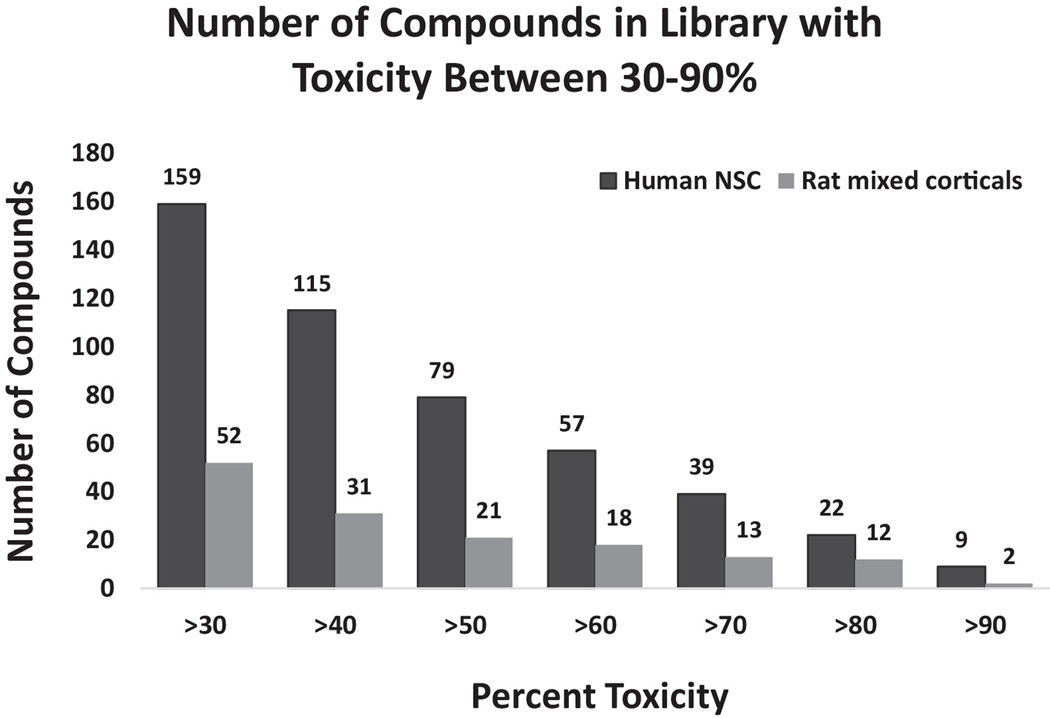
We screened human NSCs that have been previously described and characterized and mixed cultures of rat cortical neurons with a 2,000 compound Spectrum Collection library (Efthymiou et al., 2014). Rat cortical neurons were chosen for comparison because of our previous experience in obtaining robust and reproducible results with these cells in these types of screens (Nath et al., 2012; Kehn-Hall et al., 2011; Xu et al. 2011; Steiner et al, 2007; Caporello et al., 2006). Briefly, an automated primary screen was performed in triplicate in 96 well plates with drug treatment at 10 µM for 24 hours and viability was assessed by an MTT assay. When examining toxicity in 10% windows between 30–90% there were a greater number of compounds toxic to human NSCs than mixed rat cortical cultures in all the windows. We identified 79 compounds that had >50% toxicity against NSCs and 21 with the same level of toxicity against rat neuronal cultures (Figure 2). When the stringency for toxicity was decreased to >40% or >30%, 115 and 159 compounds, respectively, were identified for NSCs and 31 and 52, respectively, for mixed rat cultures (Figure 2). These data suggest that human NSCs were much more sensitive to the compounds found in this library than mixed rat cortical cultures. The variability, measured by standard error, in the screen was also low ranging from 0.5–4% for human NSCs and 1–5% for mixed rat cortical cells. Forty-three compounds from the library were selected for re-evaluation in a secondary screen. In the primary screen 18 of these were toxic to human NSCs only, 14 to human NSCs and mixed rat cortical neurons, 1 to rat cortical neurons only and 10 to neither (Supplementary Table 3).The validation rate for these compounds was 82.6% suggesting that the findings of the primary screen were reliable (Table 1).
Figure 2.
Toxicity window at 10% intervals from 30–90% displaying cumulative number of compounds that are toxic to human NSCs and rat mixed corticals in each interval.
Table 1.
The percent of selected compounds from the primary screen that were validated upon re-evaluation in NSCs and rat mixed cortical cultures.
| Primary screen | %Validated |
|---|---|
| Positive NSC | 74.2 |
| Negative NSC | 83.3 |
| Positive rat mixed cortical | 81.3 |
| Negative rat mixed cortical | 92.6 |
| Overall | 82.6 |
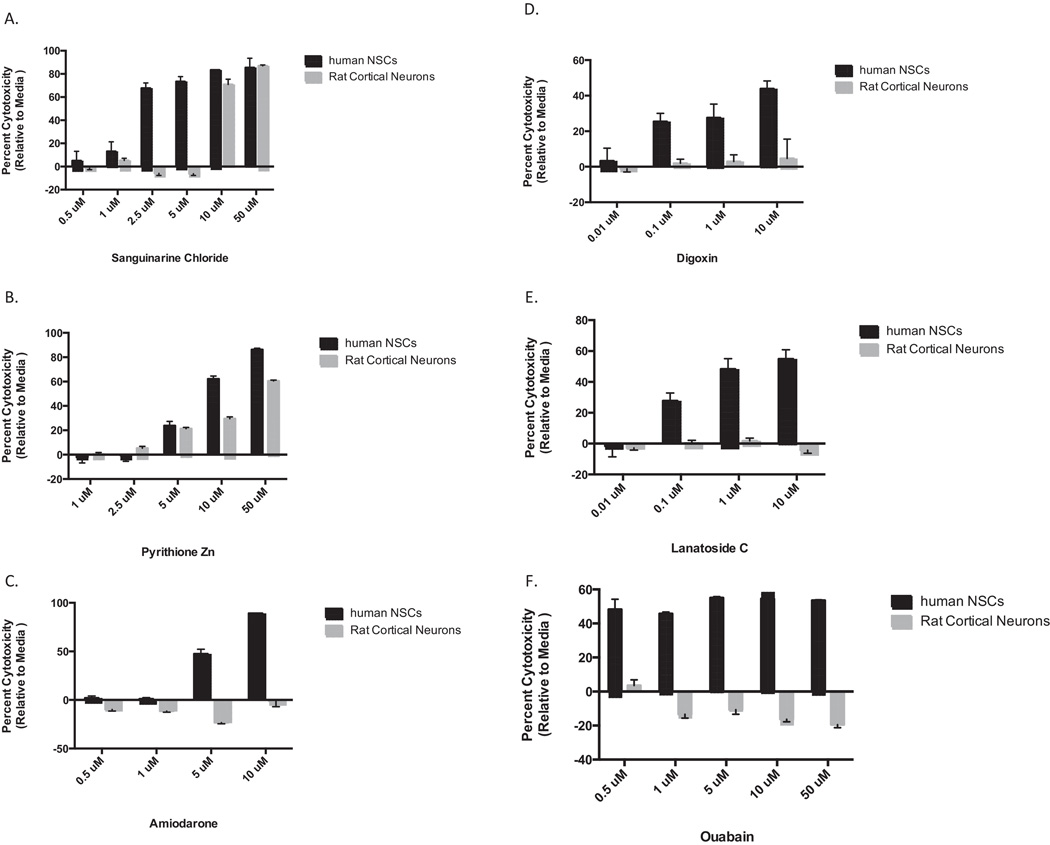
We performed dose responses on several compounds found to be toxic to further validate the results of the primary screen. The results of six of these: sanguinarine, pyrithione zinc, amiodarone, digoxin, lanatoside C, and ouabain are shown in Fig 3A–F. Digoxin, lanatoside C, and ouabain are all cardiac glycosides and were found to be toxic to human NSCs but not rat mixed cultures in the primary screen. It should be noted that the there was a total of 17 cardiac glycosides or aglycones in the library and all had similar toxicity profiles against human NSCs and rat cells. Of the six drugs for which dose responses are shown all showed toxicity to NSCs at doses of 10 µM or above, with only sanguinarine and pyrithione zinc being toxic to the rat cells. Although the cardiac glycosides were less toxic to NSCs at higher doses than the non-cardiac glycosides they were the only compounds to exhibit toxicity at sub-micromolar doses with a relatively flat dose response curve across the tested concentrations. A 2-way ANOVA analysis found a statistically significant effect of dose on each compound which differentiated between human NSCs and rat mixed cortical cultures (Supplementary Table 4).
Figure 3.
Dose response curves for selected compounds that were toxic to NSCs in primary screen. A range of concentrations was tested in triplicate and percent viability calculated relative to a DMSO control for (A) sanguinarine, (B) pyrithione zinc, (C) amiodarone, (D) digoxin, (E) lanatoside C, and (F) ouabain.
3.2. Toxic compounds affect NSC morphology in different ways
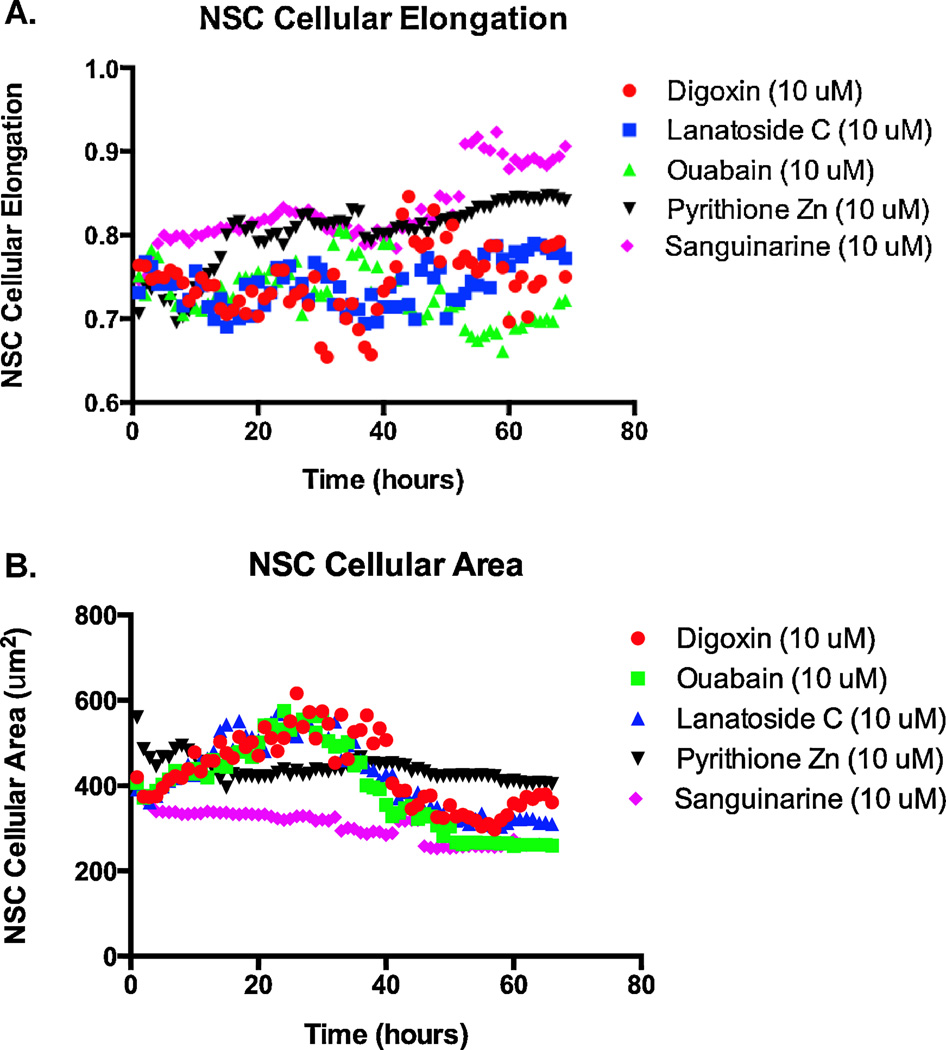
In an effort to observe changes to cellular morphology upon drug treatment we performed live cell imaging of human NSCs that ubiquitously express tdTomato. These NSCs were cultured in the presence of 10 µM of pyrithione zinc, sanguinarine or the cardiac glycosides digoxin, lanatoside C, and ouabain over a 72-hour time period. NSCs exposed to these compounds showed distinctive changes in morphology leading to cell death. Live cell imaging revealed that NSCs treated with pyrithione zinc or sanguinarine appeared to elongate and then die whereas treatment with cardiac glycosides resulted in the swelling and subsequent bursting of NSCs. Quantitative analysis confirmed these observations as pyrithione zinc and sanguinarine treated NSCs begin to elongate ~40 hours after treatment while cardiac glycoside-treated cells showed no signs of elongation (Figure 4A). Conversely, cardiac glycoside treated NSCs unlike those exposed to non-cardiac glycosides showed a spike in cellular area ~25 hours after treatment and continued to do so for ~15 hours before showing a rapid decrease in area (Figure 4B).
Figure 4.
Comparison of cellular (A) elongation and (B) area between cardiac glycosides and pyrithione zinc and sanguinarine. Total NSC cellular elongation and area was calculated for three cardiac glycoside and two non-cardiac glycoside drugs each hour and plotted for ~70 hours.
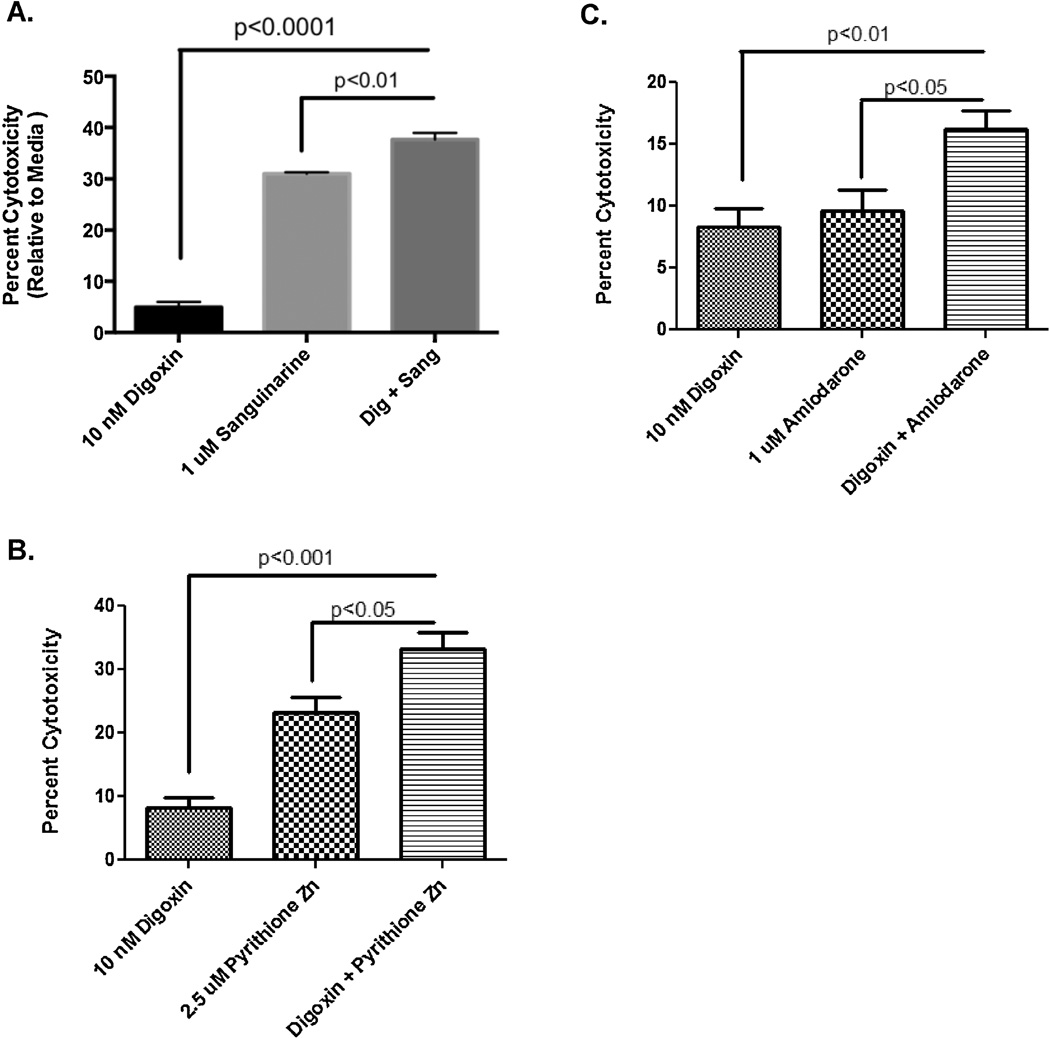
As live imaging indicated different morphological responses for the NSC treated with cardiac glycosides compared to sanguinarine and pyrithione zinc, we hypothesized that co-application of subtoxic doses of digoxin and sanguinarine, pyrithione zinc, or amiodarone as determined by the dose response would result in increased toxicity to human NSCs. We found that there were indeed statistically significant differences to cell death when compounds are co-administered compared to addition of only one compound (Figure 5). This is not surprising given that the live imaging data suggested that cardiac glycosides initiated cell death by different means than the other drugs. These data imply that toxic compounds can be combined at subtoxic doses to enhance NSC death.
Figure 5.
Combination treatment of NSCs and rat cortical mixed cultures with a cardiac glycoside and non-cardiac glycoside. NSCs were treated for 48 hours either alone or in combination with (A) digoxin and sanguinarine, (B) digoxin and pyrithione zinc, (C) digoxin and amiodarone. There were statistically significant differences between lone and co-administration for each of the three combinations.
3.3. Toxicity of selected compounds against other cell types
In an effort to determine the toxicity of selected compounds against other cell types, the 43 compounds that were validated on NSCs and mixed rat cortical cultures were re-tested against human cells from an iPSC line, NSC-derived neurons, and commercially purchased fetal astrocytes (Supplementary Table 5). Compounds were selected across the toxicity spectrum against human NSC in the primary screen ranging from high toxicity (>90%) to little or no toxicity (1–30%). As not all compounds are sold by vendors, commercial availability also played a role in which compounds were re-tested. The compounds were tested under the same conditions as the primary screen, at 10 µM for 24 hours. A summary of whether these compounds were toxic to iPSCs, NSC-derived neurons, fetal astrocytes, or mixed rat cortical cultures can be found in Table 2. The only compound that was toxic to only one cell type was thyroxin, which selectively killed astrocytes at a modest rate (35.2%). We found that 13 compounds were toxic to all individual cell culture types tested, eight of which were also toxic to mixed rat cortical cells. Interestingly, the five compounds that were non-toxic to rat cells were all cardiac glycosides, suggesting this family has the ability to kill pluripotent cells and a broad range of human neural cells. Additionally, eight compounds were toxic to all the human cells tested except neurons and five of these were also toxic to the rat cells. These compounds could be useful reagents for the purification of neuronal cells in neuronal differentiation protocols from NSCs or iPSCs. Amiodarone, a compound we previously found to be toxic to PSCs and NSCs but not dopaminergic neurons, was validated in this screen as being toxic to PSCs and NSCs but not cortical neurons differentiated from NSCs. Additionally, we found that amiodarone was also toxic to fetal astrocytes. By a literature search we found that 29 of 43 of the compounds have been shown to cross the blood-brain-barrier and 13 compounds have shown some neurotoxicity to human cells (Supplementary Table 5). Of these 13 our screen identified 11 as being neurotoxic to human NSCs with only norcantharidin and erythromycin not exhibiting toxicity to human NSCs. Overall these results suggest that there is a degree of cell specificity in toxicity amongst some of the drugs in this subset of compounds and that the screen performs well in recognizing compounds that have previously been shown to have neurotoxicity in humans.
Table 2.
Compounds that upon rescreening were >30% toxic to the listed human and rat cell types.
| Compound | iPSCs | NCRM-1 Neurons |
Fetal Astrocytes |
NSC | Mixed Rat Corticals |
|---|---|---|---|---|---|
| Actinonin | No | No | No | No | No |
| B-Estradiol | No | No | No | No | No |
| Cinchonidine | No | No | No | No | No |
| Colchicine | No | No | No | No | No |
| D-(−)-Salicin | No | No | No | No | No |
| Erythromycin Esolate | No | No | No | No | No |
| Imperatorin | No | No | No | No | No |
| Kynurenic Acid | No | No | No | No | No |
| Mechlorethamine | No | No | No | No | No |
| Myricetin | No | No | No | No | No |
| Neostigmine | No | No | No | No | No |
| Nilutamide | No | No | No | No | No |
| Norcantharidin | No | No | No | No | No |
| Piperine | No | No | No | No | No |
| Quinidine | No | No | No | No | No |
| Quinine Sulfate | No | No | No | No | No |
| Thyroxin | No | No | Yes | No | No |
| Chlorhexidine | No | No | Yes | Yes | Yes |
| Amiodarone | Yes | No | No | Yes | No |
| Androsterone Acetate | Yes | No | Yes | Yes | No |
| Astemizole | Yes | No | No | Yes | No |
| Embelin | Yes | No | Yes | Yes | No |
| Mebendazole | Yes | No | Yes | Yes | No |
| Triptolide | Yes | No | No | Yes | No |
| Acriflavinium Cl | Yes | No | Yes | Yes | Yes |
| Benzalkonium Cl | Yes | No | Yes | Yes | Yes |
| Benzethonium Cl | Yes | No | Yes | Yes | Yes |
| Celastrol | Yes | No | Yes | Yes | Yes |
| Dequalinium Cl | Yes | No | No | Yes | Yes |
| Thiomersal | Yes | No | Yes | Yes | Yes |
| Digitoxin | Yes | Yes | Yes | Yes | No |
| Digoxin | Yes | Yes | Yes | Yes | No |
| Lanatoside C | Yes | Yes | Yes | Yes | No |
| Oleandrin | Yes | Yes | Yes | Yes | No |
| Ouabain | Yes | Yes | Yes | Yes | No |
| Anisomycin | Yes | Yes | Yes | Yes | Yes |
| Cetylpyridinium Cl | Yes | Yes | Yes | Yes | Yes |
| Gambogic Acid | Yes | Yes | Yes | Yes | Yes |
| Gentian Violet | Yes | Yes | Yes | Yes | Yes |
| Gramicidin | Yes | Yes | Yes | Yes | Yes |
| Phenylmercuric acetate | Yes | Yes | Yes | Yes | Yes |
| Pyrithione Zn | Yes | Yes | Yes | Yes | Yes |
| Sanguinarine | Yes | Yes | Yes | Yes | Yes |
4. Discussion
High-throughput screens with pluripotent stem cells (PSCs) and their differentiated derivatives offer great promise for the identification of new compounds that can be used in therapies and serve as effective tools in differentiation protocols. Recently published screens in normal and patient PSCs and motor neurons differentiated from ALS patient iPSCs have identified compounds that could make a large impact in both translational and basic research (Discords et al, 2008; Lee et al, 2012; Ben-David et al, 2013; Yang et al., 2013). We have been interested in performing screens in human neural stem cells (NSCs) to determine if they are amenable to high-throughput neurotoxicity screens and whether they can be used to identify compounds that selectively kill NSCs.
Since our initial screen involving PSCs and dopaminergic neurons, we have developed an improved platform to conduct high-throughput and phenotypic screens in NSCs and NSC-derived cortical neurons (Efthymiou et al., 2014). We have now utilized this platform to screen a 2,000 compound library against human NSCs and a mixed culture of rat cortical neurons. The resulting screen was able to identify ~100 drugs that were toxic to human NSCs but not mixed rat cortical cultures. Some of these compounds, including amiodarone, were also identified in our earlier screen (Han et al., 2009). The data in the primary screen were highly reproducible as indicated by the small variation in triplicate treatments with standard error ranging from 0.5–4% for human NSCs and 1–5% for mixed rat cortical cultures, and a high degree of validation for selected compounds in secondary screens (>80%). We were also able to test the toxicity of hits against an iPSC line, fetal astrocytes, and neurons differentiated from the same NSC line that was tested in the primary assay to determine if drugs toxic to NSCs also killed these cells. Furthermore, we were able to perform live cell imaging on a reporter NSC line expressing tdTomato ubiquitously and assess the morphological changes preceding cell death. Of the 43 compounds we retested 13 have been shown to have some neurotoxicity in humans and 11 of these were toxic to neural cells used in our screen suggesting the screen has an excellent capacity to identify neurotoxic compounds. Although human neurotoxicity data is available for only a small subset of the compounds that were screened, the ability to recognize 11/13 (85%) known neurotoxicants indicates that the screen has good sensitivity in detecting compounds that are toxic to neural cells. The specificity of this screen (correct identification of negatives) is harder to assess because the published literature is heavily focused on compounds that are toxic often not reporting those that have been shown conclusively to be non-toxic. As there is a great need to develop new models for rapidly and efficiently assessing neurotoxicity, these results demonstrate that this system is a good alternative to the models that are currently being used for high-throughput viability screens. The potential to generate reporter lines also gives this system the flexibility to perform high content screens to further understand mechanisms of cell death.
Current neurotoxicity screens rely heavily on animal models or immortalized tumor cells of neural origin. The animal models are expensive, do not work for high-throughput screens, and do not always faithfully recapitulate human nervous system pathology. While screens with tumor cells bypass the first two of these issues, they also do not represent a normal human neural cell and so do not solve the third issue (National Research Council 2007; Breier et al., 2012; Llorens et al., 2012). A recent study has proposed a different model using neurons differentiated from umbilical stem cells to assess the toxicity of pesticides (Kashyap et al., 2014). While the model described in that study is a viable one for neurotoxicity testing, we believe our system has several additional advantages including the ability to generate a stable NSC state that has longer term cryopreservation and passaging potential as well as the capacity to be differentiated to multiple neuronal and glial types (Efthymiou et al., 2014). As such, our NSCs are more amenable for higher throughput assays and can be the starting source material for testing a broader array of cell types. The availability of a reporter NSC line also makes it possible to perform large scale high content screens with this platform. As no one cell culture model can perfectly predict toxicity in vivo, positive hits will have to be followed up in other in vitro models and in animals but because the platform we describe can be scaled up greatly it offers an opportunity to screen thousands of compounds and greatly narrow the list of those that can be further validated in these other systems.
There are several important findings from this screen. One of these is the presence of species differences in toxicity amongst certain compounds within this library: ~100 compounds exhibited significant toxicity to human NSCs without affecting rat cortical mixed cultures. The library contains a broad list of compounds that affect many cellular processes, so this is unlikely to be due to a library bias towards compounds that kill proliferative cells like NSCs. The rat cells used in this study are mixed cultures consisting of both neurons and astrocytes, and it is possible that the differential toxicity is more attributable to cell type than species differences. However, we tested 43 of the hits against human fetal astrocytes and neurons differentiated from the human NSCs and found that nine of the compounds toxic to human NSCs but not rat cells were also toxic to neurons and/or fetal astrocytes.
One class of compounds that shows toxicity against all the human NSCs but not the rat cells is cardiac glycosides. Seventeen compounds in the library belong to this family and all fit this toxicity profile. Four of these were validated in human NSCs (digitoxin, digoxin, lanatoside C, oleandrin, and ouabain) and also found to exhibit toxicity against NSC-derived neurons and fetal astrocytes. An iPSC toxicity screen identified lanatoside C as a drug that could be a potential PSC toxin and our screen verified that finding in iPSCs and extended these findings to include additional cardiac glycosides that have toxicity to several neural cell types (Ben-David et al., 2013). In retrospect, this result that the cardiac glycosides affect human and not rat cells is not surprising as they act through the Na, K-ATPase alpha subunit and it has long been known that rat brain predominantly expresses an alpha subunit gene that is resistant to cardiac glycosides (Herrera et al., 1987; Jewell and Lingrel, 1991). The fact that our screen was able to identify these compounds as human-specific demonstrates the potential of human NSC based neurotoxicity assays to identify compounds with species specific toxicity profiles and conversely the possibility that screening compounds against non-human cells may result in initially labeling potentially toxic compounds as non-toxic. It is also interesting to note that these compounds have the capacity to cross the blood-brain-barrier and have been shown to kill cultured glioblastoma cells, suggesting that there is a high probability that the results of our screen represent an accurate assessment of the potential toxicity of cardiac glycosides to neural cell types in general and NSCs more specifically.
A second important finding of this study is the identification of compounds that have cell-type selective toxicities. Eight compounds were identified that spared neurons but were toxic to iPSCs, NSCs, and fetal astrocytes. As such, these compounds might be good candidates to use in neuronal differentiation protocols to obtain homogeneous populations of neurons by killing astrocytes and NSCs that are present to varying degrees as byproducts of the protocol. Future work with both differentiation and co-culture paradigms will address these questions.
The ability to selectively kill NSCs could have broad therapeutic implications for cancers confined to the central nervous system In this regard it should be noted that cardiac glycosides have been shown to be effective against glioblastoma in vitro and considered as a therapeutic option for this tumor type (Badr et al., 2011; Joshi et al., 2011). Additionally, there is growing evidence that NSCs are cells of origin for glioblastoma with these cells being highly refractory to all currently used treatment regimens (Chen et al., 2012). As such glycosides could be attractive candidates for treatment of recurrent glioblastoma but our study also suggests that these drugs have toxicity against neurons and fetal astrocytes so future potential use will have to be carefully weighed against unwanted side effects. In this context it is possible that other compounds that were found to be toxic to NSCs in our initial screen but not yet followed up may be more cell type selective and, therefore, better options for glioblastoma therapy. Alternatively, it may be possible to synthesize cardiac glycoside derivatives that show toxicity against only NSCs. Future studies will be needed to further address these issues.
5. Conclusions
The work described in this study demonstrates that human NSCs can serve as the starting material for conducting effective high-throughput neurotoxicity screens. One of the most important findings uncovered by our human NSC-based neurotoxicity screen is the ability to identify compounds that specifically affect human but not rodent cells. Our described screen has the capacity to examine multiple neural cell types differentiated from the same NSC line and can also identify compounds that only kill specific cell types. Although human neurotoxicity data is available for only a small subset of compounds in the library it appears that this platform is able to identify a high percentage of the compounds that have been shown to have neurotoxic effects in humans. The ability to identify compounds only affecting NSCs could have broad implications for the treatment of tumors of neural origin. Of course, the results of this screen will be need to be validated in in vivo models and additional pharmacokinetic data will have to be gathered on potentially neurotoxic compounds including blood-brain-barrier permeability to assess their true neurotoxicity in the human nervous system. Nevertheless, easy access to large quantities of previously difficult to obtain human neural cells type will have a profound effect on how neurotoxicity testing is conducted in the future.
Supplementary Material
Acknowledgements
This work was funded by the NIH Common Fund.
References
- Badr CE, Wurdinger T, Nilsson J, Niers JM, Whalen M, Degterev A, Tannous BA. Lanatoside C sensitizes glioblastoma cells to tumor necrosis factor-related apoptosis-inducing ligand and induces an alternative cell death pathway. Neuro Oncol. 2011;13:1213–1224. doi: 10.1093/neuonc/nor067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Gan Q-F, Golan-Lev T, Arora P, Yanuk O, Oren YS, Leikin-Frenkel A, Graf M, Garippa R, Boehringer M, Gromo G, Benvenisty N. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell. 2013;12:167–179. doi: 10.1016/j.stem.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Breier JM, Gassmann K, Kayser R, Stegeman H, De Groot D, Fritsche E, Shafer TJ. Neural progenitors as models for high-througput screens of developmental neurotoxicity: state of the science. Neurotoxicol Teratol. 2010;32:4–15. doi: 10.1016/j.ntt.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Caporello E, Nath A, Slevin J, Galey D, Hamilton G, Williams L, Steiner JP, Haughey NJ. The immunophilin ligand GPI 1046 protects neurons from the lethal effects of the HIV-1 proteins gp120 and Tat by modulating endoplasmic reticulum calcium load. J Neurochem. 2006;98:146–155. doi: 10.1111/j.1471-4159.2006.03863.x. [DOI] [PubMed] [Google Scholar]
- Cavalloro M, Marianni J, Lancini C, Latorre E, Caccia R, Gullo F, Valotta M, DeBiasi S, Spinardi L, Ronchi A, et al. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox 2 mutants. Development. 2008;135:541–557. doi: 10.1242/dev.010801. [DOI] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DY, Lin SZ, Yang WK, Lee HC, Hsu DM, Lin HL, Chen CC, Liu CL, Lee WY, Ho LH. Targeting cancer stem cells for treatment of glioblastoma multiforme. Cell Transplant. 2013;22:731–739. doi: 10.3727/096368912X655136. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbordes SC, Placantonakis DG, Ciro A, Socci ND, Lee G, Djaballah H, Studer L. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2:602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;28:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efthymiou A, Shaltouki A, Steiner JP, Jha B, Heman-Ackha SM, Swistowski A, Zeng X, Rao MS, Malik N. Functional screening assays with neurons generated from pluripotent stem cell-derived neural stem cells. J Biomol Screen. 2014;19:32–43. doi: 10.1177/1087057113501869. [DOI] [PubMed] [Google Scholar]
- Han Y, Miller A, Mangada J, Liu Y, Swistowski A, Zhan M, Rao MS, Zeng X. Identification by automated screening of a small molecule that selectively eliminates neural stem cells derived from hESCs but not dopamine neurons. PLoS ONE. 2009;4:e7155. doi: 10.1371/journal.pone.0007155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughey NJ, Cutler RG, Tamara A, McArthur JC, Vargas DL, Pardo CA, Turchan J, Nath A, Mattson MP. Perturbation of sphingolipid metabolism and ceramide production in HIV-dementia. Ann Neurol. 2004;55:257–267. doi: 10.1002/ana.10828. [DOI] [PubMed] [Google Scholar]
- Hendrickson ML, Rao AJ, Demerdash ON, Kalil RE. Expression of nestin by neural cells in the adult rat and human brain. PLoS One. 2011;6:e18535. doi: 10.1371/journal.pone.0018535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera VLM, Emanuel JR, Ruiz-Opazo N, Levenson R, Nadal-Ginard B. Three differentially expressed Na,K-ATPase subunit isoforms: structural and functional implications. J Cell Biol. 1987;105:1855–1865. doi: 10.1083/jcb.105.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BY, Du ZW, Li XJ, Ayala M, Zhang SC. Human oligodendrocytes from embryonic stem cells: conserved SHH signaling networks and divergent FGF effects. Development. 2009;136:1443–1452. doi: 10.1242/dev.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell EA, Lingrel JB. Comparison of the substrate dependence properties of the rat Na,K-ATPase a1, a2, and a3 isoforms expressed in HeLa cells. J Biol Chem. 1991;266:16925–16930. [PubMed] [Google Scholar]
- Joshi AD, Parsons DW, Velculescu VE, Riggins GJ. Sodium ion channel mutations in glioblastoma patients correlate with shorter survival. Mol Cancer. 2011;10:17. doi: 10.1186/1476-4598-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehn-Hall K, Guendel I, Carpio L, Skaltounis L, Meijer L, Al-Harthi L, Steiner JP, Nath A, Kutsch O, Kashanchi F. Inhibition of Tat-mediated HIV-1 replication and neurotoxicity by novel GSK3-beta inhibitors. Virology. 2011;415:56–68. doi: 10.1016/j.virol.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap MP, Kumar V, Singh AK, Tripathi VK, Jahan S, Pandey A, Srivastava RK, Khanna VK, Pant AB. Differentiating neurons derived from human umbilical cord blood stem cells work as a test for developmental neurotoxicity. Mol Neurobiol. doi: 10.1007/s12035-014-8716-7. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Lee DH, Lee CS, Kim DW, Ae JE, Lee TH. Digitoxin sensitizes glioma cells to TRAIL-mediated apoptosis by upregulation of death receptor 5 and downregulation of surviving. Anticancer Drugs. 2014;25:44–52. doi: 10.1097/CAD.0000000000000015. [DOI] [PubMed] [Google Scholar]
- Lee G, Ramirez CN, Kim CN, Zeltner N, Liu B, Radu C, Bhinder B, Kim YJ, Choi IY, Mukherjee-Clavin B, Djaballah H, Studer L. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds than rescue IKBKAP expression. Nat Biotechnol. 2012;30:1244–1248. doi: 10.1038/nbt.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens J, Li AA, Ceccatelli S, Sunol C. Strategies and tools for preventing neurotoxicity: to test, to predict, and how to do it. Neurotoxicology. 2012;33:796–804. doi: 10.1016/j.neuro.2012.01.019. [DOI] [PubMed] [Google Scholar]
- Ludolph AC, He F, Spencer PS, Hammerstad J, Sabri M. 3-Nitropropionic acidexogenous animal neurotoxin and possible human striatal toxin. Canad J Neurol Sci. 1991;18:492–498. doi: 10.1017/s0317167100032212. [DOI] [PubMed] [Google Scholar]
- Luo Y, Liu C, Cerbini T, San H, Lin Y, Chen G, Rao MS, Zou J. Stable enhanced green fluorescent protein expression after differentiation and transplantation of reporter human induced pluripotent stem cells generated by AAVS1 transcription activator-like effector nucleases. Stem Cells Trans Med. 2014;3:821–835. doi: 10.5966/sctm.2013-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik N, Wang X, Shah S, Efthymiou AG, Yan B, Heman-Ackah S, Zhan M, Rao M. Comparison of gene expression profiles of human fetal cortical astrocytes with pluripotent stem cell derived neural stem cells identifies human astrocyte markers and signaling pathways and transcription factors active in human astrocytes. PLoS One. 2014;9:e96139. doi: 10.1371/journal.pone.0096139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assay. J Immunol Methods. 1983;16:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- National Research Council. Toxicity testingin the 21st century: a vision and a strategy. Washington, DC: 2007. [Google Scholar]
- Nath S, Bachani M, Harshavardhana D, Steiner JP. Catechins protect neurons against mitochondrial toxins and HIV proteins via activation of the BDNF pathway. J Neurovirol. 2012;18:445–455. doi: 10.1007/s13365-012-0122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingerland M, Cerella C, Guchelaar HJ, Diedrich M, Gelderblom H. Cardiac glycosides in cancer therapy: from preclinical investigations towards clinical trials. Invest New Drugs. 2013;31:1087–1094. doi: 10.1007/s10637-013-9984-1. [DOI] [PubMed] [Google Scholar]
- Shaltouki A, Peng J, Liu Q, Rao MS, Zeng X. Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells. 2013;31:941–952. doi: 10.1002/stem.1334. [DOI] [PubMed] [Google Scholar]
- Swistowski A, Peng J, Han Y, Swistowska AM, Rao MS, Zeng X. Xeno-free defined conditions for culture of human embryonic stem cells, neural stem cells and dopaminergic neurons derived from them. PLoS One. 2009;14:e6233. doi: 10.1371/journal.pone.0006233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner JP, Galey D, Haughey NJ, Asch D, Nath A. Neuroprotective and antiretroviral effects of the immunophilin ligand GPI 1046. J Neuroimmune Pharmacol. 2:49–57. doi: 10.1007/s11481-006-9060-0. [DOI] [PubMed] [Google Scholar]
- Yan Y, Shin S, Jha BS, Liu Q, Sheng J, Li F, Zhan M, Davis J, Bharti K, Zeng X, Rao M, Malik N, Vemuri MC. Efficient and rapid derivation of primitive neural stem cells and generation of brain subtype neurons from human pluripotent stem cells. Stem Cells Transl Med. 2013;2:862–870. doi: 10.5966/sctm.2013-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YM, Gupta SK, Kim KJ, Powers BE, Cerqueira A, Wainger BJ, Ngo HD, Rosowski KA, Schein PA, Ackeifi CA, Arvanites AC, Davidow LS, Woolf CJ, Rubin LL. A small molecule screen in stem-cell-derived motor neurons identifies a kinase inhibitor as a candidate therapeutic for ALS. Cell Stem Cell. 2013;12:713–726. doi: 10.1016/j.stem.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao AY, Wei L, Xia S, Rothman S, Yu SP. Ionic mechanism of ouabain-induced concurrent apoptosis and necrosis in individual cultured cortical neurons. J Neurosci. 2002;22:1350–1362. doi: 10.1523/JNEUROSCI.22-04-01350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Bae M, Tovar-y-Romo LB, Patel N, Bandaru VV, Pomerantz D, Steiner JP, Haughey NJ. The HIV coat protein gp120 promotes forward trafficking and surface of clustering of NMDA receptors in membrane microdomains. J Neurosci. 2011;31:17074–17090. doi: 10.1523/JNEUROSCI.4072-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.