Monoclonal B-cell lymphocytosis (MBL) is defined as an asymptomatic expansion of clonal B cells with less than 5 × 109/L cells in the peripheral blood and without other manifestations of chronic lymphocytic leukemia (CLL; for example, lymphadenopathy, cytopenias, constitutional symptoms).1 Approximately 1% of the MBL cohort develops CLL per year. Evidence suggests that nearly all CLL cases are preceded by an MBL state.2 Our understanding of the genetic basis, clonal architecture and evolution in CLL pathogenesis has undergone significant improvements in the last few years.3–8 In contrast to CLL, our knowledge of the genetics in MBL is still very fragmented, with few prior studies focused on the status of selected genetic abnormalities.1,9,10 In addition, previous reports have demonstrated the co-existence of two B-cell subclones in a small subset of MBL cases,11 and clonal changes in sequential MBL samples measured by IGHV mutational analysis.12
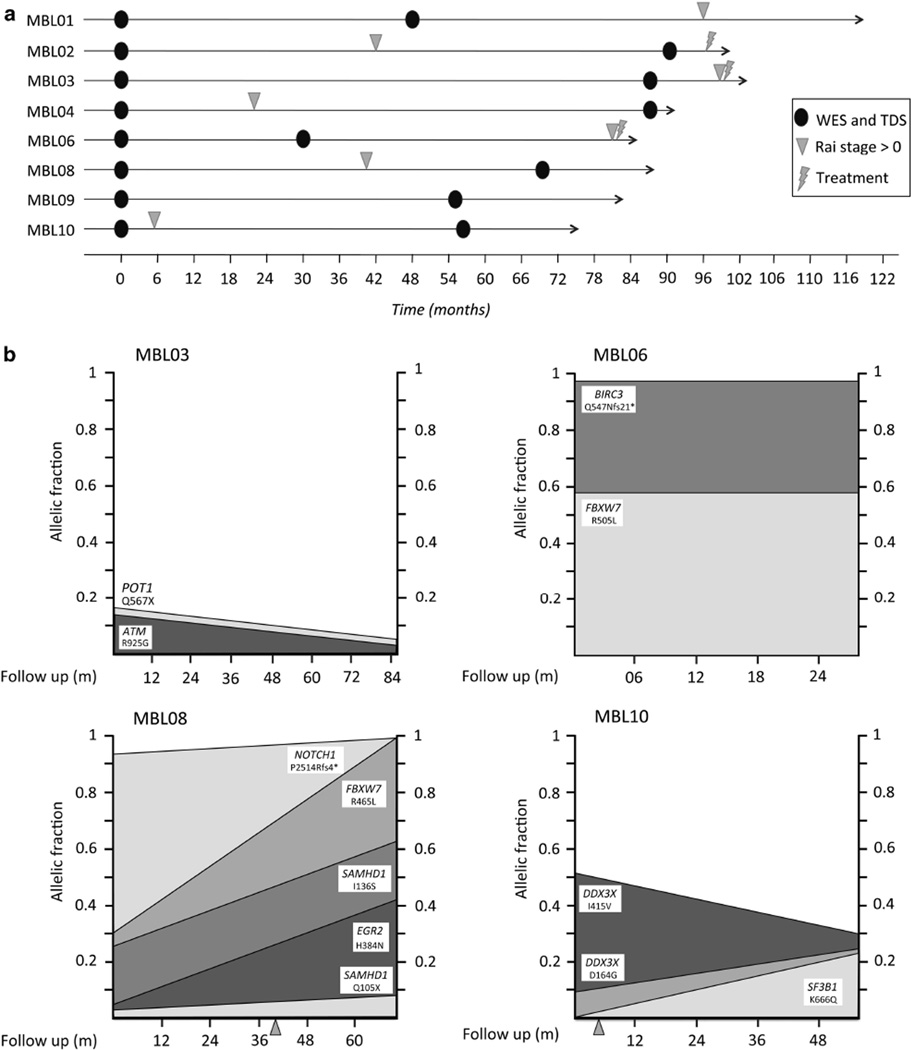
To better understand the genomic landscape of MBL and clonal evolution overtime, we performed a longitudinal analysis in a cohort of eight MBL cases, analyzed by whole-exome sequencing (WES) at two time points. The criteria for sample selection were that the individual with MBL had a sample collected at diagnosis of MBL and had one additional sample collected more than 30 months apart that had stored cells for bead selection of clonal B cells and normal non-B cells for DNA extraction. Samples for WES were collected on average 65 months apart (median 62.5 months, range 30–91). At the second time point analyzed, four cases were still MBL, whereas the remaining four cases had progressed to detectable lymphadenopathy by physical exam but had not yet required treatment. After the second sample analyzed, cases were followed up for an average of 26 months (median 24 months, range 3–72). Thus, the total median follow-up of the cohort from MBL diagnosis to the final visit was 88 months (average 91, range 75–120). Overall, one case remained as an MBL and the remaining seven cases had now progressed to detectable lymphadenopathy, and three of which required treatment by IWCLL criteria. The follow-up information and time points analyzed are summarized in Figure 1a.
Figure 1.
(a) Time points analyzed in each MBL case. Changes in Rai stage (Rai > 0) and start of treatment are indicated for each case. (b) Changes in allelic fractions of the mutations found in driver genes between time points analyzed. Y-axis shows the tumor fraction with each mutation. MBL8 and MBL10 changed Rai stage between time points (indicated with an arrowhead).
The MBL individuals provided written informed consent for the collection and use of samples for research purposes according to the Declaration of Helsinki. Clinical information of cases analyzed is provided in Supplementary Table S1. B lymphocytes were enriched from peripheral blood mononuclear cells using the EasySep Human CD19+ Cell Enrichment Kit without CD43 Depletion. T cells were enriched using the EasySep Human CD3 Positive Selection Kit (Stemcell Technologies, Vancouver, BC, Canada) and subsequently were used as germline samples in the sequencing genetic studies. After cell enrichment, all fractions were stained by four-color immunophenotypic analysis to assess sample purity. The following antibody conjugates were used: anti-CD5-fluorescein isothiocyanate, anti-CD16-phycoerythrin, anti-CD19-allophycocyanin and anti-CD3-peridinin chlorophyll protein (Beckton Dickinson, Franklin Lakes, NJ, USA). Flow cytometry analysis was performed using the FACSCalibur (Beckton Dickinson) and data analyzed using Cell Quest software. On the basis of FACS (fluorescence-activated cell sorting) analysis, we observed after enrichment an average of 91% of CD19+ cells (range 76–99%) and 91% of the CD19+ fraction were CD19+/CD5+ cells (range 66–99%). We used the values of the CD19+/CD5+ fraction to calculate the leukemic B-cell fraction and reduce any significant contamination of non-clonal B cells in each biopsy. DNA was extracted from the clonal B cells and non-clonal (that is, T cells) cells using the Gentra Puregene Cell Kit (Qiagen, Hilden, Germany). Extracted DNAs were fingerprinted to confirm the relationship between samples of the same MBL individual and to rule out sample cross-contamination between individuals. In addition, the molecular analysis of IGHV gene family was performed to confirm the existence of one or more B-cell clones (Supplementary Table S2). Mononuclear cells were used for this analysis. One microgram of RNA was converted to cDNA using the BioRad iScript cDNA Kit (Hercules, CA, USA). cDNA (2 µl) were amplified in separate PCR reactions for each of the seven IGHV family genes using a consensus sense primer and an IgM antisense primer. The reactions were carried out using the HotStarTaq PCR kit (Qiagen) in a total volume of 50 µl with 20 pmol of each primer. Cycling conditions were 94 °C for 15 min followed by 35 cycles of 30 s at 94 °C; 1 min at 60 °C; 1 min at 72 °C; and a final cycle of 72 °C for 10 min. PCR products were electrophoresed and visualized with ethidium bromide, purified using the Wizard PCR Prep kit (Promega, Madison, WI, USA) and sequenced using an automated sequencer (Applied Biosystems, Foster City, CA, USA). In those cases where gDNA was used (MBL04 and MBL09), the conditions were the same except that a consensus antisense primer to the IGHJ region was used instead of the IgM antisense primer. Resulting sequences were aligned to germline sequences using ImMunoGeneTics Information (IMGT) System reference sets and IMGT/V-Quest software (http://imgt.cines.fr).
Genomic DNA from each sample was sheared and used for the construction of paired end sequencing library as described in the protocol provided by Illumina. The exome was captured using the Sure Select 50 Mb Exome Enrichment kit (Agilent, Santa Clara, CA, USA) following the manufacturer’s instructions. Samples were sequenced using Illumina HiSeq2000. 100 bp paired end reads were aligned to human genome hg19 using Novoalign (Novocraft Technologies, Selangor, Malaysia). Realignment and recalibration was done to take advantage of Best Practice Variant Detection v3 recommendations implemented in the GATK. Somatic single-nucleotide variations (SNVs) were genotyped using SomaticSniper, whereas insertions and deletions were called by GATK Somatic Indel Detector. Each variant in coding regions was analyzed with PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) and MutationTaster (http://www.mutationtaster.org/) to predict biological effects. Pair-wise analyses were performed comparing each tumor time point with the germline sample. Variants with read depth less than 10× were excluded from further analysis. To validate the findings obtained from WES and increase the sensitivity of the screening, a panel of 24 genes relevant to CLL (Supplementary Table S3) was sequenced in all MBL samples at both time points. Targeted deep sequencing (TDS) was performed in a Ion Torrent PGM (Life Technologies, Carlsbad, CA, USA) sequencer with an average coverage depth of 630 × (range 506 × –1180 ×). Finally, SNV, insertions and deletions (indels) were visually inspected using Integrative Genomics Viewer (IGV, Broad Institute). Sequencing findings were then integrated with FISH panel data previously performed in these cases (Table 1).
Table 1.
Summary of FISH and sequencing results in the MBL cases analyzed
| Case no. | FISH | % Positive cells | Gene | Mutation | Polyphen prediction |
Mutation taster prediction |
Time point 1 | Time point 2 | P-value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time point 1 |
Time point 2 |
Coverage depth |
Allelic fractiona |
Coverage depth |
Allelic fractiona |
|||||||
| MBL01 | Normal | — | — | PRDM1 | D344N | Moderate | Disease causing | 342 | 0.467 | 410 | 0.303 | 0.0001 |
| MBL02 | 13q − | 41 | 94 | — | ||||||||
| MBL03 | 13q − | 31 | 95 | POT1 | Q567X | High | Disease causing | 377 | 0.18 | 359 | 0.04 | 0.0002 |
| ATM | R925G | Moderate | Disease causing | 641 | 0.140 | 837 | 0.020 | 0.0001 | ||||
| MBL04 | 13q − | 56 | NA | — | ||||||||
| MBL06 | Trisomy 12 | 51 | NA | BIRC3 | Q547Nfs21* | High | Disease causing | 1457 | 1.000 | 2756 | 1.000 | NS |
| FBXW7 | R505L | Moderate | Disease causing | 154 | 0.60 | 418 | 0.59 | NS | ||||
| MBL08 | Trisomy 12 | 16 | NA | NOTCH1 | P2514Rfs4* | High | Disease causing | 244 | 0.941 | 220 | 1.000 | 0.007 |
| FBXW7 | R465L | Moderate | Disease causing | 368 | 0.306 | 418 | 1.000 | 0.0001 | ||||
| EGR2 | H384N | Moderate | Disease causing | 263 | 0.038 | 301 | 0.420 | 0.0001 | ||||
| SAMHD1 | Q105X | High | Disease causing | 736 | 0.003 | 730 | 0.080 | 0.0001 | ||||
| SAMHD1 | I136S | Moderate | Disease causing | 702 | 0.260 | 679 | 0.620 | 0.0001 | ||||
| MBL09 | Trisomy 12 | 46 | NA | — | ||||||||
| MBL10 | 11q − | 11 | NA | SF3B1 | K666Q | Moderate | Disease causing | 624 | 0.000 | 528 | 0.220 | 0.0001 |
| DDX3X | D164G | Moderate | Disease causing | 530 | 0.09 | 382 | 0.23 | 0.0009 | ||||
| DDX3X | I415V | Moderate | Disease causing | 927 | 0.51 | 933 | 0.30 | 0.0001 | ||||
Abbreviations: FISH, fluorescence in situ hybridization; MBL, monoclonal B-cell lymphocytosis; NA, not applicable; NS, not significant.
Only putative tumorigenic genes were included. All mutations are somatic. All mutations excepting PRDM1 (D344N) were validated using targeted sequencing. P-values indicate whether the allelic fraction differences between time points are significant.
The values correspond to allelic fractions after tumor content correction.
Overall, an average of 122-fold depth coverage was obtained by WES (range 92 × –178 ×) with an average of 80% of the targeted regions being covered by at least 30 ×. We found an average of 6.6 and 7.5 nonsynonymous SNVs and indels at the first and second time points, respectively. The complete list of mutations and the respective allelic fractions is shown in Supplementary Table S4. In four cases, we found mutations in driver genes previously identified in CLL, including ATM, DDX3X, EGR2, FBXW7, SAMHD1 and SF3B1. TDS not only validated the WES results, but also identified additional mutations in BIRC3, POT1 and NOTCH1 in regions that were initially missed due to low coverage obtained from WES in those specific regions. We found a fifth case, MBL01, with a damaging mutation affecting PRDM1, a gene frequently inactivated in diffuse large B-cell lymphomas.13 Of note, FBXW7 was the only gene recurrently mutated in our cohort. All mutations were found in highly evolutionarily conserved regions and considered damaging by PolyPhen and MutationTaster.
In all four cases, we found the mutations in two or more driver genes (Table 1). Thus, MBL03 has mutations in ATM and POT1; MBL06 has mutations in BIRC3 and FBWX7, MBL08 has mutations in EGR2, FBXW7, NOTCH1 and two independent mutations in SAMHD1, whereas MBL10 has a mutation in SF3B1 and two independent mutations in DDX3X. We analyzed sequential time points to recreate the clonal architecture and evolution. The number of mutations found per case was insufficient to run comprehensive clustering analysis. However, we focused the analysis on the dynamics of mutations found in putative driver genes. In three of the four cases harboring mutations in putative driver genes (MBL03, MBL06 and MBL08), all the mutations were already detected at the first time point analyzed, but mostly found in subclones that either remained stable (MBL03 and MBL06) or become more prevalent overtime (MBL08; Figure 1b). In the remaining case (MBL10), the first time point analyzed was characterized by the presence of DDX3X (p.I415V) in nearly 50% allelic fraction and a second DDX3X mutation (p.D164G) in 10% of allelic fraction. In the second time point, an increased abundance of p.D164G and reduced abundance of p.I415V were observed. This opposite trend in prevalence suggests the presence of driver mutations in different subclones with alternated dominance between time points. Furthermore, at the second time point, a novel mutation in SF3B1 was found (p.K666Q). This mutation was not identified in the first time point even using deep sequencing. All four cases with putative CLL mutations eventually developed lymphadenopathy and/or required treatment on average 56 months after clinical recognition of MBL compared with an average of 61 months in the remaining cases. In all four samples, the existence of a single B-cell clone was confirmed by molecular analysis of IGHV gene (Supplementary Table S2).
Previous studies have identified genetic abnormalities in a subset of MBL, including deletion 13q14 and NOTCH1 mutations.2,10 Furthermore, ATM mutations have been found in linkage studies of familial CLL.14 Our study brings new insights regarding the genomic landscape and clonal architecture of MBL, the precursor to CLL,1,15 by identifying mutations in one or more driver genes and confirming the existence of genetic subclones in a subset of MBL cases. Furthermore, our study brings new data providing insight into the temporal succession of genetic events in MBL and CLL pathogenesis. Thus, a recent study has shown the subclonal nature of FBXW7 in all four CLL cases harboring mutations in the gene, suggesting this mutation is a later event in CLL pathogenesis.5 However, our data demonstrate the existence of recurrent mutations of FBXW7 in the premalignant stages of the disease, and suggest that FBXW7 mutations can be an early genetic event in CLL pathogenesis.
Remarkably, we identified tumorigenic mutations on average 60 months before clear disease progression occurred in MBL cases. These findings support the implementation of a targeted sequencing strategy including the subset of putative cancer genes now known to be recurrently mutated in CLL (~20 genes). Using available sequencing technologies, this subset of genes could be easily screened in very high depth coverage, thus providing early identification of oncogenic mutations in small subclones at the MBL phase.
Supplementary Material
ACKNOWLEDGEMENTS
EB is a recipient of the Marriott Specialized Workforce Development Awards in Individualized Medicine and the Henry Predolin Foundation Career Development Award. This work was supported by the Henry Predolin Foundation and grant NIH-CA95241.
RF has received a patent for the prognostication of MM based on genetic categorization of the disease. He has received consulting fees from Medtronic, Otsuka, Celgene, Genzyme, BMS, Lilly, Onyx, Binding Site, Millenium and AMGEN. He also has sponsored research from Cylene and Onyx. TS was supported by fundings from Hospira, Genentech, Glaxo-Smith-Kline, Jannsen, Celgene and Cephalon.
Footnotes
CONFLICT OF INTEREST
The remaining authors declare no conflcit of interest.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
REFERENCES
- 1.Rawstron AC, Bennett FL, O'Connor SJ, Kwok M, Fenton JA, Plummer M, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359:575–583. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 2.Landgren O, Albitar M, Ma W, Abbasi F, Hayes RB, Ghia P, et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009;360:659–667. doi: 10.1056/NEJMoa0806122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braggio E, Kay NE, VanWier S, Tschumper RC, Smoley S, Eckel-Passow JE, et al. Longitudinal genome-wide analysis of patients with chronic lymphocytic leukemia reveals complex evolution of clonal architecture at disease progression and at the time of relapse. Leukemia. 2012;26:1698–1701. doi: 10.1038/leu.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knight SJ, Yau C, Clifford R, Timbs AT, Sadighi Akha E, Dreau HM, et al. Quantification of subclonal distributions of recurrent genomic aberrations in paired pre-treatment and relapse samples from patients with B-cell chronic lymphocytic leukemia. Leukemia. 2012;26:1564–1575. doi: 10.1038/leu.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 7.Schuh A, Becq J, Humphray S, Alexa A, Burns A, Clifford R, et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 2012;120:4191–4196. doi: 10.1182/blood-2012-05-433540. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greco M, Capello D, Bruscaggin A, Spina V, Rasi S, Monti S, et al. Analysis of SF3B1 mutations in monoclonal B-cell lymphocytosis. Hemato Oncol. 2013;31:54–55. doi: 10.1002/hon.2013. [DOI] [PubMed] [Google Scholar]
- 10.Rasi S, Monti S, Spina V, Foa R, Gaidano G, Rossi D. Analysis of NOTCH1 mutations in monoclonal B-cell lymphocytosis. Haematologica. 2012;97:153–154. doi: 10.3324/haematol.2011.053090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghia P, Prato G, Scielzo C, Stella S, Geuna M, Guida G, et al. Monoclonal CD5+ and CD5− B-lymphocyte expansions are frequent in the peripheral blood of the elderly. Blood. 2004;103:2337–2342. doi: 10.1182/blood-2003-09-3277. [DOI] [PubMed] [Google Scholar]
- 12.Marti GE, Shim YK, Albitar M, Middleton D, Abbasi F, Anderson A, et al. Long-term follow-up of monoclonal B-cell lymphocytosis detected in environmental health studies. Cytometry B Clin Cytom. 2010;78:S83–S90. doi: 10.1002/cyto.b.20522. [DOI] [PubMed] [Google Scholar]
- 13.Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bevan S, Catovsky D, Marossy A, Matutes E, Popat S, Antonovic P, et al. Linkage analysis for ATM in familial B cell chronic lymphocytic leukaemia. Leukemia. 1999;13:1497–1500. doi: 10.1038/sj.leu.2401531. [DOI] [PubMed] [Google Scholar]
- 15.Shanafelt TD, Ghia P, Lanasa MC, Landgren O, Rawstron AC. Monoclonal B-cell lymphocytosis (MBL): biology, natural history and clinical management. Leukemia. 2010;24:512–520. doi: 10.1038/leu.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.