Abstract
Visual analysis is required to perform many biological experiments, from counting colonies to measuring the size or fluorescence intensity of individual cells or organisms. This unit outlines the use of CellProfiler, a free, open-source image analysis tool that extracts quantitative information from biological images. It includes a step-by-step protocol for automated analysis of the number, color, and size of yeast colonies growing on agar plates, but the methods can be adapted to identify and measure many other types of objects in images. The flexibility of the software allows experimenters to create pipelines of adjustable modules to fit different biological experiments and to generate accurate measurements from dozens or even hundreds of thousands of images.
Keywords: automatic image analysis, yeast colonies, open-source software, phenotypes, colony counting
Unit Introduction
Many experiments in a biology laboratory involve visual inspection, such as examining yeast colonies or growth patches on agar plates, or examining live or stained cell samples by microscopy. Acquiring images and analyzing them automatically with image analysis software has several advantages over simple visual inspection. It is less tedious, more objective and quantitative, and, while the set up can be time-consuming, the analysis itself is usually much faster for large sample sets.
This unit outlines a protocol for the automated counting and analysis of yeast colonies growing on agar plates; however, the methods described can be adapted to a wide variety of biological “objects” and can be used to measure a wide variety of features for each object. The protocol uses the open-source, freely downloadable software package, CellProfiler. CellProfiler has been cited in more than a thousand papers and validated for a wide variety of biological applications, including yeast colony counting and classification, cell microarray annotation, yeast patch assays, cell-cycle classification, mouse tumor quantification, wound healing assays, and tissue topology measurement, as well as analysis of fluorescence microscopy images for measurement of cell size and morphology, cell cycle distributions, fluorescence staining levels, and other features of individual cells in images (Lamprecht, Sabatini, and Carpenter 2007; Carpenter et al. 2006; Kamentsky et al. 2011).
Basic Protocol
Setting Up and Using CellProfiler
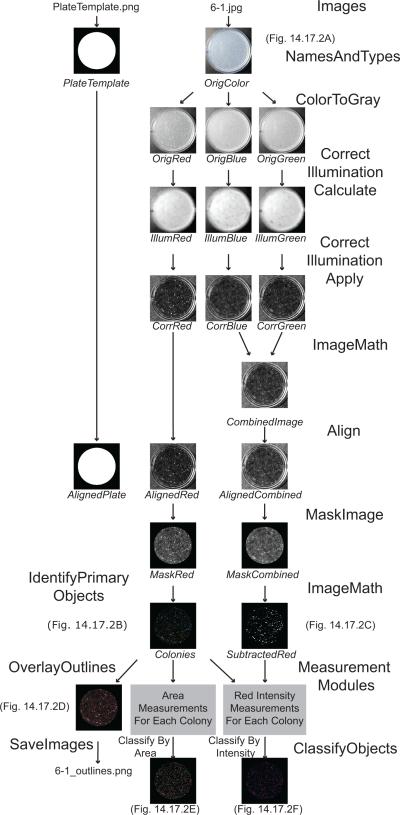
The protocol begins with instructions for downloading the CellProfiler program and an example “pipeline” file. The workflow of the pipeline is depicted in Figure 14.17.1. The pipeline is then adjusted so that it can analyze your own images. Tens of thousands of images can be routinely analyzed per experiment. In this example, CellProfiler is used to identify and count yeast colonies on each plate, and to measure each colony's size, shape, texture, and color. Lastly, instructions are given for analyzing the numerical results within CellProfiler using its built-in data tools, or by exporting the data in a comma-delimited text file for use in a spreadsheet program such as Microsoft Excel or more sophisticated analysis programs such as R (R Development Core Team 2008).
Figure 14.17.1.
An overview of the CellProfiler example pipeline. The names of the images created or objects identified appear in italics below each image, whereas the module names appear in a larger regular font.
NOTE: In addition to CellProfiler's main “Help” menu, there are many “?” buttons within the software containing more information about how to use CellProfiler. For example, clicking the “?” button below the pipeline panel will show information about the selected module within the pipeline. Additionally, the CellProfiler user manual is available in PDF format (http://www.cellprofiler.org/manuals.shtml), and a user forum is available for posting and reading questions and answers about how to use the software (http://www.cellprofiler.org/forum).
NOTE: There are several options available as preferences for modifying the appearance of the main CellProfiler window. To change your preferences, click on File > Preferences from the main menu bar.
Materials List
Images of yeast plates to be processed
Images can be taken with a flatbed scanner or digital camera (Dahle et al. 2004; Memarian et al. 2007); see Critical Parameters for guidance. The images can be located within subfolders and need not be in a particular order or follow a particular naming convention. While this example only analyzes one image, it is possible to analyze hundreds of images on a single computer, or hundreds of thousands of images using a computing cluster (see Alternate Protocol). More than 100 file formats are currently readable by CellProfiler, including BMP, GIF, JPG, PNG, TIF, DIB, LSM, and FLEX. See Critical Parameters for more information about acquiring images and image file types.
Computer with at least 4 GB of RAM and multiple processors each running at least 2 GHz. A 64-bit operating system is strongly recommended. CellProfiler is available for Macintosh, Windows, and Unix/Linux. A complete list of compatible operating systems can be found at http://www.cellprofiler.org/download.shtml. The example image pipeline demonstrated here will be processed in ~1 minute per image on a single computer with a 2.67 GHz processor and 8 GB RAM. CellProfiler is optimized to take advantage of multiple computing processors on a single computer, but large image sets (greater than ~500 images) will likely require a computing cluster (see Alternate Protocol).
Decompression software (e.g., WinZip, Stuffit) for unpacking compressed files, if not already included in your operating system.
CellProfiler software (see step 1). This protocol was written for CellProfiler version 2.1.0. Example images and corresponding CellProfiler pipeline (see step 4)
Download and install CellProfiler software
- Decide whether to use the compiled version or the developer's version.Most experimenters will use the installation package (i.e., compiled) suitable for their computing platform (Macintosh, Windows, or Unix/Linux). This version is simple to install and is free for use (GPLv2 license). However, researchers or programmers wishing to implement their own image analysis algorithms should download the developer's version, available as Python source code hosted at https://github.com/CellProfiler/CellProfiler. The developer's version is also free and open-source (GPLv2 license), but does require the installation of additional software libraries; detailed instructions are provided at https://github.com/CellProfiler/CellProfiler/wiki.
- Download the chosen version of the software from http://www.cellprofiler.org/download.shtml.CellProfiler downloads in less than one minute with a 1 Gbps internet connection.
Follow the installation instructions from the web page to install CellProfiler. If you encounter difficulties on this step, consult the installation instructions (http://www.cellprofiler.org/install.shtml), or visit the online forum (http://www.cellprofiler.org/forum) to see if the problem has been encountered and solved before.
Download the example pipeline and run on example images
-
4Download the example pipeline and images called “Yeast colony classification” from http://www.cellprofiler.org/examples.shtml; the downloaded file is called “ExampleYeastColonies_BT_Images.zip”. After downloading the ZIP file, decompress the file contents, which will yield a single folder “ExampleYeastColonies_BT_Images” containing the pipeline and images.If decompression of the downloaded file does not occur automatically, the file should be unzipped manually by double-clicking it, which should launch the decompression software. The file contents should be unzipped to a location on your computer or otherwise accessible from your computer.
-
5Double-click on the pipeline file “ExampleYeastColonies_BT.cppipe” in the unzipped folder to start CellProfiler and load the pipeline.When the example pipeline is loaded, a dialog box appears with further instructions. If the pipeline is later saved, this box will not appear when the pipeline is loaded again.
-
6Follow the instructions in the dialog box to provide example images to analyze using the Images module in CellProfiler.Setting this module tells CellProfiler the location of the images to analyze. Click on the Images module (if not already selected), which is the first module listed in the “Input modules” panel near the top left of the main CellProfiler window. You will then see the File list panel in the module settings panel on the right, which is indicated with the text “Drop files and folders here”. Using your computer's file browser, drag-and-drop the example image folder into the File list panel.
-
7Run the example pipeline by pressing the “Analyze Images” button.This step demonstrates how image processing typically proceeds. You can find more details on performing an analysis run in CellProfiler by following the instructions found in Help > Running Your Pipeline, accessible from the main menu bar. You will note that a display window opens for each module in the pipeline, showing the module results. The user can toggle whether the windows are shown or hidden by clicking the eye-shaped icon in the pipeline; this is a useful step during the testing phase when you no longer need to see the display for modules that you have already optimized. Under normal circumstances, you will be processing more than one image, and the module windows will refresh upon completing the analysis of each image.See Help > Troubleshooting Memory and Speed Issues if you obtain an “Out of memory” error.
Select a sampling of images for testing
-
8Using your computer's file browser, create a test image folder and copy several test images into it.These images will be used to optimize the module settings in your pipeline. To preview your settings thoroughly and ensure accurate results from your entire experiment, be sure to select a variety of images from the entire collection of images. For example, choose one or two images from the beginning, middle, and end of the experimental run, rather than choosing images that were collected near each other. Alternately, choose samples of positive and negative control images in order to span the range of visible characteristics.
-
9
Using your computer's file browser interface, create a test output folder.
-
10
In CellProfiler, set the Default Output Folder by pressing the “View output settings” button and in the resulting module settings panel, pointing the “Default Output Folder” setting to the test output folder you created.
Adjust example pipeline for test images
-
11Adjust the plate template image for your particular test images (if needed).This pipeline is flexible regarding the placement of each plate within the image, in that specific modules allow for CellProfiler to find the plate anywhere within the image, even if the position of the plate within the image varies from sample to sample. This is accomplished by using a single template image, “PlateTemplate.png”, to represent the image region corresponding to the interior of the plastic plate.The template image will be used later in the pipeline to remove the edges and exterior region of the plastic plate, operating on the assumption that the plates are the same size from image to image. If your test plates are not the same size as those in the example images, you will need to create your own plate template. To do this, use Adobe Photoshop (or an alternative image modification program) to modify and save one of your images to use as a template, making the center of the plate pure white, and the surrounding background pure black. Alternatively, resize the PlateTemplate.png image in Photoshop or in CellProfiler (using a pipeline consisting of the analysis modules Resize and SaveImages).
-
12Select the desired images using the Images module (see Figs. 14.17.1).Setting this module tells CellProfiler where to retrieve images. Click on the Images module (if not already selected), which is the first module listed in the “Input modules” panel, to display the File list panel on the right. If files are already listed in the File list panel, you can clear this list by right-clicking in the panel and selecting “Clear File list” from the menu that appears. The panel will then display the text “Drop files and folders here”. Using your computer's file browser, drag-and-drop the test images into the File list panel.You can drag-and-drop as many or as few images as you want to test, and you can drag-and-drop individual image files or entire folders of images. The images need not be named or organized in a particular way in order to use CellProfiler.
-
13Describe the yeast plate images using the NamesAndTypes module.This module is used to assign a user-defined name to particular images or channels, and define their relationship to one another.When analyzing images of yeast plates, or other samples in which there is only one image per plate, all that is needed is to change the rule criteria setting to look for text that all of the images have in common (e.g., a file extension such as .tif). The current example pipeline looks for an exact match between “6-1.jpg” and the file listing you provided in the Images module. If the image file names do not have precise text in common, the “Contain regular expressions” operation in the rule criteria might be useful.When there are pairs of images from the same plate (for example, one brightfield and one fluorescence image of the same biological sample), the typical method to denote the image types is to indicate the particular piece of text in the file name that is unique to that image type. For example, if all of the light images contain “LT” and all of the fluorescence images contain “FL” in the file names, you can create one rule criteria to match files that contain “LT” in the filename, and another to match “FL”; assuming that the same number of images exist for each type, these two channels will be automatically paired up. Any number of channels can be analyzed; for example, multiple brightfield and fluorescence images. See the help for the NamesAndTypes module for more information.
-
14Specify the template image using the NamesAndTypes module.The example pipeline is configured to use a single binary (black/white) plate template image to align a single yeast plate image. Ideally for an analysis run, a single template image (from step 11) will be used to align multiple yeast plate images. To configure the pipeline to do this, remove the current PlateTemplate setting by clicking the “Remove this image” button under it, then click the “Add a single image” button. Click the “Browse” button to show a listing of the current image files, and select the plate template image you used or modified from step 11. Assign this image the name “PlateTemplate” to maintain consistency with the rest of the pipeline.
-
15Set the image type to “Binary image”, that is, black-and-white. Even if your original template image is saved as grayscale instead, this setting will convert it into the proper binary type by rendering any non-zero pixels as white. Split images using the ColorToGray module.ColorToGray splits the original color images of yeast plates into three separate images: red, blue, and green. Each of these images is then converted to an image with varying grayscale intensities. The images are used for separate purposes later in the pipeline. For example, the red channel is used to identify all colonies (white and red) in the example pipeline. A different channel or combination of channels might be better suited to your own images. This can be decided by running the pipeline (pressing the Analyze Images button) and examining the output from the ColorToGray module. Your decision can be based on a visual inspection of which image channel (red, blue, or green) shows the best contrast for all colonies as compared to background, or you can check the contrast in each channel numerically by hovering the mouse over each channel in the display window to show the pixel intensity in the bottom-right of the window. Note: If the original images are collected in grayscale rather than color, you will not need to use the Color To Gray module. Simply delete this module from your pipeline, change the image type for OrigColor from “Color image” to “Grayscale image” in Names And Types, and adjust the downstream image names to allow the Names And Types module to directly feed to the next module.
-
16Calculate corrections for uneven illumination using the CorrectIlluminationCalculate module.Because most images are taken with uneven lighting across the image (or uneven thickness of the agar, resulting in a similar effect), it is important to correct the images prior to further processing. Three CorrectIllumination Calculate modules are used, one for each channel of the original image (red, green, and blue). The goal is to produce an image (called the “illumination correction function”) for each channel that represents smooth shading across the plate; this image will be subtracted from the image in the next step.There are several options for calculating the illumination correction function. The “Background” option calculates the illumination correction function across each color channel while ignoring the colonies, so that background can be subtracted in the next step. The background option finds the minimum pixel intensities across the image within blocks of a given block size.Depending on the image, it may be necessary to adjust the block size before calculating the optimal illumination correction function; the block size should be slightly larger than the diameter of the largest colony expected in the experiment. Note also that within this module, a smoothing function is applied so that the illumination correction function resembles the uneven illumination pattern present in the image. The smoothing size is set automatically and displayed in the figure window. You can adjust this setting if the smoothing size does not seem appropriate upon visual inspection. The smoothing should be set high enough so that individual colonies are no longer visible in the illumination correction function. Once this decision is made for the setting, it will apply to all images analyzed in the set.The uneven illumination pattern is likely to change when images are acquired on different days, under different conditions, or when the thickness of the agar plate varies. Therefore, for yeast plates, it is usually appropriate to use the “Each” option so that the illumination correction function is calculated for each individual plate. The “All” option should only be used if the entire set of images is well aligned and shows the identical shading pattern. Refer to Critical Parameters for further information.
-
17Apply the illumination correction function using CorrectIllumination Apply.This module applies the illumination correction functions, thus normalizing the red, green, and blue channels. The option to “Divide” or “Subtract” depends on the method used in the CorrectIllumination Calculate module. When the “Background” option is used in the CorrectIllumination Calculate module, “Subtract” is used in the CorrectIllumination Apply module as described in the module help. The resulting illumination-corrected images should no longer show an uneven illumination pattern across the background of the plate: they will have a darker background, with the colonies nicely visible.
-
18Combine the corrected blue and green images into one image using the ImageMath module.This module is used to add the pixel intensities from the two images together to produce a new combined image. This image will be used later in the pipeline so that the blue and green contributions to the red channel can be subtracted in the ImageMath module. This is needed for measuring the “redness” of each colony.
-
19Align the PlateTemplate within the plate images using the Align module.The Align module allows the interior of the plate to be found in an image, even if there is experimental variation in the plate placement.
-
20Mask the images using the MaskImage module.The term “masking” refers to using a binary (i.e., black-and-white) image to define which pixels in another image to keep and which to ignore. In each of the MaskImage modules in this example, the “Aligned Plate” image is used to mask the plate edges from each of the plate images, thereby effectively removing them from consideration. This prevents erroneous detection of colonies outside the plate and at the plate rim.
-
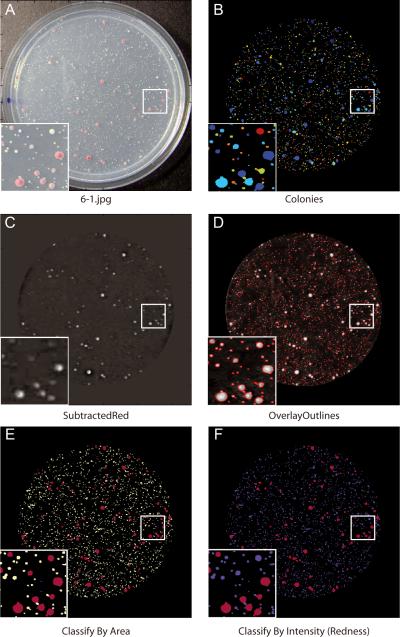
21Subtract the MaskedCombined image from the Masked Red Plate image using another ImageMath module to create an image called “SubtractedRed” (Fig. 14.17.2C).The resulting image accurately displays the “redness” of each colony. This is because white colonies have high pixel intensity values in all three channels (red, green, and blue), but red colonies have high pixel intensity values in the red channel only.
-
22Use the Identify Primary Objects module to identify all yeast colonies (white and red) within the plate (Fig. 14.17.2B). Use the red channel image, since both red and white colonies are bright in this image. Adjust the minimum and maximum diameter (in pixel units) depending on the expected colony size in your own images.It may also be necessary to adjust the maximum suppression neighborhood, which controls the distance allowed between the centers of the colonies, and is important for determining whether an object is an individual colony or a clump of colonies. CellProfiler is usually capable of separating clumped colonies provided the Identify Primary Objects settings are appropriate for your images. In some cases, (such as in the example images), you will notice that colonies are inappropriately clumped together - this is unfortunately unavoidable due to the poor resolution and the lossy JPG format of these example images.In this pipeline, Identify Primary Objects separates the clumped colonies in a two-step process. First, the number of colonies in a clump is identified, and second, Cell Profiler decides where to draw the boundaries between the clumped objects. For the first step (identifying the number of colonies in a clump), two commonly used options are “Intensity” and “Shape”. “Intensity” tends to work well if the objects are brighter in the center and dimmer at the edges, whereas “Shape” works well when the objects have definite indentations where clumped objects touch each other (especially if the objects are round). Since the yeast colonies are fairly circular, they are therefore best analyzed with the “Shape” option. Once the number of colonies in a clump is identified, Cell Profiler carries out the second step of deciding where to draw the boundaries between clumped objects. Here, the options include “Distance” and “Intensity”, where the “Distance” option draws boundary lines midway between the object centers, and the “Intensity” option draws boundary lines at the dimmest line between objects. Yeast colonies usually do not have dim lines separating them, so the “Distance” option is preferred.As shown in Figure 14.17.2 B, the identified colonies appear as arbitrary colors. These colors help you to determine if each colony is identified and separated from its neighbors properly. When two colonies are touching, but identified separately using the declumping settings, each object will appear as a distinct color. The color scheme can be changed using File > Preferences. If you wish to include objects identified at the edge of the plate in the analysis, change “Discard objects touching the border of the image?” to “No.”
-
23Use Measure Object Intensity to measure the intensity of each colony in the “Subtracted Red” image.Adjustments should not be necessary, unless you have added more Identify modules to detect other objects in the images, or if you want to measure the intensity of a different color for the colonies. The measurements displayed in the figure window are the average measurements of the colonies, but the individual colony measurements are available for export by using the Export To Spread sheet module (step 28).
-
24Use Measure Object Size Shape to measure morphological features.Several features can be measured for each colony. The average measurements for all colonies in the image are displayed in the figure window. The individual colony measurements are available for export by using the ExportToSpreadsheet module (step 28).
-
25Use the ClassifyObjects modules to classify each colony for the desired parameters.Objects can be classified by any feature that has been measured upstream in the pipeline, in any number of bins.There are two groups of settings in this module for classifying each colony in the example pipeline. The first group of settings classifies the colonies based on area (Fig. 14.17.2E) in a histogram with three bins. You might wish to adjust the thresholds for distinguishing tiny, small, and large colonies. In figure 14.17.2E, colonies are classified and labeled with different colors: tiny (blue), small (aqua) and large (yellow). The second group of settings classifies the intensity of the colonies (Fig. 14.17.2F) into two bins, for distinguishing white and red colonies, which are shown as blue and red, respectively.
-
26
Use OverlayOutlines to overlay the colony outlines on the “Masked Red Plate” image (Fig. 14.17.2D).
-
27Use the SaveImages module to save the image with the overlaid outlines to the output folder.Because there are many intermediate image processing steps, Cell Profiler never saves processed images unless specifically requested via a Save Images module. In Save Images modules, you can adjust Cell Profiler to save any of the images produced in the pipeline, such as the results of the Classify Objects module.
-
28Using the Export To Spread sheet module, the data can be exported to a comma-delimited text file that can be opened in Microsoft Excel.You can export measurements for each individual colony, and/or export the means, medians, or standard deviations of the colony measurements within each image by selecting the setting for each aggregate statistic.
-
29Add additional modules to adjust your pipeline as needed.There are dozens of optional modules that can be added to customize your pipeline. These include additional image processing steps, saving processed images to the hard drive, making additional types of measurements, defining sub-regions of each colony for analysis, measuring neighbor relationships between colonies, etc. For a detailed description and instructions, see the CellProfiler manual. Modules are added, removed, and rearranged in the pipeline using the [+], [−], [^] and [v] buttons below the pipeline.
-
30If you would like to use Cell Profiler's built-in data tools to explore your data after the analysis run (see step 31), you will need to produce an output file in a compatible format. Press the “View output settings” button and select either “HDF5” or “MATLAB” from the Output file format setting.The output file is a file where all information about the analysis as well as any measurements will be stored to the hard drive. It can either be stored as (a) a .mat file; or (b) a .h5 file in the HDF5 format. Both file formats are readable by CellProfiler or MATLAB.Note that these output files are distinct from the files produced by the export modules such as Export To Spread sheet or Export To Database; selecting “Do not write MATLAB or HDF5 files” will not affect the output from the export modules.
Figure 14.17.2.
(A) The original plate image. (B) The colonies identified. (C) The Subtracted Red image (or “the red channel”). (D) All identified colonies outlined. (E) The colonies classified by area. (F) The colonies classified by “redness”.
Run adjusted pipeline on your images
-
31
Once you have tested the pipeline with your test images, you are ready to run the pipeline to process all of your images. If the number of images is manageable for a single computer, do this by selecting the Images module, clearing the File list panel (by selecting Edit > Clear File List from the main menu bar), and drag-and-dropping the folder(s) containing the entire set of images you want to process.
-
32Hide the display windows during the analysis run, by selecting Window > Hide All Windows on Run from the main menu bar.You can monitor progress using the display windows that open for each module (see step 7). However, having CellProfiler display these windows during the analysis run will slow processing as the computer refreshes each one. This is especially the case if a large number of images are being analyzed, so it recommended that these windows remain closed.
-
33
Change the Default Output Folder to a different location, if desired.
-
34Change the output file name in SaveImages, if desired.If you are using a SaveImages module, Cell Profiler will save the processed images to the Default Output Folder during each cycle. Even before processing has completed on the entire set, you can open these processed images to check whether the processing is accurate by examining whether the outlines properly identify colonies.
-
35Click the “Analyze images” button to begin the analysis run.If not, you can cancel the pipeline using the “Stop Analysis” button and adjust settings in the pipeline appropriately, using the guidance for each module above, before beginning processing again.For sets of images too large for a single computer, see the Alternate Protocol to run images on a computing cluster.
Explore data
-
36The comma-delimited text file created by Export To Spread sheet can be opened in a spreadsheet program such as Microsoft Excel or more sophisticated analysis programs such as R, GraphPad Prism or MATLAB. See http://www.cellprofiler.org/interfaces.shtml for a partial listing of compatible programs.Note that Microsoft Excel 2013 has a spreadsheet limit of 1,048,576 rows by 16,384 columns, with a column width limit of 255 characters; other programs such as Open Office's Calc and GraphPad's Prism may have different limits. If exporting a large dataset, exporting the data to a database may be a better option; see the Alternate Protocol, part 3.
-
37Cell Profiler has several data tools for analysis, including tools for plotting histograms and scatter plots. To use the tools after analysis completes, select “Data Tools” from the main menu bar, then select one of the following:
- Display Histogram: To display your analyzed data in a histogram, the tool will prompt you to choose the output file (.mat or .h5) from your analysis. Follow the prompts to select the data to be displayed.
- Display Scatter Plot: To visualize your data as a one- or two-dimensional scatter plot, the Display Scatter Plot tool will prompt you to choose the output file (.mat or .h5) from your analysis and the features you would like to visualize.
Alternate Protocol
Analyzing Images on a Computing Cluster
Depending on the number of images and size of the pipeline, it may be necessary to use a computing cluster. While a few hundred image sets can usually be run on a standalone desktop within a few hours, users should consider running larger image sets on a computing cluster in batch mode to speed the processing. Cell Profiler can create batch files to run any pipeline on a Linux cluster.
- Download the appropriate version of Cell Profiler for Linux and install it on a computing cluster. See the installation instructions as well as Help > Other Features > Batch Processing and the online forum (http://www.cellprofiler.org/forum).You will need to decide between the developer's version and the compiled version for Linux installation. There are a wide variety of computing clusters in existence; one compiled version of Cell Profiler for 64-bit cluster computers running CentOS 6 and similar distributions is available for download at http://cellprofiler.org/download.shtml. If this version is not compatible with a particular cluster, the developer's version (source code) can be downloaded from the GIT repository at https://github.com/CellProfiler/CellProfiler (you should clone the branch corresponding to the current release version) and re-compiled on a representative cluster computer.
- Create the batch files for running your analyses on a computing cluster.
- Add the Create Batch Files module (in the “File Processing” category) to the end of the pipeline (using the + button below the pipeline panel) and configure it appropriately, according to the Help for the module.
- Add the Export To Database module if your dataset will be large and require analysis in a database environment. It should be added after all other modules in your pipeline, but before the Create Batch Files module.
- Click on the “Analyze Images” button. Cell Profiler will not analyze all images but instead will produce a file “Batch_data.h5” containing the necessary information for batch processing.
- Submit the batches to your cluster for processing using this file. See Help > Other Features > Batch Processing for details.
- Manage data processed on a computing cluster.When processing your images in batches on a cluster, the resulting measurements will be written to separate data files for each batch. There are two options to access your results. (A) If the resulting data files are not over whelmingly large, you can merge the output files into a single output file using the Data Tool “Merge Output Files.” Then, Cell Profiler's built-in Data Tools can be used to visualize the data from this output file (see step 30), or the Data Tool “Export To Spreadsheet” can be used to export the data into a comma-delimited text file. (B) Most often for large image sets, you will prefer to export the resulting data to a MySQL database for further analysis and exploration. In this case, be sure to use the Export To Database module in the pipeline, as described in step 2b.
Commentary
Background Information
As research laboratories continue to adopt high-throughput sample preparation and data acquisition, visual inspection of images becomes less desirable. Traditionally, biologists visually inspected images and drew meaningful conclusions, but these conclusions were usually qualitative and, because measuring more than a few metrics was rarely possible, valuable information was often overlooked. Using automated image analysis programs like CellProfiler, visual assays can be scaled up from a few samples to hundreds of thousands of samples. By analyzing the size, shape, texture, and color intensity of every object in each image quantitatively, new types of experiments can be quickly and accurately accomplished. Unlike more user-interactive programs like Adobe Photoshop or ImageJ/Fiji, Cell Profiler contains modules designed to be mixed and matched for automated high-throughput image analysis.
Critical Parameters
It is absolutely critical that images be acquired using a uniform protocol that is followed as strictly as any traditional biochemical procedure.
The lighting and image acquisition apparatus (camera or scanner) should be kept as constant as possible throughout the entire sample set, including parameters like exposure time, shutter speed, focus, lighting conditions, and sensitivity.
Air bubbles and noticeable imperfections on the agar plate should be minimized. Large imperfections might be subtracted effectively with the illumination correction steps built into the pipeline, but—depending on the severity of the imperfections—Cell Profiler may incorrectly identify imperfections as colonies.
When capturing images to be analyzed by Cell Profiler, it is best to use lossless image file formats when possible, such as BMP, GIF, PNG, or TIF. Although JPG images are commonly used for photography, the file compression for JPG files results in artifacts that can hinder accurate image processing and measurement. Thus, even though the example images are JPG files, this format should be avoided when acquiring experimental images. If the JPG format must be used, be sure to set the quality to maximum. For further information, see Internet References. More tips on image acquisition have been published (Pearson 2007).
When you are designing your plate template, it is best to make as much of the interior of the plate white as possible. The plastic plate edges, and the remaining parts of the image, should be black.
Troubleshooting
If a module fails, an error message will appear. In addition to the user manual, Cell Profiler has a forum (http://www.cellprofiler.org/forum) for posting questions and reporting problems, which is frequently monitored by the developers.
If your computer does not have adequate memory, you will receive an “Out of Memory” error. This can often be ameliorated by reducing the number of display windows shown during processing by selecting Window > Hide All Windows on Run from the main menu bar (see step 32).
Anticipated Results
Once the pipeline from the standard protocol is completed, the measurements will be saved in output files (comma-delimited spreadsheet files, if selected in step 28, or .mat or .h5, if selected in step 30). In addition, a processed image will be saved to the hard drive for each input image, showing the cropped plate with the colonies outlined.
Time Considerations
Downloading and installing the software should take less than 10 minutes, and running the example pipeline only a few minutes more. Depending on how much your images differ from the examples, a day should be allotted to adjust the pipeline to your images and learn the basics of how to operate Cell Profiler before proceeding to analyze all of your images. The setup time for an analysis is the same whether a handful or hundreds of thousands of images are processed. Tens of thousands of images can be routinely analyzed per experiment.
Once the pipeline has begun to cycle through your images, Cell Profiler will run until all images are analyzed, at a rate of roughly one image per minute. Version 2.1 of Cell Profiler is optimized to take advantage of multiple processing cores, so the total analysis time will be reduced in proportion to the number of processors your computer has. After completing your first analysis on a set of your own images, it usually takes only 15 minutes to double check the settings on a few test images and begin running a new batch of images.
Acknowledgements
The authors thank Matthew Veneskey for writing assistance and acknowledge funding from the National Institutes of Health (R01 GM089652 to AEC).
Footnotes
Internet Resources
The Cell Profiler home page allows free access to the software, example pipelines, and the discussion forum.
http://en.wikipedia.org/wiki/Lossy_data_compression
A detailed web page relevant to the choice of image file formats which discusses lossless versus lossy image compression. Example images are available that demonstrate the difference in quality in lossless images versus lossy images.
Literature Cited
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahle J, Kakar M, Steen HB, Kaalhus O. Automated counting of mammalian cell colonies by means of a flat bed scanner and image processing. Cytometry A. 2004;60(2):182–8. doi: 10.1002/cyto.a.20038. [DOI] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, Bray M-A, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Improved structure, function, and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42(1):71–5. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- Memarian N, Jessulat M, Alirezaie J, Mir-Rashed N, Xu J, Zareie M, Smith M, Golshani A. Colony size measurement of the yeast gene deletion strains for functional genomics. BMC Bioinformatics. 2007;8:117. doi: 10.1186/1471-2105-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson H. The good, the bad and the ugly. Nature. 2007;447(7141):138–40. doi: 10.1038/447138a. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria.: 2008. [Google Scholar]