Abstract
Studying how genetic predispositions come together with environmental factors to contribute to complex behavioral outcomes has great potential for advancing our understanding of the development of psychopathology. It represents a clear theoretical advance over studying these factors in isolation. However, research at the intersection of multiple fields creates many challenges. We review several reasons why the rapidly expanding candidate gene-environment interaction (cGxE) literature should be considered with a degree of caution. We discuss lessons learned about candidate gene main effects from the evolving genetics literature and how these inform the study of cGxE. We review the importance of the measurement of the gene and environment of interest in cGxE studies. We discuss statistical concerns with modeling cGxE that are frequently overlooked. And we review other challenges that have likely contributed to the cGxE literature being difficult to interpret, including low power and publication bias. Many of these issues are similar to other concerns about research integrity (e.g., high false positive rates) that have received increasing attention in the social sciences. We provide recommendations for rigorous research practices for cGxE studies that we believe will advance its potential to contribute more robustly to the understanding of complex behavioral phenotypes.
Keywords: genetics, candidate genes, GxE, gene-environment interaction
Background of this Article
There have been radical shifts as to belief about whether human behavior is more strongly determined by genes or by environment over the course of scientific history. Different fields seemingly advocated for the importance of one over the other (the so-called nature versus nurture debate), with some camps studying genetic influence and others studying environmental factors. It is now widely accepted that both genetic and environmental influences are important, and characterizing how these influences come together to impact outcome, ie the study of gene-environment interaction (GxE) has become an important area of study across multiple disciplines. That said, few research topics have generated more controversy and less clarity than the study of candidate gene by environment interaction (cGxE) in complex behavioral outcomes. Following the publication of cGxE studies in high profile scientific journals (Caspi et al., 2002; Caspi et al., 2003), the last decade has witnessed an explosion of interest in this area. There has been an exponential increase in the number of cGxE studies published, with researchers from diverse backgrounds routinely incorporating cGxE components into their studies. However, there has been growing skepticism about the replicability of many of these findings (e.g., Risch, Herrell, Lehner, Liang, Eaves, Hoh, Griem, Kovacs, et al., 2009) and increasing concern about the quality of this rapidly expanding literature.
This concern led the National Institute on Alcohol Abuse and Alcoholism (NIAAA) to sponsor a workshop in January 2013 that brought together a small group of researchers to discuss these challenges and provide recommendations for how to move the field forward. Those discussions formed the foundation for this paper, in which we review a number of reasons why the existing cGxE literature should be considered with a degree of caution. This is not to imply that true discoveries are absent in the literature. However, there are reasons to be concerned about the methods employed and the conclusions drawn from many cGxE studies. Drawing from accumulating findings in psychiatric genomics1, we consider potential pitfalls and logical inconsistencies with some of the extant cGxE literature. We discuss ways of refining the development of cGxE hypotheses, conducting statistically rigorous analyses, and interpreting findings within the broader context of genetics research – all directions that we believe hold promise for advancing the potential of cGxE studies to contribute more robustly to the understanding of complex behavioral phenotypes.
History
The idea that genetic or biological predispositions are likely to interact with environmental factors to contribute to psychiatric and substance use disorders has been entertained for quite some time (Whytt, 1765). Long before it was feasible and cost-efficient to measure specific genes, twin studies documented that the importance of overall genetic influences (i.e., heritability) could vary considerably as a function of measured environmental factors (K. S. Kendler & Eaves, 1986). For instance, Kendler and colleagues found that people at highest genetic risk for depression (i.e., individuals with an identical twin with a history of depression) were significantly more likely than individuals not carrying a genetic predisposition to have an onset of the disorder in the presence of exposure to a severe stressful life event, suggesting that genetic factors influence the risk for major depression in part by altering individual sensitivity to the depression-inducing effects of stressful life events (K. S. Kendler et al., 1995). With methodological advances that allowed twin researchers to model how genetic influences change as a function of the environment (Button et al., 2009), studying gene-environment interaction (GxE) became a popular area of research in behavior genetics (Button, Lau, Maughan, & Eley, 2008; Dick, Bernard, et al., 2009; Dick, Pagan, Holliday, et al., 2007; Dick, Pagan, Viken, et al., 2007; Dick, Rose, Viken, Kaprio, & Koskenvuo, 2001; Dick, Viken, et al., 2007; Harden, Hill, Turkheimer, & Emery, 2008; Purcell, 2002; R. J. Rose, Dick, Viken, & Kaprio, 2001; South & Krueger, 2008).
The accumulating body of research has demonstrated that the importance of genetic influences can vary dramatically as a function of environmental context; importantly and alternatively phrased, the importance of environmental influences can vary dramatically as a function of genetic factors. For example, it has been demonstrated that genetic influences on adolescent substance use and externalizing behavior are far stronger under conditions of low parental monitoring (Dick, Pagan, Viken, et al., 2007; Dick, Viken, et al., 2007), high peer deviance (Button et al., 2009; Dick, Pagan, Holliday, et al., 2007; Dick, Pagan, Viken, et al., 2007; Harden et al., 2008), and state, school and neighborhood conditions that provide reduced social monitoring and enhanced opportunity to use (J. D. Boardman, 2009; Dick, Bernard, et al., 2009; Dick et al., 2001; R. J. Rose et al., 2001). However, this body of GxE research did not gain widespread recognition outside the field of twin research. It was not until the influential Science publication by Caspi and colleagues (Caspi et al., 2003) attributing part of the genetic sensitivity to the depressogenic effects of stressful life events to variations in a specific DNA sequence (a polymorphism in the serotonin-transporter-linked polymorphic region [5-HTTLPR]) that gene-environment interaction research became a widely-recognized area of study outside the field of behavior genetics. However, an important distinction arose between the GxE work conducted in the field of behavior genetics, and the widespread adoption of GxE by other fields, particularly the social sciences. Historically, the research conducted by behavior geneticists focused on “latent” genetic influences. This means that the importance of genetic factors is estimated statistically by phenotypic similarity across individuals with different degrees of genetic and environmental sharing, using methodologies such as family, twin, and adoption studies (Bergeman & Plomin, 1989). This method estimates the overall importance of genetic effects on a phenotype, i.e., the total contribution of all genes influencing the phenotype. Gene-environment interaction in this context means that the overall importance of genetic variance differs across environments. In contrast, most GxE research in fields outside behavior genetics has studied measured candidate genes. These studies test whether the association of a specific genetic variant with a given outcome varies across different environments. We refer to these studies as cGxE (where cG refers to candidate gene), and they are the focus of this review.
The publication of several high profile cGxE studies (e.g., MAOA × maltreatment in antisociality; Caspi et al., 2002), as well as the technological advances in genetics that made genotyping accessible and cost-efficient, likely contributed to the dramatic increase in cGxE research. Regardless of the validity and reproducibility of those initial efforts (which continues to be debated; Brown & Harris, 2008; Clarke, Flint, Attwood, & Munafo, 2010; Culverhouse et al., 2013; Karg, Burmeister, Shedden, & Sen, 2011; Munafo, Durrant, Lewis, & Flint, 2009; Risch, Herrell, Lehner, Liang, Eaves, Hoh, Griem, Kovas, et al., 2009), they left their mark on the field by creating widespread recognition of the potential importance of the interplay between genetic and environmental factors in developmental pathways underlying the etiology of behavioral outcomes in ways that the latent GxE work of behavior geneticists had failed to do. In the excitement surrounding the initial cGxE findings, and spurred by funding initiatives that encouraged research in this area, investigators from disparate backgrounds incorporated measured genotypes into their studies. In the wake of historical tension surrounding the relative importance of genetic versus environmental effects (the so-called nature versus nurture debate), GxE provided a conciliatory framework that could facilitate a synthesis of scientific fields that had historically been at odds with one another.
However, early enthusiasm for cGxE findings has waned as the number of failures to replicate original findings mounted. In many ways, the progression of cGxE studies has closely paralleled the trajectory of studies of the main effects of candidate genes, with early enthusiasm and adoption of genotyping candidate genes giving way to a literature plagued by small studies, failures to replicate, and a proliferation of novel findings with effect sizes that appeared at odds with what was subsequently found with well-powered studies (Neiswanger, Kaplan, & Hill, 1995). However, the study of genetic main effects has advanced dramatically since the early days of candidate gene research. We believe that what has been learned about the genetics of complex behavior from studies of genetic main effects yields insights into previous cGxE studies and ways to improve such research in the future. We begin by providing a broad review of developments in the field of psychiatric genetics over the past decade and discuss how this knowledge can inform studies of cGxE.
Early Statistical Genetics: Linkage and Candidate Gene Studies
The field of statistical genetics, focused on finding genes that contribute to behavioral outcomes and disorders, has undergone rapid advances over the past decade. As our knowledge about genetics has progressed, so too have the methodologies favored for gene identification (See Figure 1 for an overview of common gene finding strategies). Linkage and candidate gene studies were early gene identification strategies, believed to have complementary strengths.
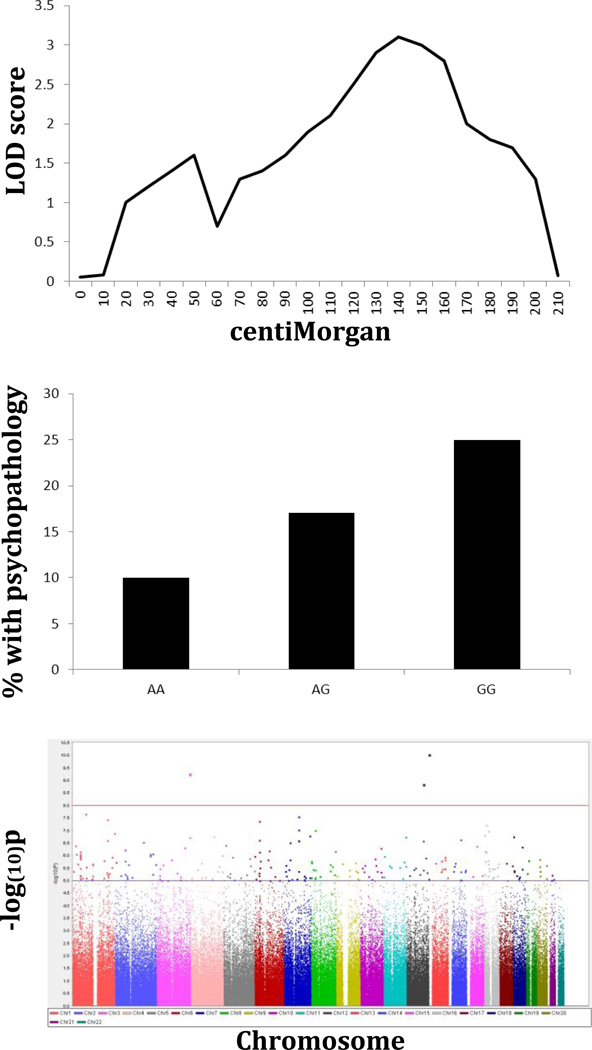
Figure 1.
Overview of common gene finding strategies.
Results of a linkage analysis are often depicted using a Logarithm of Odds (LOD) score plot that depicts the genomic region(s) (measured in genetic distance, or centiMorgans, where 1cM roughly equals 1,000,000 base pairs) with the linkage peak(s), or the highest LOD scores. The LOD is the ratio of the likelihood that there is excess allele sharing to the null hypothesis of no excess allele sharing. The adjacent diagram illustrates these results for one chromosome. Elevated LOD scores indicate that the genomic region is shared by affected relative pairs more often than expected by chance alone, suggesting there is a gene in the region contributing to the outcome under study.
Results from a classical candidate gene study are illustrated. The prevalence of psychopathology increases in an additive fashion with increasing copies of the risk allele “G”.
The adjacent figure illustrates the results of a genome-wide association study (GWAS). Each dot represents the negative logarithm (base 10) of the p-value for an individual association test (usually hundreds of thousands or millions of SNPs tested across the genome). Therefore, a p-value of 5 × 10−8, or the threshold for genome-wide significance, as denoted by the horizontal solid line, is noted at the midway point between 7 and 8. The dotted line reflects a p-value of 1 × 10−5, indicating SNPs of interest. The x-axis denotes physical positions (in base pairs) across each of the 22 autosomal chromosomes. In the hypothetical example, there are three “hits” (SNPs) that surpass the genome-wide significance threshold, and many more SNPs that are suggestive.
Linkage studies agnostically scanned the mapped genome by looking for chromosomal regions that were shared among affected family members (suggesting there was a gene in that region that contributed to the disorder). The advantage of linkage was that it did not require any a priori knowledge of the underlying biology of the outcome, in theory making it possible to discover new genes involved in the outcome that could expand our understanding of the biology of the disorder. Linkage studies were used to successfully identify many genes that contributed to Mendelian disorders, where a single gene following a straightforward inheritance pattern with a major impact on outcome was present (Gusella et al., 1983; Murray et al., 1982; Tsui et al., 1985). However, linkage methods were less successful when applied to complex behavioral outcomes where many genes are likely to be involved, each having just a small effect on the behavior, along with the environment.
In contrast to the hypothesis-free linkage approach, candidate gene studies focused on genes, and variants within those genes, that were hypothesized to have biological relevance to the outcome of interest. In this way, candidate gene studies had the advantage of being more precise than linkage studies in that they had the potential to pinpoint specific genes or genetic variants, rather than just specific chromosomal regions. However, they relied on the investigator to correctly “guess” what genes were biologically relevant to the outcome. Despite thousands of candidate gene publications on behavioral phenotypes, the approach remains controversial, and very few candidate gene findings are widely accepted within the genetics community. Over time, it has become clear that the genetic architecture of behavioral traits is highly complex, that effect sizes of genetic polymorphisms are likely to be small (see below), and that scientists’ ability to predict a priori which genes are likely to be relevant to a behavioral outcome has been very poor (F.J. Bosker et al., 2011; Colhoun, McKeigue, & Davey Smith, 2003; Need et al., 2009; P.F. Sullivan et al., 2008).
Later Statistical Genetics: Genome-Wide Association Studies (GWAS)
As the cost of genotyping dropped during the 2000s, it became possible to conduct association tests as are done in candidate gene studies, but across the entire genome. Such genome-wide association studies (GWAS) are hypothesis-free as with linkage studies, but have much higher power to detect small effects of common variants. In GWAS, hundreds of thousands to millions of genetic markers known as single nucleotide polymorphisms (SNPs) are genotyped across the genome in an attempt to identify common variants that are associated with a particular outcome (disorder, behavior, etc.), suggesting that a particular genetic variant (or one very nearby) contributes to the outcome. In a sense, GWAS combines the advantages of linkage studies (agnostic screening across the genome) and candidate gene studies (more precise localization of the gene/genetic variant).
GWAS were made possible by the rapid discovery of many more genetic variants through the International HapMap Project (hapmap.ncbi.nlm.nih.gov), which had the goal of developing a public resource to catalogue normally occurring genome-wide variation in SNPs by annotating the sample genomes of small groups of ethnically homogenous individuals (e.g., Caucasians of European descent, or CEU, are represented by Utah residents with ancestry from northern and western Europe collected in 1980) to create what is known as a reference panel. In addition to providing a database for identification and comparison of the polymorphic nature of SNPs, HapMap also allows for the examination of the extent to which neighboring SNPs are correlated with each other via a population genetics process called linkage disequilibrium (LD). When two or more SNPs are in high LD (e.g., correlations > 0.8), if one of the SNPs is genotyped then the genotype at the others can be probabilistically inferred via imputation. The ability to infer surrounding genotypes based on LD patterns means that we can now cost-efficiently scan the genome with a much smaller subset of markers than previously needed (e.g., in the range of 370,000–600,000) and impute the remaining commonly occurring markers across the genome. Whereas early attempts to impute variation were restricted to common SNPs (>5% minor allele frequency), the landmark 1000 Genomes Project (http://www.1000genomes.org/) used a similar strategy to identify less common SNPs (≤1%). Our growing knowledge of genetic variants across the genome, coupled with exponential decreases in costs of high density GWAS arrays (>1 million SNPs at <100 USD per participant currently), now allows investigators to interrogate over 10 million polymorphisms in relation to outcomes.
Two primary points have become apparent over the last several years from GWAS: (1) the effect sizes associated with individual genetic variants are very small, usually with odds ratios (ORs) on the order of 1.1, and (2) our ability to select a priori which genes are viable candidates for psychiatric and substance use disorders has been poor (K. S. Kendler, 2013; P. F. Sullivan, Daly, & O'Donovan, 2012). There are rare exceptions, such as the role of alcohol dehydrogenase genes in alcohol dependence (Shen et al., 1997). We now realize that early candidate gene studies, as well as early atheoretical systematic gene finding efforts (such as linkage studies), were underpowered to detect genes with the small effect sizes that more recent studies suggest are likely to be realistic. Although GWAS were more successful for some conditions (Crohn’s, diabetes, macular degeneration; Manolio & Collins, 2009), like linkage studies, early GWAS were largely unsuccessful in the area of psychiatric and substance use disorders (K. S. Kendler, 2013). Few SNPs were detected that met genome-wide levels of significance. As increasingly large sample sizes have been procured, we now know that most early GWAS were simply underpowered (Visscher, Brown, McCarthy, & Yang, 2012). Amassing sample sizes through large consortia on the order of tens of thousands or more has revealed that the number of significant findings increases as the sample sizes increase.
The discoveries from these large GWAS studies are robust and replicable, and for certain phenotypes, the amount of variance explained in total from genome-wide significant SNPs is becoming non-trivial (e.g., 60% for type-I diabetes; 10% for height; Visscher et al., 2012). In the area of psychiatric genetics, studies of the genetic basis of schizophrenia are currently enjoying the most success, where sample sizes on the order of >13,800 cases and >18,000 controls have now been accumulated, leading to the detection of >3500 loci in 12 genomic regions that contribute to the disorder (Ripke et al., 2013). It is noteworthy, however, that even with such impressive sample sizes, the Ripke et al. (2013) landmark study (which is undergoing a further growth in sample size) acknowledged the lack of power to detect genotype relative risks less than 1.1.
Reasons to be Concerned about the Published cGxE Literature
The emerging genetics literature suggests that the small sample sizes (e.g., n<1000), typical of candidate gene studies to date, are likely to be grossly underpowered for detecting genetic influences with small effect sizes. (The reason that many, perhaps most, candidate gene studies report positive associations despite such lack of power is discussed below.) Some of the confusion about expected effect sizes and the sample sizes needed for cGxE studies may surround differences in the conceptualization of GxE effects across different fields (see Figure 2 for an illustration of this). Candidate GxE studies can be conceptualized in two ways: (1) as a genotype moderating the association between an environmental factor and an outcome (i.e., increasing exposure to major stressful life events is more strongly associated with an increased risk for depression in the presence of the short allele of 5HTTLPR; often the default conceptualization for psychologists) or (2) as an environment moderating the association between a genotype and an outcome (i.e., the association between the short allele of 5HTTLPR and depression is strongest in individuals experiencing major stressful life events; often the default conceptualization for geneticists). Statistically, these are equivalent and indistinguishable, but they can lead to different interpretations of the same data and different expectations about the likelihood of detecting a gene-environment interaction effect. Conceptualizing GxE as a genetic effect (on an environment-behavior association) may lead one to assume small effect sizes based on the growing GWAS literature demonstrating that genetic effects on complex outcomes generally have very small effect sizes. Conceptualizing GxE as an environmental effect on a gene-behavior association may lead one to assume larger effects sizes. Nevertheless, under either interpretation, the effect size of the candidate gene is critical. Recognizing the modest main effect sizes produced by individual candidate gene polymorphisms, it may be overly optimistic to presume that the effect sizes associated with cGxE will be systematically larger. This is clearly a matter of some debate, as if certain types of cross-over interactions were prevalent, it would be possible. Nevertheless, the lessons we have learned from the history of gene finding underscore the need for researchers to be cognizant of the strong possibility that one is dealing with small effect sizes (or to provide strong justification for why larger effect sizes are expected), and to demonstrate that their samples are adequately powered.
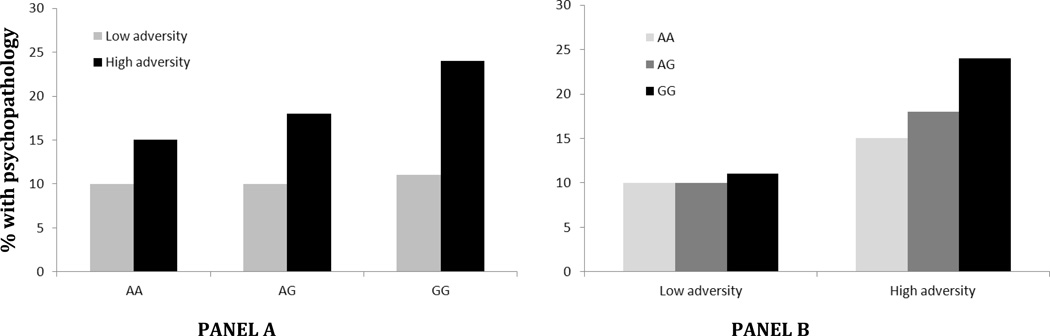
Figure 2.
Alternative portrayals of a candidate genotype (in this example a single nucleotide polymorphism, SNP) × environment (in this example high versus low adversity) interaction. Panel A emphasizes that the strongest impact of high adversity on psychopathology is found for individuals with the GG genotype. Alternatively, Panel B emphasizes that the strongest association of the genotype with psychopathology is among those experiencing high adversity. These are alternate presentations of the same interaction.
In addition to what GWAS has taught us about genetic effect sizes, GWAS has also been informative as to the likelihood that a hypothesized candidate gene will be associated with the hypothesized outcome. Robust and replicable GWAS signals (e.g., CACNA1C for Schizophrenia; Ripke et al., 2013 and Bipolar Disorder; Ruderfer et al., 2013) have tended to be distinct from those that were routinely hypothesized in candidate gene studies (e.g., COMT, MAOA; Craddock, Dave, & Greening, 2001) casting considerable doubt regarding the burden of a priori evidence for these selections. With rare exceptions (e.g., rs16969968 in the CHRNA5-CHRNA3-CHRNB4 cluster, associated at p < 10–70 across several GWAS was initially posited as a candidate gene; Tobacco_and_Genetics_Consortium, 2010), widely studied candidate genes were not found to be significant when studied systematically across the backdrop of the genome in well-powered studies (F. J. Bosker et al., 2011; Collins et al., 2012; Lasky-Su et al., 2008). In fact, some of the findings to emerge from GWAS studies in other areas, such as Crohn’s disease, have suggested new pathways that were not previously suspected to play a role in the disorder and that drastically changed the presumed understanding of the underlying biology of the disorder (Manolio & Collins, 2009). Past experience would suggest that best guesses for candidate genes affecting environmental sensitivity (i.e., cGxE) are unlikely to fare better than they have for other phenotypes investigated to date in GWAS. The combination of low prior likelihood of a given candidate being correct, compounded by likely small effect sizes and low power for any given truly associated variant, suggests that the false discovery rate—the proportion of “discoveries” in candidate gene main effect and cGxE studies that are actually false—may be unacceptably high (Duncan & Keller, 2011).
In addition to the potential for low power and low prior probabilities associated with the study of candidate genes (Munafo, 2009), it is also likely that there is insufficient correction for and/or underreporting of multiple testing. Publication bias is also probable, whereby authors are more likely to submit, and editors are more likely to accept, cGxE findings that are statistically significant. A recent report notes that problems contributing to and a consequence of such bias are rampant in the cognitive sciences (J.P. Ioannidis, Munafo, Fusar-Poll, Nosek, & David, 2014) and there is similar evidence in the cGxE literature (Duncan & Keller, 2011). Specifically, the vast majority of first-reports of a given cGxE finding were positive, whereas a much lower proportion of attempted replications were positive. Ioannidis and colleagues (2014) refer to this as the “Proteus phenomenon” whereby the rapid publication of positive findings might temporarily create a halo in which negative findings might be more readily entertained by journals for a short period of time. Furthermore, and inconsistent with expectations based on statistical power, Duncan and Keller found that the larger the cGxE sample, the less likely it was to be significant. This trend would not be expected if cGxE findings were valid. These observations are consistent with widespread publication bias in the cGxE literature. Publication bias can arise when authors, editors, and reviewers believe that positive findings are more worthy of publication than are negative or null results. There are many uninteresting ways for an empirical test of a hypothesis to fail to support it---the hypothesis was implausible to start with, the power of the test is inadequate, the operationalization of the variables is not valid, etc. As a consequence, the greater interest in positive results is understandable, especially when they bring an insightful increment to, or even transformation of, our understanding. Yet, interest in positive findings is clearly misplaced if they are false, as they can (mis)guide research efforts and funding priorities.
These problems are not unique to the study of cGxE (J.P. Ioannidis et al., 2014; Spellman, 2012). Concern about unacceptably high false positive rates in the social sciences has garnered growing attention over the past several years. In a provocative paper by Iaonnidis (J. P. Ioannidis, 2005) entitled “Why Most Published Research Findings are False,” a number of conditions are outlined that contribute to why a novel research finding ultimately may be in error. These conditions include smaller studies; smaller effect sizes; greater number and lesser preselection of tested relationships; greater flexibility in designs, definitions, outcomes, and analytic models; greater interests and prejudices surrounding the area of research; and situations in which there are more scientists in a field involved in chase of statistical significance. We believe that all of these conditions are likely to contribute to findings in the study of cGxE.
A more recent paper compellingly demonstrated how flexibility in data collection, analysis, and reporting can dramatically increase false positive rates (Simmons, Nelson, & Simonsohn, 2011). This recognition has led to a growing movement in the social sciences to adopt new practices to promote research integrity (Cumming, 2014), including prespecification of studies and hypotheses, avoidance of selection and other questionable data-analytic practices, complete reporting of analyses and variables, and encouragement of replication. A “new statistics” has been proposed that includes recommended statistical practices such as estimation based on effect sizes, confidence intervals, and meta-analyses (Cumming, 2014). These are practices that have already become more widespread in the genetics field (A. Agrawal et al., 2012; Boraska et al., 2014; Ripke et al., 2013; Steinberg et al., 2014; Stephens et al., 2013; Thompson et al., 2014) where a plague of inconsistent and nonreplicable genetic main effects has led to the adoption of more rigorous statistical practices, which have proven successful in advancing the field (Corvin, Craddock, & Sullivan, 2010; P. F. Sullivan et al., 2012; van Assen, van Aert, Nuijten, & Wicherts, 2014). Following these guidelines would go a long way toward improving the quality and trustworthiness of the cGxE literature as well.
In the following sections, we focus on practical problems as they relate to cGxE. We focus this discussion on two major components of cGxE research – the core ingredients of the interaction (i.e., how G and E are measured) and the recipe for combining them (i.e., statistical problems with modeling their interaction).
The Ingredients of cGxE: The Choice of Genetic and Environmental Variables
In the enthusiasm surrounding the study of cGxE, many investigators have expanded their studies to include measures of G or E, when that was not the original focus of the study. This expansion into new areas has happened in both directions: researchers who have focused on carefully characterizing environmental effects have expanded their studies to include measured genes, and researchers who have focused on gene finding have expanded their studies to include measures of the environment. In theory this expansion of cross-disciplinary science is a positive development; however, an unfortunate corollary has been that the added component does not always represent the state of the science in the other respective field.
The Choice of “G”
For example, the vast majority of the “G” that has been incorporated into cGxE studies consists of a handful of “usual suspect” candidate genes (Munafo, 2006; e.g., SLC6A4 [aka 5-HTT, MAOA, DRD2, COMT]), as discussed above. Often a single genetic marker is genotyped to represent the gene. Genotyping a single marker in a gene does not reflect the state of the science in genetics, which has moved toward more comprehensive approaches to gene finding. It may be appropriate to genotype a single genetic variant when that variant has a known functional impact on the gene, (i.e., it produces an observable alteration in the manner in which the gene encodes the protein product). However, over and above this simple annotation of function, the impact of a candidate polymorphism is very challenging to establish. At its simplest, even when modeling a single variant, the characterization of that variant (or its mode of inheritance) can profoundly impact detection of cGxE. For instance, which allele is assigned as a risk allele, and how many copies of this allele are required to quantify the diathesis, need to be determined. How to model a genotype becomes a particular concern with smaller samples, where homozygotes (individuals who carry two copies of a given allele) of the minor allele are often combined with heterozygotes (individuals who carry one copy of a given allele) in the interest of statistical power, potentially obfuscating the complexity underlying the genetic model. Consider the consequences if one applied this practice to other known genetic outcomes: for example, we know that two copies of one of several of the CFTR mutations are required for the diagnosis of cystic fibrosis (a disorder with a recessive mode of genetic inheritance), and that this is etiologically distinct from the one copy of the HTT trinucelotide expansion required for a diagnosis of Huntington’s disease (a dominant disorder). Imagine the confusion if a prenatal genetic counselor were to ascribe the same degree of vulnerability to a fetus that tests positive for one copy (a carrier, unaffected with disease) versus two copies (will manifest the disease) of the CFTR mutation! Although the action of genetic variants on complex traits is not monogenic, the technical specification of their purported mode of action is still significant.
Large-scale gene finding efforts have moved to systematic screens of the genome (e.g., Sklar et al., 2011; Treutlein et al., 2009; Wray et al., 2012), pathway and network analysis (Weng et al., 2011), and integration with model organism genetics (Zhao et al., 2012) as new avenues for increasing the probability of identifying relevant genes. Further, when a gene of interest is being studied, genetic variants across the gene are usually genotyped in order to capture the many locations across that gene that could be involved2 in altering gene regulation or function and producing different effects on behavior. For example, many early studies genotyped the Taq1A1 polymorphism in DRD2 as a hypothesized biological candidate for a variety of outcomes related to reward deficiency, for which dopamine transmission was thought to play a role (Blum et al., 1990; Meyers et al., 2013; Young, Lawford, Nutting, & Noble, 2004). Subsequently, it was discovered that the polymorphism was actually located in the neighboring gene ANKK1 (Neville, Johnstone, & Walton, 2004). More systematic studies of genetic variation across the region containing ANKK1 and DRD2 have found evidence that multiple genes may be involved (Gelernter et al., 2006; B.Z. Yang et al., 2007; B. Z. Yang et al., 2008). Differences in the way that individual genes, gene networks, and whole genomes are systematically studied has led to increasing distance between the gene finding world and studies of cGxE, which still focus largely on the “usual suspect” candidate polymorphisms. Clearly, better integration of these research areas is necessary.
The Choice of “E”
An equally important issue concerns the choice of E. The challenges within this area are illustrated in the literature examining life stress as an environmental risk factor for depression. In attempting to replicate the original cGxE results of Caspi and colleagues (2003) pertaining to life events, many investigators have incorporated a wide variety of ad hoc measures of life stress (Monroe & Reid, 2008). Almost any form of adversity or challenge, at any time in a person’s life, has been used as an alternative index of “stress.” For example, replication studies on this cGxE topic have included participant life stress occurring over a range of time, from one month through a lifetime. Other studies have adopted life event scales known to possess serious measurement deficiencies (Monroe, 2008), or simply have employed unique measures never used previously. In one review of this literature, only 5 of 18 studies reported psychometric properties of the stress measures (Monroe & Reid, 2008). Finally, even when roughly common omnibus measures of life events were adopted, how the life events were combined (over time, severity, and type) for the final index of stress varied greatly across investigations (Uher & McGuffin, 2010).
The use of measures of the environment with proven reliability, empirical precedent, and theoretical plausibility is critical for advancing the field. The elasticity and looseness of the environmental construct can present serious problems for measurement and for the likelihood of detecting a cGxE (Monroe & Reid, 2008). In the example of stress, it is highly likely that different types of “stress” are relevant for different types of cGxE interactions for particular kinds of disorder or disease. For example, chronic stress over years may be most relevant for conditions that develop over protracted periods of time (e.g., coronary heart disease), whereas acute, aversive life events may be most relevant for conditions that often appear to come on rather quickly (e.g., a major depressive episode). Additionally, the developmental timing of the stressor can be critical as the social and biological impact can be expected to vary as a function of stage of development. More specific theoretical dimensions associated with “stress” should accompany such temporal distinctions (e.g., loss or humiliation life events versus life events conveying threat and danger versus other types of adaptive challenges). In a very real sense, there should be candidate “stressors” proposed for particular conditions based on a theoretical understanding of the plausible underlying mechanisms. Viewed from an alternative perspective, no one would expect to find a ‘true’ cGxE interaction if the wrong gene was assessed for the particular outcome. In a similar manner, any potentially valid cGxE interaction will go undetected if the wrong form of environment is assessed, or if the right form of the environment is assessed poorly (Monroe & Reid, 2008).
Problems with the Recipe: Statistical Concerns in cGxE Research
There are a number of statistical considerations that influence the detection and interpretation of cGxE that have not received widespread attention in the literature. We highlight some of the most critical issues next, and many of our concerns are consistent with those of others who’ve recently written on this topic (e.g., Roisman et al., 2012; Zammit, Lewis, Dalman, & Allebeck, 2010; Zammit, Owen, & Lewis, 2010).
The importance of scale
First, evidence for interactions can depend on choice of scale as well as choice of statistical model. Despite one’s conviction in the presence or absence of cGxE, interactions are statistical phenomena and only have meaning in the context of a specific statistical and measurement model. However, many behavioral measures have no “true scale.” One might, for instance, argue that a construct like height has a true scale, and, moreover, a ratio scale with a meaningful zero point and equal intervals between data points (Stevens, 1946): a board of two feet is twice as long as a board one foot long. However, many, if not most of our constructs in the behavioral sciences, do not have meaningful zero points and, thus, are scaled somewhat arbitrarily. Quantitative scales without meaningful zero points can vary further as to whether they have meaningful intervals between measurement points or simply reflect differences in relative magnitude. This is true of measures of both behavioral outcomes (e.g., a depression score) and environments (e.g., family function, peer deviance, neighborhood disintegration). The scale of measurement matters profoundly in interaction research because evidence for an interaction can change solely depending upon arbitrary choice of scale (L. J. Eaves, 2006). For example, predictors that combine multiplicatively to influence the outcome variable will combine additively if the outcome is log-transformed. In such situations, the significance of the interaction term depends on how the outcome is scaled which, in most behavioral research, is arbitrary.
The selection of model
As with choice of transformation, choice of how to model interactions can profoundly affect evidence for them. In particular, there has long been debate in epidemiology regarding the relative utility of risk differences versus risk ratios. That is, if the rate of illness is 10 and 50 per 10,000 in groups unexposed and exposed to some risk factor, are we more interested in the risk difference (40 new cases per 10,000) or the risk ratio of 5? From a practical perspective (e.g., public heath impact, focus for possible prevention, advice to patients), the risk difference approach has much to recommend it. However, the risk ratio approach is the more dominant in large part because it is easily implemented statistically in logistic regression. This distinction is critical because it defines the baseline model from which we assess interactions. In a risk difference framework, an interaction reflects a deviation from a model in which risk factors add together. In a risk ratio framework, an interaction reflects a deviation from a model in which risk effects multiply. Accordingly, the usage of logistic regression to study cGxE for binary outcomes of interest (such as presence/absence of a disorder) fundamentally changes the nature of the relationship between two variables: multiplication on the original scale of a variable conforms to addition on the logarithmic scale. Thus, an “interaction” on the original scale can ‘disappear’ or even be of opposite sign on the logarithmic scale, and vice-versa. We refer the reader to other sources (K. Kendler & Gardner, 2010; Zammit, Lewis, et al., 2010) for further discussion of these important issues.
The use of cross-product terms
Statistical tests of cGxE effects often rely on the modeling of a cross-product term in a regression-type model. Valid detection of true interactions in these models requires that factors that could produce spurious interactions be ruled out. For example, when predictors are correlated and quadratic terms are not modeled, the cross-product term can carry the variance of the unmodeled quadratic term and generate spurious interactions (Lubinski & Humphreys, 1990). Moreover, failure to include quadratics can also result in false negative findings of interactions or the reversal of sign of true interactions (Ganzach, 1997). More generally, if the underlying relationship between G and/or E and an outcome is nonlinear (e.g., a spline or higher-order polynomial), misspecification of the analysis by failing to include a term to model the nonlinearity can generate a significant interaction term in the absence of a true interaction.
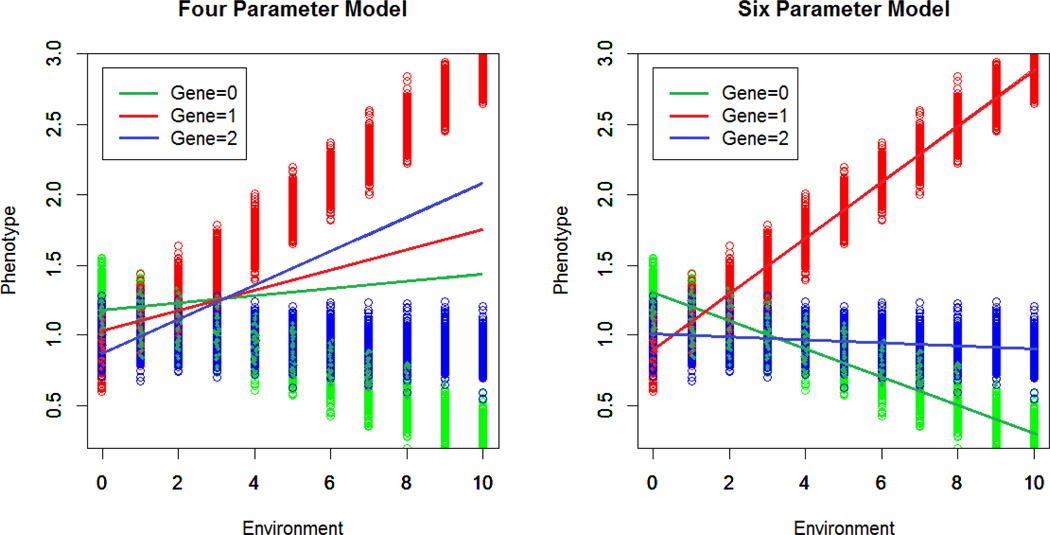
The use of a cross-product term can be particularly problematic for modeling three-level, categorical genotypes. In the standard practice of assuming an additive genetic model, the use of a cross-product term will force the slope difference to be the same between all genotypic groups (e.g., the difference in slope between people carrying 0 versus 1 copies of the risk allele is constrained to be the same as the slope difference between individuals who carry 1 versus 2 copies of the risk allele). It also forces the lines for the three genotypic groups to all cross at the same point when an interaction exists. Accordingly, an interaction will only be accurately represented by the cross-product term when these conditions are met, and there is no a priori reason to assume that these constraints are sensible. This means that the regression lines implied by the use of a cross-product term may not accurately reflect the interaction present in the data. Figure 3 illustrates the problem, and demonstrates how a reparameterization of the regression equation using parameters additional to the single cross-product term can correct it (as further delineated in Aliev, Latendresse, Bacanu, Neale, & Dick, 2014).
Figure 3.
The figure presents simulated phenotypic data for three genotypic groups (G=0, 1, 2, indicating groups of individuals who carry 0, 1, or 2 copies of a particular allele) each shown in a different color. The four parameter model corresponds to the case where the interaction term is modeled by a cross-product term only. Although a significant interaction is detected, the corresponding linear regression lines do not match the data points and the slopes are incorrectly ordered from 0 to 1 to 2, based on the constraints imposed by the use of the cross-product term to model the interaction. Thus, although the model would produce a “significant interaction”, the regression lines implied by the model inaccurately represent the data and would be misleading as to the nature of the interaction. The data can be accurately reproduced by an extended parameterization of the regression model (six parameter model) as detailed in (Aliev, Latendresse, Bacanu, Neale, & Dick, 2014).
The importance of covariates
Failure to properly control for potential confounds can also be problematic in cGxE research. In non-experimental research, researchers typically enter potential confounding variables (e.g., gender, ethnicity, socioeconomic status, genotype quality, etc.) into regression equations to control for their effects. However, this approach controls only for the additive effects of covariates; it does nothing to control for the potential confounding effects these covariates might have on the interaction itself (Keller, 2014; Yzerbyt, Muller, & Judd, 2004). To properly control for confounders in cGxE research, investigators must also evaluate all relevant gene-by-covariate and environment-by-covariate interaction terms. To date, virtually no cGxE studies have appropriately controlled for all covariate interactions (Keller, 2014). This failure to include covariates is particularly concerning in mixed-ethnicity samples, where stratification can not only produce spurious genetic main effect association to be detected (Price, Zaitlen, Reich, & Patterson, 2010), but can also cause ethnicity-by-environment interactions to appear as spurious gene-by-environment interactions. This is because the frequency of alleles naturally varies across ethnic populations and, in the presence of a coincidental excess of affected individuals belonging to one ethnic group, spurious associations and interactions with polymorphisms of no functional consequence, except a degree of natural ethnic variation, may emerge.
Power to detect and characterize different types of interactions
Yet another concern is the low power to detect most plausible forms of interactions in the first place (McClelland & Judd, 1993) in observational field studies as compared to experimental studies where independent variables can be efficiently manipulated. Under many conditions, theoretically meaningful interactions are likely to be quite small, accounting for 1% of the outcome variance and the power to detect most plausible interactions will be quite limited without large N.
Further, even if an interaction is detected, discerning the true pattern of an interaction from observed results is even more tenuous. In recent years, there has been great interest in determining the form of the observed interaction in cGxE research since the interpretation of disordinal (i.e., “cross-over”) interactions theoretically differs from ordinal interactions. Specifically, cross-over interactions lend themselves to a differential susceptibility interpretation where a given “risk” or “malleable” allele is associated with both poorer outcomes in a “bad” environment but better outcomes in a “good” environment; ordinal interactions lend themselves to a diathesis-stress interpretation where it is the combination of a risk-conferring allele and a “bad” environment that exacerbates the likelihood of manifesting the outcome (e.g., Belsky, Bakermans-Kranenburg, & van Ijzendoorn, 2007; Belsky et al., 2009). However, simulations demonstrate that, conditional upon a Type 1 error, the form of an ostensibly “significant” interaction is usually of a “cross-over” (i.e., disordinal) nature, especially when samples sizes are small (Sher & Steinley, 2013). Boardman et al. (2014) recently made a similar observation in reference to the emerging genome-wide gene-by-environment (GWGEI) approach (Cornelis et al., 2012; Mukherjee, Ahn, Gruber, & Chatterjee, 2012; Thomas, Lewinger, Murcray, & Gauderman, 2012) by demonstrating that when many interaction tests are performed, the most significant p-values will come from disordinal interactions even when such interactions are generated from random data (J.D. Boardman et al., 2014). Boardman et al. note these findings “conform to the differential susceptibility model but will not tell us anything meaningful about the way in which environments systematically moderate genetic factors…because they will likely be a statistical artifact” (p. 123).
Moreover, even “true” ordinal interactions that are statistically significant can appear to be of a cross-over type (Sher & Steinley, 2013) due to random error. All linear interactions imply a cross over at some point, even if outside the range of observed values. This is not a trivial issue since a typical practice is to plot values +1 standard deviation (SD) and −1 SD above and below the mean of the moderator (Aiken & West, 1991). However, −1 SD can represent values that rarely or never exist in nature for skewed predictors. We note that some authors (Roisman et al., 2012) have recently recommended extending the Aiken & West (1991) guidelines to + 2 SDs, in order to provide 95% coverage of the observed values. Given the highly skewed nature of many environmental exposures, attention to the underlying distribution of all study constructs is necessary so as not to generate misleading regression plots covering regions of sparse or imaginary data.
Recently, techniques for estimating the standard error of the cross-over point have been proposed which could, in principle, allow stronger inferences about the actual form of the interaction (Widaman et al., 2012). Alternatively, establishing regions of significance around each regression line using standing approaches (e.g., Johnson & Neyman, 1936) to characterize where two slopes overlap and where they do not could also be used to increase confidence that an ostensible cross-over shows a desired degree of statistical differentiation from an ordinal interaction. Such approaches, in principle, could provide greater confidence in believing a true “cross-over” has been detected. Consistent with the other points made above, such approaches are dependent upon being confident that the interaction is not an artifact of scaling, is not caused by (unmodeled) nonlinearity, and is not a Type 1 error.
cGxE versus gene-environment correlation (rGE)
Gene-environment correlation, or rGE, refers to instances where exposure to environment is non-random and correlated with genetic vulnerability3, whether through passive, active or evocative processes4. For instance, in classical behavior genetics, rGE is represented by genetic factors that influence the outcome (e.g. alcohol and tobacco use) and the environment (e.g. peer relationships; Harden et al., 2008) as indexed by a genetic correlation. Similarly, in measured gene studies, presence of rGE is indexed by variations in genotype or allelic frequency as a function of the environmental exposure. For instance, Salvatore and colleagues report an association between a polygenic score for alcohol problems and peer deviance (Salvatore et al., 2014), indicating that individuals who are at genetic risk for alcohol problems are also more likely to have deviant peer groups. It is likely that for many outcomes both rGE and cGxE may be important; however, the presence of rGE can complicates the interpretation of cGxE. Behavioral genetic and twin models implement several statistical approaches to account for and even explicitly model rGE (e.g., L. Eaves & Erkanil, 2003; Purcell, 2002; van der Sluis, Posthuma, & Dolan, 2012). In measured gene studies, the first step in testing for potential rGE involves estimation of a correlation between genotype and environment. In the absence of such a correlation, as has been noted for 5-HTTLPR and stressful life events, cGxE testing can proceed without concern. If the correlation is solely attributable to outliers in the environmental measure, removal or winsorization may eliminate rGE (e.g., Bogdan, Williamson, & Hariri, 2012). A modest correlation between genotype and environment may require more careful consideration. For instance, in revisiting the interaction between a polymorphism in the monoamine oxidase A (MAOA) gene and exposure to childhood physical abuse in the development of antisocial behaviors (Caspi et al., 2002), Kim-Cohen and colleagues examined whether MAOA genotype was correlated with not only exposure to abuse (i.e. evocative rGE) but also, maternal antisocial behavior (i.e. passive rGE) with the latter being a key correlate of transmission of risk for antisociality and for increased likelihood of exposure to abuse (Kim-Cohen et al., 2006). There was evidence for the latter, whereby maternal antisocial behavior was correlated with offspring exposure to abuse, however the effect of the interaction persisted even after accounting for this effect. Alternatively, Salvatore and colleagues accounted for rGE by residualizing both their polygenic score and their environmental measures (parental knowledge and peer deviance) for each other prior to testing for cGxE in the etiology of alcohol use problems (Salvatore et al., 2014). The interaction between parental knowledge and the polygenic scores remained significant; however, the interaction with peer deviance was no longer significant, indicating the possibility of both rGE and cGxE for the former but rGE alone for the latter. Therefore, while both mechanisms of gene-environment interplay (rGE and GxE) may be at work, testing for rGE is necessary before conclusions regarding GxE are made. When rGE is presented, methods to account for it should be implemented. In some instances, relevant data may not be available (e.g. availability of parental phenotypes to test for passive rGE) or the correlation may be complex and mediated by other unmeasured factors. In such instances, the possibility that rGE may contribute to the relationship between genotype and environment should be acknowledged.
In summary, although the statistical approaches for modeling interactions are well established, having confidence in the statistical validity of an interaction requires due diligence on the part of the investigator. These include attention to scaling issues, characterizing the underlying linearity of the relationships under investigation and determining whether nonlinear models are necessary, controlling for relevant confounders including various forms of rG-E, and insuring plotted results are not unduly influenced by the constraints imposed for rendering an easy-to-interpret graph. Perhaps the greatest challenge is to minimize the likelihood that an observed interaction is not a Type 1 error given that various data sets have a large number of candidate Gs and candidate Es, there is considerable flexibility in approach to analysis, and under most plausible conditions, power to detect GxE is likely to be low. Many of the issues described above (as well as some others) are described by Roismann et al. (2012) who provide a list of thoughtful guidelines for addressing various issues such as characterizing whether an obtained interaction is of a cross-over type (and to the extent the magnitude of the cross-over is meaningful), the problem of nonlinearity, and Type 1 errors.
Recommendations
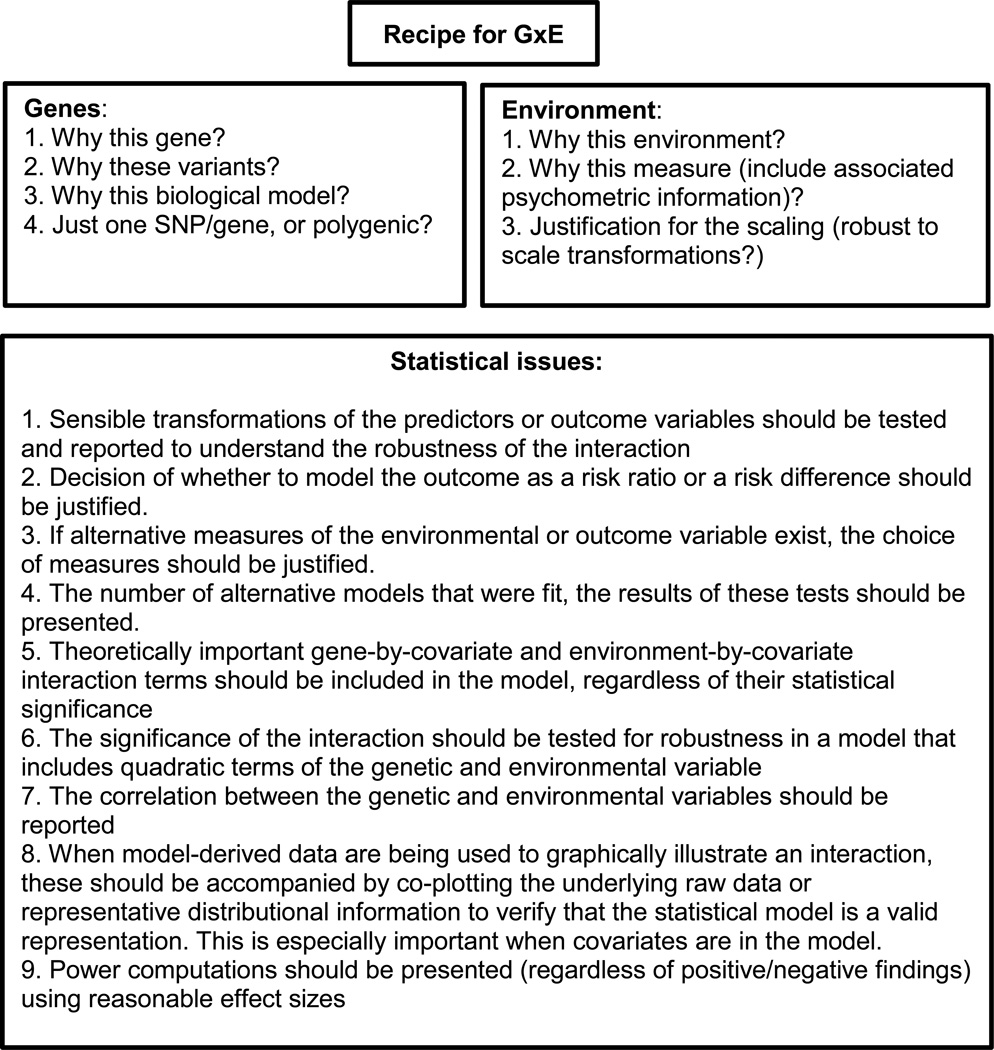
Although the list of challenges associated with characterizing cGxE is long, many of these can be addressed by adopting a handful of rigorous research practices. Below we delineate a series of recommendations that we believe will help advance the study of GxE and ensure that the literature provides meaningful progress for science. We focus at greatest length on the issues pertaining to the G (genetics) in GxE, under the assumption that this will be most unfamiliar to social scientists. Many of the other concerns, reviewed in less detail below, are not specific to the study of cGxE, but complement the broader discussion (Cumming, 2014; J. P. Ioannidis, 2005; Lakens & Evers, 2014; Simmons et al., 2011) in the social sciences about how to produce robust, replicable findings that advance science. The recommendations below are reviewed in Figure 4.
Figure 4.
Recommendations for rigorous GxE research practices.
Selection of genes
Given the prevailing skepticism surrounding candidate gene research, the burden of proof for the selection of a candidate gene is high. Such a rationale should be convincingly articulated in a manner specific to the phenotype and environment under study. The crux of the argument for selection of a particular gene to study lies in the statistical priors for the gene, i.e., based on prior evidence and the quality of the source of that evidence, how likely is it that this is a robust candidate. There is nothing inherently wrong with studying candidate genes, though the very idea of “candidate gene research” has fallen out of favor due to the historical issues with studying hypothesized biological candidates that have not held up in more systematic well-powered studies, as reviewed above. There are notable cases where hypothesized biological candidates have shown robust associations with outcome. For instance, rs1229984 in alcohol dehydrogenase (ADH1B) gene was one of the earliest candidate gene variants proposed in the etiology of alcoholism, particularly in Asians (A Agrawal & Bierut, 2012)5. The variant, which is rather rare in European-American populations, has recently been identified in adequately powered genome-wide association studies as well (Bierut et al., 2012). Similarly, early candidate gene and experimental studies implicated a SNP in a nicotinic receptor (rs16969968 in CHRNA5) as a risk factor for tobacco smoking and nicotine dependence, but the role of this variant in cigarette smoking was not widely accepted until it was identified in multiple meta-analyses of cigarettes smoked per day, the largest of which had a sample size exceeding 70,000 (J. Z. Liu et al., 2010; Thorgeirsson et al., 2010; Tobacco_and_Genetics_Consortium, 2010).
Knowing that the prior for a candidate gene selected on hypothesized biological rationale is low, there are a variety of other methods for selecting candidate genes for study that should produce more robust and reliable findings, including focusing on genes with either large main effects or stronger a priori evidence. For the latter, methods of gene selection that have a greater likelihood of producing meaningful cGxE results include focusing on candidates suggested by well-powered GWAS or meta-analysis, or by model organism work, ideally with replication. Identification of genotypes of substantial main effect is challenging and limited. For example, the ε2/ ε3/ ε4 polymorphism at the APOE locus is a known and important risk factor for Alzheimer’s and coronary heart disease (Corder et al., 1993; Ward et al., 2009). Such large-effect polymorphisms seem to be particularly compelling candidates for cGxE research because the genetic effect is already known; only an environmental modification of this effect is required for evidence for an interaction.
A systematic strategy of gene identification followed by efforts to characterize moderation of the effect associated with that gene can be found in the example of GABRA2, and moderation by parental monitoring. GABRA2 was originally identified by the Collaborative Study on the Genetics of Alcoholism, the largest gene identification project in the area of alcohol dependence (Begleiter et al., 1995), by systematically interrogating GABA receptor genes with evidence for involvement in ethanol related response (Harris, 1999) that were located in a region of linkage identified with both clinical alcohol dependence phenotypes and electrophysiological endophenotypes (Ghosh et al., 2003; Porjesz et al., 2002; Reich et al., 1998). Association between GABRA2 and alcohol dependence was subsequently reported (Edenberg et al., 2004) and replicated by multiple independent research groups around the world using a variety of research designs (Covault, Gelernter, Hesselbrock, Nellissery, & Kranzler, 2004; Enoch, Schwartz, Albaugh, Virkkunen, & Goldman, 2006; Fehr et al., 2006; Lappalainen et al., 2005; Olfson & Bierut, 2012; Soyka et al., 2008). Although there have been failures to replicate (Covault, Gelernter, Jensen, Anton, & Kranzler, 2008; Drgon, D'Addario, & Uhl, 2006; Matthews, Hoffman, Zezza, Stiffler, & Hill, 2007) a recent meta-analysis confirms the evidence for association (Li et al., 2014). In addition, translational research has found the role of GABRA2 in rodent drinking (Dixon, Walker, King, & Stephens, 2012; J. Liu et al., 2011) and in the brain’s response to alcohol-related (Kareken et al., 2010) and monetary reward cues (Villafuerte et al., 2012). While GABRA2 SNPs have not been identified via GWAS, they typically have the lowest p-values of candidate polymorphisms extracted from GWAS data (Olfson & Bierut, 2012). Interaction between GABRA2 and parental monitoring was tested based on the twin literature suggesting that parental monitoring moderates the relative importance of overall genetic effects (as inferred based on comparisons of twins, not using measured genotypes) on substance use outcomes in adolescence (Dick, Pagan, Viken, et al., 2007; Dick, Viken, et al., 2007); genetic effects assumed greater importance under conditions of lower parental monitoring, presumably because adolescents with lower monitoring have more access to the substance and opportunity to express their genetic predisposition. An interaction between GABRA2 and parental monitoring was tested in an independent sample; a stronger association between the gene and trajectories of externalizing behavior was found under conditions of lower parental monitoring, as was hypothesized based on the twin findings (Dick, Latendresse, et al., 2009). This series of studies illustrates how different literatures and study designs (linkage and association studies, twin studies, and longitudinal, developmental studies) were integrated to characterize a cGxE effect. In this case, the candidate gene under study was selected based on a series of converging pieces of evidence that indicated involvement in alcohol and externalizing outcomes before it was studied in the context of cGxE.
In short, not all candidate genes are created equal nor is there a single pathway to determining their viability in a cGxE study. The burden is on the researcher to provide a compelling argument for the study of a particular candidate. And we suggest that bar should be much higher than the field has insisted upon to date.
Selection of genetic variants
In addition to strong justification for the selection of the gene under study, a second area that should be justified is the genotyping strategy: if only a select number of polymorphisms in a gene are being genotyped, how and why were those selected? There are several methods for selecting polymorphisms – one can utilize SNP content from a preexisting GWAS array or genotype custom content individually or en masse (e.g., as offered by the Illumina Golden Gate technology; Hodgkinson et al., 2008). Prices for GWAS arrays, especially those designed to include custom content, have dropped considerably, and often it is far more expensive and less cost-efficient to genotype a small number of polymorphisms than to genotype in large scale. In some cases, the technology required to genotype a particular variant, especially one that is not a SNP (e.g., a variable number of tandem repeats) is specialized and may entail unique requirements as most commercial arrays and custom genotyping platforms may not include this. Nonetheless, if only a few SNPs can be genotyped, tagging a gene may be preferable to simply pursuing the “usual suspects”. Tagging refers to identifying all variation, regardless of function, that captures variation across the gene. This includes important regulatory regions, such as promoters and enhancers which are increasingly recognized as key contributors (Zannas & Binder, 2014). Reliance on simple annotation of “function” (i.e., typically, a nonsynonymous exonic variant, meaning that the location is known to be in a part of the gene that produces an alteration in the gene’s protein product) is short-sighted as modern annotations available via the identification of epigenetic marks along the genome suggest that even intronic variants can have a profound impact on genomic action (e.g., Ziller et al., 2013). An exciting and upcoming possibility is a highly cost efficient chip being designed by the Psychiatric Genomics Consortium (PGC) that will include custom common and rare variation and will be based on SNPs nominated by expert consensus and validated via meta-analytic methods.
Modeling genes
Another consideration is that how genotype is coded implies a biological model, as reviewed above, and, as such, the model should be specified and justified (or explicitly stated as exploratory, with appropriate corrections for multiple testing). Genotypes should not be collapsed purely to increase power and if they are, then effects should be described across all genotype groups for completeness. With increasing recognition that individual genetic polymorphisms on their own are likely to have very small effects, there should be justification provided if single polymorphisms are being studied in isolation. Further, with growing knowledge about gene networks and integrated functional pathways, there is opportunity to think more broadly about the potential for incorporating gene networks or pathways into tests of GxE, allowing one to go beyond focusing on a single gene and instead focus on sets of genes that interact biologically. This brings its own set of complications, as decisions must be made about the nature of genetic effects across the pathway or network: is a mutation in any of the genes in the network sufficient? Are all variations within the network expected to have an equal effect on outcome? Are mutations across multiple genes in the network acting cumulatively to affect outcome? Similar questions can be asked about multiple variants within any given gene of interest. Should polygenic risk scores that capture risk across the genome be used? The answer to these questions depends on the investigator’s theory behind how the environment is operating: does the investigator believe that all genes involved in outcome should be moderated by that environment in a parallel fashion or only subsets of the relevant genes? There is no straightforward answer to these questions, but what is clear is that deeper thought must be given to these issues in order to move the study of cGxE forward. Justification for the choices made in any given GxE study should be included in the publication. Because it is challenging to keep up with advances in the field of genetics, we suggest that this is an area where collaborations between geneticists and psychologists can be particularly fruitful, as connection to the latest findings from statistical and psychiatric genetics about our rapidly evolving knowledge of the underlying genetic basis for a given outcome of interest can ensure that the genetics being integrated into psychological science represents the latest advances from the field of genetics. The annual meetings of the Behavior Genetics Association (www.bga.org) and the International Society for Psychiatric Genetics (www.ispg.net) are opportunities to learn about the latest advances in psychiatric and behavioral genetics and to potentially develop collaborations with scientists working directly in those areas. In addition, Text Box 1 of this review lists a variety of genetics resources that may be useful to investigators in the social sciences that are interested in adding an informed genotyping component to their study.
Text Box 1. Genetics resources.
dbGAP
All federally funded studies with genome wide association study (GWAS) data are required to deposit genotypes and a select set of phenotypes in dbGAP. Investigators can follow procedures to access these data for analyses, including cGxE.
dbSNP
Repository of genetic variation.
Epigenome browser (Zhou et al., 2013)
Annotation of epigenetic marks in human genome.
GCTA (Yang, Lee, Goddard, & Visscher, 2011)
Uses plink-format files above to estimate the total genetic variance explained in a phenotype by GWAS data; can be used for GxE using all GWAS variants.
HapMap
Includes reference data on allele frequencies, gene assignment and linkage disequilibrium for common variants in sample populations.
Linkage disequilibrium (LD) estimation and plotting software
The following software packages offer investigators the ability to compute LD in their own data and in reference datasets.
Reference data: http://www.broadinstitute.org/mpg/snap/
Own data with Haploview (Barrett, Fry, Maller, & Daly, 2005):
Identify tagging SNPs: http://www.broadinstitute.org/mpg/tagger/
Regional association plots with LD (Johnson et al., 2008; Prium et al., 2010):
PGC (Psychiatric Genomics Consortium)
Conducts some of the largest meta-analyses of psychopathology. Results (effect sizes) are available for developing cGxE hypotheses based on strong priors. Free lectures by genetics experts available to view.
PLINK (Purcell et al., 2007)
Free and user-friendly software for analysis of measured and imputed genomic association, creation of polygenic risk scores, meta-analysis, cGxE and GWAS analyses. Allows for easy transformation of genomic data.
Power computations for genetic main effects and GxE
These user-friendly power calculators are useful in determining power given sample size and genetic model.
For a variety of study designs and genetic models (Purcell, Cherny, & Sham, 2003):
http://pngu.mgh.harvard.edu/~purcell/gpc/
More restrictive range of genetic models, includes GxE models (Gauderman, 2002):
http://biostats.usc.edu/Quanto.html
Primarily for case control association (Skol, Scott, Abecasis, & Boehnke, 2006):
UCSC Genome Browser/ENCODE
Detailed annotation of variation and features of the human genome, including expression, cross-species conservation and epigenomic marks via the Encyclopedia of DNA Elements.
Selection of the environment
Investigators should provide theoretical rationale for the selection of the environmental factor under study. Why is there reason to believe this aspect of the environment will have a moderating effect? Does the investigator believe this is an environmental factor with a time-limited or enduring effect? Justification for the scale of measurement of the environment should be provided, as well as reporting of results on biologically defensible transformations of the independent and dependent variables. The environmental measure should reflect the “state of the art” method for measuring the particular feature of the environment. If it does not, justification for why this particular measure should be relevant/adequate should be included, as well as traditional supportive psychometric information.
Accurate reporting of multiple testing
Researchers should explicitly detail how many total polymorphisms were available to them, how many were tested, and what type of guarding against inflation of type-I errors were made. In an identical vein, researchers should explicitly detail how many environmental variables, and how many different methods of operationalizing these environmental variables, were considered, as well as transformations of such, and selection of models. These types of procedures for genes are now routine in GWAS but are rare in cGxE studies. Such explicit disclosures will help increase confidence in positive findings that survived proper multiple testing corrections.
Power and small samples
Investigators should demonstrate that the sample being employed has adequate power to detect an interaction effect for the variables under study. Power computations should be presented regardless of the nature of the finding. In other words, studies with positive findings should be encouraged to present power computations as well (K. S. Button et al., 2013). Computations should be specific to the statistical technique, distributions of the variables under investigation, and the hypothesized form of the interaction. Power analyses should assume realistic cGxE effect sizes. GWAS suggest that main effects are typically of a small magnitude, most accounting for less than 1% of the variance in a psychiatric phenotype. If the investigator has reason to believe that a larger effect size is likely for their study this should be clearly spelled out and justified. For instance, one might hypothesize that genes exert stronger effects on endophenotypes (e.g., neuroimaging outcomes) that are, arguably, more proximal to their action (though see Flint & Munafo, 2007; Munafo & Flint, 2009). Or, it is possible that the use of a phenotypic measure thought to be of much greater reliability and validity than existing clinical phenotypes could enhance power. Finally, investigators should use
A question that sometimes arises is what can and should be done with studies whose small sample size is necessitated by the prohibitive costs associated with measurement of the phenotype, for example, with behavioral or neuroimaging studies, multi-wave longitudinal studies or studies of special populations. Regardless of the nature of the variables (e.g., self-report vs. neuroimaging), a small sample size nearly always implies reduced power. In fact, Button and colleagues (K.S. Button et al., 2013) reported that the median statistical power across 49 meta-analyses of neuroscience studies was typically less than 20%, with the estimate plummeting to 8% when examining neuroimaging studies alone -- and that is without estimation of G or GxE! Some have argued that genotypic effect sizes associated with endophenotypes (such as neuroimaging outcomes) are likely to be higher because the outcome is more proximal to the biological substrate (E. J. Rose & Donohoe, 2013), yet there is no demonstrable evidence that we understand the genetic underpinnings of threat-related amygdala reactivity and habituation, any better than we do the etiology of depression and anxiety. In other words, endophenotypes may be as polygenic as self-report measures (Flint & Munafo, 2007).
Replication is key for findings based on small samples, but, as Button and colleagues note, the winner’s curse often inflates initial results and unless the replication sample is substantially better powered, the ceiling placed on average sample size likely perpetuates false positive findings until enough samples have been amassed to conduct a meta-analysis. Meta-analysis and/or integrative data analysis (Hussong, Curran, & Bauer, 2013) are particularly attractive techniques to combine data across multiple studies so that small scale research can contribute to more definitive results. In instances where that is impossible, perhaps because the sample is rare and unique, the outcome under study is highly novel or because the environmental factors have been measured using superior assessments that other studies do not include, we recommend that these studies acknowledge their limitations and present a candid account of power in the study, even if it implies that their findings might be spurious. Importantly, readers of this literature should carefully consider the caveats of such small sized studies so as not to perpetuate a series of cGxE emerging from and replicated in small samples.
Manipulation/Presentation of Data
The statistical properties that surround the detection of the interaction should be specified and discussed. Is the interaction significant on an additive or multiplicative scale? Investigators should provide evidence that detected interaction effects are robust to transformation of scale and/or provide a strong defense of the scaling employed. If it is not robust to transformation, the implications of this should be discussed. Common artifacts associated with nonlinearity should also be evaluated and ruled out. Similarly, if the interaction is detected under some conditions or using some measures of the environment but not others, this should be reported and incorporated into the theory behind what the findings contribute to the literature. Perhaps most importantly, the raw data along with 95% confidence intervals should always be plotted and reported in the publication, not just the model-derived regression lines, which can be misleading, especially when some combinations of the independent variable have small sample sizes and when control covariates are included in the model. Because apparent cGxE effects can be due to stratification, genotyping artifacts, and even gender (Keller, 2014), careful attention must be paid to the variables under study in order to bolster confidence that the moderation effect is due to the specific gene/environment under study. For example, if whites are more sensitive to environmental trauma, leading to Posttraumatic Stress Disorder (PTSD), in a mixed ethnicity sample examining environmental trauma-by-gene interactions, any SNP that differentiates whites from blacks will also show an apparent cGxE interaction. In this case, the gene isn't the true moderator - race is - the gene was merely correlated with race. To increase confidence in cGxE findings and to eliminate alternative explanations for them, researchers need to include all relevant gene-by-covariate and environment-by-covariate interaction terms in their models. Researchers should consider including quadratic transformations of the environmental term as well as covariates, as these may change the interpretation of the interaction depending on multicollinearity. Finally, the assumptions that are imposed by modeling interactions using cross-product terms in linear regression and/or by the use of logistic regression (as discussed above) should be acknowledged and, when appropriate, justified.
Addressing Publication Bias
In circumstances where the proportion of false findings is high, as we suspect is the case for candidate gene main effect and cGxE studies, the issue of publication bias should be considered by both authors and editors. Authors, journal editors, and reviewers should understand that novel positive findings may often be false, and so should set higher standards of evidence, including stricter standards of significance testing, full accounting for all sources of multiple testing, and direct independent replication prior to publication. Equally, investigators should strive to submit, and editors to publish, adequately powered, independent direct replication attempts, irrespective of the outcome -- positive, negative, or null. Meta- or mega-analyses that combine data across multiple studies while accounting for across-study heterogeneity, allow smaller sized studies to contribute to hypotheses necessitating larger samples, such as cGxE. Such collaborative research should be encouraged. These kinds of recommendations, aimed at reducing the role of publication and other biases, are being adopted by journals that have an interest in cGxE; some recent examples are Behavior Genetics (Hewitt, 2012), the Journal of Abnormal Child Psychology (Johnston, Lahey, & Matthys, 2013) and Drug and Alcohol Dependence (Munafo & Gage, 2013). We would recommend that other journals dealing with substance abuse, psychiatry, and related fields adopt similar policies.
Conclusions
Studying how genetic predispositions and environmental circumstances come together to contribute to complex behavioral outcomes has great potential for advancing our understanding of the development of psychopathology. It represents a clear theoretical advance over studying these factors in isolation. However, research at the intersection of multiple fields creates many challenges. Studying cGxE requires appropriate understanding of genetic mechanisms, appropriate measurement of the environment, a conceptual framework for integrating the two with respect to a specific outcome of interest and, critically, of the statistical principles that underpin cGxE studies. It is not likely, or expected, that every investigator conducting cGxE research will have the requisite expertise in all of these areas; accordingly, we encourage cGxE studies that are collaborative efforts involving individuals with common interests but diverse expertise.
The National Institutes of Health has initiatives aimed at addressing challenges associated with genotypic research, many of which are also relevant to the study of cGxE. For example, in recognition of the fact that gene identification requires large numbers, but one of the challenges that is often encountered as researchers attempt to pool their samples is the use of different measures across studies, the PhenX initiative was launched (www.phenX.org) (Hamilton et al., 2011). PhenX brought together panels of experts across a variety of research areas to come up with recommended consensus measures (including both outcome and environmental measures) for inclusion in genetics studies to encourage the use of common measures to facilitate cross-study comparisons and analyses. There are limitations to this approach: brief, low burden measures were preferentially selected to encourage more widespread uptake, which may result in less precise or comprehensive assessments in the case of some constructs. However, it represents a step toward facilitating collaborative efforts in genetics research. The use of standardized measures across studies could also help advance the cGxE field, with greater emphasis placed on replication and combined analyses across research groups to enhance sample size and corresponding power. The Office of Behavioral and Social Sciences Research (OBSSR) at NIH also has resources on its website [http://obssr.od.nih.gov/] to help social scientists with the incorporation of genetic information into their studies. Many of these issues are not specific to the study of psychopathology. A recent paper on gene-environment interactions in cancer epidemiology, coming out of a National Cancer Institute think tank, described similar challenges (Hutter, Mechanic, Chatterjee, Kraft, & Gillanders, 2013).
As investigators who have explored cGxE hypotheses ourselves, with some of our own work not meeting the standards delineated in this review, we were compelled to ask: how can we do better? We hope we have outlined some such strategies. Through greater awareness of the challenges in conducting cGxE research, resources available to aid in conducting high quality cGxE studies, and proactive efforts to move cGxE studies in this direction, it is our hope that this growing area of research will eventually reach its potential to deeply inform to our understanding of complex behavioral outcomes.
Acknowledgments
This paper developed out of discussions from the workshop “Challenges and Opportunities in GxE Research: Creating Consensus Recommendations for Guidelines on GxE Research”, sponsored by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) in January 2013. In addition to the co-authors who contributed to the writing of this manuscript, other workshop participants included Lindon Eaves, Ph.D., Virginia Commonwealth University; Andrew Heath, Ph.D., Washington University, St. Louis; and Eric Turkheimer, Ph.D., University of Virginia. We are deeply grateful for the leadership of Marcia S. Scott, Ph.D., chair of the NIAAA Gene and Environment Team, Dionne C. Godette, Ph.D., and Mariela C. Shirley, Ph.D., Division of Epidemiology and Prevention Research, National Institute on Alcohol Abuse and Alcoholism, (Dr. Shirley now at the Office of Research on Women's Health), National Institutes of Health, in organizing support for the workshop and for their helpful comments on drafts of this manuscript. Support for co-authors involved in drafting this manuscript includes: K02AA018755, R01DA070312, and U10AA008401 to DMD and R01AA018333 to DMD and KSK; K01MH085812 and R01MH100141 to MCK; K02DA32573, R21AA21235, and R01DA23668 to AA; DA011015 to JKH; K05 AA017242 to KJS. We are also grateful to Erica Spotts, Ph.D., Health Scientist Administrator at the Office of Behavioral and Social Science Research (OBSSR) for her assistance in securing funding (R01 AA018333–04S1 to DMD) to create a website to facilitate understanding of the concepts related to conducting cGxE research, as delineated in this manuscript. This information will be hosted through the OBSSR website (anticipated 2014 release).
Glossary*
- additive
a genetic model where each increasing copy of an allele (AA vs AG vs GG) is associated with a corresponding incremental change in association with the outcome.
- allele
alternate version of a DNA sequence or variant (e.g. SNP rs1229984 has two alleles, A and G)
- carrier
an individual who is heterozygous for the risk conferring allele of a variant or mutation with a recessive pattern of inheritance. Carriers do not express recessive phenotypes but can transmit a copy of the recessive allele on to their children.
- common variants
typically, single nucleotide polymorphisms where the less common allele is present in ≥5% of the population under study; e.g. the less common allele in the widely studied Val66Met polymorphism rs6265 in BDNF (brain derived neurotrophic factor) is seen in 19–27% of those of Caucasian European descent making it a common variant.
- Cross-product term
In a regression, the coefficient/term that captures the interaction between two variables (e.g. GxE)
- dominant
an allele that is expressed when present in one copy. For alleles that follow Mendelian inheritance patterns, individuals who have one or two copies of the dominant allele will express the phenotype
- effect size
the magnitude or strength of the relationship between two variables, such as genotype and phenotype; can be reflected by a regression coefficient, an odds-ratio etc.
- enhancers
a sequence of DNA that represses or increases the rate of transcription of a gene
- epigenetic
inherited or de novo changes in gene, expression that are not directly related to a change in DNA sequence
- exonic
a genetic variant that lies within coding region of a gene, meaning a sequence of DNA that is translated into a protein product
- functional (SNP)
a single nucleotide polymorphism that changes the protein product or expression of a gene
- genetic marker
a sequence of DNA that maps to a particular (known) chromosomal location
- genomics
the study of genomes (a complete set of DNA sequence information for an organism)
- genotype relative risks
the ratio of penetrance parameters where penetrance refers to the likelihood of being affected given a certain genotype.
- GWAS
Genome-wide association study; an unbiased study looking for allele frequency differences across the genome between individuals who vary in a phenotype of interest
- Genome-wide significance
when investigators perform genome-wide association studies, the traditional p-value cut-off for significance (p<=0.05) is no longer valid as nearly one million tests are being preformed. A genome-wide significance threshold accounts for multiple testing via a Bonferroni correction for a million tests (0.05/1,000,000), shifting the significance level to 5×10–8 (usually)
- Heritability
the proportion of observable differences in a trait between individuals within a population that is due to genetic differences
- heterozygote
an individual who has two different alleles (i.e. one copy of each AG) at a particular locus (location on a chromosome)
- homozygote
an individual who has two copies of the same allele at a particular chromosomal location(i.e. AA or GG). Individuals can be homozygous dominant (two copies of the dominant allele at a locus) or homozygous recessive (two copies of the recessive allele at a locus).
- Imputation
probabilistic inference of genotype of an untyped variant in the vicinity of a typed variant, based on curated data that estimate the extent to which the typed variant is typically correlated with the untyped variant.
- intronic
a genetic variant that lies within a noncoding region of a gene, which is a sequence of DNA that is not translated into a protein
- linkage disequilibrium
the correlated nature of adjacent variants such that knowledge about genotype at one location can result in high accuracy of prediction of genotype at the other location. This is due to the nonrandom association of alleles in a population resulting from historical recombination events
- Minor allele frequency (MAF)
in a particular population, the frequency of the minor (least common) allele (variant) at a particular locus
- Mendelian disorder
a single gene disorder that follows a Mendelian inheritance pattern (i.e. dominant or recessive) where there is a direct correspondence between carrying the disease-conferring genotype and expressing the disorder.
- monogenic
a phenotype or disorder that is caused by one gene
- mutation
a DNA sequence change that results in observed differences in phenotype
- nonsynonomous
a DNA sequence change that results in an amino acid change in the gene’s protein product (see also missense)
- pleiotropy
where one gene influences multiple outcomes that may be unrelated
- polygenic risk scores
the summation of multiple alleles that are putatively associated with a phenotype to create a single index of genetic vulnerability
- polymorphism
a DNA sequence change, i.e. a location in the genome that can come in multiple forms
- population genetics
the study of allelic variation in a population characterized by certain evolutionary influences.
- power
in statistics, the probability of detecting an effect, if an effect truly exists
- Priors/prior probabilities
the probability of an event before statistical evidence from other studies/observations that favor/disprove a hypothesis is taken into account. For instance, the prior probability that a newborn in the Caucasian population has cystic fibrosis is 1/2500 but this probability is higher (25%) if both parents are carriers.
- promoter
a region of DNA that regulates transcription of a gene
- recessive
an allele that is only expressed when present in two copies. For alleles that follow Mendelian inheritance patterns, only an individual who has the homozygous recessive genotype will express the recessive phenotype
- reference panel
genomic data regarding the physical location, gene assignment and other features of a variant based on a select group of genotyped individuals (e.g. for Whites, residents of Utah of Western and Northern European descent were selected by the Centre d'Etude du Polymorphisme Humain (CEPH) – this constitutes the CEU reference panel).
- risk allele
the allele associated with increasing vulnerability to the phenotype.
- SNP
single nucleotide polymorphism a single base pair change in the DNA sequence
- Stratification
naturally occurring variation in the frequency of the minor allele of SNPs across different ethnic groups. When unaccounted for, stratification can lead to false positive associations if the outcome is correlated with ethnicity.
- Tandem repeats
DNA sequence repeats that occur side by side, e.g., a repeating pattern of the nucleotides ACGACG would be a trinucleotide tandem repeat.
- Variant
alternative DNA sequence
(*)Note: This glossary is not meant to be exhaustive or serve as a substitute for a genetics textbook. Rather, we provide a brief overview of key genetic phenomena here to assist the reader with the interpretation of this paper.
Footnotes
Authorship
All authors contributed to writing sections of the manuscript and reviewing drafts.
Words indicated in bold in the text are defined in the Glossary.
This is often called linkage disequilibrium/LD-tagging, and indicates that you can use information about the LD or correlation structure across variants within and across genes in order to know how many genetic variants you need to genotype to capture most of the variable locations in the gene that could be associated with outcome (see Text Box 1 for software).
Mendelian randomization is a related and novel conceptualization of genotype representing environmental exposure propensity and might explain cGxE in the presence of rGE. We refer the reader to (Smith, 2010) to learn more.
Passive rGE is more common during childhood and adolescence and refers to an individual being differentially exposed to an environment without their own initiative, most likely because aspects of their environment are provided by their parents with whom they also share genetic variance. For instance, antisocial parents may pass on a genetic liability to antisociality and expose their children to an abusive environment. Evocative rGE refers to environments that an individual elicits/evokes from others based on their genetic predisposition. For instance, an antisocial adolescent may evoke harsher parenting. Finally, as an individual matures into adulthood, the importance of active rGE increases. Here, an individual actively selects their environment, or niche, based on characteristics of their genetic predisposition. For example, antisocial youth may select into high risk neighborhoods or affiliate with deviant peers.
The ADH1B polymorphism (rs1229984) has been implicated in Asian populations to afford protection against the development of alcoholism, putatively via flushing (facial reddening due to accelerated conversion of ethanol to acetaldehyde). Rs1229984 was not originally detected in GWAS in European-American populations. The minor allele frequency of this variant in European-Americans is low (<5%) and, commercial GWAS arrays rely on common variation. Accordingly, the SNP was neither genotyped on these arrays nor could it be reliably imputed. However, genotyping this polymorphism resulted in genome-wide significant association signals (Bierut et al., 2012), and with the advent of more recent arrays that target this variant better, there is now evidence in a GWAS of an association between rs1229984 and alcoholism at p=1.2×10−31 (Gelernter et al., 2014). In this instance, GWAS did not initially guide identification of a logical and validated candidate gene, highlighting that all techniques have their strengths and weaknesses.
References
- Agrawal A, Bierut L. Identifying genetic variation for alcohol dependence. Alcohol Research. 2012;34(3):274–281. [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Freedman ND, Cheng YC, Lin P, Shaffer JR, Sun Q, Bierut LJ. Measuring alcohol consumption for genomic meta-analyses of alcohol intake: opportunities and challenges. Am J Clin Nutr. 2012;95(3):539–547. doi: 10.3945/ajcn.111.015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken LS, West SG. Multiple regression: Testing and interpreting interactions. Newbury Park, CA: Sage; 1991. [Google Scholar]
- Aliev F, Latendresse SJ, Bacanu S, Neale M, Dick DM. Testing for measured gene-environment interaction: Problems with the use of cross-product terms and a regression model reparameterization solution. Behavior Genetics. 2014;44(2):165–181. doi: 10.1007/s10519-014-9642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li TK, Schuckit M, Rice J. The Collaborative Study on the Genetics of Alcoholism. Alcohol Health & Research World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- Bergeman CS, Plomin R. Genotype-environment interaction. In: Bornstein MH, Bruner JS, editors. Interaction in human development. Hillsdale, NJ: Lawrence Erlbaum Associates; 1989. pp. 157–171. [Google Scholar]
- Bierut L, Goate A, Breslau N, Johnson E, Bertelsen S, Fox L, Edenberg H. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Molecular Psychiatry. 2012;17(4):445–450. doi: 10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum K, Noble EP, Sheridan PJ, Montgomery A, Ritchie T, Jagadeeswaran P, Cohn JB. Allelic association of human dopamine D2 receptor gene in alcoholism. Journal of the American Medical Association. 1990;263:2055–2060. [PubMed] [Google Scholar]
- Boardman JD. State-level moderation of genetic tendencies to smoke. Am.J.Public Health. 2009;99(3):480–486. doi: 10.2105/AJPH.2008.134932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman JD, Domingue BW, Blalock CL, Haberstick BC, Harris KM, McQueen MB. Is the gene-environment interaction paradigm relevant to genome-wide studies? The case of education and body mass index. Demography. 2014;51(1):119–139. doi: 10.1007/s13524-013-0259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan R, Williamson D, Hariri AR. Mineralocorticoid receptor Iso/Val (rs5522) genotype moderates the association between previous childhood emotional neglect and amygdala reactivity. American Journal of Psychiatry. 2012;169(5):515–522. doi: 10.1176/appi.ajp.2011.11060855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraska V, Franklin CS, Floyd JA, Thornton LM, Huckins LM, Southam L, Builk CM. A genome-wide association study of anorexia nervosa. Molecular Psychiatry. 2014 doi: 10.1038/mp.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, Posthuma D, WA N. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Molecular Psychiatry. 2011;16(5):516–532. doi: 10.1038/mp.2010.38. [DOI] [PubMed] [Google Scholar]
- Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, Posthuma D, Nolen WA. Poor replication of candidate genes for major depressive disorder using genome-wide association data. Molecular Psychiatry. 2011;16(5):516–532. doi: 10.1038/mp.2010.38. [DOI] [PubMed] [Google Scholar]
- Brown GW, Harris TO. Depression and the serotonin transporter 5-HTTLPR polymorphism: a review and a hypothesis concerning gene-environment interaction. Journal of Affective Disorders. 2008;111(1):1–12. doi: 10.1016/j.jad.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES, Munafo MR. Power failure: why small sample size undermines the reliability of neuroscience. Nature Reviews Neuroscience. 2013;14(5):365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- Button KS, Ioannidis JP, Morkrysz C, Nosek BA, Flint J, Robinson ES, Munafo MR. Power failure: Why small sample size undermines the reliability of neuroscience. Nature Reviews - Genetics. 2013;14(5):365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- Button TM, Lau JY, Maughan B, Eley TC. Parental punitive discipline, negative life events and gene-environment interplay in the development of externalizing behavior. Psychological Medicine. 2008;38(1):29–39. doi: 10.1017/S0033291707001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button TM, Stallings MC, Rhee SH, Corley RP, Boardman JD, Hewitt J. Perceived peer delinquency and the genetic predisposition for substance dependence vulnerability. Drug & Alcohol Dependence. 2009;100:1–8. doi: 10.1016/j.drugalcdep.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, Poulton R. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, Poulton R. Influence of Life Stress on Depression: Moderation by a Polymorphism in the 5-HTT Gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Clarke H, Flint J, Attwood AS, Munafo MR. Association of the 5-HTTLPR genotype and unipolar depression: a meta-analysis. Psychological Medicine. 2010;40(11):1767–1778. doi: 10.1017/S0033291710000516. [DOI] [PubMed] [Google Scholar]
- Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361(9360):865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- Collins AC, Kim Y, Sklar P, International Schizophrenia C, O'Donovan M, PF S. Hypothesis driven candidate genes for schizohrenia compared to genome-wide association results. Psychological Medicine. 2012;42(3):607–616. doi: 10.1017/S0033291711001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Pericak-Vance MA. Gene does of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Cornelis MC, Tchetgen EJ, Liang L, Qi L, Chatterjee N, Hu FB, Kraft P. Gene-environmnet interactions in genome-wide association studies: a comparative study of tests applied to empirical studies of type 2 diabetes. American Journal of Epidemiology. 2012;175(5):191–202. doi: 10.1093/aje/kwr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvin A, Craddock N, Sullivan PF. Genome-wide association studies: a primer. Psychological Medicine. 2010;40(7):1063–1077. doi: 10.1017/S0033291709991723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. American Journal of Medical Genetics (Neuropsychiatric Genetics) 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5'-region of GABRG1 associate to alcohol dependence and ar in linkage disequillibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33(4):837–848. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock N, Dave S, Greening J. Association studies of bipolar disorder. Bipolar Disorders. 2001;3(6):284–298. doi: 10.1034/j.1399-5618.2001.30604.x. [DOI] [PubMed] [Google Scholar]
- Culverhouse RC, Bowes L, Breslau N, Nurnberger J, Burmeister M, Fergusson DM 5-HTTLPR_Stress_and_Depression_Consortium. Protocol for a collaborative meta-analysis of 5-HTTLPR, stress and depression. BMC Psychiatry. 2013;13:304. doi: 10.1186/1471-244X-13-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming G. The new statistics: Why and how. Psychological Science. 2014;25(1) doi: 10.1177/0956797613504966. [DOI] [PubMed] [Google Scholar]
- Dick DM, Bernard M, Aliev F, Viken R, Pulkkinen L, Kaprio J, Rose RJ. The Role of Socioregional Factors in Moderating Genetic Influences on Early Adolescent Behavior Problems and Alcohol Use. Alcoholism: Clinical and Experimental Research. 2009;33(10):1739–1748. doi: 10.1111/j.1530-0277.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Latendresse SJ, Lansford JE, Budde J, Goate A, Dodge KA, Bates J. Role of GABRA2 in Trajectories of Externalizing Behavior Across Development and Evidence of Moderation by Parental Monitoring. Archives in General Psychiatry. 2009;66:649–657. doi: 10.1001/archgenpsychiatry.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Pagan JL, Holliday C, Viken R, Pulkkinen L, Kaprio J, Rose RJ. Gender differences in friends' influences on adolescent drinking: a genetic epidemiological study. Alcoholism: Clinical and Experimental Research. 2007;31(12):2012–2019. doi: 10.1111/j.1530-0277.2007.00523.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Pagan JL, Viken R, Purcell S, Kaprio J, Pulkkinen L, Rose RJ. Changing environmental influences on substance use across development. Twin Research and Human Genetics. 2007;10(2):315–326. doi: 10.1375/twin.10.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Rose RJ, Viken RJ, Kaprio J, Koskenvuo M. Exploring gene-environment interactions: Socioregional moderation of alcohol use. Journal of Abnormal Psychology. 2001;110:625–632. doi: 10.1037//0021-843x.110.4.625. [DOI] [PubMed] [Google Scholar]
- Dick DM, Viken R, Purcell S, Kaprio J, Pulkkinen L, Rose RJ. Parental monitoring moderates the importance of genetic and environmental influences on adolescent smoking. Journal of Abnormal Psychology. 2007;116(1):213–218. doi: 10.1037/0021-843X.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon CI, Walker SE, King SL, Stephens DN. Deletion of the gabra2 gene results in hypersensitivity to the acute effects of ethanol but does not alter ethanol self administration. PLoS One. 2012;7(10):e47135. doi: 10.1371/journal.pone.0047135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgon T, D'Addario C, Uhl GR. Linkage disequilibrium, haplotype and association studies of a chromosome 4 GABA receptor gene cluster: Candidate gene variants for addictions. American Journal of Medical Genetics Part B (Neuropsychiatric Genetics) 2006;141B(8):854–860. doi: 10.1002/ajmg.b.30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L, Keller MC. A critical review of the first ten years of measured gene-by-environment interaction research in psychiatry. American Journal of Psychiatry. 2011;168:1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves L, Erkanil A. Markov Chain Monte Carlo approaches to analysis of gentic and environmental components of human development change and G × E interaction. Behavior Genetics. 2003;33(3):279–299. doi: 10.1023/a:1023446524917. [DOI] [PubMed] [Google Scholar]
- Eaves LJ. Genotype × Environment interaction in psychopathology: fact or artifact? Twin Res Hum Genet. 2006;9(1):1–8. doi: 10.1375/183242706776403073. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Begleiter H. Variations in GABRA2, encoding the ‡2 subunit of the GABA-A receptor are associated with alcohol dependence and with brain oscillations. American Journal of Human Genetics. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Schwartz L, Albaugh B, Virkkunen M, Goldman D. Dimensional anxiety mediates linkage of GABRA2 haplotypes with alcoholism. Am. J.Med.Genet B Neuropsychiatr.Genet. 2006;141B(6):599–607. doi: 10.1002/ajmg.b.30336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatric Genetics. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychological Medicine. 2007;37:163–180. doi: 10.1017/S0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganzach Y. Misleading interaction and curvilinear terms. Psychological Methods. 1997;2:235–247. [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Farrer LA. Genome-wide association study of alcohol dependence: Significant findings in African- and European-Americans including novel risk loci. Molecular Psychiatry. 2014;19(1):41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, Farrer L. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Human Molecular Genetics. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Begleiter H, Porjesz B, Chorlian DB, Edenberg HJ, Foroud T, Reich T. Linkage mapping of beta 2 EEG waves via non-parametric regression. American Journal of Medical Genetics (Neuropsychiatric Genetics) 2003;118B:66–71. doi: 10.1002/ajmg.b.10057. [DOI] [PubMed] [Google Scholar]
- Gusella JF, Wexler NS, Conneally PM, Naylor SL, Anderson MA, Tanzi RE, et al. A polymorphic DNA marker genetically linked to Huntington's disease. Nature. 1983;306(5940):234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- Hamilton CM, Strader LC, Pratt JG, Maiese D, Hendershot T, Kwok RK, Haines J. The PhenX toolkit: Get the most from your measures. American Journal of Epidemiology. 2011;174(3):253–260. doi: 10.1093/aje/kwr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden KP, Hill JE, Turkheimer E, Emery RE. Gene-environment correlation and interaction in peer effects on adolescent alcohol and tobacco use. Behavior Genetics. 2008;38:339–347. doi: 10.1007/s10519-008-9202-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA. Ethanol actions on multiple ion channels: Which are important? Alcoholism: Clinical and Experimental Research. 1999;23(10):1563–1570. [PubMed] [Google Scholar]
- Hewitt J. Editorial policy on candidate gene association and candidate gene-by-environment interaction studies of complex traits. Behavior Genetics. 2012;42:1–2. doi: 10.1007/s10519-011-9504-z. [DOI] [PubMed] [Google Scholar]
- Hodgkinson C, Yuan Q, Xu K, Shen P, Heinz E, Lobos E, Goldman D. Addictions Biology: Haplotype-Based Analysis for 130 Candidate Genes on a Single Array. Alcohol & Alcholism. 2008;43(5):505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussong AM, Curran PJ, Bauer DJ. Integrative data analysis in clinical psychology research. Annual Review of Clinical Psychology. 2013;9:61–89. doi: 10.1146/annurev-clinpsy-050212-185522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter CM, Mechanic LE, Chatterjee N, Kraft P, Gillanders EM. Gene-environment interactions in cancer epidemiology: A National Cancer Institute think tank report. Genetic Epidemiology. 2013;37:643–657. doi: 10.1002/gepi.21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2(8):e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Munafo MR, Fusar-Poll P, Nosek BA, David SP. Pubilcation and other reporting biases in cognitive sciences: detection, prevalence, prevention. Trends in Cognitive Science. 2014;18(5):235–241. doi: 10.1016/j.tics.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PO, Neyman J. Tests of certain linear hypotheses and their applications to some educational problems. Statistical Research Memoirs. 1936;1:57–93. [Google Scholar]
- Johnston C, Lahey BB, Matthys W. Editorial policy for candidate gene studies. J Abnorm Child Psychol. 2013;41:511–514. [Google Scholar]
- Kareken DA, Liang T, Wetherill L, Dzemidzic M, Bragulat V, Cox C, Foroud T. A polymorphism in GABRA2 is associated with the medial frontal response to alcohol cues in an fMRI study. Alcoholism: Clinical and Experimental Research. 2010;34(12):2169–2178. doi: 10.1111/j.1530-0277.2010.01293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stresss, and depression meta-analysis revisited: evident of genetic moderation. Archives of General Psychiatry. 2011;68:444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MC. Gene by environment interaction studies have not properly controlled for potential confounders: The problem and the (simple) solution. Biological Psychiatry. 2014;75(1):18–24. doi: 10.1016/j.biopsych.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler K, Gardner C. Interpretation of interactions: Guide for the perplexed. British Journal of Psychiatry. 2010;197(3):170–171. doi: 10.1192/bjp.bp.110.081331. [DOI] [PubMed] [Google Scholar]
- Kendler KS. What psychiatric genetics has taught us about the nature of psychiatric illness and what is left to learn. Molecular Psychiatry. 2013;18:1058–1966. doi: 10.1038/mp.2013.50. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Eaves LJ. Models for the joint effect of genotype and environment on liability to psychiatric illness. The American Journal of Psychiatry. 1986;143:279–289. doi: 10.1176/ajp.143.3.279. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Kessler RC, Walters EE, MacLean CJ, Neale MC, Heath AC, Eaves LJ. Stressful life events, genetic liability, and onset of an episode of major depression in women. American Journal of Psychiatry. 1995;152(6):833–842. doi: 10.1176/ajp.152.6.833. [DOI] [PubMed] [Google Scholar]
- Kim-Cohen J, Caspi A, Taylor A, Williams B, Newcombe R, Craig IW, Moffitt TE. MAOA, maltreatment, and gene-environment interaction predicting children's mental health: new evidence and a meta-analysis. Mol Psychiatry. 2006;11(10):903–913. doi: 10.1038/sj.mp.4001851. [DOI] [PubMed] [Google Scholar]
- Lakens D, Evers E. Sailing from the seas of chaos into the corridor of stability: practical recommnedations to increase the informational value of studies. Perspectives on Psychological Science. 2014;9(3):278–292. doi: 10.1177/1745691614528520. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, Gelernter J. Association between alcoholism and Gamma-Amino Butyric Acid alpha2 receptor subtype in a Russian population. Alcoholism: Clinical and Experimental Research. 2005;29:493–498. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- Lasky-Su J, Neale BM, Franke B, Anney RJ, Zhou K, Maller JB, Faraone SV. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. American Journal of Medical Genetics Part B (Neuropsychiatric Genetics) 2008;147B(8):1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- Li D, Sulovari A, Cheng C, Zhao H, Kranzler HR, Gelernter J. Association of gamma-aminobutyric acid A receptor a2 (GABRA2) with alcohol use disorder. Neuropsychopharmacology. 2014;39(4):907–918. doi: 10.1038/npp.2013.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yang AR, Kelly T, Puche A, Esoga C, June JL, Aurelian L. Binge alcohol drinking is associated with GABAA alpha2-regulated Toll-like receptor 4 (TLR4) expression in the central amygdala. Proceedings of the National Academy of Sciences in the USA. 2011;108(11):4465–4470. doi: 10.1073/pnas.1019020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillal SG, Middelton L, Berrettini W, Marchini J. Meta-analysis and imputation refines the association of 12q25 with smoking quality. Nature Genetics. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski D, Humphreys LG. Assessing spurious "moderator effects": Illustrated substantively with the hypothesized ("synergistic") relation between spatial visualization and mathematical ability. Psychological Bulletin. 1990;107:385–393. doi: 10.1037/0033-2909.107.3.385. [DOI] [PubMed] [Google Scholar]
- Manolio T, Collins FS. The HapMap and Genome-Wide Association Studies in Diagnosis and Therapy. Annual Review of Medicine. 2009;60:443–456. doi: 10.1146/annurev.med.60.061907.093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Hoffman EK, Zezza N, Stiffler S, Hill SY. The role of the GABRA2 polymorphism in multiplex alcohol dependence families with minimal comorbidity: within-family association and linkage analyses. Journal of Studies on Alcohol and Drugs. 2007;68(5):625–633. doi: 10.15288/jsad.2007.68.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland GH, Judd CM. Statistical difficulties of detecting interactions and moderator effects. Psychological Bulletin. 1993;114(2):376–390. doi: 10.1037/0033-2909.114.2.376. [DOI] [PubMed] [Google Scholar]
- Meyers JL, Nyman E, Loukola A, Rose RJ, Kaprio J, Dick D. The association between DRD2/ANKK1 and genetically informed measure of alcohol use and problems. Addiction Biology. 2013;10(3):523–536. doi: 10.1111/j.1369-1600.2012.00490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe SM. Modern approaches to conceptualizing and measuring human life stress. Annual Review of Clinical Psychology. 2008;4:33–52. doi: 10.1146/annurev.clinpsy.4.022007.141207. [DOI] [PubMed] [Google Scholar]
- Monroe SM, Reid MW. Gene-environment interactions in depression: Genetic polymorphisms and life stress polyprocedures. Psychological Science. 2008;10:947–956. doi: 10.1111/j.1467-9280.2008.02181.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee B, Ahn J, Gruber SB, Chatterjee N. Testing gene-environment interaction in large-scale case-control association studies: Possible choices and comparisons. American Journal of Epidemiology. 2012;175(3):177–190. doi: 10.1093/aje/kwr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo MR. Candidate gene studies in the 21st century: meta-analysis, mediation, moderation. Genes, Brain, and Behavior. 2006;5(S1):3–5. doi: 10.1111/j.1601-183X.2006.00188.x. [DOI] [PubMed] [Google Scholar]
- Munafo MR. Reliability and replicability of genetic association studies. Addiction. 2009;104(9):1439–1440. doi: 10.1111/j.1360-0443.2009.02662.x. [DOI] [PubMed] [Google Scholar]
- Munafo MR, Durrant C, Lewis G, Flint J. Gene × environment interactions at the serotonin transporter locus. Biological Psychiatry. 2009;65(3):211–219. doi: 10.1016/j.biopsych.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Munafo MR, Flint J. Replication and heterogeneity in gene × environment interaction studies. Int J Neuropsychopharmacol. 2009;12:727–729. doi: 10.1017/S1461145709000479. [DOI] [PubMed] [Google Scholar]
- Munafo MR, Gage SH. Improving the reliability and reporting of genetic association studies. Drug and Alcohol Dependence. 2013;132(3):411–413. doi: 10.1016/j.drugalcdep.2013.03.023. [DOI] [PubMed] [Google Scholar]
- Murray JM, Davies KE, Harper PS, Meredith L, Mueller CR, Williamson R. Linkage relationship of a cloned DNA sequence on the short arm of the × chromosome to Duchenne muscular dystrophy. Nature. 1982;300(5887):69–71. doi: 10.1038/300069a0. [DOI] [PubMed] [Google Scholar]
- Need AC, Ge D, Weale ME, Maia J, Feng SHEL, Shianna KV, Goldstein DB. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genetics. 2009;5(2):e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiswanger K, Kaplan BB, Hill SY. What can the DRD2/alcoholism story teach us about association studies in psychiatric genetics? American Journal of Medical Genetics (Neuropsychiatric Genetics) 1995;60:272–275. doi: 10.1002/ajmg.1320600403. [DOI] [PubMed] [Google Scholar]
- Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANKK1: A novel kinase gene closely linked to DRD2 on chromosome band 11q23.1. Human Mutation. 2004;23(6):540–545. doi: 10.1002/humu.20039. [DOI] [PubMed] [Google Scholar]
- Olfson E, Bierut LJ. Convergence of Genome-Wide Association and Candidate Gene Studies for Alcoholism. Alcohol Clin Exp Res. 2012 doi: 10.1111/j.1530-0277.2012.01843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porjesz B, Almasy L, Edenberg HJ, Wang K, Chorlian DB, Foroud T, Begleiter H. Linkage disequilibrium between the beta frequency of the human EEG and a GABAA receptor gene locus. PNAS. 2002;99:3729–3733. doi: 10.1073/pnas.052716399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nature Reviews - Genetics. 2010;11(7):459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. Variance components models for gene-environment interaction in twin analysis. Twin Research. 2002;5(6):554–571. doi: 10.1375/136905202762342026. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. American Journal of Medical Genetics. 1998;81:207–215. [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, Sullivan PF. Genome-wide association analysis identified 13 new risk loci for schizophrenia. Nature Genetics. 2013;45(10):1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, Merikangas KR. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA. 2009;301(23):2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, Merikangas K. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. Journal of the American Medical Association. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roisman GI, Newman DA, Fraley RC, Haltigan JD, Groh AM, Haydon KC. Distinguishing differential susceptibility from diathesis-stress: Recommendations for evaluating interaction effects. Development and Psychopathology. 2012;24(2):389–409. doi: 10.1017/S0954579412000065. [DOI] [PubMed] [Google Scholar]
- Rose EJ, Donohoe G. Brain vs behavior: An effect size comparison of neuroimaging and cognitive studies of genetic risk for schizophrenia. Schizophrenia Bulletin. 2013;39(3):518–526. doi: 10.1093/schbul/sbs056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RJ, Dick DM, Viken RJ, Kaprio J. Gene-environment interaction in patterns of adolescent drinking: Regional residency moderates longitudinal influences on alcohol use. Alcoholism: Clinical and Experimental Research. 2001;25:637–643. [PubMed] [Google Scholar]
- Salvatore J, Aliev F, Edwards A, Evans D, Macleod J, Hickman M, Dick D. Polygenic scores predict alcohol problems in an independent sample and show moderation by the environment. Genes. 2014;5(2):330–346. doi: 10.3390/genes5020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YC, Fan JH, Edenberg HJ, Li TK, Cui YH, Wang YF, Xia GY. Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcoholism: Clinical and Experimental Research. 1997;21(7):1272–1277. [PubMed] [Google Scholar]
- Sher K, Steinley DS. Some issues surrounding interactions; Paper presented at the NIAAA Workshop on Gene-Environmnet Interactions; Rockville, MD. 2013. [Google Scholar]
- Simmons JP, Nelson LD, Simonsohn U. False-positive psychology: undisclosed flexibility in data collection and analysis allows presenting anything as significant. Psychological Science. 2011;22(11):1359–1366. doi: 10.1177/0956797611417632. [DOI] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Purcell SM. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature Genetics. 2011;43(10):977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. Mendelian radomization for strengthening causal interference in observational studies: Application to gene × environment interactions. Perspectives on Psychological Science. 2010;5(5):527–545. doi: 10.1177/1745691610383505. [DOI] [PubMed] [Google Scholar]
- South SC, Krueger RF. Martial quality moderates genetic and environmental influences on the internalizing spectrum. Journal of Abnormal Psychology. 2008;117(4):826–837. doi: 10.1037/a0013499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyka M, Preuss UW, Hesselbrock V, Zill P, Koller G, Bondy B. GABA-A2 receptor subunit gene (GABRA2) polymorphisms and risk for alcohol dependence. Journal of Psychiatric Research. 2008;42(3):184–191. doi: 10.1016/j.jpsychires.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Spellman BA. Introduction to the special section: Data, data, everywhere…especially in my file drawer. Perspectives on Psychological Science. 2012;7:58–59. doi: 10.1177/1745691611432124. [DOI] [PubMed] [Google Scholar]
- Steinberg S, de Jong S, Mattheisen M, Costas J, Demontis D, Jamain S, Stefansson K. Common variant 16p11.2 conferring risk of psychosis. Molecular Psychiatry. 2014;19(1):108–114. doi: 10.1038/mp.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens SH, Hartz SM, Hoft NR, Saccone NL, Corley RC, Hewitt JK, Ehringer MA. Distinct loci in the CHRNA5/CHRNA3/CHRNB4 gene cluster are associated with onset of regular smoking. Genetic Epidemiology. 2013;37(8):846–859. doi: 10.1002/gepi.21760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SS. On the theory of scales of measurement. Science. 1946;103(2684):677–680. [PubMed] [Google Scholar]
- Sullivan PF, Daly M, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nature Reviews Genetics. 2012;13(8):537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkings D, Stroup TS, Close SL. Genomewide association for schizophrenia in the CATIE study: Results of stage 1. Molecular Psychiatry. 2008;13(6):570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC, Lewinger JP, Murcray CE, Gauderman WJ. Invited commentary: GE-Whiz! Ratcheting gene-environment studies up to the whole genome and the whole exposome. American Journal of Epidemiology. 2012;175(3):203–207. doi: 10.1093/aje/kwr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME Saguenay_Youth_Study_(SYS)_Group. The ENIGMA Consortium: Large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging and Behavior. 2014;8(2):153–182. doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Stefansson K. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nature Genetics. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacco_and_Genetics_Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature Genetics. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Rietschel M. Genome-wide association study of alcohol dependence. Archives of General Psychiatry. 2009;66(7):773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui LC, Buchwald M, Barker D, Braman JC, Knowlton R, Schumm JW, et al. Cystic fibrosis locus defined by a genetically linked polymorphic DNA marker. Science. 1985;230(4729):1054–1057. doi: 10.1126/science.2997931. [DOI] [PubMed] [Google Scholar]
- Uher R, McGuffin P. The moderation by the serotonin transporter gene of environmental adversity in depression: 2009 update. Molecular Psychatry. 2010;15(1):18–22. doi: 10.1038/mp.2009.123. [DOI] [PubMed] [Google Scholar]
- van Assen MA, van Aert RC, Nuijten MB, Wicherts JM. Why publishing everything is more effective than selective publishing of statistically significant results. PLoS One. 2014;9(1):e84896. doi: 10.1371/journal.pone.0084896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluis S, Posthuma D, Dolan CV. A note on false positives and power in GxE modelling of twin data. Behavior Genetics. 2012;42:170–186. doi: 10.1007/s10519-011-9480-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villafuerte S, Heitzeg MM, Foley S, Yau WY, Majczenko K, Zubieta JK, Burmeister M. Impulsiveness and insula activation during reward anticipation are associated with genetic variants in GABRA2 in a family sample enriched for alcoholism. Molecular Psychiatry. 2012;17(5):511–519. doi: 10.1038/mp.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. American Journal of Human Genetics. 2012;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward H, Mitrou PN, Bowman R, Luben R, Wareham NJ, Khaw KT, Bingham S. APOE genotype, lipids, and coronary heart disease risk: A prospective population study. Archives of Internal Medicine. 2009;169(15):1424–1429. doi: 10.1001/archinternmed.2009.234. [DOI] [PubMed] [Google Scholar]
- Weng L, Macciardi F, Subramanian A, Guffanti G, Potkin SG, Yu Z, Xie X. SNP-based pathway enrichment analysis for genome-wide association studies. BMC Bioinformatics, Apr 15 Epub. 2011 doi: 10.1186/1471-2105-12-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whytt R. Observations on the nature, causes, and cure of those disorders which are commonly called nervous, hypochondriac, or hysteric. Edinburgh, Scotland: Becket & Du Hondt; 1765. [Google Scholar]
- Widaman KF, Helm JL, Castro-Schilo L, Pluess M, Stallings MC, Belsky J. Distinguising ordinal and disordinal interactions. Psychological Methods. 2012;17(4):615–622. doi: 10.1037/a0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DHR, Penninx BWJH, Gordon SD, Nyholt DR, Sullivan PF. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry. 2012;17(1):36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Association of haplotypic variants in DRD2, ANKK1, TTC12 and NCAM1 to alcohol dependence in independent case control and family samples. Human Molecular Genetics. 2007;16(23):2844–2853. doi: 10.1093/hmg/ddm240. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Haplotypic variants in DRD2, ANKK1, TTC12, and NCAM1 are associated with comorbid alcohol and drug dependence. Alcoholism: Clinical and Experimental Research. 2008;32(12):2117–2127. doi: 10.1111/j.1530-0277.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RM, Lawford BR, Nutting A, Noble EP. Advances in molecular genetics and the prevention and treatment of substance misuse: Implications of association studies of the A1 allele of the D2 dopamine receptor gene. Addictive Behaviors. 2004;29:1275–1294. doi: 10.1016/j.addbeh.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Yzerbyt VY, Muller D, Judd CM. Adjusting researchers' approach to adjustment: On the use of covariates when testing interactions. Journal of Experimental Social Psychology. 2004;40:424–431. [Google Scholar]
- Zammit S, Lewis G, Dalman C, Allebeck P. Examinging interactions between risk factors for psychosis. British Journal of Psychiatry. 2010;197(3):207–211. doi: 10.1192/bjp.bp.109.070904. [DOI] [PubMed] [Google Scholar]
- Zammit S, Owen MJ, Lewis G. Misconceptions about gene-environment interactions in psychiatry. Evidenced-Based Mental Health. 2010;13(3):65–68. doi: 10.1136/ebmh.13.3.65. [DOI] [PubMed] [Google Scholar]
- Zannas AS, Binder EB. Gene-environment interactions at the FKBP5 locus: Sensitive periods, mechanisms and pleiotropism. Genes, Brain and Behavior. 2014;13(1):25–37. doi: 10.1111/gbb.12104. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Guo A, van den Oordd EJCG, Aliev F, Jia P, Riley BP, Miles MF. Multi-species data integration and gene ranking enriches significant results in an alcoholism genome-wide association study. BMC Genomics. 2012;13(Suppl 8):S16. doi: 10.1186/1471-2164-13-S8-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LTY, Kohlbacher O, Meissner A. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Statistics in Medicine. 2002;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit S, Nizzari MM, O'Donnell CJ, de Bakker PIW. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prium RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Giledt TP, Willer CJ. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale BM, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Sham PC. PLINK: A toolset for whole-genome association and population-based linkage analysis. Americal Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nature Genetics. 2006;38(2):209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A toll for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Lowdon RF, Li D, Lawson HA, Madden PA, Cstello JF, Wang T. Exploring long-range genome interactions using the WashU Epigenome Browser. Nature Methods. 2013;10(5):375–376. doi: 10.1038/nmeth.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]