Abstract
Plants assimilate sulfate from the environment to synthesize biologically active sulfur-containing compounds required for growth and cellular development. The primary steps of sulfur metabolism involve sequential enzymatic reactions synthesizing adenosine 5′-phosphosulfate (APS) and 3′-phosphoadenosine 5’-phosphosulfate (PAPS). Recent finding suggests that an adenosine nucleotide transport system facilitating the exchange of PAPS and 3′-phosphoadenosine 5′-phosphate across the plastid envelope is essential for establishing an intimate connection between the plastidic and cytosolic sulfate assimilation pathways in plants. Subcellular partitioning and integration of metabolic pathways provide focal points for investigating metabolic flux regulations. This perspective article presents an integrative view of sulfur metabolic flux control mechanisms with an emphasis on subcellular partitioning of APS/PAPS biosynthetic pathways in Arabidopsis thaliana.
Keywords: sulfur metabolism, sulfate assimilation, subcellular localization, metabolic flux, metabolite distribution
SUBCELLULAR LOCALIZATION AND PATHWAY DISTRIBUTIONS
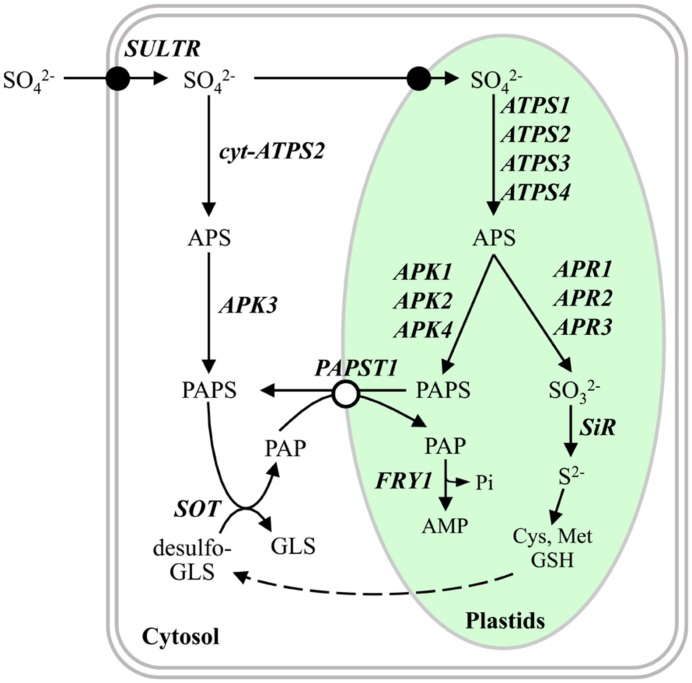
Sulfate assimilation occurs in both plastids and cytosol in vascular plants (Figure 1; reviewed in Takahashi et al., 2011; Gigolashvili and Kopriva, 2014; Koprivova and Kopriva, 2014). Sulfate imported across the plasma membrane is the primary substrate provided to the sulfate assimilation pathways, where the ATP sulfurylase (ATPS) serves as an enzyme to catalyze the initial metabolic reaction generating adenosine 5′-phosphosulfate (APS) from ATP and sulfate in both plastids and cytosol. APS is subsequently phosphorylated to 3′-phosphoadenosine 5′-phosphosulfate (PAPS) by the APS kinase (APK), or reduced to sulfite through the function of the APS reductase (APR). APK is present in both plastids and cytosol for phosphorylation, while APR and the subsequent pathway enzyme, sulfite reductase (SiR), are localized only in plastids for catalyzing the reduction steps. The sulfate assimilation pathway thus bifurcates into two directions to phosphorylate or reduce APS in plastids, whereas only the APS phosphorylation pathway is present in cytosol (Figure 1).
FIGURE 1.
Metabolic pathway of sulfur assimilation in Arabidopsis. The illustration shows subcellular partitioning of sulfate assimilation and APS/PAPS metabolic pathways in plastids and cytosol in Arabidopsis. Cys, Met, and GSH biosynthetic pathways are simplified in this diagram to describe their connections downstream of the reductive sulfate assimilation pathway. The names of enzymes and transporters are indicated in bold italicized letters. Closed and open circles indicate sulfate transporter and PAPS transporter, respectively. Cytosolic ATP sulfurylase (cyt-ATPS2) forms by alternative translational initiation (Bohrer et al., 2014). Abbreviations of metabolites: APS, adenosine-5′-phosphosulfate; Cys, cysteine; GLS, glucosinolate; GSH, glutathione; Met, methionine; PAP, 5′-phosphoadenosine 3′-phosphate; PAPS, 3′-phosphoadenosine 5′-phosphosulfate. Abbreviations of enzymes and transporters: APK, APS kinase; APR, APS reductase; ATPS, ATP sulfurylase; FRY1, inositol polyphosphate 1-phosphatase (FIERY1); PAPST1, PAPS transporter; SiR, sulfite reductase; SOT, sulfotransferase; SULTR, sulfate transporter.
Plants switch-control the APS reduction and phosphorylation pathways to change the partitioning of sulfur into the primary and secondary metabolisms (Kopriva et al., 2012). APR plays a key role in channeling APS into the sulfate reduction pathway, responding to the demands for Cys and GSH. The demand-driven flux control mechanism has been suggested based on observations of APR transcripts and proteins over-accumulating following sulfate deprivation and being repressed in the presence of reduced S sources (Takahashi et al., 1997; Vauclare et al., 2002). The significance of APR in the primary sulfur metabolism is evidenced by the accumulation of sulfate and total S in Arabidopsis accessions with less active variants of APR2 enzyme (Loudet et al., 2007; Chao et al., 2014). In contrast, the expression of plastidic APK, which is required for PAPS biosynthesis providing sulfate donors used in secondary sulfur metabolism, is repressed under sulfur-deficient conditions (Maruyama-Nakashita et al., 2006; Mugford et al., 2009). In addition to the transcriptional mechanisms, APR and APK enzyme activities are regulated by the redox status as demonstrated by Arabidopsis APR1 and APK1 gaining maximum catalytic efficiency in their oxidized and reduced forms, respectively (Bick et al., 2001; Ravilious and Jez, 2012; Ravilious et al., 2012, 2013). Thus, the metabolic flux through the bifurcate pathway for APS utilization is affected by both transcriptional and post-transcriptional mechanisms controlling enzyme activity.
INTEGRATION OF PAPS METABOLISM THROUGH PAPS TRANSPORTER
The plastidic and cytosolic pathways merge following PAPS biosynthesis as PAPS is mainly utilized in the cytosol (Figure 1) where it serves as sulfate donor for synthesizing sulfated metabolites including glucosinolates (Klein and Papenbrock, 2004; Piotrowski et al., 2004; Hirai et al., 2005). Therefore, PAPS metabolism has to include a PAPS transporter in the plastid envelopes. Indeed, a PAPS transporter (PAPST1) has been found to export PAPS from plastids to cytosol (Gigolashvili et al., 2012). When sulfotransferases (SOT) synthesize sulfated metabolites, the sulfate moieties of PAPS are transferred to the hydroxyl groups of suitable acceptors, and 3′-phosphoadenosine 5′-phosphate (PAP) is generated as a byproduct. PAP is, however, a cytotoxic compound, as it inhibits RNA metabolizing enzymes responsible for decomposing aberrant RNA (Gy et al., 2007). Since PAPST1 is capable of facilitating the plastid import of PAP to be coupled with the export of PAPS, PAP can be degraded to AMP by PAP phosphatase, FIERY1 (FRY1), in plastids (Rodríguez et al., 2010; Estavillo et al., 2011). PAPS biosynthesis, PAPS utilization, and PAP degradation are therefore connected as a sequential network of metabolic steps in both plastids and cytosol. Such functional interplays can be achieved only with the presence of a suitable transporter, such as PAPST1, that enables the PAPS/PAP exchange following the concentration gradients of substrates necessarily formed across the plastid envelope (Figure 1).
The absence of FRY1 induces plant responses to drought, salinity, cold, and excess light stresses, where ABA and jasmonate are involved in signaling (Xiong et al., 2001; Wilson et al., 2009; Rodríguez et al., 2010; Chen et al., 2011; Estavillo et al., 2011; Chan et al., 2013). The significance of the FRY1-mediated pathway lies in the fact that PAP can be a retrograde signal for inducing molecular mechanisms protecting viable cells from stresses in adverse environments. Given the pathway connections with the PAPS/PAP exchange across the plastid envelope (Gigolashvili et al., 2012), cellular PAP concentrations are likely modulated by FRY1 in plastids (Rodríguez et al., 2010; Estavillo et al., 2011). Chloroplast-mitochondrion dual localizations of FRY1 from Arabidopsis and PAPST from rice further suggest that mitochondria also serve for detoxifying PAP (Estavillo et al., 2011; Xu et al., 2013). Cross-species conservation of PAP metabolism requires further investigation.
FLUX CONTROL
In this framework of metabolic pathway connections, abundance and functions of ATPS and APK in plastids and cytosol, together with efficiency of PAPS transport, appear pivotal for controlling subcellular distributions of PAPS and PAP (Figure 1). ATPS and APK are predominant in plastids, as evident from the localization of individual isoforms, distribution of enzyme activities, and phenotypes of corresponding mutants (Rotte and Leustek, 2000; Mugford et al., 2009). Such subcellular distributions of ATPS and APK may be important to increase the concentration gradient of PAPS across the plastid envelope, leading to an increased export of PAPS from the plastids to the cytosol as well as a more efficient transport of PAP into the plastids for its detoxification to be accomplished. Since SOTs and consequently PAP production are localized in the cytosol (Figure 1), increase in PAPS synthesis in the cytosol would thus prevent PAP/PAPS shuttling and may lead to accumulation of PAP in the cytosol.
These flux control models are supported in part by evidence showing a strong requirement of plastidic APK for synthesis of sulfated metabolites including glucosinolates (Mugford et al., 2009). The phenotypes of the Arabidopsis apk1 apk2 mutant also suggest that cytosolic APK3 is not compensating for the loss of plastidic APK activity to provide sulfate donors to SOT (Mugford et al., 2009). Mutants in PAPST1 present similar phenotypes with accumulation of desulfo-precursors of sulfated compounds, pointing to the importance of the transporter, however, as these phenotypes are milder than those of apk1 apk2 plants, another PAPST1 has to be postulated (Gigolashvili et al., 2012). It is therefore conceivable that APK, PAPST1, and SOT are functionally coupled to utilize PAPS and sequester PAP to the plastids. In this metabolic cycle, the PAPS/PAP shuttling mechanism may not properly operate when the plastids are deficient in PAPS. To avoid elevation of PAP concentration in the cytosol, the sulfation reactions catalyzed by SOTs may be not only limited by low PAPS supply, but also actively inhibited when the PAPS/PAP shuttling mechanism is disabled. The coordinate induction of PAPST1, SOT, and FRY1 gene expression in the apk1 apk2 mutant suggests that a transcriptional coexpression mechanism is activated in an attempt to overcome the defect in plastidic PAPS production (Mugford et al., 2009). In contrast, PAP accumulates disproportionately in the plastids of fry1/fou8 mutants (Estavillo et al., 2011; Lee et al., 2012), leading to disturbance of the PAP gradient across plastid envelopes. Such circumstances also appear unfavorable for PAPST1 to shuttle PAPS/PAP and to cooperate with APK and SOT to produce sulfated metabolites (Lee et al., 2012).
The overexpression of a bacterial APK in Arabidopsis in either plastids or cytosol demonstrates, however, rather complex metabolic interconnections (Mugford et al., 2011). In the APK-overexpressing lines, APS is more likely used for synthesizing PAPS than producing sulfite. Limitation of PAPS availability can induce the expression of glucosinolate biosynthetic genes (Mugford et al., 2009). In contrast, an increased supply of PAPS does not seem to have an opposing effect on gene expression in glucosinolate biosynthesis but rather induces accumulation of MAM3 and SOT17 transcripts in the APK-overexpressing lines (Mugford et al., 2011). Furthermore, overexpression of APK causes no significant effect on increasing the flux of glucosinolate production, suggesting that pathways are under control of multifaceted mechanisms. In contrast, the metabolic flux of reductive sulfate assimilation appears to increase for adjustment of Cys and GSH biosynthesis in the APK-overexpressing lines (Mugford et al., 2011). It is possible to hypothesize that APS can be limiting in APK overexpressors, and its shortage may trigger an increase in metabolic flux of reductive sulfate assimilation. Metabolic regulation by APS has been described in bacteria (Bykowski et al., 2002).
APS biosynthesis is a thermodynamically unfavorable reaction and can be a bottleneck of the sulfate assimilation pathway. With regard to the metabolic flux control through the function of ATPS, plastidic ATPS1 makes substantial contribution to the reductive sulfate assimilation pathway (i.e., Cys and GSH biosynthesis) in Arabidopsis (Kawashima et al., 2011). In support of this evidence, ATPS1 is found as a genetic locus that significantly affects sulfate accumulation among the Arabidopsis natural variations (Koprivova et al., 2013). Furthermore, a reaction mechanism of substrate-enzyme interaction is proposed based on structural and kinetic analyzes of ATPS1 (Herrmann et al., 2014). In contrast to a wealth of information documenting the function and regulation of plastidic ATPS1, the genetic identity of a cytosolic isoform has remained elusive until recently, when alternative translational initiation of ATPS2 has been identified as a potential mechanism underlying the cytosolic ATPS activity in Arabidopsis (Bohrer et al., 2014). It has been reported that cytosolic ATPS activity becomes relatively abundant in matured Arabidopsis leaves (Rotte and Leustek, 2000), suggesting that the presence of cytosolic isoform may be conditional. Identification of mechanisms involved in regulation of cytosolic ATPS isoform will open a way to altering the function of this key enzyme and engineering metabolic flux partitioning of sulfate assimilation.
CONCLUSION
Sulfur metabolic enzymes are not equally expressed in all plant cell types and organelles. ATPS, APK, APR, and PAPST focused on in this article, represent a sub-network of sulfur assimilation pathway, in which partitioning between cytosol and plastids is particularly important. Details of the subcellular localizations of individual isoforms have only recently been acquired and although the identity of at least one additional PAPST1 is still not known, this knowledge will facilitate the dissection of isoform-dependent and compartment-specific functions of these enzymes and transporters and provide new insights into their contributions to control of flux through sulfate assimilation. Given the biological significance of metabolites synthesized in this pathway, the flux regulation of APS/PAPS biosynthesis in specific cell-types and compartments may be associated with physiological adaptations.
Conflict of Interest Statement
The Reviewer Stephan Krueger declares that, despite having collaborated in the past with the author Stanislav Kopriva, the review process was handled objectively and no conflict of interest exists. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Hideki Takahashi is supported by the National Science Foundation (MCB 1244300) and the AgBioResearch. Stanislav Kopriva is supported by Deutsche Forschungsgemeinschaft (EXC 1028).
REFERENCES
- Bick J. A., Setterdahl A. T., Knaff D. B., Chen Y., Pitcher L. H., Zilinskas B. A., et al. (2001). Regulation of the plant-type 5’-adenylyl sulfate reductase by oxidative stress. Biochemistry 40 9040–9048 10.1021/bi010518v [DOI] [PubMed] [Google Scholar]
- Bohrer A.-S., Yoshimoto N., Sekiguchi A., Rykulski N., Saito K., Takahashi H. (2014). Alternative translational initiation of ATP sulfurylase underlying dual localization of sulfate assimilation pathways in plastids and cytosol in Arabidopsis thaliana. Front. Plant Sci. 5:750 10.3389/fpls.2014.00750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykowski T., van der Ploeg J. R., Iwanicka-Nowicka R., Hryniewicz M. M. (2002). The switch from inorganic to organic sulphur assimilation in Escherichia coli: adenosine 5’-phosphosulphate (APS) as a signalling molecule for sulphate excess. Mol. Microbiol. 43 1347–1358 10.1046/j.1365-2958.2002.02846.x [DOI] [PubMed] [Google Scholar]
- Chan K. X., Wirtz M., Phua S. Y., Estavillo G. M., Pogson B. J. (2013). Balancing metabolites in drought: the sulfur assimilation conundrum. Trends Plant Sci. 18 18–29 10.1016/j.tplants.2012.07.005 [DOI] [PubMed] [Google Scholar]
- Chao D.-Y., Baraniecka P., Danku J., Koprivova A., Lahner B., Luo H., et al. (2014). Variation in sulfur and selenium accumulation is controlled by naturally occurring isoforms of the key sulfur assimilation enzyme APR2 across the Arabidopsis thaliana species range. Plant Physiol. 166 1593–608 10.1104/pp.114.247825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhang B., Hicks L. M., Xiong L. (2011). A nucleotide metabolite controls stress-responsive gene expression and plant development. PLoS ONE 6:e26661 10.1371/journal.pone.0026661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo G. M., Crisp P. A., Pornsiriwong W., Wirtz M., Collinge D., Carrie C., et al. (2011). Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23 3992–4012 10.1105/tpc.111.091033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T., Geier M., Ashykhmina N., Frerigmann H., Wulfert S., Krueger S., et al. (2012). The Arabidopsis thylakoid ADP/ATP carrier TAAC has an additional role in supplying plastidic phosphoadenosine 5’-phosphosulfate to the cytosol. Plant Cell 24 4187–4204 10.1105/tpc.112.101964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T., Kopriva S. (2014). Transporters in plant sulfur metabolism. Front. Plant Sci. 5:442 10.3389/fpls.2014.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gy I., Gasciolli V., Lauressergues D., Morel J.-B., Gombert J., Proux F., et al. (2007). Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19 3451–3461 10.1105/tpc.107.055319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J., Ravilious G. E., McKinney S. E., Westfall C. S., Lee S. G., Baraniecka P., et al. (2014). Structure and mechanism of soybean ATP sulfurylase and the committed step in plant sulfur assimilation. J. Biol. Chem. 289 10919–10929 10.1074/jbc.M113.540401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M. Y., Klein M., Fujikawa Y., Yano M., Goodenowe D. B., Yamazaki Y., et al. (2005). Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 280 25590–25595 10.1074/jbc.M502332200 [DOI] [PubMed] [Google Scholar]
- Kawashima C. G., Matthewman C. A., Huang S., Lee B.-R., Yoshimoto N., Koprivova A., et al. (2011). Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J. 66 863–876 10.1111/j.1365-313X.2011.04547.x [DOI] [PubMed] [Google Scholar]
- Klein M., Papenbrock J. (2004). The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J. Exp. Bot. 55 1809–1820 10.1093/jxb/erh183 [DOI] [PubMed] [Google Scholar]
- Kopriva S., Mugford S. G., Baraniecka P., Lee B.-R., Matthewman C. A., Koprivova A. (2012). Control of sulfur partitioning between primary and secondary metabolism in Arabidopsis. Front. Plant Sci. 3:163 10.3389/fpls.2012.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A., Giovannetti M., Baraniecka P., Lee B.-R., Grondin C., Loudet O., et al. (2013). Natural variation in the ATPS1 isoform of ATP sulfurylase contributes to the control of sulfate levels in Arabidopsis. Plant Physiol. 163 1133–1141 10.1104/pp.113.225748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A., Kopriva S. (2014). Molecular mechanisms of regulation of sulfate assimilation: first steps on a long road. Front. Plant Sci. 5:589 10.3389/fpls.2014.00589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.-R., Huseby S., Koprivova A., Chételat A., Wirtz M., Mugford S. T., et al. (2012). Effects of fou8/fry1 mutation on sulfur metabolism: is decreased internal sulfate the trigger of sulfate starvation response? PLoS ONE 7:e39425 10.1371/journal.pone.0039425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O., Saliba-Colombani V., Camilleri C., Calenge F., Gaudon V., Koprivova A., et al. (2007). Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat. Genet. 39 896–900 10.1038/ng2050 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Tohge T., Saito K., Takahashi H. (2006). Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18 3235–3251 10.1105/tpc.106.046458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford S. G., Lee B.-R., Koprivova A., Matthewman C., Kopriva S. (2011). Control of sulfur partitioning between primary and secondary metabolism. Plant J. 65 96–105 10.1111/j.1365-313X.2010.04410.x [DOI] [PubMed] [Google Scholar]
- Mugford S. G., Yoshimoto N., Reichelt M., Wirtz M., Hill L., Mugford S. T., et al. (2009). Disruption of adenosine-5’-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21 910–927 10.1105/tpc.109.065581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M., Schemenewitz A., Lopukhina A., Müller A., Janowitz T., Weiler E. W., et al. (2004). Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 279 50717–50725 10.1074/jbc.M407681200 [DOI] [PubMed] [Google Scholar]
- Ravilious G. E., Jez J. M. (2012). Nucleotide binding site communication in Arabidopsis thaliana adenosine 5’-phosphosulfate kinase. J. Biol. Chem. 287 30385–30394 10.1074/jbc.M112.387001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravilious G. E., Nguyen A., Francois J. A., Jez J. M. (2012). Structural basis and evolution of redox regulation in plant adenosine-5’-phosphosulfate kinase. Proc. Natl. Acad. Sci. U.S.A. 109 309–314 10.1073/pnas.1115772108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravilious G. E., Westfall C. S., Jez J. M. (2013). Redox-linked gating of nucleotide binding by the N-terminal domain of adenosine 5’-phosphosulfate kinase. J. Biol. Chem. 288 6107–6115 10.1074/jbc.M112.439414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez V. M., Chételat A., Majcherczyk P., Farmer E. E. (2010). Chloroplastic phosphoadenosine phosphosulfate metabolism regulates basal levels of the prohormone jasmonic acid in Arabidopsis leaves. Plant Physiol. 152 1335–1345 10.1104/pp.109.150474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotte C., Leustek T. (2000). Differential subcellular localization and expression of ATP sulfurylase and 5’-adenylylsulfate reductase during ontogenesis of Arabidopsis leaves indicates that cytosolic and plastid forms of ATP sulfurylase may have specialized functions. Plant Physiol. 124 715–724 10.1104/pp.124.2.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Kopriva S., Giordano M., Saito K., Hell R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62 157–184 10.1146/annurev-arplant-042110-103921 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Yamazaki M., Sasakura N., Watanabe A., Leustek T., Engler J. A., et al. (1997). Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 94 11102–11107 10.1073/pnas.94.20.11102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauclare P., Kopriva S., Fell D., Suter M., Sticher L., von Ballmoos P., et al. (2002). Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5’-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J. 31 729–740 10.1046/j.1365-313X.2002.01391.x [DOI] [PubMed] [Google Scholar]
- Wilson P. B., Estavillo G. M., Field K. J., Pornsiriwong W., Carroll A. J., Howell K. A., et al. (2009). The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 58 299–317 10.1111/j.1365-313X.2008.03780.x [DOI] [PubMed] [Google Scholar]
- Xiong L., Lee B., Ishitani M., Lee H., Zhang C., Zhu J. K. (2001). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15 1971–1984 10.1101/gad.891901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang J., Wu Z., Liu H., Huang F., Wu Y., et al. (2013). Identification of a dual-targeted protein belonging to the mitochondrial carrier family that is required for early leaf development in rice. Plant Physiol. 161 2036–2048 10.1104/pp.112.210831 [DOI] [PMC free article] [PubMed] [Google Scholar]