Abstract
After completion of the Schizosaccharomyces pombe genome sequence, we have carried out a pilot gene deletion project to assess the feasibility of a genome-wide deletion project and to estimate the percentage of essential genes. Using a PCR-based gene deletion procedure, we investigated 100 genes within a 253-kb region of chromosome II. Eight of nine genes located within a region of 18 kb could not be deleted, suggesting that systematic deletion of all fission yeast genes may be difficult to achieve using this PCR approach. The percentage of essential genes was found to be 17.5%. Further deletion of selected S. pombe genes revealed that whether a gene is essential or not is correlated with the timing of its appearance on the tree of life and its conservation within all branches of the tree. None of the investigated ancient genes in fission yeast that have been lost in the Saccharomyces cerevisiae lineage are essential. In agreement with S. cerevisiae and Caenorhabditis elegans genome analyses, our data suggest that natural selection has preferentially kept the genes required for vital functions. We propose that many of the essential eukaryotic genes appeared with the first eukaryotic cell and have remained conserved in all species.
The fission yeast Schizosaccharomyces pombe was the sixth eukaryote to be sequenced (Wood et al. 2002), following the budding yeast (Goffeau et al. 1996) and four multicellular organisms (C. elegans [The C. elegans Sequencing Consortium 1998], Drosophila. melanogaster [Adams et al. 2000], Arabidopsis thaliana [The Arabidopsis Genome Initiative 2000], and Homo sapiens [International Human Genome Sequencing Consortium 2001; Venter et al. 2001]). S. pombe is predicted to have a maximum of 4940 protein-coding genes, the smallest number of open reading frames (ORFs) in a eukaryote to date (Wood et al. 2002). In comparison, there are 5300 to 5400 ORFs predicted for the budding yeast Saccharomyces cerevisiae (Mackiewicz et al. 2002). The genomes of multicellular organisms contain more ORFs, with ∼15,000 for worm and fly and at least twice as many for human and Arabidopsis. Up to 100 genomes of both eubacteria and archaebacteria are now also publicly available at http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html.
The plethora of ORF sequences from organisms located in various branches of the tree of life has allowed the development of new organism classification systems and the elaboration of novel genomic trees (Tekaia et al. 1999; Korbel et al. 2002). Satisfyingly, these trees are similar to the more traditional analyses based on molecular phylogeny (Tekaia et al. 1999; Korbel et al. 2002). Paleontological work suggests that the first prokaryotic cell may have arisen ∼3800 million years ago, whereas the acquisition of a closed nucleus may have occurred ∼2000 million years ago (Feng et al. 1997). It has been estimated that fungi separated from metazoa and plants ∼1000–1200 million years ago, and that fission yeast diverged from budding yeast ∼400 million years ago (Sipiczki 2000), although older time estimates have been proposed for divergence of these yeasts by Heckman et al. (2001).
The availability of a variety of genome sequences provides a powerful tool to follow the history of a protein or protein family. Comparisons of the S. pombe, S. cerevisiae, and C. elegans gene sets led Wood et al. (2002) to conclude that 14% of the S. pombe ORFs are found exclusively in that yeast and therefore, are absent from S.cerevisiae, whereas 3% of the S. pombe ORFs have homologs in C. elegans, which appear to have disappeared from the S. cerevisiae lineage. Therefore, it appears that both acquisition and loss of genes have occurred since the divergence of S. pombe and S. cerevisiae from their common ancestor (Aravind et al. 2000). In this paper, we carry out a pilot gene deletion project in S. pombe using a PCR-based procedure (Bähler et al. 1998), to assess the feasibility of a genome-wide project and to determine the percentage of essential genes in S. pombe. Included in this analysis are genes that have been gained recently in the S. pombe and S. cerevisiae lineages, ancient genes that have been lost in the budding yeast lineage, and genes that have remained conserved throughout evolution.
RESULTS
The availability of the S. pombe genome sequence and methods for PCR-mediated deletion of ORFs (Bähler et al. 1998) has allowed us to carry out a pilot gene deletion project to assess the number of essential S. pombe genes and to address several evolutionary questions. The first step of the work was the classification of fission yeast ORFs based on phylogenetic criteria (Fig. 1). We performed BlastP analysis (Altschul and Lipman 1990) on 450 S. pombe proteins out of the 4929 predicted by PombePD (see Wood et al. 2002). These proteins were encoded by three sets of 150 successive protein-encoding genes located on each of the three chromosomes. We searched for the presence of homologs in prokaryotes, S. cerevisiae, metazoa, and plants using a threshold value for E of 10−5, and classified the fission yeast proteins into eight different classes according to the distribution of homologs within these organisms (Ia, Ib, Ic, Id, II, III, IV, and V) (classes defined in legend of Fig. 1). A similar analysis was performed on 450 predicted proteins of S. cerevisiae on chromosomes I, IV, and VIII (Fig. 1).
Figure 1.
Comparison of S. pombe and S. cerevisiae proteome. (A) A consensus phylogeny of fission yeast and budding yeast adapted from Sipiczki (2000). (B) 450 proteins from S. pombe and S. cerevisiae (YPD and PombePD) were compared to proteins from prokaryotes, metazoa, plants, S. pombe, and S. cerevisiae using BLASTP (Altschul and Lipman 1990) with a cutoff E value of 10−5. The proteins from both yeasts were classified into eight different classes, according to the distribution of homologous proteins in other species. Class I: homologous proteins are found in both S. pombe and S. cerevisiae and in either metazoa + plants + prokaryotes (Ia), or metazoa + plants with no homolog in prokaryotes (Ib), or metazoa + prokaryotes (Ic), or only metazoa (Id). Class II: homologous proteins are found in both yeasts and in plants or prokaryotes, but there is no homolog in metazoa. Class III: homologous proteins are present in both S. cerevisiae and S. pombe, but there is no homolog outside the fungal branch. Class IV: homologous proteins of S. pombe are not found in S. cerevisae and S. cerevisiae proteins do not have an homolog in S. pombe, but homologous proteins of both yeasts are found at least in the metazoa branch. Class V: there is no homologous protein of one yeast in the other yeast, and no homolog in other branches. Numbers of genes in each category are shown for both yeasts. Percentages are given in the table. The third column gives the gene distribution in the 253-kb region that we selected for systematic gene deletion in this study.
The distribution of proteins in the eight classes is very similar in the two yeasts. In both organisms, 28% of the total proteins have homologs in prokaroytes and eukaryotes (S. cerevisiae, plants, and metazoa) (class Ia, Fig. 1), whereas 21.5%–24 % are specific to eukaryotes with no homologous protein being detected in prokaryotes (class Ib, Fig. 1). About 10% of the proteins do not have an homolog within the metazoa but do have an homolog in other eukaryotes and prokaryotes (classes II and III, Fig. 1). The class IV proteins share homology with proteins from other species (with at least an homolog in the metazoa branch) but do not have homologs in the other yeast; this class accounts for 4.4% of the S. pombe proteins and 3.5% of the S. cerevisiae proteins. The only significant difference between the two yeasts is found in class V, which consists of proteins specific to either S. pombe or S. cerevisiae. This group comprises 19% of total fission yeast proteins and up to 28% of total budding yeast proteins (class V, Fig. 1). The budding yeast class V proteins may be too high by ∼10% because of overestimation of the total number of proteins in S. cerevisiae, especially those that are not conserved (Mackiewicz et al. 1999, 2002; Blandin et al. 2000; Wood et al. 2001).
Fission Yeast Genes Essential for Vegetative Growth
To estimate the percentage of S. pombe genes required for vegetative growth, we screened a contiguous region of chromosome II containing 100 ORFs. The 253-kb region, selected at random, is likely to be appropriate to estimate the percentage of essential genes. First, according to phylogenetic criteria, the gene distribution within this region containing 100 genes is similar to our estimated distribution for the S. pombe genome (Fig. 1B). Second, assuming that fission yeast genes play a similar role than their budding yeast homologs, the genes appear to be organized randomly relative to their function, with the exception of four genes encoding putative ribosomal proteins (for an estimated total of ∼55 on the entire genome). Interestingly, cytoplasmic ribosomal subunit-encoding genes also appear to cluster in S. cerevisiae (CYGD, http://mips.gsf.de/proj/yeast/CYGD/db/index.html).
We designed 85 pairs of primers for targeted PCR-based gene deletion (Bähler et al. 1998) because deletion of the other 15 genes had already been published (Table 1). After transformation of a fission yeast diploid strain with the PCR-amplified deletion cassette, geneticin-resistant clones were selected and the gene deletion was checked by colony PCR (see Methods). Diploids were sporulated and four-spored asci dissected on rich medium. When tetrads did not contain spores that could form geneticin-resistant colonies, the gene deletion was classified as giving a lethal phenotype (Table 1). We deleted 65 of the 85 genes by this procedure. Of the 20 remaining genes, the deletion cassette could not be PCR amplified for 3, and correctly deleted geneticin-resistant colonies could not be obtained for the other 17, despite repeating the deletion procedure up to six times. Eight of these undeletable 17 genes were located within a string of 9 contiguous genes that lie between SPBC106.10 and SPBC106.20 ORFs (Table 1), suggesting that this whole chromosomal segment of 18 kb was refractory to gene deletion. We measured the recombination frequency between two markers (cut 4 and cdc13) flanking this segment to check whether the region was a cold spot for recombination. Analysis of random spores generated by a cross between cut4-533 and cdc13-117 indicated that the genetic distance between the two markers was 5 cM, compatible with their physical separation of 30 kb, suggesting that this region is not a recombination cold spot. The efficiency with which genes were correctly deleted varied from 5%–100% with an average of 51% based on 650 geneticin-resistant clones analyzed. Including the genes for which the deletion phenotype had already been published, 14 of the 80 genes analyzed were essential for fission yeast vegetative growth (Table 1). This suggests that the percentage of essential genes in S. pombe is 17.5%, compared with 17.8% in budding yeast (YPD; see Garrels 2002), with an interval of confidence (P90) for S. pombe essential genes of 9.5%–25.5%.
Table 1.
Deletion Phenotype of S. pombe Genes and Their Closest Homolog in S. cerevisiae
| Gene name | Class | Deletion | Deletion of closest S. cerevisiae homolog17 | |
| SPBC1271.15c | Ia | viable | viable | YOL023w/FM1 |
| SPBC1271.14 | II | viable | viable | YMR062c/ECM40 |
| SPBC1271.13 | Ia | lethal | viable | YJL063c/MRPL8 |
| SPBC1271.12 | Ib | viable | viable | YPLI45c/KES1 |
| SPBC1271.11 | Ib | viable | viable | YNL003c/PET8 |
| SPBC1271.10c | III | viable | viable | YNR055c/HOL1 |
| SPBC1271.09 | Ia | viable | viable | YCR098c/GIT1 |
| SPBC1271.08c | V | viable | — | |
| SPBC1271.07c | IV | viable | — | |
| SPBC1271.06c | V | viable | — | |
| SPBC1271.05c | III | viable | viable | YOR052c |
| SPBC1271.04c | Ia | no deletion | lethal | YHR068w/DYS1 |
| SPBC1271.03c | V | viable | — | |
| SPBC1271.02/stt3 | Ic | lethal | lethal | YGL022w/STT3 |
| SPBC1271.01c | V | viable | — | |
| SPBC106.02c | Ib | no deletion | viable | YKL086w |
| SPBC106.03 | V | viable | — | |
| SPBC106.04 | Ib | viable | viable | YML035c/AMD1 |
| SPBC106.05c | V | viable | — | |
| SPBC106.06 | Id | lethal | lethal | YDL143w/CCT4 |
| SPBC106.07c | Ic | viable | lethal | YGR147c |
| SPBC106.08c | V | viable | — | |
| SPBC106.09/cut4 | Id | lethal2 | lethal | YNL172w/APC1 |
| SPBC106.10/pka1 | Ic | viable3 | viable | YPL203w/TPK2 |
| SPBC106.11c | IV | no deletion | — | |
| SPBC106.12c | IV | no deletion | — | |
| SPBC106.13 | Ib | no deletion | viable | YIL097w |
| SPBC106.14c | Ib | no deletion | lethal | YGR245c/SDA1 |
| SPBC106.15 | Ia | no deletion | lethal | YPL117c/IDI1 |
| SPBC106.16 | Ia | no deletion | lethal | YOL038w/PRE6 |
| SPBC106.17c | II | viable | viable | YNL277w/MET2 |
| SPBC106.18 | Ia | no deletion | lethal | YOL127w/RPL25 |
| SPBC106.19 | V | no deletion | — | |
| SPBC106.20 | Id | viable | lethal | YJL085w/EXO70 |
| SPBC582.03/cdc13 | Ib | lethal4 | viable | YPR119w/CLB2 |
| SPBC582.04c | V | viable | — | |
| SPBC582.05c/brc1 | Ib | viable5 | viable | YHR154w/ESC4 |
| SPBC582.06c | V | viable | — | |
| SPBC582.07c | Ib | lethal | lethal | YPR108w/RPN7 |
| SPBC582.08 | Ia | viable | viable | YDR111c |
| SPBC582.09 | IV | no deletion | — | |
| SPBC582.10c | Ia | viable | viable | YBR114w/RAD16 |
| SPBC582.11c | Ib | lethal | viable | YDL116w/NUP84 |
| SPBC582.12c | Ia | viable | viable | YJR148w/BAT2 |
| SPBC428.03c | Ib | viable | viable | YAR071w/PHO11 |
| SPBC428.04 | V | viable | — | |
| SPBC428.05c | Ia | viable | viable | YOL058w/ARG1 |
| SPBC428.06c | V | viable | — | |
| SPBC428.07 | V | no PCR* | — | |
| SPBC428.08c/clr4 | Ia | viable6 | viable | YJL168c/SET2 |
| SPBHC428.09c | V | viable | — | |
| SPBC428.10 | Ic | viable | viable | YIR019c/MUC1 |
| SPBC428.11 | Ia | viable | viable | YLR303w/MET17 |
| SPBC428.12c | Ib | viable | lethal | YER165w/PAB1 |
| SPBC428.13c/mob1 | Ib | lethal7 | lethal | YIL106w/MOB1 |
| SPBC428.14 | Ib | viable | viable | YBR042c |
| SPBC428.15 | Ia | no deletion | viable | YGR210c |
| SPBC428.16c/rhb1 | Ib | lethal8 | viable | YCR027c/RSG1 |
| SPBC428.17c | V | viable | — | |
| SPBC428.18/cdt1 | IV | lethal9 | — | |
| SPBC428.19c | Ib | viable | lethal | YMR093w |
| SPBC902.01c/alp6 | Ib | lethal10 | lethal | YNL126w/SPC98 |
| SPBC902.02c | Ia | viable | viable | YMR078c/CTF18 |
| SPBC902.03 | III | viable | viable | YAL009w/SPO7 |
| SPBC902.04 | IV | viable | — | |
| SPBC902.05c/glu2 | Ia | viable11 | viable | YOR136w/IDH2 |
| SPBC902.06 | V | viable | — | |
| SPBC1685.01/pmp1 | Ib | viable12 | viable | YIL113w |
| SPBC1685.02c | Ib | viable | viable | YOR369c/RPS12 |
| SPBC1685.03 | Ia | lethal | lethal | YIR022w/SEC11 |
| SPBC1685.04 | V | viable | — | |
| SPBC1685.05 | II | viable | viable | YNL123w |
| SPBC1685.06 | Ib | viable | viable | YNL299w/TRF5 |
| SPBC1685.07c | Ib | viable | viable | YBL089w |
| SPBC1685.08 | Ib | viable | viable | YPL181w |
| SPBC1685.09 | Ia | viable | viable | YLR388w/RPS29A |
| SPBC1685.10 | Ia | no deletion | viable | YHR021c/RPS27B |
| SPBC1685.11 | V | viable | — | |
| SPBC1685.12c | non- | coding ? | ||
| SPBC1685.13 | III | viable | viable | YPR149w/NCE102 |
| SPBC1685.14c | Ib | viable | viable | YNL212w/VID27 |
| SPBC649.01c/klp6 | Ib | viable13 | viable | YGL216w/KIP3 |
| SPBC649.02 | Ia | viable | viable | YNL302c/RPS19B |
| SPBC649.03 | Id | viable | viable | YMR201c/RAD14 |
| SPBC649.04/uvi15 | V | viable14 | — | |
| SPBC649.05/cut12 | V | lethal15 | — | |
| SPBC649.06 | Ia | viable | viable | YGR173w |
| SPBC354.02c/sec61 | Ia | lethal16 | lethal | YLR378c/SEC61 |
| SPBC354.03 | Ib | no PCR* | lethal | YBR198c/TAF90 |
| SPBC354.04 | V | no PCR* | — | |
| SPBC354.05c | Id | no deletion | viable | YOR032c/HMS1 |
| SPBC354.06 | Ia | no deletion | viable | YPL013c |
| SPBC354.07c | Ib | viable | viable | YPL145c/KES1 |
| SPBC354.08c | Ib | viable | viable | YMR266w |
| SPBC354.09c | Ib | viable | viable | YPL176c |
| SPBC354.10 | Ia | viable | viable | YIR019c/MUC1 |
| SPBC354.11c | V | viable | — | |
| SPBC354.12 | Ia | no deletion | viable | YJL052w/TDH1 |
| SPBC354.13 | Id | no deletion | viable | YDR389w/SAC7 |
| SPBC354.14c | Ib | viable | viable | YEL013w/VAC8 |
| SPBC354.15 | IV | viable | — | |
Fission yeast genes from chromosome II were deleted in a diploid strain using targeted PCR-based gene deletion procedure (Bähler et al., 1998). Diploids were sportulated and the spore viability was assessed on rich glucose medium. Deletion phenotype of the closest homolog in S. cerevisiae (if any) is given.
Yoshida et al. (1999), 2Yamashita et al. (1996), 3Fernandez et al. (1997), 4Booher and Beach (1988), 5Verkade et al. (1999), 6Bannister et al. (2001) and Nakayama et al. (2001), 7Hou et al. (2000), 8Mach et al. (2000), 9Hofmann and Beach (1994), 10Vardy and Toda (2000), 11Barel and MacDonald (1993), 12Sugiura et al. (1998) 13our study and West et al. (2002), 14Lee et al. (1995), 15Bridge et al. (1998), 16Broughton et al. (1997), 17YPD (see Garrels, 2002) and EUROSCARF (http://www.uni-frankfurt.de/fb15/mikro/euroscarf/). *No amplification of the deletion cassette.
Nine of the 14 S. pombe essential genes of Table 1 have been previously described and can be classified into the functional categories of genes described by MIPS (CYGD, http://mips.gsf.de/proj/yeast/CYGD/db/index.html). For the other five fission yeast essential genes, a putative function can be assigned by homology with S. cerevisae. Classification into functional categories reveals that among the 14 S. pombe essential genes of Table 1, 6 belong to the so-called Protein Fate functional category that includes genes involved in protein folding, modification, and targeting. They include stt3, SPBC106.06, cut4, SPBC582.07c SPBC1685.03, and sec61, with cut4, and possibly SPBC106.06 and SPBC582.07c, being also required for completion of mitosis. Another four genes (cdc13, mob1, alp6, cut12) are essential for mitosis. SPBC582.11c is likely to encode the fission yeast homolog of Nup84p nucleoporin; cdt1 is required for DNA replication initiation (Nishitani et al. 2000); rhb1 encodes a Rheb-related GTPase that putatively regulates alternative responses to limiting nutrients (Mach et al. 2000); and SPBC1271.13 probably encodes a ribosomal protein.
Essential Genes: Comparison Between S. pombe and S. cerevisiae
We then tested whether deletion of homologous genes in the two yeasts showed the same deletion phenotype (whether essential or not), choosing the closest S. cerevisiae homolog of each of the S. pombe genes from Table 1 and Figure 2B (see below). Among the 81 S. pombe genes with an homolog in budding yeast, 88% (71 genes) show the same deletion phenotype in both yeasts, 6% (5 genes) are essential for S. cerevisiae but not for S. pombe, and 6% (5 genes) are essential for S. pombe but not S. cerevisiae (Table 2). This means that of the 15 fission yeast essential genes included in our study, only 10 (67%) are also essential for budding yeast growth. This number is similar to that calculated from previously published data (PombePD), which reveals that 135 of the 198 (68%) S. pombe essential genes with S. cerevisiae homologs are also essential for S. cerevisiae. These data indicate that, although the absolute percentage of essential genes is similar between S. cerevisiae and S. pombe, surprisingly only two-thirds of the essential genes in one yeast have essential homologous genes in the other yeast.
Figure 2.
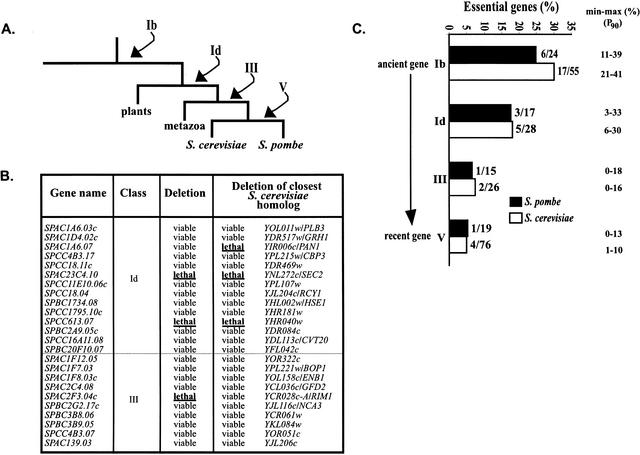
Correlation between age and “essentiability” of the S. pombe genes. (A) A simplified phylogenetic tree of the eukaryotic kingdom (Sipiczki 2000) is given. The arrows indicate the point of appearance of S. pombe class Ib, Id, III, and V genes on the tree. (B) Additional S. pombe genes from class Id and class III were deleted using the procedure described in Methods. The phenotype of the deletion is given in the third column. The deletion phenotype of the S. cerevisiae closest homolog of each S. pombe gene is given in the last column. (C) Lethality in gene classes Ib, Id, III, and V of S. pombe and S. cerevisiae is compared. Intervals of confidence (P90) are given for each class of genes.
Table 2.
Comparison of Deletion Phenotype Between S. pombe and S. cerevisiae Homologs
| S. pombe | S. cerevisiae closest homolog | |
| viable | viable | 61 |
| lethal | lethal | 10 |
| viable | lethal | 5 |
| lethal | viable | 5 |
The table compares the deletion phenotype, on rich glucose medium, of the 81 S. pombe genes with an homolog in S. cerevisiae that are included in our study.
One hypothesis would be that, in budding yeast, the genes have been duplicated to compensate for essential gene loss. However, analysis of Table 1 genes reveals that it is unlikely in fission yeast. The SPBC1271.13 and SPBC582.11c ORFs, which are required for S. pombe growth but do not have essential homologous genes in budding yeast, have not been duplicated in S. cerevisiae. Similarly, the YGR147c, EXO70, and YMR093w ORFs of S. cerevisiae, which do not have essential homologs in fission yeast, do not belong to gene families in S. pombe. On the other hand, CCT4, which is required for growth of S. cerevisiae and S. pombe, belongs to gene families in both yeasts. Moreover, most of the paralogous genes of budding yeast CCL4 are also essential, suggesting the existence of essential gene families. Other gene families, like the MFS superfamily of permeases, which includes the nonessential SPBC1271.10c ORF (Table 1), are very unlikely to comprise a high percentage of essential genes, as revealed by S. cerevisiae studies (for review, see Sa-Correia and Tenreiro 2002).
Age of S. pombe Genes and Whether They Are Essential
We then tested whether a gene was essential and correlated it with the time of appearance of a gene on the life tree. Sipiczki (2000) has proposed a consensus tree for eukaryotes based on molecular phylogeny of both 18s rRNA and HMG-CoA reductase sequences. If we refer to this tree shown in Figure 2A, we can postulate that our gene classes appeared in the following order: Ib > Id > III > V. To estimate the percentage of essential genes in each of these classes, we deleted another 24 genes from both class Id and class III (Fig. 2B). Together with the data from Table 1, we estimate that lethality in the classes is as follows: Ib, 25% (6/24); Id, 18% (3/17); III, 7% (1/15), and V, 5% (1/19). From these data, if we consider only the fission yeast genes that do not have homologs within the prokaryotic branch, the more ancient the gene is, the more likely it is to be essential (Fig. 2C). Data from the S. cerevisiae genome (YPD, see Garrels 2002) give a similar profile (Fig. 2C). Focusing on the genes with homologs in both eukaryotic and prokaryotic cells (Table 1, class Ia), we find that 22.5% (4/18) are essential for S. pombe. Data in S. cerevisiae are similar as we estimate that 26% of class Ia genes are required for budding yeast growth. Comparing this data with the average of 17.5% of essential genes for the whole genome, we conclude that ancient genes maintained in all eukaryotic species or in both eukaryotic and prokaryotic species, are more likely to be essential. In contrast, yeast-specific genes (class V), which have appeared recently, are less likely to be essential.
We then focused on S. pombe class IV genes, which have an homolog in the metazoa branch but do not have an homolog in the S. cerevisae lineage, deleting another 36 genes within this class (Table 3). Of these 40 genes, only 1 (cdt1) was found to be essential (Hofmann and Beach 1994). However, in this case, a functionally equivalent gene to cdt1 has been reported in S. cerevisiae, which has very low sequence similarity (Tanaka and Diffley 2002). This may be a highly diverged gene derived from a common ancestor or may be an example of nonorthologous gene replacement (Koonin et al. 1996), when a gene is functionally replaced by another that is unrelated by descent. The two genes may, however, share limited sequence similarity acquired by convergent evolution. Assuming that class IV genes are more likely to be the result of gene loss instead of lateral transfer from plants/animals to S. pombe, we conclude that a fission yeast gene for which the homolog has been lost in the budding yeast lineage is very unlikely to be essential, although its origin may be ancient.
Table 3.
Deletion of Class IV S. pombe Genes
| Gene name | Deletion |
| MLO2 | viable |
| SPAC30.03c | viable |
| SPAC13G6.09 | viable |
| SPAC17A5.04c | viable |
| SPAC1805.08 | viable |
| SPAC343.18 | viable |
| SPAC22F3.02 | viable |
| SPAC3A11.10c | viable |
| SPAC630.13c | viable |
| SPAC1565.02c | viable |
| SPAC15A10.10 | viable |
| SPAC1952.06c | viable |
| SPAC1687.17c | viable |
| SPAC25H1.02 | viable |
| SPAC22A12.03c | viable |
| SPAP8A3.12c | viable |
| SPBC1271.07c | viable |
| SPBC428.18.cdt1 | lethal2 |
| SPBC902.04 | viable |
| SPBC19F8.02 | viable |
| SPBC354.15 | viable |
| SPBC13G1.04c | viable |
| SPBC31F10.02 | viable |
| SPBC146.06c | viable |
| SPBC2F12.12c | viable |
| SPBC249.10 | viable |
| SPBC3B9.08c | viable |
| SPBC725.10 | viable |
| SPBC20F10.03 | viable |
| SPBC16G5.07c | viable |
| SPBC342.04 | viable |
| SPBC577.03c | viable |
| SPBP23A10.12 | viable |
| SPBP35G2.02 | viable |
| SPCC736.08 | viable |
| SPCC736.09c | viable |
| SPCC645.08c | viable |
| SPCC1020.09 | viable |
| SPCC14G10.06 | viable |
| SPCC965.12 | viable |
DISCUSSION
This pilot work has shown that a systematic deletion of all S. pombe ORFs using a PCR-based gene deletion procedure may be difficult to achieve compared with the equivalent work in S. cerevisiae because the efficiency of homologous recombination is lower in S. pombe than in S. cerevisiae (Kaur et al. 1997). Bähler et al. (1998) have shown that the use of longer flanking sequences (60–80 bp instead of the 40 bp used for budding yeast) increases the efficiency of homologous integration in the PCR-based procedure. However, increasing the length of flanking sequences up to 80 bp as used in our study was not sufficient to delete all target genes. We identified one region of 18 kb in length on chromosome II containing 9 genes, within which we were unable to delete 8 of the genes. Meiotic recombination frequency was normal between two markers flanking this region, indicating that it is not a cold spot region for meiotic recombination. One possibility is that the chromatin structure may be different and gene transcription low in this region. Because S. pombe transcription factors have been shown to alter local chromatin structure and to activate meiotic recombination hotspots (for review, see Davis and Smith 2001), we speculate that some regions of the genome may contain poorly transcribed genes with “closed” chromatin structure, resulting in a low efficiency of targeted gene deletion. Alternatively, integration of the deletion cassette may have occurred but, because of the silent chromatin, the kanamycin resistance gene was insufficiently expressed, reminiscent of transcriptional silencing observed at mating-type, telomeric and centromeric regions of S. pombe chromosomes (for review, see Huang 2002).
From our data, we calculate that the percentage of essential genes in fission yeast growing on a rich medium is 17.5%, similar to the 17.8% of genes that are essential for S. cerevisiae growth on rich medium. However, taking into account our failure to delete six genes with essential homologs in budding yeast, the percentage of essential genes in fission yeast may be higher (between 18% and 20%). The probability of an unknown gene being essential for S. pombe is dependent on whether homologs are found within other branches of the life tree. We estimate that 27% of the proteins conserved in prokaryotes, mammals, plants, and S. cerevisiae (our class Ia), and 27% of the proteins conserved in mammals, plants, and S. cerevisiae, but not found in prokaryotes (class Ib), are essential in S. pombe. If we calculate the absolute number of essential genes in each of these two classes of genes, we estimate that class Ia and Ib genes account, respectively, for 43% and 38% of the total number of essential S. pombe genes (Fig. 3). This means that 80% of the S. pombe essential genes are found in highly conserved gene classes (Ia and Ib), although they account for only 50% of the total number of protein-encoding genes. A lower fraction of fungal-specific proteins (III and V) is essential for S. pombe growth (Fig. 3); these account for 10% of the essential genes, although they form 26% of the total number of protein-encoding genes (Fig. 3).
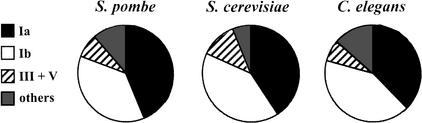
Figure 3.
Distribution of essential genes in S. pombe, S. cerevisiae and C. elegans. From this study, we estimated the percentage of essential genes in the S. pombe gene classes Ia (both yeasts + metazoa + plants + prokaryotes), Ib (both yeasts + metazoa + plants), and III + V (one or both yeasts). The distribution of S. cerevisiae essential genes was calculated from data in YPD (see Garrels 2002). The figures for C. elegans were adapted from Gönczy et al. (2000) who used RNA-mediated interference to investigate the percentage of essential genes in chromosome III: “multi-kingdom” genes (our class Ia), eukaryotic genes (Ib), and nematode-specific genes (that we classified under III + V). The essential genes that do not belong to any of the above classes were classified under “others.”
Analysis of data contained in the YPD database (see Garrels 2002) revealed similar conclusions for budding yeast. Classes Ia and Ib contain 82% of S. cerevisiae essential genes, whereas fungal-specific proteins only comprise 12% of the essential proteins (Fig. 3). Using RNA-mediated interference in C. elegans, Gönczy et al. (2000) have shown that the essential genes in this nematode worm are mainly those conserved in other organisms. They found that genes conserved in both eukaryotes and prokaryotes (our class Ia) account for 40% of the total essential genes in C. elegans embryos, whereas genes conserved in eukaryotes but not prokaryotes (our class Ib) account for 45%. Nematode-specific genes form only 7.5% of the essential genes, although they account for 40% of the total genome (Gönczy et al. 2000). Therefore, a general consensus emerges suggesting that genes essential for eukaryotic cells are mainly found in gene classes that have homologs within all the eukaryotic branches.
One interpretation of this data is that essential genes evolve more slowly as proposed by the adaptive theory of mutation rates, which argues that essential protein-encoding genes evolve at lower rates than nonessential ones. However, Hurst and Smith (1999) have shown that, in rat and mouse, mutation rate is not correlated with the severity of the knockout phenotype, suggesting that, for these organisms, essential and nonessential genes evolve at the same rate. Moreover, Jordan et al. (2001) have shown that relative rates of amino acid sequence evolution are very similar in all three domains of life (eubacteria, archaebacteria, and eukaryotes). Therefore, we interprete the data gathered from the S. pombe, S. cerevisiae, and C. elegans genomes as indicating that natural selection has preferentially kept genes that are required for essential functions. This interpretation gains further support from the observation that S. pombe conserved genes, which have been lost in the S. cerevisiae lineage (class IV), are unlikely to be essential. We conclude that genes that have been maintained throughout evolution are more likely to be essential.
However, what about genes that appeared late in the tree of life? We have established that the more ancient a gene is, the more likely it is to be essential. Therefore, organism-specific genes (class III and V) that have arisen more recently appear to be less likely to be required for vital functions of the cell. It has been suggested that these proteins may be required for more specialized functions (Chervitz et al. 1998; Rubin et al. 2000). In C. elegans, for example, processes that are unique to the metazoa and have arisen more recently are carried out by proteins that do not have homologs in yeast, whereas core biological functions use orthologous proteins (Chervitz et al. 1998). We propose that generally in eukaryotes, many of the essential genes are those that appeared with the origin of eukaryotic life and have remained conserved within all branches of the tree of life.
METHODS
Protein Sequence Comparison
S. pombe proteins from PombePD (see Wood et al. 2002) were compared to nr database using BLASTP (Altschul and Lipman 1990) with a cutoff E value of 10−5.
Gene Deletion
PCR-based gene deletion, using the kanR marker, was performed according to Bähler et al. (1998) with flanking sequences of 80 bp. An h+/h− ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-M210/ade6-M216 S. pombe diploid strain was transformed with the PCR product, and geneticin-resistant colonies were selected on YES medium containing 100 μg/mL G418 (Life Technologies). Gene deletion was checked by colony PCR. Diploid strains were sporulated and tetrads were dissected as described in http://www.bio.uva.nl/pombe/. The deletion phenotype was assessed on YES medium.
WEB SITE REFERENCES
http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html; microbial genomes.
http://mips.gsf.de/proj/yeast/CYGD/db/index.html; MIPS Comprehensive Yeast Genome Database with annotation for all yeast ORFs.
http://www.bio.uva.nl/pombe; S. pombe site with an online fission yeast handbook.
http://www.uni-frankfurt.de/fb15/mikro/euroscarf/; collection of S. cerevisiae deletion strains.
Acknowledgments
We are grateful to Adam Bermange and Damien Hermand for their help. We thank Pierre Tilquin for help with statistical analysis of the data. We thank all members of the Cancer Research UK Cell Cycle Laboratory for their support. We thank Jacky Hayles, John Sgouros, André Goffeau, and Françoise Foury for critical reading of the manuscript. This work was supported by the European Commission, Biotechnology Programme (A.D.), Comunidad Autonoma de Madrid (I. S.-P.), and Cancer Research UK.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL anabelle.decottignies@gece.ucl.ac.be; FAX 32-2-762-9405.
E-MAIL Paul.Nurse@cancer.org.uk; FAX 44-20-72693610.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.636103.
REFERENCES
- 1.Adams M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., and Galle, R.F. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F. and Lipman, D.J. 1990. Protein databases searches for multiple alignments. Proc. Natl. Acad. Sci. 87: 5509-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Arabidopsis Genome Initiative 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. [DOI] [PubMed] [Google Scholar]
- 4.Aravind L., Watanabe, H., Lipman, D.J., and Koonin, E.V. 2000. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl. Acad. Sci. 97: 11319-11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bähler J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie, A., III, Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943-951. [DOI] [PubMed] [Google Scholar]
- 6.Bannister A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120-124. [DOI] [PubMed] [Google Scholar]
- 7.Barel I. and MacDonald, D.W. 1993. Enzyme defects in glutamate-requiring strains of Schizosaccharomyces pombe. FEMS Microbiol. Lett. 113: 267-272. [DOI] [PubMed] [Google Scholar]
- 8.Blandin G., Durrens, P., Tekaia, F., Aigle, M., Bolotin-Fukuhara, M., Bon, E., Casaregola, S., de Montigny, J., Gaillardin, C., Lepingle, A., et al. 2000. Genomic exploration of the hemiascomycetous yeasts: 4. The genome of Saccharomyces cerevisiae revisited. FEBS Lett. 487: 31-36. [DOI] [PubMed] [Google Scholar]
- 9.Booher R. and Beach, D. 1988. Involvement of cdc13+ in mitotic control in Schizosaccharomyces pombe: Possible interaction of the gene product with microtubules. EMBO J. 7: 2321-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bridge A.J., Morphew, M., Bartlett, R., and Hagan, I.M. 1998. The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes & Dev. 12: 927-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broughton J., Swennen, D., Wilkinson, B.M., Joyet, P., Gaillardin, C., and Stirling, C.J. 1997. Cloning of SEC61 homologues from Schizosaccharomyces pombe and Yarrowia lipolytica reveals the extent of functional conservation within this core component of the ER translocation machinery. J. Cell Sci. 110: 2715-2727. [DOI] [PubMed] [Google Scholar]
- 12.The C. elegans Sequencing Consortium 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- 13.Chervitz S.A., Aravind, L., Sherlock, G., Ball, C.A., Koonin, E.V., Dwight, S.S., Harris, M.A., Dolinski, K., Mohr, S., Smith, T., et al. 1998. Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science 282: 2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis L. and Smith, G.R. 2001. Meiotic recombination and chromosome segregation in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. 98: 8395-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng D.-F., Cho, G., and Doolittle, R.F. 1997. Determining divergence times with a protein clock: Update and reevaluation. Proc. Natl. Acad. Sci. 94: 13028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez J., Soto, T., Vicente-Soler, J., Cansado, J., and Gacto, M. 1997. Heat-shock response in Schizosaccharomyces pombe cells lacking cyclic AMP-dependent phosphorylation. Curr. Genet. 31: 112-118. [DOI] [PubMed] [Google Scholar]
- 17.Garrels J.I. 2002. Yeast genomic databases and the challenge of the post-genomic area. Funct. Integr. Genomics 2: 212-237. [DOI] [PubMed] [Google Scholar]
- 18.Goffeau A., Barrell, B.G., Bussey, H., Davis, R.W., Dujon, B., Feldmann, H., Galibert, F., Hoheisel, J.D., Jacq, C., Johnston, M., et al. 1996. Life with 6000 genes. Science 275: 1051-1052. [DOI] [PubMed] [Google Scholar]
- 19.Gönczy P., Echeverri, G., Oegema, K., Coulson, A., Jones, S.J., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331-336. [DOI] [PubMed] [Google Scholar]
- 20.Heckman D.S., Geiser, D.M., Eidell, B.R., Stauffer, R.L., Kardos, N.L., and Hedges, S.B. 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293: 1129-1133. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann J.F. and Beach, D. 1994. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: Requirement for DNA replication and inhibition of mitosis. EMBO J. 13: 425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou M.C., Salek, J., and McCollum, D. 2000. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr. Biol. 10: 619-622. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y. 2002. Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 30: 1465-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurst L.D. and Smith, N.G.C. 1999. Do essential genes evolve slowly? Curr. Biol. 9: 747-750. [DOI] [PubMed] [Google Scholar]
- 25.International Human Genome Sequencing Consortium 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- 26.Jordan I.K., Kondrashov, F.A., Rogozin, I.B., Tatusov, R.L., Wolf, Y.I., and Koonin, E.V. 2001. Constant relative rate of protein evolution and detection of functional diversification among bacterial, archaeal and eukaryotic proteins. Genome Biol. 2: RESEARCH0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaur R., Ingavale, S.S., and Bachhawat, A.K. 1997. PCR-mediated direct gene disruption in Schizosaccharomyces pombe. Nucleic Acids Res. 25: 1080-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koonin E.V., Mushegian, A.R., and Bork, P. 1996. Non-orthologous gene displacement. Trends Genet. 12: 334-336. [PubMed] [Google Scholar]
- 29.Korbel J.O., Snel, B., Huynen, M.A., and Bork, P. 2002. SHOT: A web server for the construction of genome phylogenies. Trends Genet. 18: 158-162. [DOI] [PubMed] [Google Scholar]
- 30.Lee J.K., Kim, M., Choe, J., Seong, R.H., Hong, S.H., and Park, S.D. 1995. Characterization of uvi15+, a stress-inducible gene from Schizosaccharomyces pombe. Mol. Gen. Genet. 246: 663-670. [DOI] [PubMed] [Google Scholar]
- 31.Mach K.E., Furge, K.A., and Albright, C.F. 2000. Loss of Rhb1, a Rheb-related GTPase in fission yeast, causes growth arrest with a terminal phenotype similar to that caused by nitrogen starvation. Genetics 155: 611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackiewicz P., Kowalczuk, M., Gierlik, A., Dudek, M.R., and Cebrat, S. 1999. Origin and properties of non-coding ORFs in the yeast genome. Nucleic Acids Res. 27: 3503-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackiewicz P., Kowalczuk, M., Mackiewicz, D., Nowicka, A., Dudkiewicz, M., Laszkiewicz, A., Dudek, M.R., and Cebrat, S. 2002. How many protein-coding genes are there in the Saccharomyces cerevisiae genome? Yeast 19: 619-629. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama J., Rice, J.C., Strahl, B.D., Allis, C.D., and Grewal, S.I. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110-113. [DOI] [PubMed] [Google Scholar]
- 35.Nishitani H., Lygerou, Z., Nishimoto, T., and Nurse, P. 2000. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404: 625-628. [DOI] [PubMed] [Google Scholar]
- 36.Rubin G.M., Yandell, M.D., Wortman, J.R., Gabor Miklos, G.L., Nelson, C.R., Hariharan, I.K., Fortini, M.E., Li, P.W., Apweiler, R., Fleischmann, W., et al. 2000. Comparative genomics of the eukaryotes. Science 287: 2204-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sa-Correia I. and Tenreiro, S. 2002. The multidrug resistance transporters of the major facilitator superfamily, 6 years after disclosure of Saccharomyces cerevisiae genome sequence. J. Biotechnol. 98: 215-226. [DOI] [PubMed] [Google Scholar]
- 38.Sipiczki M. 2000. Where does fission yeast sit on the tree of life? Genome Biol. 1: 1011.1-1011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugiura R., Toda, T., Shuntoh, H., Yanagida, M., and Kuno, T. 1998. pmp1+, a suppressor of calcineurin deficiency, encodes a novel MAP kinase phosphatase in fission yeast. EMBO J. 17: 140-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka S. and Diffley, J.F. 2002. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2—7 during G1 phase. Nat. Cell Biol. 4: 198-207. [DOI] [PubMed] [Google Scholar]
- 41.Tekaia F., Lazcano, A., and Dujon, B. 1999. The genomic tree as revealed from whole proteome comparisons. Genome Res. 9: 550-557. [PMC free article] [PubMed] [Google Scholar]
- 42.Vardy L. and Toda, T. 2000. The fission yeast γ-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 19: 6098-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venter J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- 44.Verkade H.M., Bugg, S.J., Lindsay, H.D., Carr, A.M., and O'Connell, M.J. 1999. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 10: 2905-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West R.R., Malmstrom, T., and McIntosh, J.R. 2002. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J. Cell Sci. 115: 931-940. [DOI] [PubMed] [Google Scholar]
- 46.Wood V., Rutherford, K.M., Ivens, A., Rajandream, M.-A., and Barrell, B. 2001. A re-annotation of the Saccharomyces cerevisiae genome. Comp. Funct. Genome 2: 143-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood V., Gwilliam, R., Rajandream, M.-A., Lyne, M., Stewart, A., Sgouros, J., Peat, N., Hayles, J., Baker, S., Basham, D., et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871-880. [DOI] [PubMed] [Google Scholar]
- 48.Yamashita Y.M., Nakaseko, Y., Samejima, I., Kumada, K., Yamada, H., Michaelson, D., and Yanagida, M. 1996. 20S cyclosome complex formation and proteolytic activity inhibited by the cAMP/PKA pathway. Nature 384: 276-279. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida S., Matsuura, A., Merregaert, J., and Anraku, Y. 1999. Schizosaccharomyces pombe stt3(+) is a functional homologue of Saccharomyces cerevisiae STT3 which regulates oligosaccharyltransferase activity. Yeast 15: 497-505. [DOI] [PubMed] [Google Scholar]
- 50.Zhou C., Seibert, V., Geyer, R., Rhee, E., Lyapina, S., Cope, G., Deshaies, R.J., and Wolf, D.A. 2001. The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem. 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]