Abstract
The evolution of metabolic enzymes and pathways has been a subject of intense study for more than half a century. Yet, so far, previous studies have focused on a small number of enzyme families or biochemical pathways. Here, we examine the phylogenetic distribution of the full-known metabolic complement of Escherichia coli, using sequence comparison against taxa-specific databases. Half of the metabolic enzymes have homologs in all domains of life, representing families involved in some of the most fundamental cellular processes. We thus show for the first time and in a comprehensive way that metabolism is conserved at the enzyme level. In addition, our analysis suggests that despite the sequence conservation and the extensive phylogenetic distribution of metabolic enzymes, their groupings into biochemical pathways are much more variable than previously thought.
One of the fundamental tenets in molecular biology was expressed by Monod, in his famous phrase “What is true for Escherichia coli is true for the elephant” (Jacob 1988). For a long time, this statement has inspired generations of molecular biologists, who have used Bacteria as model organisms to understand the basic principles of life. The discovery of the three domains of life (Woese and Fox 1977) testified that there exist some pronounced differences between organisms, for example in transcription regulation (Struhl 1999). Paradoxically, metabolism has always been considered as one of the most conserved cellular processes (Lehninger 1979), that remains invariable from Bacteria to Eucarya, but no quantification of this view has been provided.
Instead, the phylogenetic extent of metabolism has been assessed by experimental case studies of individual biochemical pathways (Crawford 1989) and, more recently, by comparative genomics. Entire genome sequences from a wide variety of species offered the possibility of performing metabolic reconstruction, based on known metabolic pathways and genome sequence comparison (Karp et al. 1996). Case studies have suggested that even some of the most central pathways in biochemistry such as the citric acid cycle (Huynen et al. 1999), glycolysis (Dandekar et al. 1999), and amino acid biosynthetic pathways (Forst and Schulten 2001) may vary significantly over large phylogenetic distances.
A comprehensive analysis of metabolism has not been performed until now, possibly due to the scarcity of systematically collected information on genome sequences and metabolic pathways. Metabolic enzyme families are considered to be highly conserved and have been used to reconstruct the deep branching patterns of the tree of life (Doolittle et al. 1996). Yet, it remains unclear which enzymes are represented in all major taxa, what pathways they participate in, and which ones are most conserved at the sequence level.
We set out to address the phylogenetic extent and conservation of enzymes and pathways by using a highly curated, reliable source of metabolic information. The EcoCyc database holds information about the full genome and all known metabolic pathways of Escherichia coli (Karp et al. 2000). Recently, the database has been used to represent computational predictions of other organisms (Karp 2001).
RESULTS
We have searched the nonredundant protein sequence database, previously partitioned in seven major taxonomic groups, with all 548 enzymes from the known metabolic complement of Escherichia coli. Whenever a homolog of each query enzyme is found in the corresponding taxonomic group, this is recorded into a binary vector (see Methods). Conceptually, this approach is similar to a low-resolution version of the phylogenetic profile method (Pellegrini et al. 1999). Instead of searching individual species, however, we focus on major taxonomic groups and we seek enzymes that “travel together” within or across these groups. No assumptions about functional roles or associations are made (Pellegrini et al. 1999): We only examine the phylogenetic extent of the query set, in this case the entire known metabolic complement of E. coli. The end result is a matrix of 548 genes across all the taxonomic combinations; in all, 37 (out of 128 possible) such combinations can be observed. Genes that have the same distribution pattern (i.e., identical binary vectors) in each taxonomic category are collected accordingly.
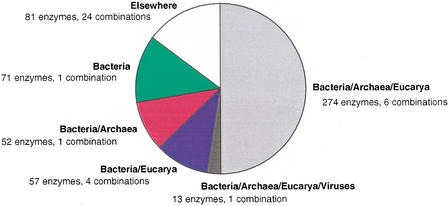
The majority of E. coli enzymes (274 of them, or 50%) have homologs in all domains of life, Bacteria, Archaea, and Eucarya, covering six taxonomic combinations (Fig. 1). Furthermore, there are an additional 13 enzymes (2%) which are universally present in all seven taxa, including the viruses (Table 1). This universal set represents enzyme families involved in various biochemical processes, including amino acid, cofactor, and nucleotide biosynthesis (Kyrpides et al. 1999). It is worth noting that the presence of all these enzymes in viruses is not fully understood yet (Kaiser et al. 1999). Enzymes present in Bacteria and (1) Eucarya are 57, covering four taxonomic combinations (10%; e.g., glucosamine-6-phosphate isomerase) or (2) Archaea are 52 (9%) (e.g., cytochrome D ubiquinol oxidase). It is interesting that 71 enzymes are present only in Bacteria (13%; e.g., L-fucose isomerase), possibly representing unique metabolic capabilities of this taxon. Notably, we have not observed any metabolic enzymes that are species-specific to E. coli. Finally, the remaining 81 enzymes (15%) have homologs in various taxonomic combinations (24 in total), with very low counts that are not statistically significant (see below). Overall, 52% of known metabolic enzymes from E. coli are found to be present in all three domains, a fact indicating that metabolism is highly conserved during evolution (Fig. 1).
Figure 1.
Distribution of the 548 known E. coli metabolic enzymes into 37 taxonomic combinations (see Methods). The seven taxonomic groups correspond to domains of life in the case of Archaea and Bacteria or the four major eukaryotic groups (Fungi, Metazoa, Protista, Viridiplantae), while Viruses are considered as an additional group (see Methods). Universal enzymes are colored in gray (dark gray for those with viral homologs), those with homologs in Eucarya in blue, with homologs in Archaea in red, and in Bacteria only in green. Other combinations of the seven taxonomic groups are shown in white (see Methods).
Table 1.
The 13 Metabolic Enzymes Which Have Homologs in All Major Taxonomic Partitions, Including Viruses
| Accession | Description | Pathway |
| P00379 | Dihydrofolate reductase (EC 1.5.1.3) | FormylTHF biosynthesis, folic acid biosynthesis |
| P00470 | Thymidylate synthase (EC 2.1.1.45) | FormylTHF biosynthesis, deoxypyrimidine nucleotide/side metabolism |
| P00479 | Aspartate carbamoyltransferase catalytic chain (EC 2.1.3.2) | Pyrimidine biosynthesis |
| P00861 | Diaminopimelate decarboxylase (EC 4.1.1.20) | Lysine and diaminopimelate biosynthesis |
| P04391 | Ornithine carbamoyltransferase chain I (EC 2.1.3.3) | Arginine biosythesis II |
| P05459 | Erythronate-4-phosphate dehydrogenase (EC 1.1.1.-) | Pyridoxal 5′-phosphate biosynthesis |
| P06960 | Ornithine carbamoyltransferase chain F (EC 2.1.3.3) | Arginine biosynthesis II |
| P08328 | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) | Serine biosynthesis |
| P17169 | Glucosamine-fructose-6-phosphate aminotransferase [isomerizing] (EC 2.6.1.16) | Dissimilation of N-acetylglucosamine, N-acetylmannosamine and N-acetylneuraminic acid, UDP-N-acetylglucosamine biosynthesis |
| P22939 | Geranyltranstransferase (EC 2.5.1.10) | Polyisoprenoid biosynthesis |
| P27830 | DTDP-glucose 4,6-dehydratase (EC 4.2.1.46) | dTDP-rhamnose biosynthesis, enterobacterial common antigen biosynthesis |
| P37759 | DTDP-glucose 4,6-dehydratase (EC 4.2.1.46) | dTDP-rhamnose biosynthesis, enterobacterial common antigen biosynthesis |
| P52643 | D-lactate dehydrogenase (EC 1.1.1.28) | Fermentation |
Column names: database Accession number, database Description line, both from SwissProt (Bairoch and Apweiler 2000) and the corresponding Pathway name, according to EcoCyc (Karp et al. 2000). Note that an enzyme may participate in more than one pathway (Ouzounis and Karp 2000). Table is sorted by Accession number.
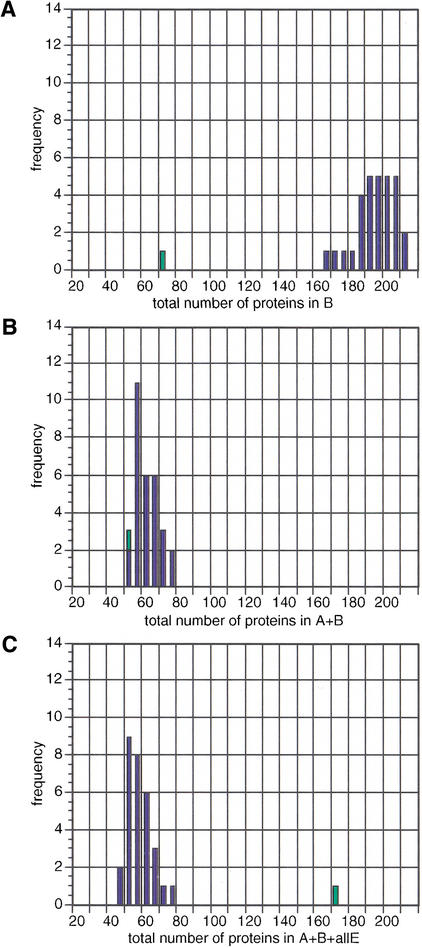
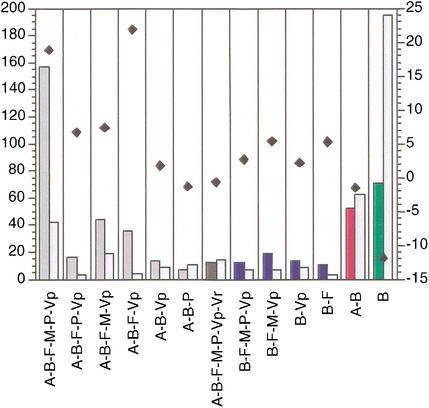
To assess whether the observed patterns of phylogenetic distribution for metabolic enzymes are different from any other proteins, we have performed simulations of this analysis using protein sets of equal size, randomly selected from the E. coli genome (see Methods). Because some of the eukaryotic taxa (e.g., Protista or Fungi) may be significantly underrepresented in terms of the amount of available sequence data, we have assessed the gross pattern of taxonomic distributions by combining all four eukaryotic taxa (Protista, Fungi, Plants, Metazoa) into one group. It is striking that the 71 Bacteria-specific enzymes are significantly underrepresented compared to the control set (with an average of 195 proteins that are Bacteria-specific; Fig. 2A). The 52 enzymes with homologs in Archaea but not Eucarya are slightly underrepresented (with an average of 63 proteins in the control sets for this taxonomic grouping; Fig. 2B). Finally, if we take enzymes with homologs in Archaea, Bacteria, all four eukaryotic taxa, and viruses (170 in total, or 31%), it is evident that this set of proteins stands out as being overrepresented in this universal taxonomic pattern compared with random (Fig. 2C). It is thus evident that randomly selected proteins from a bacterial genome tend to be confined within the corresponding taxonomic group, while metabolic enzymes are expected to be present across a much wider phylogenetic spectrum (Fig. 2). Enzymes with homologs in other taxonomic group combinations exhibit similarly strong deviations from a random background being either over- or underrepresented in the corresponding combination (Fig. 3).
Figure 2.
Frequency distributions of E. coli metabolic enzymes (green bars) versus 30 control sets of equal size with randomly selected E. coli proteins (blue bars, see Methods) in (A) Bacteria, (B) Bacteria and Archaea or (C) Bacteria, Archaea and all eukaryotic groups (including viruses). Counts are shown on the x-axis and the frequency of the sets on the y-axis. The set of E. coli enzymes are less likely to be present only in Bacteria than the control sets (A); conversely, the set of E. coli enzymes are more likely to be present in all phylogenetic groups than the control sets (C).
Figure 3.
Statistics of phylogenetic distribution of E. coli enzymes. The 13 statistically significant taxonomic combinations are shown on the x-axis (color coding as in Fig. 1; for taxonomic abbreviations see Methods). Frequency of enzymes belonging to the corresponding taxonomic combination (left bars, colored), compared to the mean value of 30 control runs (right bars, hatched), is shown on the left y-axis. Variance estimates for the mean values are not shown, for clarity. Black diamonds represent Z-score values (see Methods for details); scale is shown on the right y-axis.
To examine which enzymes are actually most conserved at the sequence level, we recorded all pairwise sequence identity values between the E. coli enzymes and their homologs from Homo sapiens (Table 2), as an indicative measure of protein sequence conservation. There are 11 E. coli metabolic enzymes which have similar lengths (±10%) and have sequence identity ≥55% to the corresponding human proteins (Table 2). The most conserved known metabolic enzyme appears to be guanosine 5′ monophosphate oxidoreductase, with 68% identity shared between the E. coli and H. sapiens sequences, followed by glyceraldehyde 3-phosphate dehydrogenase and succinyl-CoA synthetase α chain (Table 2). This is the first time, to our knowledge, that such a simple comparison has been performed.
Table 2.
The 11 Most Conserved Metabolic Enzymes in E. coli According to Their Sequence Identity to Homologs in Homo sapiens
| Accession | Description | Human homologs | Identity |
| P15344 | GMP reductase (guanosine 5′-monophosphate oxidoreductase) | Q9P2T1 AAH08021AAH08281P36959 | 68% 68% 65% 65% |
| P06977 | Glyceraldehyde 3-phosphate dehydrogenase A (GAPDH-A) | P04406P00354 Q9HCU6 O14556 | 66% 65% 63% 63% |
| P07459 | Succinyl-coa synthetase alpha chain (SCS-ALPHA) | Q9BWB0 P53597 | 66% 66% |
| P11537 | Glucose-6-phosphate isomerase (GPI) | Q9UHE6 Q9BSK5 P06744 | 64% 64% 64% |
| P05042 | Fumarate hydratase Class II (fumarase) | AAH03108P07954 | 60% 60% |
| P06994 | Malate dehydrogenase | AAH01917P40926 | 58% 58% |
| P09148 | Galactose-1-phosphate uridylyltransferase | P07902 | 56% |
| P07912 | 2-amino-3-ketobutyrate coenzyme A ligase | O75600 | 55% |
| P08324 | Enolase (2-phosphoglycerate dehydratase) | P13929 | 55% |
| P10444 | Succinate dehydrogenase flavoprotein subunit | AAH01380P31040 | 55% 55% |
| P25526 | Succinate-semialdehyde dehydrogenase [NADP+] (SSDH) | P51649 | 55% |
Selected by two criteria: difference in length ≤10% and sequence identity ≥55%. Column names: Accession and Description as in Table 1; “Human homologs” contains the accession numbers for the corresponding human proteins; Identity is the percent identity from pairwise sequence comparison. Note that there may be multiple human homologs for a single E. coli enzyme. Table is sorted by sequence identity of the closest homolog.
Finally, we have examined the conservation of pathways using the detection of enzyme homologs in the corresponding taxonomic partitions. Interestingly, only five of 87 pathways (less than 6%) whose enzymes have homologs in all seven taxa are completely conserved. In fact, these correspond only to short interconversion reactions defined as pathways (not shown). If we relax this strict criterion and admit enzymes with homologs in all three domains of life, that is, in any eukaryotic group, 23 pathways with more than four enzymes and 70% coverage are detected (Table 3). The three invariant pathways correspond to biosynthetic pathways for tryptophan, leucine, and arginine (Table 3). Other pathways in this list include metabolic reactions for cofactor biosynthesis and central metabolism (Table 3). In general, conserved pathways appear to be involved in energy metabolism, central intermediary metabolism, sugar degradation, cofactor biosynthesis, and the processing of amino acids and nucleotides. This observation lends support to the hypothesis that this set of processes is one of the most ancient aspects of cellular physiology, possibly present in the universal ancestor (Woese 1998).
Table 3.
The Most Conserved Metabolic Pathways in E. coli Whose Enzymes Are Universally Present in the Three Domains of Life, Archaea (A), Bacteria (B) and Eucarya (E)
| Pathway name | Pathway superclass | A-B-E | A-B-E-Vr | Enzymes | Coverage |
| Tryptophan biosynthesis | Individual amino acids | 5 | 0 | 5 | 1.00 |
| Leucine biosynthesis | Individual amino acids | 6 | 0 | 6 | 1.00 |
| Arginine biosynthesis II | Individual amino acids | 9 | 2 | 11 | 1.00 |
| Valine biosynthesis | Individual amino acids | 8 | 0 | 9 | 0.89 |
| Nonoxidative branch of the pentose phosphate pathway | Energy-metabolism | 7 | 0 | 8 | 0.88 |
| Histidine biosynthesis | Individual amino acids | 7 | 0 | 8 | 0.88 |
| Purine biosynthesis | Purines and pyrimidines | 12 | 0 | 14 | 0.86 |
| Enterobactin synthesis | Cofactor-biosynthesis | 5 | 0 | 6 | 0.83 |
| Riboflavin, FMN and FAD biosynthesis | Cofactor-biosynthesis | 5 | 0 | 6 | 0.83 |
| Threonine biosynthesis | Individual amino acids | 5 | 0 | 6 | 0.83 |
| FormylTHF biosynthesis | Intermediary-metabolism | 7 | 2 | 11 | 0.82 |
| Methylglyoxal metabolism | Central-metabolism | 4 | 0 | 5 | 0.80 |
| Ribose catabolism | Carbon-degradation | 4 | 0 | 5 | 0.80 |
| Thiamin biosynthesis | Cofactor-biosynthesis | 8 | 0 | 10 | 0.80 |
| Folic acid biosynthesis | Cofactor-biosynthesis | 7 | 1 | 10 | 0.80 |
| Gluconeogenesis | Central-metabolism | 10 | 0 | 13 | 0.77 |
| Isoleucine biosynthesis | Individual amino acids | 10 | 0 | 13 | 0.77 |
| Glycolysis | Energy-metabolism | 10 | 0 | 13 | 0.77 |
| Aerobic respiration, electron donors reaction list | Energy-metabolism | 22 | 0 | 29 | 0.76 |
| Glyoxylate cycle | Energy-metabolism | 5 | 0 | 7 | 0.71 |
| Glycogen catabolism | Carbon-degradation | 5 | 0 | 7 | 0.71 |
| Biosynthesis of proto- and siroheme | Cofactor-biosynthesis | 7 | 0 | 10 | 0.70 |
| TCA cycle, aerobic respiration | Energy-metabolism | 14 | 0 | 20 | 0.70 |
Column names: Pathway name and superclass according to EcoCyc (Karp et al. 2000); ABE: number of enzymes in the three phylogenetic domains; ABEVr: number of enzymes in the three phylogenetic domains plus Viruses (Vr); Enzymes: total number of enzymes in the corresponding pathway; Coverage: ratio of the sum of universal enzymes over total, per pathway. Table is sorted by coverage.
In conclusion, it appears that although metabolic enzymes are widely distributed and highly conserved during evolution, their corresponding participation as groupings in pathways might vary significantly. In that sense, pathways appear to be more variable than previously thought, and pathway evolution exhibits a primarily “mosaic” pattern (Teichmann et al. 2001; Tsoka and Ouzounis 2001), with homologous enzymes being reused in different cellular roles (Jensen 1976).
DISCUSSION
We have shown that the known metabolic complement of E. coli is highly conserved, with half of the enzymes being present in at least one species from the three domains of life, thus considered universal. With statistical simulations, we have also shown that the universal enzymes are much more conserved than the ones absent from Eucarya. The high coverage of the database and the presence of homologs in multiple species eliminates to a significant extent the possibility of detecting lateral gene transfers, also supported by recent, independent analyses (Salzberg et al. 2001; Stanhope et al. 2001). Thus, the observed taxonomic patterns of enzyme distribution are most likely to reflect genuine divergent relationships, tracing the origins of metabolic enzymes and pathways back in time. In particular, the set of universal enzymes may be used for deep phylogeny studies to assess robustness of phylogenetic trees (Huelsenbeck et al. 2001), the determination of divergence times (Feng et al. 1997), and hypotheses for lateral gene transfer (Brown et al. 2001). Finally, we have examined the conservation of biochemical pathways across species and have found that the pathways in which the universal enzymes participate correspond to reactions metabolizing small molecules, such as sugars, amino acids, and nucleotides, as previously suggested (Kyrpides et al. 1999).
The topology and functional diversification of biochemical pathways is yet to be explored, given that the experimentally determined information available is confined to only a few model species, such as E. coli, and processes, such as metabolism. It remains to be seen how representative the metabolism of E. coli is and whether our conclusions will hold for other species, or indeed for other cellular processes. Much more work is necessary to validate the metabolic reconstructions currently based on sequence homology (Tsoka and Ouzounis 2000b).
METHODS
We have extracted the sequences of all enzymes (548 in total) from EcoCyc (Karp et al. 2000), both the monomeric enzymes (208 in total) and the components of enzyme complexes (348 in total; some monomeric enzymes can also be found as enzyme complex components; Tsoka and Ouzounis 2000a). To address the generic phylogenetic distribution of enzymes, we have subsequently divided the nonredundant protein sequence database SwAll (SwissProt+TrEMBL; Bairoch and Apweiler 2000)—(602,333 entries in total) into seven major taxonomic groups: Archaea (A), Bacteria (B), Fungi (F), Metazoa (M), Protista (P), Viridiplantae (Vp), and Viruses (Vr). The section of the database containing these groups covers 582,638 sequences or 97% of the total number of entries (the 3% remainder represents vector data, artificial sequences, insertion sequences, transposons, and other unclassified entries). The partitioning was greatly facilitated by SRS (Etzold and Argos 1993), using the taxonomic records from the SwAll database (SwissProt+TrEMBL; Bairoch and Apweiler 2000). All E. coli entries were excluded from this target database.
The E. coli enzymes were subsequently used as queries to search against the seven taxonomic partitions using BLAST (Altschul et al. 1997) at an E-value threshold 10−06, as previously (Tsoka and Ouzounis 2001), filtered for compositional bias with CAST (Promponas et al. 2000; score threshold 40). The detected homologs were used to classify the query enzyme dataset into the seven taxonomic groups. For each query enzyme, we recorded the distribution pattern of all homologs in a binary vector, representing its taxonomic distribution.
To assess the statistical significance of these measurements, 30 samples of equal size were taken from the E. coli genome as control sets and were subjected to an identical analysis—as previously described (Tsoka and Ouzounis 2000a). In total, 16,988 runs (548 queries * 31 runs) were performed against the nonredundant protein sequence database. Total computation time was approximately 120 h on a 4-CPU Sun E450 with 2GB of RAM. Of the observed (62, out of 128 possible) combinations of taxonomic partitions, only 16 sets contain more than ten counts either in enzymes or in the control sets and were further tested for significance. We have used a 2-tailed t-test, which is robust for skewed nonnormal distributions, at 99% confidence level. Of the 16 sets, only 13 show a highly significant t-test value and were considered as meaningful (Figs. 1,3).
Acknowledgments
We thank Peter D. Karp (SRI International) for comments and members of the Computational Genomics Group for discussions. This work was supported by the European Molecular Biology Laboratory, the TMR Programme of the European Commission (DGXII—Science, Research and Development) and the Ministry of Science and Technology, Spain. S.T. acknowledges support from the UK Medical Research Council. C.A.O. thanks IBM Research for additional support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL ouzounis@ebi.ac.uk; FAX 44-1223-494471.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.246903.
REFERENCES
- 1.Altschul S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bairoch A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28 45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown J.R., Douady, C.J., Italia, M.J., Marshall, W.E., and Stanhope, M.J. 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28 281-285. [DOI] [PubMed] [Google Scholar]
- 4.Crawford I.P. 1989. Evolution of a biosynthetic pathway: The tryptophan paradigm. Annu. Rev. Microbiol. 43 567-600. [DOI] [PubMed] [Google Scholar]
- 5.Dandekar T., Schuster, S., Snel, B., Huynen, M., and Bork, P. 1999. Pathway alignment: Application to the comparative analysis of glycolytic enzymes. Biochem. J. 343 115-124. [PMC free article] [PubMed] [Google Scholar]
- 6.Doolittle R.F., Feng, D.F., Tsang, S., Cho, G., and Little, E. 1996. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science 271 470-477. [DOI] [PubMed] [Google Scholar]
- 7.Etzold T. and Argos, P. 1993. SRS—an indexing and retrieval tool for flat file data libraries. Comput. Appl. Biosci. 9 49-57. [DOI] [PubMed] [Google Scholar]
- 8.Feng D.F., Cho, G., and Doolittle, R.F. 1997. Determining divergence times with a protein clock: Update and reevaluation. Proc. Natl. Acad. Sci. 94 13028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forst C.V. and Schulten, K. 2001. Phylogenetic analysis of metabolic pathways. J. Mol. Evol. 52 471-489. [DOI] [PubMed] [Google Scholar]
- 10.Huelsenbeck J.P., Ronquist, F., Nielsen, R., and Bollback, J.P. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294 2310-2314. [DOI] [PubMed] [Google Scholar]
- 11.Huynen M.A., Dandekar, T., and Bork, P. 1999. Variation and evolution of the citric-acid cycle: A genomic perspective. Trends Microbiol. 7 281-291. [DOI] [PubMed] [Google Scholar]
- 12.Jacob F., 1988. A statue within. Basic Books, New York, NY.
- 13.Jensen R.A. 1976. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30 409-425. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser A., Vollmert, M., Tholl, D., Graves, M.V., Gurnon, J.R., Xing, W., Lisec, A.D., Nickerson, K.W., and Van Etten, J.L. 1999. Chlorella virus PBCV-1 encodes a functional homospermidine synthase. Virology 263 254-262. [DOI] [PubMed] [Google Scholar]
- 15.Karp P.D. 2001. Pathway databases: A case study in computational symbolic theories. Science 293 2040-2044. [DOI] [PubMed] [Google Scholar]
- 16.Karp P.D., Ouzounis, C., and Paley, S. 1996. HinCyc: A knowledge base of the complete genome and metabolic pathways of H. influenzae. Proc. Int. Conf. Intell. Syst. Mol. Biol. 4 116-124. [PubMed] [Google Scholar]
- 17.Karp P.D., Riley, M., Saier, M., Paulsen, I.T., Paley, S.M., and Pellegrini-Toole, A. 2000. The EcoCyc and MetaCyc databases. Nucleic Acids Res. 28 56-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyrpides N., Overbeek, R., and Ouzounis, C. 1999. Universal protein families and the functional content of the last universal common ancestor. J. Mol. Evol. 49 413-423. [DOI] [PubMed] [Google Scholar]
- 19.Lehninger A.L., 1979. Biochemistry, pp. 363 Worth Publishers, Inc., New York, NY.
- 20.Ouzounis C.A. and Karp, P.D. 2000. Global properties of the metabolic map of Escherichia coli. Genome Res. 10 568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pellegrini M., Marcotte, E.M., Thompson, M.J., Eisenberg, D., and Yeates, T.O. 1999. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. 96 4285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Promponas V.J., Enright, A.J., Tsoka, S., Kreil, D.P., Leroy, C., Hamodrakas, S., Sander, C., and Ouzounis, C.A. 2000. CAST: An iterative algorithm for the complexity analysis of sequence tracts. Bioinformatics 16 915-922. [DOI] [PubMed] [Google Scholar]
- 23.Salzberg S.L., White, O., Peterson, J., and Eisen, J.A. 2001. Microbial genes in the human genome: Lateral transfer or gene loss? Science 292 1903-1906. [DOI] [PubMed] [Google Scholar]
- 24.Stanhope M.J., Lupas, A., Italia, M.J., Koretke, K.K., Volker, C., and Brown, J.R. 2001. Phylogenetic analyses do not support horizontal gene transfers from bacteria to vertebrates. Nature 411 940-944. [DOI] [PubMed] [Google Scholar]
- 25.Struhl K. 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98 1-4. [DOI] [PubMed] [Google Scholar]
- 26.Teichmann S.A., Rison, S.C., Thornton, J.M., Riley, M., Gough, J., and Chothia, C. 2001. The evolution and structural anatomy of the small molecule metabolic pathways in Escherichia coli. J. Mol. Biol. 311 693-708. [DOI] [PubMed] [Google Scholar]
- 27.Tsoka S. and Ouzounis, C.A. 2000a. Prediction of protein interactions: Metabolic enzymes are frequently involved in gene fusion. Nat. Genet. 26 141-142. [DOI] [PubMed] [Google Scholar]
- 28.Tsoka S. and Ouzounis, C.A. 2000b. Recent developments and future directions in computational genomics. FEBS Lett. 480 42-48. [DOI] [PubMed] [Google Scholar]
- 29.Tsoka S. and Ouzounis, C.A. 2001. Functional versatility and molecular diversity of the metabolic map of Escherichia coli. Genome Res. 11 1503-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woese C. 1998. The universal ancestor. Proc. Natl. Acad. Sci. 95 6854-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woese C.R. and Fox, G.E. 1977. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. 74 5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]