Abstract
The corpus callosum connects cerebral hemispheres and is the largest axon tract in the mammalian brain. Callosal malformations are among the most common congenital brain anomalies and are associated with a wide range of neuropsychological deficits. Crossing of the midline by callosal axons relies on a proper midline environment that harbors guidepost cells emitting guidance cues to instruct callosal axon navigation. Little is known about what controls the formation of the midline environment. We find that two components of the Hippo pathway, the tumor suppressor Nf2 (Merlin) and the transcriptional coactivator Yap (Yap1), regulate guidepost development and expression of the guidance cue Slit2 in mouse. During normal brain development, Nf2 suppresses Yap activity in neural progenitor cells to promote guidepost cell differentiation and prevent ectopic Slit2 expression. Loss of Nf2 causes malformation of midline guideposts and Slit2 upregulation, resulting in callosal agenesis. Slit2 heterozygosity and Yap deletion both restore callosal formation in Nf2 mutants. Furthermore, selectively elevating Yap activity in midline neural progenitors is sufficient to disrupt guidepost formation, upregulate Slit2 and prevent midline crossing. The Hippo pathway is known for its role in controlling organ growth and tumorigenesis. Our study identifies a novel role of this pathway in axon guidance. Moreover, by linking axon pathfinding and neural progenitor behaviors, our results provide an example of the intricate coordination between growth and wiring during brain development.
Keywords: Commissure, Radial glia, Glial wedge, Indusium griseum, Guidepost neurons, Nervous system
INTRODUCTION
The corpus callosum is the largest commissural axon tract in the mammalian brain. It transfers sensory, motor and cognitive information between cerebral hemispheres. An innovation during eutherian evolution, the corpus callosum is believed to be involved in higher order brain functions. Callosal malformations are among the most common congenital brain anomalies and are associated with cognitive, behavioral and neurological deficits (Paul et al., 2007).
Callosal formation requires a cascade of dynamic events to be precisely coordinated spatially and temporally (Donahoo and Richards, 2009; Fame et al., 2011; Nishikimi et al., 2013). Callosal projection neurons mainly reside in cortical layers II and III and, to a lesser extent, layers V and VI. In the mouse, axons from the cingulate cortex approach and cross the midline first at around embryonic day (E) 16 and are later joined by axons from the neocortex (Koester and O'Leary, 1994; Rash and Richards, 2001). To reach their targets in the contralateral hemisphere, callosal axons follow the guidance cues emitted by several glial and neuronal guidepost structures at the corticoseptal boundary region. Defects in callosal neuron specification and production, midline patterning, guidepost formation or guidance cue expression and reception might result in complete or partial agenesis of the corpus callosum. Although it has long been recognized that the environment which callosal axons traverse is crucial for their pathfinding, how this environment is shaped is not well understood. Despite recent progress (Amaniti et al., 2013; Benadiba et al., 2012; Chinn et al., 2014; Magnani et al., 2014; Piper et al., 2009a; Shu et al., 2003a,b; Smith et al., 2006; Unni et al., 2012), our knowledge of what regulates the development of guideposts and expression of guidance molecules remains limited.
Neurofibromatosis type 2 is an inherited syndrome in which individuals develop nervous system tumors (Li et al., 2012). It is caused by inactivating mutations of the neurofibromatosis 2 (NF2) tumor suppressor gene, which encodes a Four-point-one, Ezrin, Radixin, Moesin (FERM) domain-containing protein that is also known as Merlin. Nf2 (Merlin) is widely expressed and engages in many signaling pathways, including the Rac-PAK, mTORC1, EGFR, PI3K-Akt and Hippo pathways. It regulates diverse cellular processes, such as the establishment of cell polarity, formation of cell-cell junctions and proliferation. Nf2 plays numerous roles in the nervous system, including promoting cell-cell adhesion during neural tube closure (McLaughlin et al., 2007), inhibiting glial cell proliferation (Giovannini et al., 2000; Houshmandi et al., 2009) and maintaining axonal integrity (Schulz et al., 2013).
Using Nf2 conditional knockout mouse models, we have previously shown that Nf2 limits the expansion of neural progenitor cell (NPC) populations during brain development by inhibiting the transcriptional coactivators Yap (Yap1 – Mouse Genome Informatics) and its paralog Taz (here referred to as Yap/Taz) (Lavado et al., 2013). Nf2 loss and YAP hyperactivation also render the dentate gyrus radial glial scaffold over-exuberant and impair hippocampus morphogenesis. We unexpectedly discovered that Nf2 mutants lack the corpus callosum. However, it was unclear mechanistically how Nf2 regulates callosal development.
Here, we investigate the underlying cellular and molecular basis for the role of Nf2 in callosal development. We find that Nf2 is not required in callosal neurons or their progenitors in order for the corpus callosum to form, but it is required in midline NPCs for proper formation of the midline environment that callosal axons encounter. We further demonstrate that Nf2 functions by suppressing Yap, which in turn regulates guidepost development and expression of the guidance cue Slit2. Our study uncovers the mechanistic basis of how the tumor suppressor Nf2 regulates callosal formation and provides novel insights into the molecular mechanisms that shape the callosal midline environment.
RESULTS
Nf2 is required for corpus callosum and hippocampal commissure development
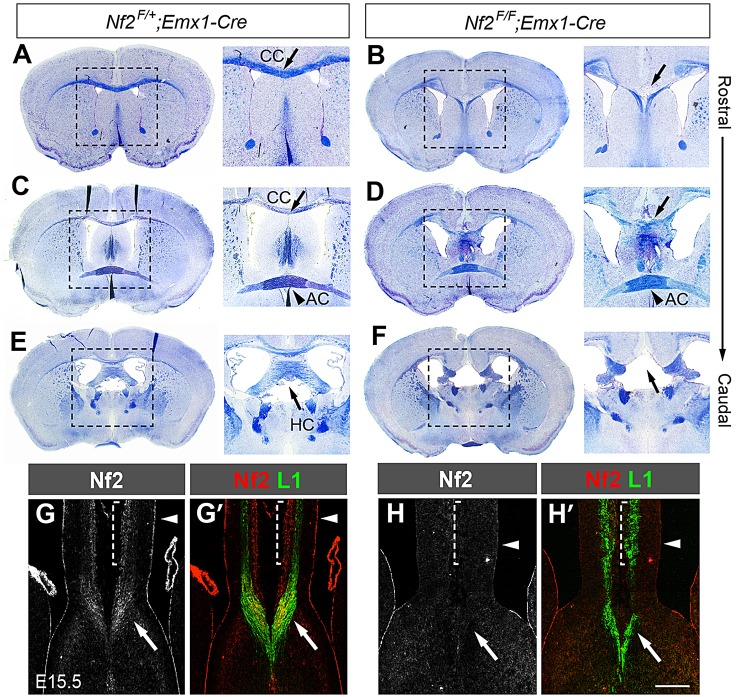
We previously found that deleting floxed alleles of Nf2 (Nf2F/F) (Giovannini et al., 2000) with Emx1-cre, which is expressed in the dorsal telencephalon by E10.5 (Gorski et al., 2002), results in callosal agenesis (Lavado et al., 2013). Luxol Blue staining of adult brain sections to detect myelinated axons showed that, in Nf2 mutants (Nf2F/F;Emx1-Cre), the corpus callosum was absent along the rostrocaudal axis with few, if any, fibers crossing the midline (Fig. 1A-D, arrow). The hippocampal commissure was also lacking (Fig. 1E,F, arrow), whereas the anterior commissure formed normally (Fig. 1C,D, arrowhead). The acallosal phenotype was never observed in Nf2F/+;Emx1-Cre animals, suggesting that the defect was not due to strain background. Deleting Nf2 using Nestin-Cre (Tronche et al., 1999), which is expressed in NPCs throughout the central nervous system, also led to agenesis of the corpus callosum and hippocampal commissure (supplementary material Fig. S1). Here, we focused on understanding the role of Nf2 in callosal development.
Fig. 1.
Agenesis of the corpus callosum and hippocampal commissure in Nf2 mutants. (A-F) Luxol Blue staining of myelinated axons (blue) and Cresyl Violet staining of cell bodies (purple) showing agenesis of the corpus callosum (CC) (A-D, arrow) and hippocampal commissure (HC) (E,F, arrow) but normal anterior commissure (AC) (C,D, arrowhead) in 2-month-old Nf2F/F;Emx1-Cre mice (n=3). A magnified view of the boxed region is shown in the image to the right. (G,G′) Immunostaining showing Nf2 localization at the ventricular surface (arrowhead), cortical plate (dashed bracket) and L1+ callosal axons (arrow) in control brains at E15.5. (H,H′) Nf2 immunoreactivity in these regions is eliminated in Nf2F/F;Emx1-Cre dorsal telencephalon. Arrowhead, ventricular surface; arrow, callosal axons. Scale bar: 200 µm.
Nf2 transcripts have been detected in NPCs and cortical neurons during mouse brain development (McLaughlin et al., 2007). We examined Nf2 protein localization at E15.5, when pioneer axons reach the midline. As reported previously (Lavado et al., 2013), Nf2 localized at the apical region of NPCs, highlighting the ventricular surface (Fig. 1G, arrowhead). Nf2 was also detected in the cortical plate (Fig. 1G, dashed bracket) and in axons reaching the midline (Fig. 1G, arrow). Co-staining with an antibody against L1 cell adhesion molecule (L1), which labels axons, confirmed that Nf2 was present in callosal axons (Fig. 1G′, arrow). All Nf2 immunoreactivity in the dorsal telencephalon was eliminated in Nf2F/F;Emx1-Cre embryos (Fig. 1H,H′), indicating that the observed signals were specific. Thus, Nf2 is expressed in callosal neurons and the NPCs that give rise to callosal neurons and midline structures. This expression pattern suggests that the acallosal phenotype caused by loss of Nf2 might arise from (1) defects in callosal neurons, including their specification, production and ability to project axons and respond to guidance cues, or (2) defects in the environment that callosal axons encounter, including midline patterning, guidepost formation and the expression of guidance cues. We investigated all of these possibilities.
A defective midline is responsible for callosal agenesis in Nf2 mutants
First we examined whether callosal neurons were generated properly in Nf2 mutants. The expression patterns of Satb2, a transcription factor required for callosal neuron specification (Alcamo et al., 2008; Britanova et al., 2008), and cortical layer-specific markers Ctip2 (layers V and VI) and Tbr1 (layers II, III, V and VI) were normal in Nf2 mutants at E15.5 and E17.5 (supplementary material Fig. S2), demonstrating that callosal neuron production and cortical laminar organization are largely unperturbed upon Nf2 deletion.
To determine whether Nf2 is required in callosal neurons for callosal formation, we specifically deleted Nf2 in callosal neurons using a Satb2-Cre line. Using the Cre-reporter line Rosa-CAG-LSL-tdTomato (R26R-tdTomato) (Madisen et al., 2010), which expresses tdTomato upon Cre-mediated deletion of the LoxP-Stop-LoxP (LSL) cassette, we confirmed that Cre activity was turned on at around E13.5 and was mostly restricted to callosal neurons (supplementary material Fig. S3A-C). No Cre activity was detected in progenitor cells at the ventricular zone and subventricular zone (supplementary material Fig. S3A,B). In E15.5 Nf2F/F;Satb2-Cre embryos, Nf2 immunoreactivity was preserved at the ventricular surface (supplementary material Fig. S3D-E′, arrowhead) but lost in callosal axons (supplementary material Fig. S3D-E′, arrow). The corpus callosum formed normally in these mutants (supplementary material Fig. S3F,G), suggesting that Nf2 is not required in callosal neurons for callosal formation.
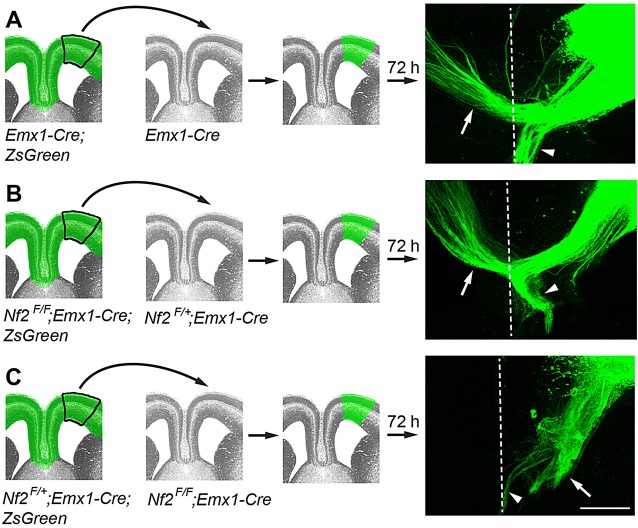
To further test whether Nf2-deficient callosal neurons were able to project axons across the midline, we performed ex vivo transplantation experiments. We introduced a Cre-reporter gene, Rosa-CAG-LSL-ZsGreen (ZsGreen) (Madisen et al., 2010), into the Nf2F/F background to visualize axons. In control experiments, homotopic transplantation of the frontal cortex of Emx1-Cre;ZsGreen embryos at E16.5 into cortical sections of Emx1-Cre hosts resulted in midline crossing of ZsGreen+ callosal axons after 72 h in culture (n=7 out of 7; Fig. 2A, white arrow). Axons from Nf2 mutant (Nf2F/F;Emx1-Cre;ZsGreen) cortices were also able to cross the midline of Nf2F/+;Emx1-Cre hosts (n=5 out of 5; Fig. 2B, white arrow). By contrast, axons from Nf2F/+;Emx1-Cre;ZsGreen cortices failed to cross the midline of Nf2 mutant hosts (n=0 out of 10; Fig. 2C, white arrow). We often observed that, in all three transplantation scenarios, some axons mis-projected ventrally into the septum (Fig. 2A-C, arrowheads). This is probably an artifact caused by, for example, imperfect sectioning or grafting. Nevertheless, the stark contrast between Nf2 mutant axons being able to cross the midline of control hosts (Fig. 2B) and control axons unable to cross the midline of Nf2 mutant hosts (Fig. 2C) strongly suggests that Nf2 is not required for the production of callosal neurons capable of projecting axons across the midline, but is required to generate the midline environment.
Fig. 2.
Callosal axons from Nf2 mutant brains can cross the midline of control forebrains. Tissues from the frontal cortex of E16.5 ZsGreen+ donor embryos were transplanted into cortical sections of ZsGreen− host embryos and then cultured for 72 h. Axon trajectory was examined using ZsGreen fluorescence. Panels show transplantation schemes with the genotypes of donor and host embryos indicated, and examples of transplants are shown on the right. Dashed line, the midline; white arrow, callosal axons; arrowhead, mis-projected axons. (A) Control callosal axons cross the midline of control hosts. (B) Callosal axons from Nf2F/F;Emx1-Cre embryos cross the midline of Nf2F/+;Emx1-Cre hosts. (C) Callosal axons from Nf2F/+;Emx1-Cre embryos fail to cross the midline of Nf2F/F;Emx1-Cre hosts. Scale bar: 200 µm.
Nf2 is required for glial wedge development
Upon determining that midline defects are responsible for callosal agenesis in Nf2 mutants, we first examined early patterning at the corticoseptal boundary where midline guideposts later form. At E12.5 and E15.5, the expression patterns of Six3, Zic2 and Nfia, transcription factors delineating subdomains of the commissural plate (Moldrich et al., 2010), were unaffected upon Nf2 deletion (supplementary material Fig. S4A-F′). Gli3 and Fgf8 are required for corticoseptal boundary patterning and callosal formation (Amaniti et al., 2013; Magnani et al., 2014; Moldrich et al., 2010). Their expression and that of Sprouty1, a target of Fgf signaling, appeared normal in Nf2 mutants (supplementary material Fig. S4A-D′,G-J′). Interestingly, quantitative western blot analyses of Gli3 using E13.5 cortex and E15.5 medial-cortex tissues showed that the amounts of the full-length activator form (Gli3FL) and cleaved repressor form (Gli3R) and the ratio of Gli3FL to Gi3R were increased in Nf2 mutants (supplementary material Fig. S5), although the functional consequence of these changes is unclear. Wnt8b and the Wnt target gene Axin2 were expressed normally in Nf2 mutants except for an elongation of the Wnt8b+ domain at E15.5 (supplementary material Fig. S4K-N′), which correlated with the elongation of the glial wedge (see below, Fig. 3A-B′). Axin2 in situ hybridization signals were very weak at E15.5 (supplementary material Fig. S4N,N′). We performed quantitative RT-PCR (qRT-PCR) analysis using rostral medial-cortex tissues at E15.5, which also showed similar Axin2 mRNA levels in control and Nf2 mutants (n=3 embryos per genotype, P=0.8). Taken together, these results suggest that patterning of the corticoseptal boundary is grossly normal in Nf2 mutants.
Fig. 3.
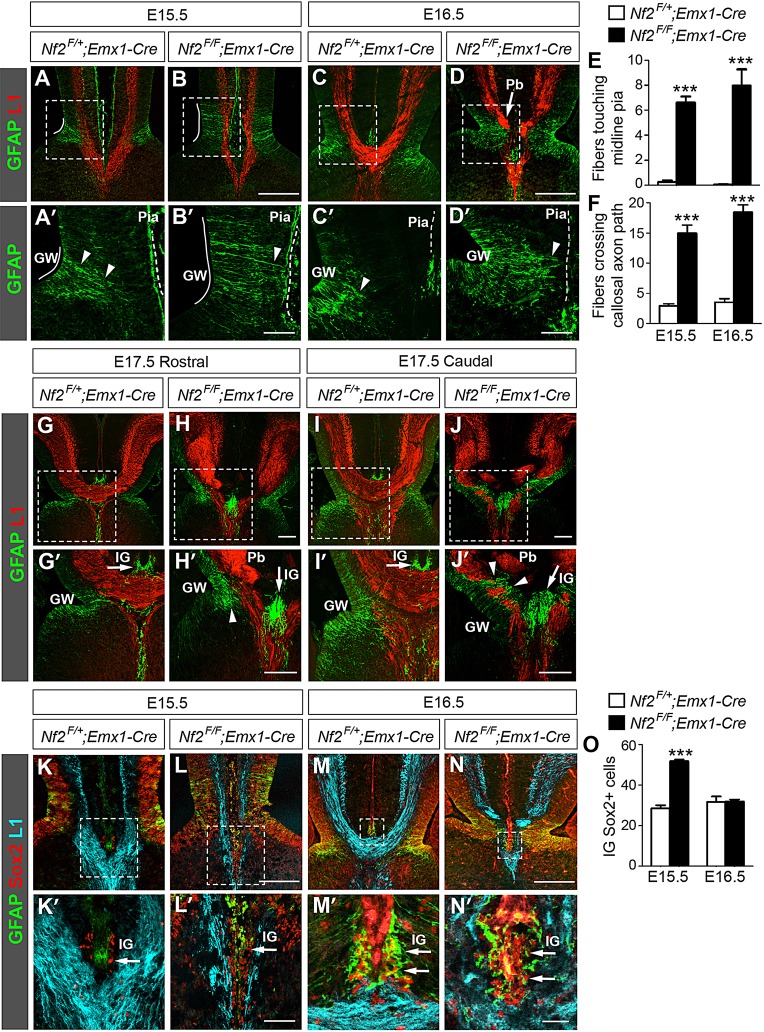
Nf2 loss impairs glial wedge development. (A,A′) In control embryos at E15.5, GFAP+ processes (arrowhead) from the glial wedge (GW, white line) have detached from the pia (dashed line). At this stage, the GFAP antibody often stains the pia nonspecifically. Panels labeled with primes are the magnified view of the boxed areas in corresponding panels. (B,B′) In Nf2F/F;Emx1-Cre embryos, the GW is elongated and many GW processes remain attached to the pia and intersect L1+ callosal axons (B′, arrowhead). (C-D′) In E16.5 Nf2F/F;Emx1-Cre embryos, GW fibers still extend close to the pia (D′, arrowhead) and Probst bundles (Pb) are formed (D, arrow). (E,F) More GFAP+ GW fibers touch the midline pia (E, n=4) and more cross callosal axon path (F, n=6) in Nf2F/F;Emx1-Cre embryos than in controls. ***P<0.001. (G-J′) In E17.5 Nf2F/F;Emx1-Cre brains, GW fibers have lost pial attachment in rostral regions (H′, arrowhead) but still project close to the pia and intersect axons of the corpus callosum or hippocampal commissure in caudal regions (J′, arrowhead). Arrow: indusium griseum (IG). (K-L′) At E15.5, Sox2+ cells are present in the IG primordium in control and Nf2F/F;Emx1-Cre embryos (arrow). These cells are GFAP− at this stage. (M-N′) GFAP+ IG astrocytes (arrow) are present in control and Nf2F/F;Emx1-Cre embryos at E16.5. Most of these cells also express Sox2. (O) The number of IG Sox2+ cells is increased in Nf2F/F;Emx1-Cre embryos at E15.5 (n=5) but is similar to that in controls at E16.5 (n=4). Scale bars: 200 µm in B,D,H,H′,J,J′,L,N; 50 µm in B′,D′,L′; 10 µm in N′.
Next we examined the development of midline guideposts between E15.5 and E17.5, before and during callosal formation. The glial wedge (GW) is located at the ventricular zone on either side of the midline and deflects callosal axons toward the midline (Shu and Richards, 2001). At these stages, the GW is composed of NPCs, which are also known as radial glia (Kriegstein and Alvarez-Buylla, 2009; Shu et al., 2003b; Smith et al., 2006). GW cells initially extend long GFAP+ radial processes to the midline pia, but lose their pial attachment by E15.5, when pioneer axons reach the midline (Shu et al., 2003b) (Fig. 3A,A′, arrowhead). In Nf2 mutants, the GW was formed but dorsoventrally spanned a broader region than that in controls (Fig. 3A-B′, white line). Moreover, many GFAP+ processes remained attached to the midline pia, crossing the path of callosal axons (Fig. 3B,B′, arrowhead; Fig. 3E,F). At E16.5, when many callosal axons have crossed the midline in controls (Fig. 3C), some GW fibers in Nf2 mutants still extended close to the pia (Fig. 3D,D′, arrowhead; Fig. 3E,F) and callosal axons, failing to cross the midline, accumulated into abnormal tangles known as Probst bundles (Fig. 3D, arrow). By E17.5, GW fibers in Nf2 mutants no longer projected close to the pia in rostral regions (Fig. 3H,H′, arrowhead), but still transected the path of callosal or hippocampal commissural axons in caudal regions (Fig. 3J,J′, arrowhead). Thus, GW formation was perturbed in Nf2 mutants, manifesting as elongation of the GW region and overextension of radial processes.
The indusium griseum (IG) comprises neurons and astrocytes underneath the midline pia, above the corpus callosum. Because IG astrocytes do not express GFAP until E17 (Shu et al., 2003b), we searched for other IG markers to examine the earlier stages of its development. At E15.5, at least a fraction of cells beneath the midline pia expressed Sox2 (Fig. 3K,K′, arrow) and some Sox2+ cells were also immunopositive for GFAP at E16.5 (Fig. 3M,M′, arrow), suggesting that Sox2 is an earlier marker for IG cells. The immunostaining pattern of Sox2 overlapped with that of Sox9 (supplementary material Fig. S6A-B′), another IG glia marker (Clegg et al., 2014). Similar to controls, Sox2+, GFAP+ and Sox9+ cells were present beneath the midline pia in Nf2 mutants from E15.5 to E17.5 (Fig. 3K-N′,G-J′, arrows; supplementary material Fig. S6C-F′). We noted that the numbers of Sox2+ cells and Sox9+ cells were increased in Nf2 mutants at E15.5 but returned to control levels at E16.5 (Fig. 3O; supplementary material Fig. S6G). The functional significance of this change is unclear. We also noticed that the IG in Nf2 mutants was mis-positioned at the same dorsoventral level as the GW, instead of dorsal to it as in controls (Fig. 3G-J′). This defect, however, might be secondary to the lack of crossing callosal axons. Taken together, we conclude that IG development is largely unaffected in Nf2 mutants, except for a transient increase in IG glia at E15.5.
Nf2 promotes differentiation of glutamatergic guidepost neurons
Next we investigated the development of midline guidepost neurons, which comprise GABAergic interneurons and Calretinin-positive glutamatergic neurons and attract callosal axons (Niquille et al., 2009). Because GABAergic neurons are produced by NPCs at the ventral telencephalon (Niquille et al., 2009), Nf2 deletion using Emx1-Cre is unlikely to directly affect GABAergic neurons. We therefore focused on Calretinin+ glutamatergic neurons. At E15.5, these neurons resided at the midline cortical plate, forming a band between the pia and GW processes (Fig. 4A,A′). In Nf2 mutants, Calretinin+ neurons were significantly reduced [310±4 per section in controls versus 225±4 (mean±s.e.m.) in Nf2 mutants, n=4, P<0.0001], and the band of these cells was bisected by a gap (Fig. 4B,B′, arrowhead). This gap coincided precisely with the GW fibers that remained attached to the pia (Fig. 4B′, arrowheads) and was occupied by Tbr2+ intermediate progenitors (Fig. 4D,D′, arrowheads), which normally reside at the subventricular zone (Fig. 4C,C′). The presence of Tbr2+ cells in the cortical plate indicated that differentiation of Calretinin+ neurons might be affected. To test this, we performed birth-dating experiments. We injected BrdU into pregnant mice at E13.5 to label S-phase progenitor cells. Two days later, many BrdU-labeled cells in controls had differentiated into Calretinin+ neurons (Fig. 4E,E′, yellow cells, arrowheads). In Nf2 mutants, BrdU-labeled cells predominantly remained as Tbr2+ intermediate progenitors or ventricular zone NPCs, and significantly fewer of them differentiated into Calretinin+ neurons (Fig. 4F,F′; 84±9 BrdU+Calretinin+ neurons per section in controls versus 33±2 in Nf2 mutants, n=3, P=0.0056). These results demonstrate that Nf2 promotes differentiation of Calretinin+ guidepost neurons.
Fig. 4.
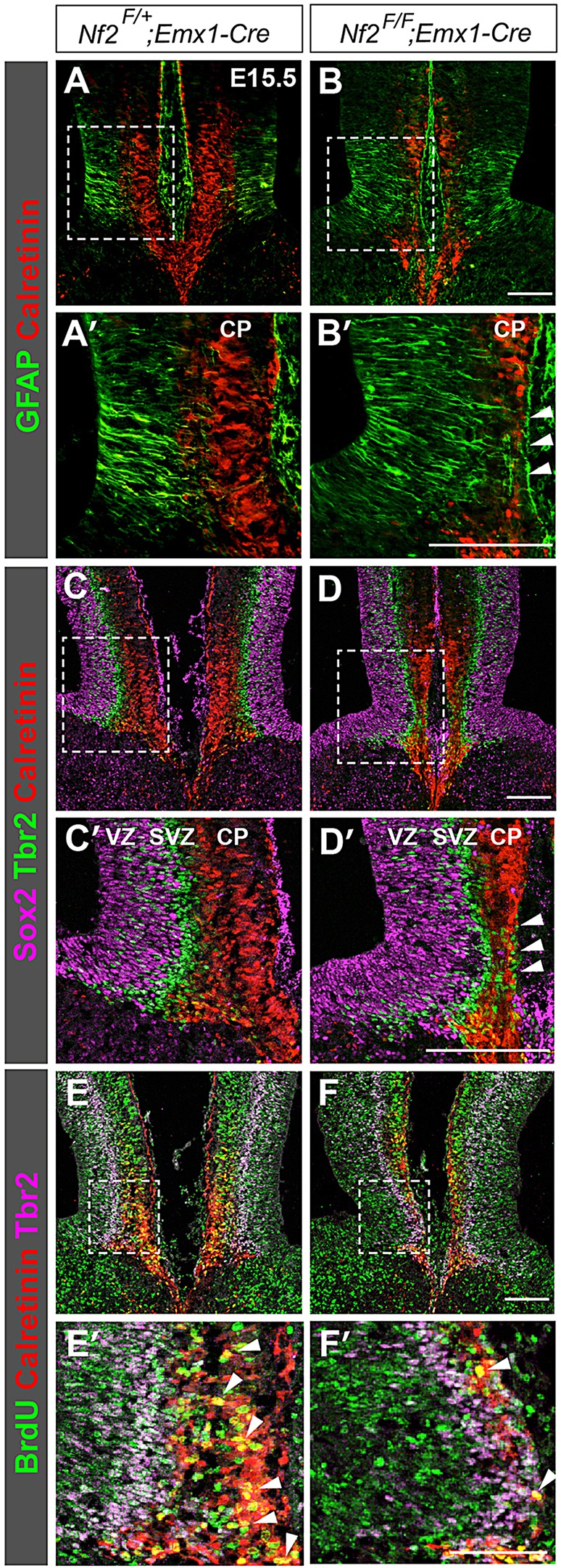
Loss of Nf2 reduces glutamatergic guidepost neurons. (A,A′) In control embryos at E15.5, Calretinin+ guidepost neurons reside at the midline cortical plate (CP). (B,B′) Fewer Calretinin+ neurons are found in Nf2F/F;Emx1-Cre midline, especially at the region where GW fibers remain attached to the pia (B′, arrowhead). (C,C′) In controls, Sox2+ neural progenitor cells (NPCs), Tbr2+ intermediate progenitors and Calretinin+ neurons reside at the ventricular zone (VZ), subventricular zone (SVZ) and CP, respectively. (D,D′) In the midline of Nf2F/F;Emx1-Cre mice, Tbr2+ intermediate progenitors intermingle with Calretinin+ neurons (arrowheads). (E-F′) BrdU was administered at E13.5, and embryos were harvested 48 h later. Many BrdU-labeled cells in controls are Calretinin+ (E,E′, yellow cells, arrowhead). In Nf2F/F;Emx1-Cre embryos, most BrdU-labeled cells remain as VZ NPCs or Tbr2+ intermediate progenitors (F,F′) and fewer of them have differentiated into Calretinin+ neurons (F′, yellow cells, arrowheads). Panels labelled with primes are the magnified view of the boxed areas in corresponding panels. Scale bars: 100 µm.
Upregulation of Slit2 contributes to callosal agenesis in Nf2 mutants
Next, we examined whether Nf2 deletion affects the expression of guidance molecules that are important for callosal formation. Similar to E15.5-16.5 controls, callosal axons in Nf2 mutants were immunopositive for guidance receptors Neuropilin-1 (Piper et al., 2009,b) and Dcc (Fazeli et al., 1997), and Robo1 (Andrews et al., 2006) transcripts were detected in the cell bodies of cingulate neurons (supplementary material Fig. S7A-F). Midline guidance cues Sema3c (Niquille et al., 2009; Piper et al., 2009,b), Netrin1 (Serafini et al., 1996) and Draxin (Islam et al., 2009) were expressed normally in Nf2 mutants (supplementary material Fig. S7G-L). Robo1, Sema3c and Netrin1 transcripts are usually present in the region where Calretinin+ neurons reside (supplementary material Fig. S7E,G,I, dashed bracket). In Nf2 mutants, the apparent lack or downregulation of these transcripts in that region is probably due to fewer Calretinin+ neurons (see Fig. 4) rather than to reduced transcription. Interestingly, Slit2, which is expressed in the midline ventricular zone at E15.5 and is required for callosal formation (Bagri et al., 2002; Shu and Richards, 2001), was induced in Nf2 mutants (Fig. 5A,B). To confirm the change in the level of Slit2, we performed qRT-PCR analysis using E14.5 medial-cortex tissues. The Slit2 mRNA level was increased 1.7±0.1 fold in Nf2 mutants (n=3, P=0.0087).
Fig. 5.
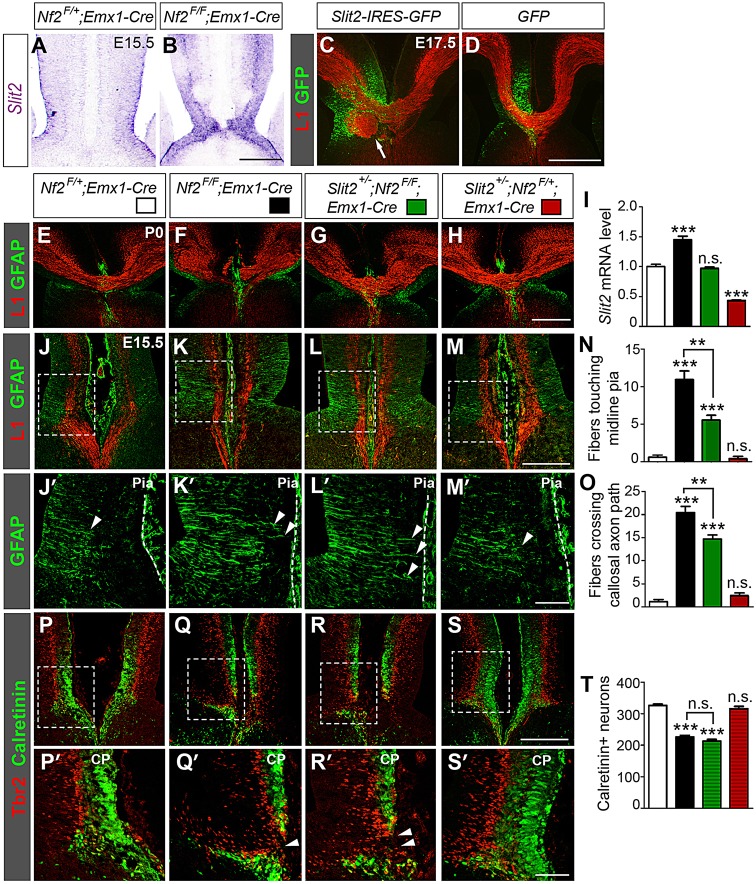
Slit2 upregulation contributes to callosal agenesis in Nf2 mutants. (A,B) In situ hybridization shows higher Slit2 levels in E15.5 Nf2F/F;Emx1-Cre midline than in control. (C,D) Overexpressing Slit2 by using in utero electroporation at E13.5 results in formation of axon tangles at E17.5 (C, arrow). GFP overexpression does not cause this defect (D). (E-H) Sections of littermates show that Slit2 heterozygosity in the Nf2F/F;Emx1-Cre background restores the corpus callosum (G). (I) qRT-PCR analyses of E15.5 rostral medial-cortex tissues show that although the Slit2 mRNA level is upregulated in Nf2F/F;Emx1-Cre embryos, it is returned to control (Nf2F/+;Emx1-Cre) levels in Slit2+/−;Nf2F/F;Emx1-Cre embryos (n=4). (J-O) Slit2 heterozygosity does not fully rescue the GW defects of Nf2 mutants. More GFAP+ GW fibers (arrowheads) touch the midline pia (dashed line) and more cross the callosal axon path in Slit2+/−;Nf2F/F;Emx1-Cre embryos than in littermate Nf2F/+;Emx1-Cre controls. These defects are similar to those in Nf2F/F;Emx1-Cre embryos, albeit less severe. Slit2+/−;Nf2F/+;Emx1-Cre, n=3; the rest, n=5. (P-T) Calretinin+ neurons are reduced in Slit2+/−;Nf2F/F;Emx1-Cre embryos compared to that in Nf2F/+;Emx1-Cre controls and Tbr2+ intermediate progenitors are present in the midline cortical plate (CP) (arrowheads). These defects are similar to those in Nf2F/F;Emx1-Cre embryos. Slit2+/−;Nf2F/+;Emx1-Cre, n=3; the rest, n=4. Scale bars: 200 µm (B,M,S); 500 µm (D,H); 50 µm (M′,S′). Panels labeled with primes are magnified views of the boxed areas in corresponding panels. In panels I,N,O,T, statistical significance scores are evaluated against Nf2F/+;Emx1-Cre embryos unless specifically indicated. **P<0.01; ***P<0.001; n.s., not significant.
Previous in vitro assays show that Slit2 repels cortical axons (Shu and Richards, 2001). To test whether Slit2 upregulation in Nf2 mutants contributes to callosal agenesis, we overexpressed Slit2 in vivo at the corticoseptal boundary by performing in utero electroporation at E13.5. At E17.5, Slit2 overexpression resulted in the accumulation of abnormal axon tangles in the side that had been transfected (n=4 out of 4; Fig. 5C, arrow), although it did not prevent callosal formation. Axon tangles were never observed after overexpression of green fluorescent protein (GFP) (n=0 out of 4, Fig. 5D). These results suggest that Slit2 upregulation could contribute to callosal agenesis in Nf2 mutants.
To further confirm the involvement of Slit2 upregulation, we tested whether deleting Slit2 in Nf2 mutants suppresses the acallosal phenotype. Remarkably, deleting one allele of Slit2 fully restored the corpus callosum in neonatal (P0) Nf2 mutants (n=5 out of 5, Fig. 5G), whereas it had no discernible effect in the Nf2 heterozygous background (Fig. 5H). qRT-PCR analysis of E15.5 rostral medial-cortex tissues showed that although the Slit2 mRNA level was increased in Nf2F/F;Emx1-Cre embryos, it was returned to littermate control (Nf2F/+;Emx1-Cre) levels in Slit2+/−;Nf2F/F;Emx1-Cre embryos (Fig. 5I). Slit2 heterozygosity, however, did not fully rescue guidepost development. In Slit2+/−;Nf2F/F;Emx1-Cre embryos, more GFAP+ GW processes extended close to the pia than those in littermate controls (Nf2F/+;Emx1-Cre and Slit2+/−;Nf2F/+;Emx1-Cre) (Fig. 5J-O, arrowheads), although the defects were not as severe as they were in Nf2F/F;Emx1-Cre embryos. Fewer Calretinin+ guidepost neurons were found in Slit2+/−;Nf2F/F;Emx1-Cre embryos than in controls (Fig. 5P-T), and the bands of Calretinin+ neurons were interrupted by Tbr2+ intermediate progenitors (Fig. 5R,R′, arrowhead), again resembling the phenotypes of Nf2F/F;Emx1-Cre embryos. Taken together, these results suggest that Slit2 upregulation contributes to callosal agenesis in Nf2 mutants.
Yap overactivation is responsible for callosal agenesis in Nf2 mutants
Next, we sought to understand how Nf2 deletion leads to guidepost malformation and Slit2 upregulation. Two lines of evidence suggested that overactivation of the Yap/Taz transcriptional coactivators might be responsible. First, we have previously shown that Yap/Taz activities are increased in Nf2 mutant brains and that overexpressing YAP in NPCs increases the abundance of GFAP+ radial processes in the developing hippocampus (Lavado et al., 2013), a phenotype resembling that of Nf2 mutant GW. Second, SLIT2 is induced by YAP overexpression in several cell types and is considered a signature gene of enhanced YAP activity (Dupont et al., 2011; Mohseni et al., 2014; Zhao et al., 2008).
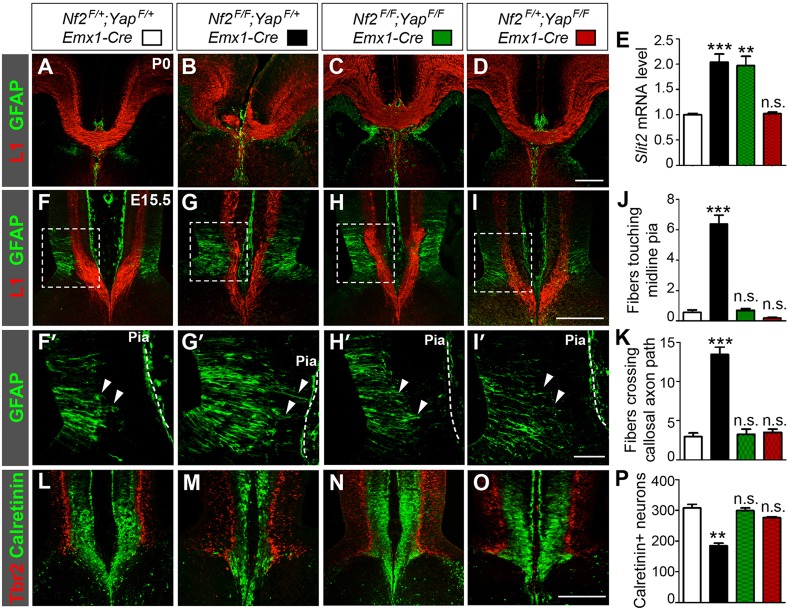
If Yap overactivation is responsible for callosal agenesis in Nf2 mutants, then deleting Yap should restore the corpus callosum. To test this, we generated Nf2;Yap double conditional mutants. Indeed, every double mutant examined between stages E17.5 and P0 developed the corpus callosum (n=10, Fig. 6C). Moreover, the formation of midline guideposts was completely restored in E15.5 Nf2;Yap double mutants. As in littermate Nf2F/+;YapF/+;Emx1-Cre controls, few GW processes remained attached to the pia in Nf2;Yap double mutants (Fig. 6F-K, arrowheads). The number of Calretinin+ neurons in Nf2;Yap double mutants was similar to that in Nf2F/+;YapF/+;Emx1-Cre controls (Fig. 6L-P). However, qRT-PCR analysis of E15.5 rostral medial-cortex tissues showed that, in Nf2;Yap double mutants, the Slit2 mRNA level remained upregulated compared with that of littermate Nf2F/+;YapF/+;Emx1-Cre controls and was similar to that in Nf2F/F;YapF/+;Emx1-Cre mutants (Fig. 6E). Yap deletion in the Nf2 heterozygous background did not have obvious effects on guidepost development, Slit2 level, or callosal formation (Fig. 6D,E,I-K,O,P). Taken together, these results strongly suggest that Yap overactivation, which disrupts guidepost development, contributes to callosal agenesis in Nf2 mutants. Because Yap expression is restricted to NPCs (Milewski et al., 2004) (Fig. 7A), these results also suggest that Nf2 functions within NPCs to regulate callosal development. That Slit2 remains upregulated in Nf2;Yap double mutants indicates that Taz overactivation might also contribute to Slit2 upregulation.
Fig. 6.
Yap deletion restores the corpus callosum in Nf2 mutants. (A-D) Sections of littermates show that deleting Yap in the Nf2F/F;Emx1-Cre background restores the corpus callosum at P0 (C). (E) qRT-PCR analyses of E15.5 rostral medial-cortex tissues show that, compared with Nf2F/+;YapF/+;Emx1-Cre controls, the Slit2 mRNA level is upregulated in Nf2F/F;YapF/F;Emx1-Cre animals, similar to that in Nf2F/F;YapF/+;Emx1-Cre embryos. Nf2F/+;YapF/+;Emx1-Cre, n=4; the rest, n=3. (F-K) Yap deletion rescues the GW defects of Nf2 mutants. At E15.5, the number of GFAP+ GW fibers (arrowheads) touching the midline pia and that crossing callosal axon path in Nf2F/F;YapF/F;Emx1-Cre embryos are similar to those in littermate Nf2F/+;YapF/+;Emx1-Cre controls. n=4. (L-P) The number of Calretinin+ neurons is restored to control levels in the midline of Nf2F/F;YapF/F;Emx1-Cre animals. n=3. Panels labeled with primes are magnified views of the boxed areas in corresponding panels. In panels E,J,K,P, statistical significance scores are evaluated against Nf2F/+;YapF/+;Emx1-Cre embryos. **P<0.01; ***P<0.001; n.s., not significant. Scale bars: 200 µm, except 50 µm in I′.
Fig. 7.
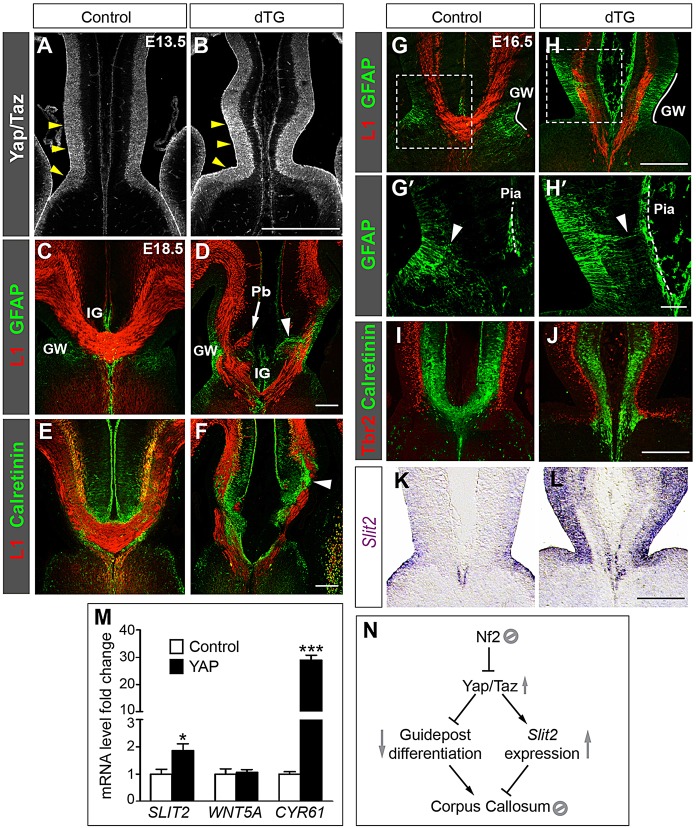
Overexpressing YAP in midline NPCs causes callosal agenesis. (A,B) Immunostaining with an antibody recognizing Yap and Taz shows Yap/Taz expression in ventricular zone NPCs of control brains and increased expression specifically in the midline NPCs of TetO-YAP1;Axin2-rtTA double-transgenic (dTG) embryos (arrowheads). (C-F) dTG embryos at E18.5 exhibit callosal agenesis, with Probst bundles (Pb) accumulating at the midline (D, arrow), GW fibers protruding to the pia (D, arrowhead) and Calretinin+ guidepost neurons aggregating within the callosal axon path (F, arrowhead). n=4. (G-L) dTG embryos at E16.5 exhibit midline defects, including elongation of the GW (G,H, white line), extension of GW fibers (arrowhead) to the pia (dashed line) (G′,H′), fewer Calretinin+ neurons (I,J) and Slit2 upregulation (using in situ hybridization) (K,L). Panels labeled with primes are magnified views of the boxed areas in corresponding panels. (M) qRT-PCR assays show that YAP overexpression in HEK293T cells increases the mRNA levels of SLIT2 and CYR61 but not WNT5A. n=3, *P<0.05, ***P<0.001. (N) Model of the signaling cascade delineated in this study. In midline neural progenitor cells, Nf2 suppresses Yap/Taz to promote guidepost differentiation and to prevent ectopic Slit2 expression, thus allowing the formation of the corpus callosum. See text for details. Scale bars: 200 µm, except 50 µm in H′.
Elevating YAP activity in midline NPCs causes callosal agenesis
To further confirm that aberrant Yap/Taz activation underlies callosal agenesis in Nf2 mutants, we tested whether increasing their activity was sufficient to prevent callosal formation. Based on our finding that Nf2 is required to generate the proper midline environment, we predicted that elevating Yap/Taz activity selectively in midline NPCs would block callosal formation. To test this, we crossed mice carrying a doxycycline-inducible allele of human YAP1, TetO-YAP1 (Camargo et al., 2007), with an Axin2-rtTA line that expresses the doxycycline-dependent transactivator rtTA from the Axin2 promoter, which is active in the midline of the dorsal neural tube, including the dorsal telencephalic midline (Yu et al., 2007). Immunostaining confirmed that, in TetO-YAP1;Axin2-rtTA double-transgenic (dTG) embryos, the level of YAP was specifically increased in the midline ventricular zone but not in the rest of the brain (Fig. 7A,B, arrowheads). By measuring the fluorescence intensity of Yap/Taz immunostaining signals (supplementary material Fig. S8A,B), we estimated that the YAP protein level at the midline of E13.5 dTG embryos was 1.934±0.007 times greater than that in controls (n=3, P<0.0001). Quantitative western blot analysis of E16.5 medial-cortex tissues showed a 2.5±0.1 fold increase of YAP protein amount in dTG embryos (n=3, P<0.0001; supplementary material Fig. S8C).
The phenotypes of these dTG animals were strikingly similar to those of Nf2 mutants – at E18.5, callosal axons in dTG brains approached the midline but few crossed it, instead forming Probst bundles (Fig. 7D, arrow). Some GW fibers penetrated the callosal axon path (Fig. 7D, arrowhead). Calretinin+ guidepost neurons, instead of forming two bands dorsal and ventral to the corpus callosum as they did in controls (Fig. 7E), became disorganized, many invading the callosal axon path (Fig. 7F, arrowhead). Defects in midline development were evident at E16.5 and closely resembled those in Nf2 mutants. The GW expanded dorsoventrally (Fig. 7G,H, white line) and numerous radial processes overextended to the pia (Fig. 7G′,H′, arrowhead). (The number of GFAP+ processes touching the midline pia was 0.083±0.083 in controls and 6.67±0.08 in dTG; n=3, P<0.0001. The number of GFAP+ processes crossing the callosal axon path was 1.7±0.3 in controls and 20.0±1.2 in dTG; n=3, P=0.0001.) Fewer Calretinin+ guidepost neurons were present (Fig. 7I,J; 335±20 Calretinin+ neurons in controls, 212±14 in dTG; n=3, P=0.007). Moreover, Slit2 expression was markedly upregulated in dTG embryos, as shown by using in situ hybridization (Fig. 7K,L) and qRT-PCR analyses of rostral medial-cortex tissues at E16.5 (1.5±0.1 fold increase in dTG, n=3, P=0.013), supporting the hypothesis that elevated Yap/Taz activities upregulate Slit2 transcription. In summary, overexpressing YAP selectively in midline NPCs is sufficient to phenocopy the defects caused by Nf2 deletion, further strengthening the hypothesis that Nf2 regulates callosal development by suppressing Yap/Taz activities in midline NPCs.
Next, we wanted to test whether Yap/Taz activate transcription of Slit2. The Slit2 gene lies within a 2.5 Mb region (chr5:45860000-48370000) containing hundreds of evolutionarily conserved regions, which are potential enhancer elements (http://ecrbrowser.dcode.org/), making it very difficult to identify cis-regulatory elements and determine whether Yap/Taz directly act on them. We therefore took an indirect approach. We overexpressed YAP in HEK293T cells and then measured SLIT2 mRNA levels using qRT-PCR analyses. Compared with vector-transfected cells, YAP-transfected cells expressed higher levels of SLIT2, as well as CYR61, a known YAP target gene (Fig. 7M). In comparison, YAP overexpression had no effect on the level of WNT5A (Fig. 7M), another callosal guidance cue that is expressed at the midline (Keeble et al., 2006). That YAP overexpression increases SLIT2 transcript levels in HEK293T cells, a cell type unrelated to midline NPCs, strongly suggests that Yap/Taz regulate Slit2 transcription and argues against the possibility that Slit2 upregulation in Nf2-deletion or YAP-overexpression mutants was secondary to changes in the general properties of midline cells.
DISCUSSION
By dissecting the mechanism underlying the callosal agenesis caused by Nf2 deletion, we discovered a signaling cascade operating within NPCs to control callosal axon pathfinding. We found that the Nf2 tumor suppressor inhibits Yap/Taz transcriptional coactivators in NPCs, which in turn regulate midline guidepost development and expression of the guidance cue Slit2, hence influencing the midline environment that callosal axons travel through and affecting their navigation. Nf2 loss leads to aberrant Yap/Taz activation, which impairs guidepost formation and induces Slit2 expression, preventing callosal formation (Fig. 7N). We provide compelling genetic evidence for this signaling cascade by showing that Yap deletion and Slit2 heterozygosity both restore the corpus callosum in Nf2 mutants and that Yap gain-of-function selectively in midline NPCs prevents callosal formation. Nf2 and Yap/Taz are crucial components of the Hippo pathway, a conserved signaling pathway governing tissue growth and homeostasis (Yu and Guan, 2013), including mammalian brain growth (Lavado et al., 2013). In addition to offering new insights into the molecular regulation of midline development, our study demonstrates that perturbations in neural progenitor properties affect not only brain growth but also its wiring, highlighting that growth and wiring are intricately coordinated during brain development.
Nf2 loss causes guidepost defects and Slit2 upregulation. Slit2 heterozygosity and Yap deletion both re-establish the corpus callosum in Nf2 mutants. Interestingly, the former suppresses Slit2 upregulation but does not fully rescue guidepost development at E15.5 (before midline crossing), whereas the latter rescues guidepost development but does not mitigate Slit2 upregulation. These results suggest that both the guidepost defects and Slit2 upregulation contribute to, but neither is sufficient to cause, callosal agenesis in Nf2 mutants. Overexpressing YAP in midline NPCs, however, is sufficient to cause callosal agenesis and, importantly, both the guidepost defects and Slit2 upregulation, closely mimicking the effects of Nf2 loss. These results strongly suggest that aberrant Yap/Taz activation is the cause of callosal agenesis in Nf2 mutants, being responsible for both the guidepost defects and Slit2 upregulation, and that elevated Taz activity accounts for Slit2 upregulation in Nf2;Yap double mutants. This interpretation is consistent with our previous finding that the nuclear levels of both Yap and Taz proteins are increased in Nf2 mutant brains (Lavado et al., 2013). Our results do not preclude that defects other than guidepost malformation and Slit2 upregulation also contribute to callosal agenesis in Nf2-deletion and YAP-overexpression mutants. Furthermore, the apparent contradiction between the overexpression of Slit2 by using in utero electroporation resulting in abnormal axon tangles and the elevated levels of Slit2 in Nf2;Yap double mutants having little effect on callosal axon pathfinding is probably because in utero electroporation leads to higher levels of Slit2 than that found in Nf2;Yap double mutants.
Nf2 and Yap regulate the development of midline guideposts
Guidepost cells associated with the developing corpus callosum were first observed decades ago (Silver et al., 1982). Recent research using mouse mutants began to uncover the molecular mechanisms governing their development. A comparison of Nf2 mutant phenotypes to those described previously reveals interesting similarities and differences that help us to better understand the role of Nf2 during guidepost development.
Nf2 mutants closely resemble Gli3 hypomorphic and conditional knockout mutants, which have overextended GW processes, fewer Calretinin+ guidepost neurons and increased Slit2 levels (Amaniti et al., 2013; Magnani et al., 2014). However, it does not seem that Nf2 mutant phenotypes are caused by altered Gli3 function. First, Gli3 mutations lead to altered Fgf and Wnt signaling at the corticoseptal boundary and defective midline patterning (Amaniti et al., 2013; Magnani et al., 2014), whereas Fgf, Wnt signaling and midline patterning are unperturbed in Nf2 mutants. Second, unlike in Nf2 mutants, Slit2 deletion in Gli3 hypomorphic mutants does not restore the corpus callosum, consistent with Gli3 hypomorphic mutants having an earlier and broader midline patterning defect. However, the levels of Gli3FL and Gli3R and the ratio of Gli3FL to Gli3R were increased in Nf2 mutants, suggesting that Gli3 activator activities might be higher in Nf2 mutants. This change in Gli3 activity would be in the same direction as that found in Gli3 mutants because Gli3 is thought to mainly act in its repressor form in the developing cortex (Rallu et al., 2002). More molecular and genetic analyses are required to determine whether Nf2 and Gli3 functionally interact during callosal development.
We found that Nf2 promotes the transition of GW radial glia from those extending long radial processes to the pia to those having short processes. Whether this transition involves the former retracting their long processes or being replaced by radial glia born with short processes needs to be addressed through live-imaging experiments. In mice that lack Fgf receptor 1 in radial glia, GW radial glia fail to translocate from the ventricular zone to the IG. Consequently, in these mutants, GW processes remain attached to the pia at P0 and IG astrocytes are lacking (Smith et al., 2006). Fgf signaling-mediated radial glia translocation is unlikely to be involved in the GW phenotype of Nf2 mutants because IG astrocytes are present and Fgf signaling is unperturbed in Nf2 mutants.
Transcription factors Nfia and Nfib (Nfia,b) are also required for GW radial glia maturation (Shu et al., 2003a; Steele-Perkins et al., 2005). However, the GW defects of Nfia−/− and Nfib−/− mice are opposite to those of Nf2 mutants – GFAP+ processes are fewer and shorter in Nfia−/− mice than in wild-type animals and are mostly absent in Nfib−/− mice. We postulate that Nf2 and Nfia,b. act at different levels during GW radial glia development: Nf2, by inhibiting Yap, promotes the transition to later cellular states, whereas Nfia,b promote astroglial structural characteristics. Supporting this idea, it has been shown that Yap promotes stem cell and progenitor cell self-renewal in many systems (Barry and Camargo, 2013), whereas Nfi factors directly activate the expression of astroglial genes (Namihira et al., 2009).
We show that Nf2 promotes the differentiation of Calretinin+ guidepost neurons. It is unclear which progenitor population gives rise to these neurons. The juxtaposition of these neurons with GW radial glia and the phenotypic correlation between the reduction in Calretinin+ neuron production and delayed developmental transition of GW radial glia in Nf2 mutants suggest that at least some Calretinin+ guidepost neurons are derived from GW radial glia. Lineage-tracing experiments are required to determine whether this is the case. Intriguingly, these guidepost neurons share several features with Cajal–Retzius cells: both express Calretinin but are glutamatergic, whereas most if not all other cortical Calretinin+ neurons are GABAergic; both exist transiently during development and both guide axonal navigation (Borrell et al., 2007; Del Rio et al., 1997). A source of Cajal–Retzius cells is the cortical hem, residing along the caudal telencephalic midline immediately posterior to the GW (Grove et al., 1998). It is plausible that these two anatomically and ontogenetically associated structures give rise to functionally similar progenies.
Deleting Nf2 using Emx1-Cre does not affect anterior commissure formation, although Emx1-Cre is active in the NPCs that give rise to neurons projecting through the anterior commissure (Falk et al., 2005; Gorski et al., 2002). Interestingly, formation of the anterior commissure is abolished when Nf2 is removed throughout the nervous system by Nestin-Cre (supplementary material Fig. S1D). These contrasting results suggest that Nf2 is likely to be required for the development of the ventral midline environment where the anterior commissure forms. GFAP+ glia are found surrounding the developing anterior commissure (Lent et al., 2005; Pires-Neto et al., 1998; Silver et al., 1993), although their origin and function have not been established. Future studies are required to determine whether Nf2-Yap signaling regulates the development of these putative anterior commissure guidepost cells and those involved in the formation of other axon tracts (Chedotal and Richards, 2010).
Nf2 and Yap/Taz regulate Slit2 expression
Remarkable progress has been made in identifying axon guidance molecules and elucidating their functions. Although it is self-evident that these molecules must be expressed at the correct place and time for proper wiring, how their expression is regulated is poorly understood. Here, we show that Nf2 suppresses Yap/Taz to prevent aberrant Slit2 expression. Our finding is clearly just a small part of Slit2 transcriptional control. The Slit2 gene exhibits complex expression patterns during brain development (Borrell et al., 2012; Unni et al., 2012). The large and conserved non-coding region of the gene suggests multifaceted transcriptional regulation. A recent study has elegantly demonstrated that a small difference in Slit2 expression patterns in mammals and birds leads to different positioning of guidepost neurons and ultimately divergent trajectories of thalamic axons in these two species (Bielle et al., 2011). Future studies on the transcriptional regulation of guidance molecules will provide deeper understanding of how the nervous system is wired during organism development and species evolution.
MATERIALS AND METHODS
Animals
Animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of St. Jude Children's Research Hospital (SJCRH). Emx1IRES-Cre (stock: 005628), Nestin-Cre (003771), Rosa-CAG-LSL-tdTomato (007909), Rosa-CAG-LSL-ZsGreen (007906) and Axin2-rtTA (016997) lines were from the Jackson Laboratory. Nf2F/F, YapF/F (Xin et al., 2011) and TetO-YAP1 lines were provided by Marco Giovannini (University of California Los Angeles, CA, USA), Eric Olson (UT Southwestern Medical Center, Dallas, TX, USA) and Fernando Camargo (Boston Children's Hospital, MA, USA), respectively. Satb2-Cre (032908-UCD) and Slit2+/− (Plump et al., 2002) lines were obtained from the Mutant Mouse Regional Resource Center (MMRRC) at University of California Davis (Davis, CA, USA) and University of Missouri (Columbia, MO, USA), respectively. All experimental animals were in mixed background. Littermates were used for comparison studies. Doxycycline was administered in drinking water (E9.5-14.5, 100 mg/l; E14.5-18.5, 200 mg/l). For BrdU labeling, timed pregnant mice were intraperitoneally injected with BrdU at 100 mg/kg.
Histology, immunostaining and in situ hybridization
Luxol Blue and Cresyl Violet staining was performed on 8 µm thick paraffin sections using the Kluver–Barrera method (EMS no. 26681). Immunostaining was performed on 10 to 14 µm thick cryosections. Details are provided in supplementary Materials and Methods. Primary antibodies are listed in supplementary material Table S1. Fluorescent secondary antibodies were from Jackson ImmunoResearch. In situ hybridization was performed as described previously (Schaeren-Wiemers and Gerfin-Moser, 1993) using the following probes: Slit2 and Robo1 (Brose et al., 1999), Netrin1 (Serafini et al., 1996), Sema3c (Bagnard et al., 1998), Draxin (Islam et al., 2009), Fgf8 (Crossley and Martin, 1995), Sprouty1 (Minowada et al., 1999), Wnt8b (Liu et al., 2010) and Axin2 (Andoniadou et al., 2007) (supplementary material Table S2). Low-magnification images were acquired with a Zeiss AxioImager M2 microscope. The remaining images were acquired with Zeiss LSM510 or LSM780 confocal microscopes. For measuring the fluorescence intensities of Yap/Taz immunostaining signals, single-plane confocal images with signals below saturation levels were captured using the same settings for control and dTG samples. Quantification was performed on sections of comparable levels. The values shown are mean±s.e.m. per section of the indicated number of animals, and the value of each animal is the average of six sections. Statistical significance was evaluated by using two-tailed unpaired t-tests.
Transplantation and slice culture
Transplantation and slice culture procedures were performed as described previously (Niquille et al., 2009). Briefly, brains at E16.5 were dissected in cold Hank's balanced salt solution (HBSS) and embedded in 3% low-melting agarose (Invitrogen). Using a Vibratome, 300 μm thick coronal sections were obtained. After transplantation, slices were cultured for 72 h on Nucleopore Track-Etch Membranes (1-μm pore size, Whatman) in a 3:1 mixture of basal medium Eagle (BME) and HBSS supplemented with glucose, glutamine, 5% horse serum and penicillin-streptomycin (Invitrogen). Transplants showing axon growth toward the midline were included in the analysis.
In utero electroporation
In utero electroporation was performed as described previously (Saito, 2006). Briefly, the plasmid solution was microinjected into forebrain ventricles of embryos at E13.5 in utero, and square-wave electric pulses (40 V, five pulses with durations of 50 ms/pulse and at 1-second intervals) were delivered with Tweezertrodes connected to an ECM 830 electroporator (Harvard Apparatus). Embryos were returned to the dam and harvested 4 days later.
Western blot, qRT-PCR, cell culture and transfection
For western blot analyses, tissues were homogenized in 20 mM HEPES (pH 7.4), 150 mM NaCl, 5% glycerol, 2% SDS supplemented with Halt protease and phosphatase inhibitors (Thermo Scientific). Quantitative western blot was performed using IRDye 600LT- and 800CW-conjugated secondary antibodies, which were then detected with the ODYSSEY infrared imaging system (LI-COR). Primary antibodies are listed in supplementary material Table S1. HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (Invitrogen), transfected with Lipofectamine 2000 (Invitrogen) and then harvested 48 h later. RNA was isolated using Trizol (Invitrogen). cDNA was prepared with SuperScript III reverse transcriptase (Life Technologies). qRT-PCR reactions were performed with Fast SYBR Master Mix (Life Technologies) using the primers listed in supplementary material Table S3. Values are mean±s.e.m. Statistical significance was evaluated by using two-tailed unpaired t-tests.
Supplementary Material
Acknowledgements
We thank Marco Giovannini, Eric Olson and Fernando Camargo for sharing mouse lines; Marc Tessier-Lavigne, Jean Hébert, Avraham Yaron, Richard Lang, Gail Martin and Kun-Liang Guan for plasmids; Guillermo Oliver for an antibody against Six3; MMRRC, GENSAT and INSERM (France) for mouse lines; Addgene for plasmids; SJCRH Imaging Center for assistance with image acquisition; Jim Morgan, David Solecki, Young-Goo Han and Ge Bai for suggestions on the manuscript and Cherise Guess for editorial assistance.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
X.C. conceived the project. X.C., A.L. and M.W. designed the experiments and analyzed the results. A.L. performed most of the experiments. M.W. performed initial studies. J.P. performed Luxol Blue staining, western blot, qRT-PCR and HEK293T cell experiments. X.C. collected tissues for western blot and qRT-PCR assays and performed in utero electroporation. X.C. wrote and A.L. edited the manuscript.
Funding
This work was supported by American Lebanese Syrian Associated Charities; Basil O'Connor Award [5-FY10-488] from March of Dimes; the Whitehall Foundation [2012-05-106]; and the National Institutes of Health [R01NS086938]. The Imaging Center is supported by SJCRH and the National Cancer Institute [grant P30 CA021765-34]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.111260/-/DC1
References
- Alcamo E. A., Chirivella L., Dautzenberg M., Dobreva G., Fariñas I., Grosschedl R. and McConnell S. K. (2008). Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 57, 364-377 10.1016/j.neuron.2007.12.012 [DOI] [PubMed] [Google Scholar]
- Amaniti E.-M., Hasenpusch-Theil K., Li Z., Magnani D., Kessaris N., Mason J. O. and Theil T. (2013). Gli3 is required in Emx1(+) progenitors for the development of the corpus callosum. Dev. Biol. 376, 113-124 10.1016/j.ydbio.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Andoniadou C. L., Signore M., Sajedi E., Gaston-Massuet C., Kelberman D., Burns A. J., Itasaki N., Dattani M. and Martinez-Barbera J. P. (2007). Lack of the murine homeobox gene Hesx1 leads to a posterior transformation of the anterior forebrain. Development 134, 1499-1508 10.1242/dev.02829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews W., Liapi A., Plachez C., Camurri L., Zhang J., Mori S., Murakami F., Parnavelas J. G., Sundaresan V. and Richards L. J. (2006). Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development 133, 2243-2252 10.1242/dev.02379 [DOI] [PubMed] [Google Scholar]
- Bagnard D., Lohrum M., Uziel D., Puschel A. W. and Bolz J. (1998). Semaphorins act as attractive and repulsive guidance signals during the development of cortical projections. Development 125, 5043-5053. [DOI] [PubMed] [Google Scholar]
- Bagri A., Marín O., Plump A. S., Mak J., Pleasure S. J., Rubenstein J. L. R. and Tessier-Lavigne M. (2002). Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron 33, 233-248 10.1016/S0896-6273(02)00561-5 [DOI] [PubMed] [Google Scholar]
- Barry E. R. and Camargo F. D. (2013). The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr. Opin. Cell Biol. 9, 6. [DOI] [PubMed] [Google Scholar]
- Benadiba C., Magnani D., Niquille M., Morlé L., Valloton D., Nawabi H., Ait-Lounis A., Otsmane B., Reith W., Theil T. et al. (2012). The ciliogenic transcription factor RFX3 regulates early midline distribution of guidepost neurons required for corpus callosum development. PLoS Genet. 8, e1002606 10.1371/journal.pgen.1002606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielle F., Marcos-Mondejar P., Keita M., Mailhes C., Verney C., Nguyen Ba-Charvet K., Tessier-Lavigne M., Lopez-Bendito G. and Garel S. (2011). Slit2 activity in the migration of guidepost neurons shapes thalamic projections during development and evolution. Neuron 69, 1085-1098 10.1016/j.neuron.2011.02.026 [DOI] [PubMed] [Google Scholar]
- Borrell V., Pujadas L., Simó S., Durà D., Solé M., Cooper J. A., Del Río J. A. and Soriano E. (2007). Reelin and mDab1 regulate the development of hippocampal connections. Mol. Cell. Neurosci. 36, 158-173 10.1016/j.mcn.2007.06.006 [DOI] [PubMed] [Google Scholar]
- Borrell V., Cárdenas A., Ciceri G., Galcerán J., Flames N., Pla R., Nóbrega-Pereira S., García-Frigola C., Peregrín S., Zhao Z. et al. (2012). Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron 76, 338-352 10.1016/j.neuron.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O., de Juan Romero C., Cheung A., Kwan K. Y., Schwark M., Gyorgy A., Vogel T., Akopov S., Mitkovski M., Agoston D. et al. (2008). Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57, 378-392 10.1016/j.neuron.2007.12.028 [DOI] [PubMed] [Google Scholar]
- Brose K., Bland K. S., Wang K. H., Arnott D., Henzel W., Goodman C. S., Tessier-Lavigne M. and Kidd T. (1999). Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 96, 795-806 10.1016/S0092-8674(00)80590-5 [DOI] [PubMed] [Google Scholar]
- Camargo F. D., Gokhale S., Johnnidis J. B., Fu D., Bell G. W., Jaenisch R. and Brummelkamp T. R. (2007). YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054-2060 10.1016/j.cub.2007.11.016 [DOI] [PubMed] [Google Scholar]
- Chedotal A. and Richards L. J. (2010). Wiring the brain: the biology of neuronal guidance. Cold Spring Harb. Perspect. Biol. 2, a001917 10.1101/cshperspect.a001917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinn G. A., Hirokawa K. E., Chuang T. M., Urbina C., Patel F., Fong J., Funatsu N. and Monuki E. S. (2014). Agenesis of the corpus callosum due to defective glial wedge formation in Lhx2 mutant mice. Cereb. Cortex (in press). 10.1093/cercor/bhu067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg J. M., Conway C. D., Howe K. M., Price D. J., Mason J. O., Turnbull J. E., Basson M. A. and Pratt T. (2014). Heparan sulfotransferases Hs6st1 and Hs2st keep Erk in check for mouse corpus callosum development. J. Neurosci. 34, 2389-2401 10.1523/JNEUROSCI.3157-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley P. H. and Martin G. R. (1995). The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121, 439-451. [DOI] [PubMed] [Google Scholar]
- Del Río J. A., Heimrich B., Borrell V., Förster E., Drakew A., Alcántara S., Nakajima K., Miyata T., Ogawa M., Mikoshiba K. et al. (1997). A role for Cajal-Retzius cells and reelin in the development of hippocampal connections. Nature 385, 70-74 10.1038/385070a0 [DOI] [PubMed] [Google Scholar]
- Donahoo A.-L. S. and Richards L. J. (2009). Understanding the mechanisms of callosal development through the use of transgenic mouse models. Semin. Pediatr. Neurol. 16, 127-142 10.1016/j.spen.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S. et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474, 179-183 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Falk J., Bechara A., Fiore R., Nawabi H., Zhou H., Hoyo-Becerra C., Bozon M., Rougon G., Grumet M., Püschel A. W. et al. (2005). Dual functional activity of semaphorin 3B is required for positioning the anterior commissure. Neuron 48, 63-75 10.1016/j.neuron.2005.10.024 [DOI] [PubMed] [Google Scholar]
- Fame R. M., MacDonald J. L. and Macklis J. D. (2011). Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 34, 41-50 10.1016/j.tins.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazeli A., Dickinson S. L., Hermiston M. L., Tighe R. V., Steen R. G., Small C. G., Stoeckli E. T., Keino-Masu K., Masu M., Rayburn H. et al. (1997). Phenotype of mice lacking functional Deleted in colorectal cancer (Dcc) gene. Nature 386, 796-804 10.1038/386796a0 [DOI] [PubMed] [Google Scholar]
- Giovannini M., Robanus-Maandag E., van der Valk M., Niwa-Kawakita M., Abramowski V., Goutebroze L., Woodruff J. M., Berns A. and Thomas G. (2000). Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes Dev. 14, 1617-1630. [PMC free article] [PubMed] [Google Scholar]
- Gorski J. A., Talley T., Qiu M., Puelles L., Rubenstein J. L. and Jones K. R. (2002). Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove E. A., Tole S., Limon J., Yip L. and Ragsdale C. W. (1998). The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development 125, 2315-2325. [DOI] [PubMed] [Google Scholar]
- Houshmandi S. S., Emnett R. J., Giovannini M. and Gutmann D. H. (2009). The neurofibromatosis 2 protein, merlin, regulates glial cell growth in an ErbB2- and Src-dependent manner. Mol. Cell. Biol. 29, 1472-1486 10.1128/MCB.01392-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S. M., Shinmyo Y., Okafuji T., Su Y., Naser I. B., Ahmed G., Zhang S., Chen S., Ohta K., Kiyonari H. et al. (2009). Draxin, a repulsive guidance protein for spinal cord and forebrain commissures. Science 323, 388-393 10.1126/science.1165187 [DOI] [PubMed] [Google Scholar]
- Keeble T. R., Halford M. M., Seaman C., Kee N., Macheda M., Anderson R. B., Stacker S. A. and Cooper H. M. (2006). The Wnt receptor Ryk is required for Wnt5a-mediated axon guidance on the contralateral side of the corpus callosum. J. Neurosci. 26, 5840-5848 10.1523/JNEUROSCI.1175-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester S. E. and O'Leary D. D. (1994). Axons of early generated neurons in cingulate cortex pioneer the corpus callosum. J. Neurosci. 14, 6608-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A. and Alvarez-Buylla A. (2009). The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149-184 10.1146/annurev.neuro.051508.135600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavado A., He Y., Pare J., Neale G., Olson E. N., Giovannini M. and Cao X. (2013). Tumor suppressor Nf2 limits expansion of the neural progenitor pool by inhibiting Yap/Taz transcriptional coactivators. Development 140, 3323-3334 10.1242/dev.096537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lent R., Uziel D., Baudrimont M. and Fallet C. (2005). Cellular and molecular tunnels surrounding the forebrain commissures of human fetuses. J. Comp. Neurol. 483, 375-382 10.1002/cne.20427 [DOI] [PubMed] [Google Scholar]
- Li W., Cooper J., Karajannis M. A. and Giancotti F. G. (2012). Merlin: a tumour suppressor with functions at the cell cortex and in the nucleus. EMBO Rep. 13, 204-215 10.1038/embor.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Lagutin O., Swindell E., Jamrich M. and Oliver G. (2010). Neuroretina specification in mouse embryos requires Six3-mediated suppression of Wnt8b in the anterior neural plate. J. Clin. Invest. 120, 3568-3577 10.1172/JCI43219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R. et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133-140 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani D., Hasenpusch-Theil K., Benadiba C., Yu T., Basson M. A., Price D. J., Lebrand C. and Theil T. (2014). Gli3 controls corpus callosum formation by positioning midline guideposts during telencephalic patterning. Cereb. Cortex 24, 186-198 10.1093/cercor/bhs303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin M. E., Kruger G. M., Slocum K. L., Crowley D., Michaud N. A., Huang J., Magendantz M. and Jacks T. (2007). The Nf2 tumor suppressor regulates cell-cell adhesion during tissue fusion. Proc. Natl. Acad. Sci. USA 104, 3261-3266 10.1073/pnas.0700044104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski R. C., Chi N. C., Li J., Brown C., Lu M. M. and Epstein J. A. (2004). Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development 131, 829-837 10.1242/dev.00975 [DOI] [PubMed] [Google Scholar]
- Minowada G., Jarvis L. A., Chi C. L., Neubuser A., Sun X., Hacohen N., Krasnow M. A. and Martin G. R. (1999). Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development 126, 4465-4475. [DOI] [PubMed] [Google Scholar]
- Mohseni M., Sun J., Lau A., Curtis S., Goldsmith J., Fox V. L., Wei C., Frazier M., Samson O., Wong K.-K. et al. (2014). A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat. Cell Biol. 16, 108-117 10.1038/ncb2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldrich R. X., Gobius I., Pollak T., Zhang J., Ren T., Brown L., Mori S., De Juan Romero C., Britanova O., Tarabykin V. et al. (2010). Molecular regulation of the developing commissural plate. J. Comp. Neurol. 518, 3645-3661 10.1002/cne.22445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namihira M., Kohyama J., Semi K., Sanosaka T., Deneen B., Taga T. and Nakashima K. (2009). Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev. Cell 16, 245-255 10.1016/j.devcel.2008.12.014 [DOI] [PubMed] [Google Scholar]
- Niquille M., Garel S., Mann F., Hornung J.-P., Otsmane B., Chevalley S., Parras C., Guillemot F., Gaspar P., Yanagawa Y. et al. (2009). Transient neuronal populations are required to guide callosal axons: a role for semaphorin 3C. PLoS Biol. 7, e1000230 10.1371/journal.pbio.1000230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikimi M., Oishi K. and Nakajima K. (2013). Axon guidance mechanisms for establishment of callosal connections. Neural Plast. 2013, 149060 10.1155/2013/149060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul L. K., Brown W. S., Adolphs R., Tyszka J. M., Richards L. J., Mukherjee P. and Sherr E. H. (2007). Agenesis of the corpus callosum: genetic, developmental and functional aspects of connectivity. Nat. Rev. Neurosci. 8, 287-299 10.1038/nrn2107 [DOI] [PubMed] [Google Scholar]
- Piper M., Moldrich R. X., Lindwall C., Little E., Barry G., Mason S., Sunn N., Kurniawan N. D., Gronostajski R. M. and Richards L. J. (2009a). Multiple non-cell-autonomous defects underlie neocortical callosal dysgenesis in Nfib-deficient mice. Neural Dev. 4, 43 10.1186/1749-8104-4-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M., Plachez C., Zalucki O., Fothergill T., Goudreau G., Erzurumlu R., Gu C. and Richards L. J. (2009b). Neuropilin 1-Sema signaling regulates crossing of cingulate pioneering axons during development of the corpus callosum. Cereb. Cortex 19Suppl. 1, i11-i21 10.1093/cercor/bhp027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires-Neto M. A., Braga-De-Souza S. and Lent R. (1998). Molecular tunnels and boundaries for growing axons in the anterior commissure of hamster embryos. J. Comp. Neurol. 399, 176-188 [DOI] [PubMed] [Google Scholar]
- Plump A. S., Erskine L., Sabatier C., Brose K., Epstein C. J., Goodman C. S., Mason C. A. and Tessier-Lavigne M. (2002). Slit1 and Slit2 cooperate to prevent premature midline crossing of retinal axons in the mouse visual system. Neuron 33, 219-232 10.1016/S0896-6273(01)00586-4 [DOI] [PubMed] [Google Scholar]
- Rallu M., Machold R., Gaiano N., Corbin J. G., McMahon A. P. and Fishell G. (2002). Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and Hedgehog signaling. Development 129, 4963-4974. [DOI] [PubMed] [Google Scholar]
- Rash B. G. and Richards L. J. (2001). A role for cingulate pioneering axons in the development of the corpus callosum. J. Comp. Neurol. 434, 147-157 10.1002/cne.1170 [DOI] [PubMed] [Google Scholar]
- Saito T. (2006). In vivo electroporation in the embryonic mouse central nervous system. Nat. Protoc. 1, 1552-1558 10.1038/nprot.2006.276 [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N. and Gerfin-Moser A. (1993). A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry 100, 431-440 10.1007/BF00267823 [DOI] [PubMed] [Google Scholar]
- Schulz A., Baader S. L., Niwa-Kawakita M., Jung M. J., Bauer R., Garcia C., Zoch A., Schacke S., Hagel C., Mautner V.-F. et al. (2013). Merlin isoform 2 in neurofibromatosis type 2-associated polyneuropathy. Nat. Neurosci. 16, 426-433 10.1038/nn.3348 [DOI] [PubMed] [Google Scholar]
- Serafini T., Colamarino S. A., Leonardo E. D., Wang H., Beddington R., Skarnes W. C. and Tessier-Lavigne M. (1996). Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell 87, 1001-1014 10.1016/S0092-8674(00)81795-X [DOI] [PubMed] [Google Scholar]
- Shu T. and Richards L. J. (2001). Cortical axon guidance by the glial wedge during the development of the corpus callosum. J. Neurosci. 21, 2749-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T., Butz K. G., Plachez C., Gronostajski R. M. and Richards L. J. (2003a). Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J. Neurosci. 23, 203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T., Puche A. C. and Richards L. J. (2003b). Development of midline glial populations at the corticoseptal boundary. J. Neurobiol. 57, 81-94 10.1002/neu.10252 [DOI] [PubMed] [Google Scholar]
- Silver J., Lorenz S. E., Wahlsten D. and Coughlin J. (1982). Axonal guidance during development of the great cerebral commissures: descriptive and experimental studies, in vivo, on the role of preformed glial pathways. J. Comp. Neurol. 210, 10-29 10.1002/cne.902100103 [DOI] [PubMed] [Google Scholar]
- Silver J., Edwards M. A. and Levitt P. (1993). Immunocytochemical demonstration of early appearing astroglial structures that form boundaries and pathways along axon tracts in the fetal brain. J. Comp. Neurol. 328, 415-436 10.1002/cne.903280308 [DOI] [PubMed] [Google Scholar]
- Smith K. M., Ohkubo Y., Maragnoli M. E., Rasin M.-R., Schwartz M. L., Sestan N. and Vaccarino F. M. (2006). Midline radial glia translocation and corpus callosum formation require FGF signaling. Nat. Neurosci. 9, 787-797 10.1038/nn1705 [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G., Plachez C., Butz K. G., Yang G., Bachurski C. J., Kinsman S. L., Litwack E. D., Richards L. J. and Gronostajski R. M. (2005). The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol. Cell. Biol. 25, 685-698 10.1128/MCB.25.2.685-698.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P. C., Bock R., Klein R. and Schutz G. (1999). Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99-103 10.1038/12703 [DOI] [PubMed] [Google Scholar]
- Unni D. K., Piper M., Moldrich R. X., Gobius I., Liu S., Fothergill T., Donahoo A.-L. S., Baisden J. M., Cooper H. M. and Richards L. J. (2012). Multiple Slits regulate the development of midline glial populations and the corpus callosum. Dev. Biol. 365, 36-49 10.1016/j.ydbio.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Xin M., Kim Y., Sutherland L. B., Qi X., McAnally J., Schwartz R. J., Richardson J. A., Bassel-Duby R. and Olson E. N. (2011). Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci. Signal. 4, ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.-X. and Guan K.-L. (2013). The Hippo pathway: regulators and regulations. Genes Dev. 27, 355-371 10.1101/gad.210773.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.-M. I., Liu B., Costantini F. and Hsu W. (2007). Impaired neural development caused by inducible expression of Axin in transgenic mice. Mech. Dev. 124, 146-156 10.1016/j.mod.2006.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C.-Y., Chinnaiyan A. M. et al. (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962-1971 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.