Abstract
Various combinations of cardiogenic transcription factors, including Gata4 (G), Hand2 (H), Mef2c (M) and Tbx5 (T), can reprogram fibroblasts into induced cardiac-like myocytes (iCLMs) in vitro and in vivo. Given that optimal cardiac function relies on distinct yet functionally interconnected atrial, ventricular and pacemaker (PM) cardiomyocytes (CMs), it remains to be seen which subtypes are generated by direct reprogramming and whether this process can be harnessed to produce a specific CM of interest. Here, we employ a PM-specific Hcn4-GFP reporter mouse and a spectrum of CM subtype-specific markers to investigate the range of cellular phenotypes generated by reprogramming of primary fibroblasts. Unexpectedly, we find that a combination of four transcription factors (4F) optimized for Hcn4-GFP expression does not generate beating PM cells due to inadequate sarcomeric protein expression and organization. However, applying strict single-cell criteria to GHMT-reprogrammed cells, we observe induction of diverse cellular phenotypes, including those resembling immature forms of all three major cardiac subtypes (i.e. atrial, ventricular and pacemaker). In addition, we demonstrate that cells induced by GHMT are directly reprogrammed and do not arise from an Nxk2.5+ progenitor cell intermediate. Taken together, our results suggest a remarkable degree of plasticity inherent to GHMT reprogramming and provide a starting point for optimization of CM subtype-specific reprogramming protocols.
Keywords: Direct reprogramming, Pacemaker, Cardiomyocyte, Mouse
INTRODUCTION
The canonical view of development depicts lineage specification as a unidirectional process, beginning with pluripotent progenitor cells that progressively differentiate into specialized cell types with restricted potential. Recent studies, however, have directly refuted a simple linear model of lineage specification. Yamanaka and colleagues demonstrated that overexpression of four transcription factors (Oct4/Sox2/Klf4/Myc; OSKM) was sufficient to unlock the restricted potential of committed cells by directly reprogramming differentiated fibroblasts into induced pluripotent stem (iPS) cells (Takahashi and Yamanaka, 2006).
Building on this reprogramming paradigm, Ieda and colleagues showed that the combination of Gata4, Mef2c and Tbx5 (GMT) can generate cardiomyocyte-like cells, albeit with low efficiency (Ieda et al., 2010). These cells display many of the key features of bona fide cardiomyocytes (CMs), including expression of sarcomeric genes, spontaneous beating activity, calcium oscillations and characteristic action potentials. Interestingly, follow-up studies have revealed that many different combinations of transcription factors, including GHMT (H; Hand2), can function to reprogram fibroblasts into CM-like cells in vitro (Addis et al., 2013; Christoforou et al., 2013; Fu et al., 2013; Nam et al., 2013; Protze et al., 2012; Song et al., 2012; Wada et al., 2013). Based on these results, several groups subsequently demonstrated direct reprogramming of activated fibroblasts toward a cardiac phenotype in vivo (Inagawa et al., 2012; Jayawardena et al., 2012; Mathison et al., 2012; Qian et al., 2012; Song et al., 2012). Taken together, these studies provide a basis for converting fibroblasts into CMs in vivo to treat a variety of cardiovascular disorders.
In addition to potential therapeutic applications, cardiac reprogramming represents a platform to dissect the molecular details of cardiomyogenesis. Although direct cardiac reprogramming is intriguing as a model system, several important hurdles remain to be addressed. Currently, the estimated efficiency of generating reprogrammed CMs is <1% based on spontaneous beating activity as a measure of functionality (Ieda et al., 2010). Given that reprogrammed CMs rapidly exit the cell cycle and thus cannot be expanded in culture, the generation of adequate numbers of cells will be crucial for both investigational and therapeutic applications. Furthermore, as most successful CM reprogramming protocols generate immature cell types, this system is currently more suitable for studies aimed at understanding lineage specification rather than the acquisition of mature CM-like properties. Specifically, the question of whether CM reprogramming can be modulated to generate specific cardiac cell types (i.e. atrial, ventricular and pacemaker) remains to be explored. To address this issue, however, we must possess expedient and robust methods to identify and quantify specific cardiac cell types.
Here, we utilize a pacemaker (PM)-specific reporter mouse to investigate the range of CMs generated by direct reprogramming of fibroblasts. Using primary fibroblasts derived from this transgenic line, we identified a four-transcription factor combination (4F) that robustly activates Hcn4-GFP expression. However, 4F-mediated reprogramming does not generate cells with spontaneous beating activity, a cardinal feature of PM cells. By analyzing endogenous CMs, we uncover that sarcomeric protein expression is a key property of PM cells, and we identify a panel of CM subtype-specific markers that reliably distinguish individual endogenous cell types – atrial, ventricular and PM. Applying these immunostaining criteria to GHMT-reprogrammed fibroblasts, we find that immature forms of each CM subtype are induced. Based on our observation that spontaneously beating cells possess well-organized sarcomere structures, we re-calculate the reprogramming efficiency of GHMT and quantitate individual cardiac cell types generated during this process. Finally, we demonstrate that individual reprogrammed beating cells display unique action potentials that correlate retrospectively with subtype-specific immunostaining characteristics. Taken together, our results suggest an unanticipated degree of plasticity inherent to GHMT reprogramming and provide a method for assessing directed efforts to generate individual cardiac subtypes selectively.
RESULTS
Selected reprogramming factors activate Hcn4 reporter expression but fail to generate PM cells
Based on anatomical positions, gene expression patterns and unique electrical properties, there are three major types of CMs in the heart: atrial, ventricular and PM. PM CMs can be found in the sinoatrial node (SAN), which is located at the junction of the superior vena cava and right atrium (Munshi, 2012). PM CMs generate spontaneous action potentials that sequentially activate atrial and ventricular myocardium to optimize the timing of cardiac contraction. Thus, highly coordinated activity of all three CM subtypes is required for effective circulation. Previous studies have clearly demonstrated that the core cardiac transcription factors can reprogram fibroblasts into CM-like cells. It is unclear, however, which cardiac subtype is preferentially induced by current protocols or whether a specific cardiac subtype can be directed by a direct reprogramming approach. Thus, we aimed to generate induced PM (iPM) myocytes by forced expression of selected lineage-specifying transcription factors in primary fibroblasts rather than mature atrial or ventricular myocytes (Bakker et al., 2012; Kapoor et al., 2013). As a first step toward this goal, we sought to develop a reliable reporter system that faithfully marks PM cells, thereby allowing us to perform initial large-scale screening experiments. In vivo, the SAN and AVN function as the dominant sites of PM activity, due in large part to expression of the Hcn4 potassium channel. Given the importance of Hcn4 for PM function, we therefore obtained Hcn4-GFP reporter mice created for the GENSAT Project (Gong et al., 2003) and confirmed that GFP is robustly expressed in the SAN of neonatal and adult mice (Fig. 1A). In order to characterize presumptive Hcn4+ SAN cells, we isolated CMs from the right atrium of Hcn4-GFP reporter mice at P0-P1. Importantly, we found by immunostaining that Hcn4-GFP co-localized with endogenous Hcn4 (Fig. 1B), thus confirming that Hcn4-GFP faithfully recapitulates endogenous Hcn4 expression.
Fig. 1.
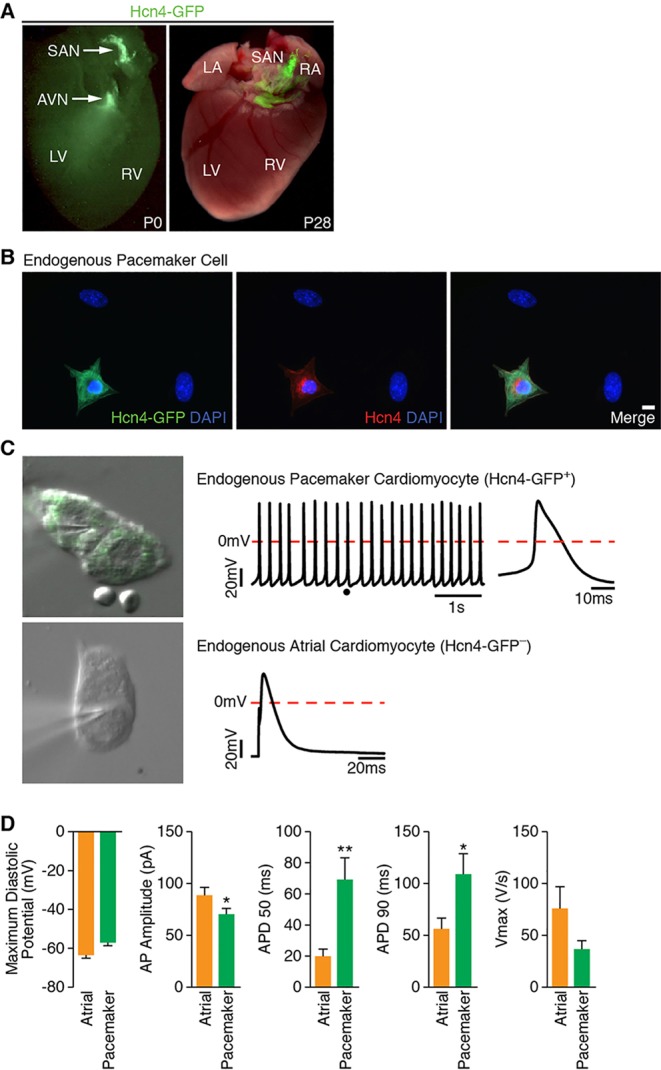
Hcn4-GFP reporter identifies bona fide PM cells. (A) Fluorescent image of Hcn4-GFP reporter mouse heart at P0 and P28, demonstrating restricted expression of Hcn4-GFP within the SAN. (B) Immunofluorescence staining of Hcn4-GFP+ atrial myocytes isolated from Hcn4-GFP reporter mouse for Hcn4-GFP reporter (green) and endogenous Hcn4 (red). Scale bar: 20 µm. (C) A representative image, action potential tracing and enlarged view of single action potential of whole-cell patch-clamped Hcn4-GFP+ atrial myocytes (top). The dot in the action potential tracing indicates the location in the tracing where a single action potential (top, right) is zoomed. A representative image and a single action potential of whole-cell patch-clamped Hcn4-GFP− atrial myocytes are shown at the bottom. (D) Comparison of Hcn4-GFP+ (n=14) and Hcn4-GFP− (n=10) atrial myocytes by multiple action potential parameters, including maximum diastolic potential, AP (action potential) amplitude, AP durations (APD50 and APD90) and Vmax. AVN, atrioventricular node; LV, left ventricle; RV, right ventricle; SAN, sinoatrial node. *P<0.05; **P<0.01.
To determine the electrical identity of individual cell types, we performed patch clamping on Hcn4-GFP+ and Hcn4-GFP− atrial CMs isolated from Hcn4-GFP reporter mice. All Hcn4-GFP+ CMs exhibited unstimulated action potentials with typical features of PM cells, including spontaneous diastolic depolarization (Fig. 1C, top). By contrast, most Hcn4-GFP− CMs required an electrical stimulus to elicit an action potential, with characteristics consistent with working atrial CMs (Fig. 1C, bottom). Furthermore, quantitative analysis of individual action potential parameters demonstrated that Hcn4-GFP+ cells conform to PM CM characteristics, whereas Hcn4-GFP− cells resemble atrial CMs (Fig. 1D). Taken together, these results clearly show that Hcn4-GFP marks genuine PM CMs that can be readily distinguished from surrounding Hcn4-GFP− atrial CMs.
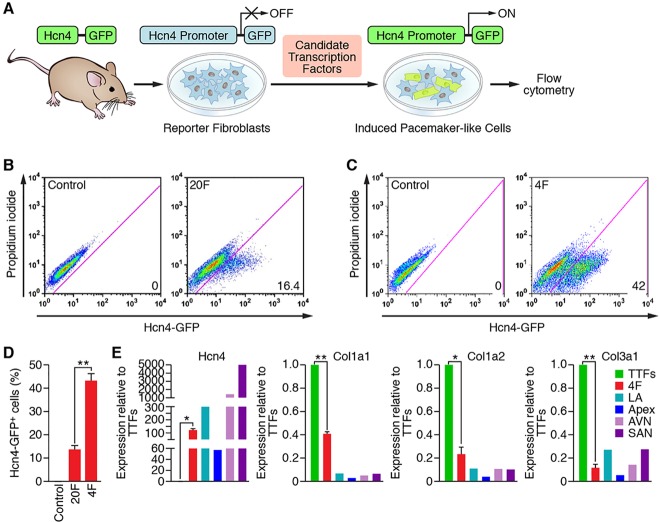
Given that Hcn4-GFP faithfully reports Hcn4 expression in bona fide PM cells in vivo, we sought to define the optimal combination of transcription factors that would robustly activate the Hcn4-GFP reporter in transgenic fibroblasts as a first step towards reprogramming PM CMs (Fig. 2A). Thus, we constructed retroviral expression vectors for 20 candidate transcription factors based on previous literature (Munshi, 2012) and our own observations (data not shown). When all 20 factors (20F) were introduced into Hcn4-GFP fibroblasts, we found that ∼15% were GFP+, whereas uninfected or empty vector-infected controls did not give rise to GFP+ cells (Fig. 2B,D). Subsequently, we performed an extensive single-factor subtraction screening to exclude inhibitory factors for PM reprogramming (data not shown). Through such reiterative screening experiments, we found an optimal combination of four factors (4F: Gata6, Tbx3, Tbx5 and Rarg or Rxra) that activated Hcn4-GFP reporter expression in up to 40% of transduced cells (Fig. 2C,D).
Fig. 2.
Reprogramming with selected transcription factors activates Hcn4-GFP expression. (A) Transgenic mice harboring a PM-specific Hcn4-GFP transgene were used as a source of fibroblasts for infection with retroviruses encoding candidate transcription factors. Cells were analyzed for expression of GFP by flow cytometry. (B,C) Representative flow cytometry plots for analyses of Hcn4-GFP+ cells after 7 days of transduction with retroviruses expressing 20F (B) and 4F (C). The indicated combinations of transcription factors were transduced into tail tip fibroblasts (TTFs) isolated from Hcn4-GFP reporter mice. The numbers in each plot indicate the percentage of Hcn4-GFP+ cells. Dead cells were excluded by propidium iodide staining. Cells transduced with empty vector or uninfected cells were used as a control. (D) Summary of flow cytometry analyses for 20F and 4F. Percentage of Hcn4-GFP+ cells following infection of TTFs with retroviruses expressing 20F and 4F is shown. Seven (for 20F) and 21 (for 4F) independent experiments are presented as mean±s.d. **P<0.0001. (E) Gene expression of endogenous Hcn4 and fibroblast markers (Col1a1, Col1a2 and Col3a1) were quantified by RT-qPCR. Gene expression induced by 4F transduction relative to TTFs was compared with specific regions of P0 mouse heart including LA (left atrium), apex, AVN (atrioventricular node) and SAN (sinoatrial node). n=3 for TTFs and 4F. Three individual heart tissues from indicated specific regions were pooled for RNA extraction. *P<0.0005; **P<0.0001.
To ensure that Hcn4-GFP fibroblasts underwent genuine reprogramming, we performed qPCR analysis on cells that were transduced with 4F. These experiments demonstrated that 4F transduction of fibroblasts resulted in robust expression of the endogenous Hcn4 gene and concomitant repression of multiple fibroblast-specific genes (Fig. 2E), suggesting that Hcn4-GFP expression reflects lineage reprogramming rather than merely reporter gene activation. Although Hcn4 activation is consistent with reprogramming towards a PM-like phenotype, the ultimate measure of PM functionality in vitro is spontaneous beating activity, as observed in endogenous Hcn4-GFP+ PM cells and subsequently confirmed by intracellular recordings (Fig. 1C). In this regard, we were unable to identify a single, spontaneously beating cell following 4F transduction, regardless of the type of fibroblast (i.e. mouse tail tip, cardiac and embryonic) or duration in culture (up to 12 weeks). Moreover, we performed patch-clamping on individual Hcn4-GFP+ cells reprogrammed by 4F (n=6), and we were unable to detect any electrical activity, even after providing a depolarizing stimulus (supplementary material Fig. S1). Thus, we conclude that, whereas 4F induction efficiently reprograms Hcn4 gene expression, this combination of transcription factors is insufficient to generate functional PM cells.
Endogenous PM cells express sarcomeric proteins
As we observed robust expression of Hcn4 and gene expression changes consistent with genuine reprogramming in 4F-transduced cells, we reasoned that certain properties of endogenous PM cells remained uncaptured by relying on the Hcn4-GFP reporter gene alone. Therefore, we performed detailed analysis of endogenous PM cells to identify key characteristics that we could incorporate into future screening assays. From a functional standpoint, PM cells are uniquely specialized for spontaneous depolarization rather than contraction. Given this specialization, however, previous studies have not clearly addressed whether PM cells exhibit a highly organized contractile apparatus similar to working atrial and ventricular CMs (Bleeker et al., 1980; Kapoor et al., 2013). Thus, in order to develop rigorous criteria for defining the PM phenotype, we first sought to determine whether PM cells express contractile proteins and to evaluate the extent to which they form organized sarcomeric structures.
To examine whether sarcomere proteins are expressed and organized properly in PM cells, we conducted immunocytochemistry experiments on right atrial cells isolated from Hcn4-GFP reporter mice. Both Hcn4-GFP− and Hcn4-GFP+ right atrial cells displayed well-organized sarcomeric structures as determined by expression of the sarcomeric proteins α-actinin and cardiac troponin-T (cTnT) (Fig. 3A-C). Hcn4-GFP+ atrial cells, which we defined as PM CMs by direct intracellular recordings (Fig. 1C), unequivocally showed sarcomere structures labeled by α-actinin and cTnT.
Fig. 3.
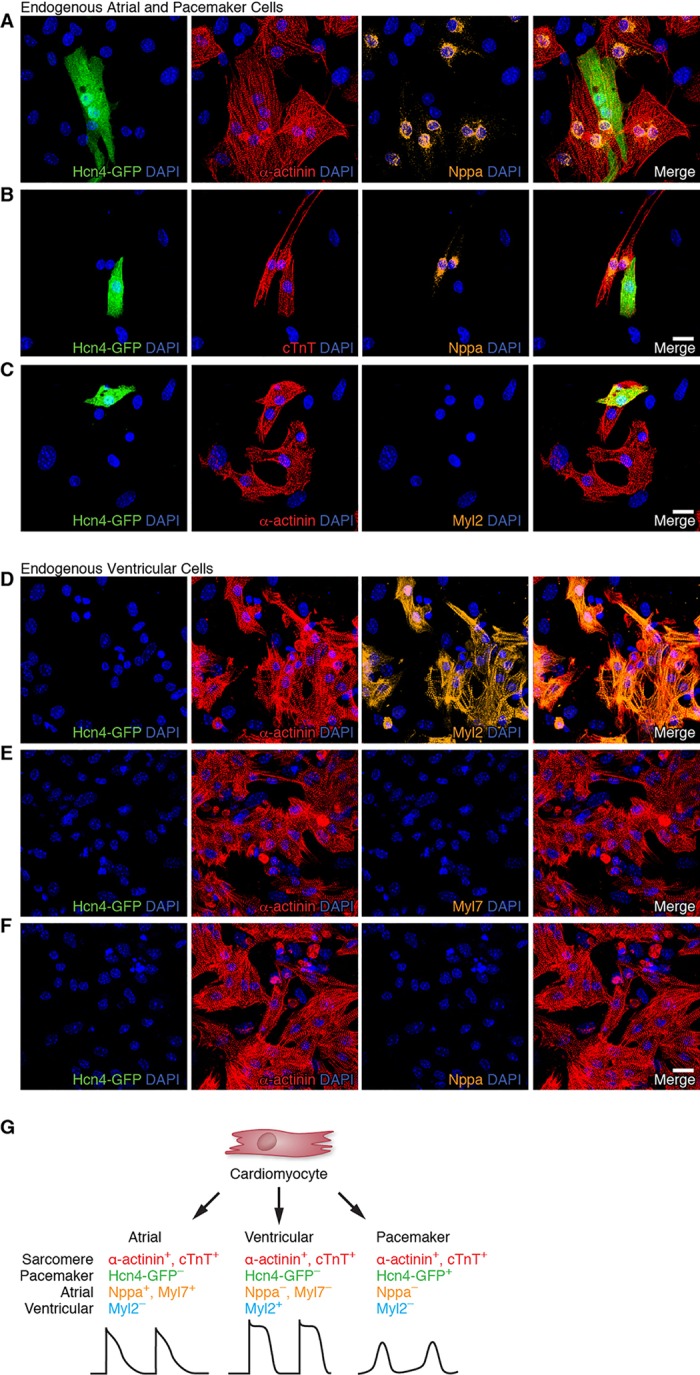
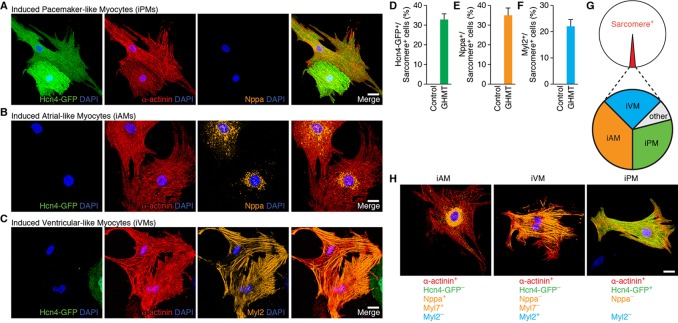
Multiplex immunostaining identifies individual CM subtypes. (A-C) Immunofluorescence staining of neonatal atrial cells isolated from Hcn4-GFP reporter mice for Hcn4-GFP (PM marker, green), α-actinin (sarcomere marker, red) and Nppa (atrial marker, A,B) or Myl2 (ventricular marker, C) (orange). (D-F) Immunofluorescence staining of neonatal ventricular cells isolated from Hcn4-GFP reporter mice for Hcn4-GFP (green), α-actinin (red) and Myl2 (D) or Myl7 atrial markers (E,F) (orange). (G) Schematic of multiplex immunostaining approach for defining individual cardiac subtypes. Scale bars: 20 µm.
We further analyzed endogenous PM cells by immunostaining with other cardiac markers. We confirmed that the atrial marker Nppa was excluded from Hcn4-GFP+ cells (Mommersteeg et al., 2007), although it was strongly expressed in Hcn4-GFP− atrial cells (Fig. 3A,B). By contrast, the atrial marker Myl7 did not clearly distinguish Hcn4-GFP− atrial myocytes from Hcn4-GFP+ SAN cells (supplementary material Fig. S2), although both atrial markers (Nppa and Myl7) co-localized in Hcn4− atrial myocytes (supplementary material Fig. S3). As expected, the ventricular marker Myl2 was not expressed in either Hcn4-GFP+ or Hcn4-GFP− atrial cells (Fig. 3C). From these experiments, we conclude that PM CMs exhibit organized sarcomeric structure, and immunostaining for atrial (Nppa) and PM (Hcn4-GFP) markers readily distinguishes these two CM subtypes. In order to develop a similar immunostaining strategy for ventricular CMs, we isolated neonatal ventricular CMs from Hcn4-GFP reporter mice. As expected, these cells did not express atrial markers (Myl7 and Nppa) or the PM marker Hcn4-GFP (Fig. 3D-F). However, we found that ventricular CMs strongly expressed the ventricular marker Myl2 (Fig. 3D), confirming that Myl2 can specifically identify ventricular myocytes (He et al., 2003).
Taken together, we conclude that the development of sarcomeric structure is a common feature of CMs regardless of subtype (i.e. atrial, ventricular and PM). In addition, combining the results of these experiments, we can derive a multiplex immunostaining strategy to prospectively identify individual CM subtypes using sarcomeric (α-actinin or cTnT), atrial (Nppa or Myl7), ventricular (Myl2) and PM (Hcn4-GFP) markers (Fig. 3G). Applying this approach, we can assign a unique identity to each electrophysiologically defined CM subtype: (1) PM, α-actinin+/cTnT+/Hcn4-GFP+/Nppa−/Myl2−; (2) atrial, α-actinin+/cTnT+/Hcn4-GFP−/Nppa+/Myl7+/Myl2−; and (3) ventricular, α-actinin+/cTnT+/Hcn4-GFP−/Nppa−/Myl7−/Myl2+. These experiments establish a rigorous method for distinguishing individual CM subtypes and suggest that such an approach can be readily applied to CMs generated by direct reprogramming.
GHMT reprograms fibroblasts into Hcn4-GFP+/cTnT+ cells with varying degrees of sarcomere organization
Based on our findings that endogenous PM cells express both Hcn4 and sarcomeric proteins, we re-evaluated how 4F, the transcription factor combination optimized for Hcn4-GFP expression, performs with these revised criteria, using two-color flow cytometry on reprogrammed cells labeled with anti-GFP and anti-cTnT antibodies. Interestingly, whereas fibroblasts expressed Hcn4-GFP following 4F transduction, they did not display measurable levels of cTnT (Fig. 4A,C). As a comparison, we performed similar two-color flow cytometry on GHMT-transduced fibroblasts and unexpectedly found a significant fraction of reprogrammed cells that expressed both Hcn4-GFP and cTnT (Fig. 4B,C). Combined with our prior observation that 4F transduction does not form spontaneously beating cells, we conclude that the absence of sarcomeric gene expression during reprogramming precludes complete lineage conversion to a PM phenotype. In addition, this result suggested an unexpected potential for GHMT to induce a PM phenotype.
Fig. 4.
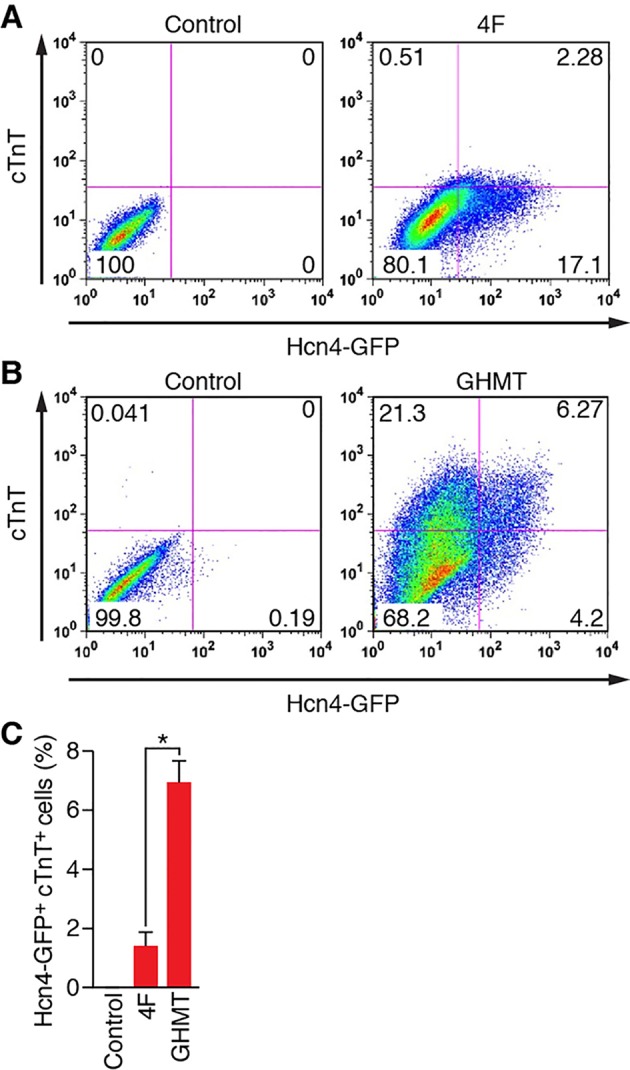
GHMT, but not 4F, reprograms fibroblasts into Hcn4-GFP+/cTnT+ cells. (A,B) Representative flow cytometry plots for analyses of both Hcn4-GFP and cTnT, induced by 4F (A) and GHMT (B) in MEFs after 10 days of transduction. The indicated combination of transcription factors were transduced into MEFs isolated from Hcn4-GFP reporter mice. Cells transduced with empty vector or uninfected cells were used as a control. Percentages of cells expressing both markers are indicated in the right upper quadrants of plots. (C) Summary of flow cytometry experiments for 4F and GHMT. Three (for 4F) and ten (for GHMT) independent experiments are presented as mean±s.d. *P<0.0005.
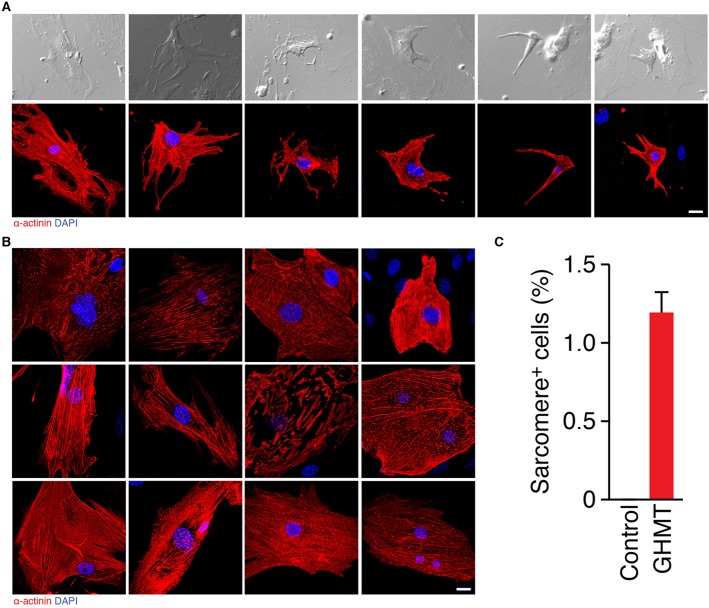
Consistent with the finding that GHMT transduction results in sarcomere protein expression, we observed that a small subset of reprogrammed cells (0.0-0.16%) displayed spontaneous beating activity as shown in previous studies (Addis et al., 2013; Ieda et al., 2010; Song et al., 2012). To assess whether sarcomere structure correlates with spontaneous beating activity, we performed retrospective immunostaining with α-actinin on these rare, beating cells. In each spontaneously beating cell for which retrospective immunostaining was technically feasible and the originating cell was readily identifiable, we observed well-organized sarcomere structure (Fig. 5A; supplementary material Movies 1-6). Although not unexpected, a single retrospective property of reprogrammed CMs that correlates with 100% spontaneous beating activity has, to our knowledge, not been described previously. When we prospectively immunostained GHMT-transduced fibroblasts, we found a striking range of sarcomeric organization, with only a small fraction of reprogrammed cells displaying well-organized sarcomere structures (Fig. 5B). In other words, sarcomeric protein expression and organization are not strictly correlated with one another. This suggests that current estimates of reprogramming efficiency, based on sarcomere protein expression derived from flow cytometry analysis, are probably vastly overestimated. Collectively, these experiments indicate that functional CM reprogramming relies upon proper sarcomere organization and that subtype specificity is likely to require the additional expression of a specialized cadre of unique proteins.
Fig. 5.
Well-organized sarcomere structure correlates with spontaneous beating in reprogrammed cells. (A) Bright-field images of beating cells after 2-3 weeks of GHMT transduction into MEFs (top). Immunostaining of corresponding beating cells with α-actinin (red), demonstrating sarcomere structure (bottom). The movies for spontaneous contraction of these cells are shown in supplementary material Movies 1-6. Nuclei were stained with DAPI (blue). (B) Immunofluorescence staining of MEFs 2 weeks after GHMT transduction for α-actinin (red) displayed a wide spectrum of sarcomere development. The degree of organization in sarcomere structures increases from top to bottom and from left to right. (C) Quantitation of well-organized sarcomere+ cells after GHMT transduction. The number of α-actinin+ cells that developed organized sarcomere structures were counted on a 20× field of a confocal microscope and then divided by the number of MEFs initially plated [sarcomere+ cells (%)]. A total of 3252 α-actinin+ cells in 16 independent experiments were counted. Data are presented as mean±s.d. Scale bars: 20 µm.
Given the remarkable degree of sarcomeric structural diversity induced by GHMT transduction, we sought to re-assess reprogramming efficiency based on a rigorous definition of sarcomere organization. Using this stringent approach, we found that ∼1.2% of fibroblasts transduced with GHMT developed an organized sarcomeric structure (Fig. 5C). Although ∼10-30% of GHMT-reprogrammed fibroblasts were shown by flow cytometry or immunocytochemistry to express cTnT (Ieda et al., 2010; Nam et al., 2013; Song et al., 2012), only a small subset of these cells demonstrate well-organized sarcomeric structures (Fig. 5B), suggesting that the overwhelming majority of fibroblasts either fail to undergo complete reprogramming or remain in an immature state.
Reprogramming of fibroblasts with GHMT generates diverse cardiomyocyte cell types
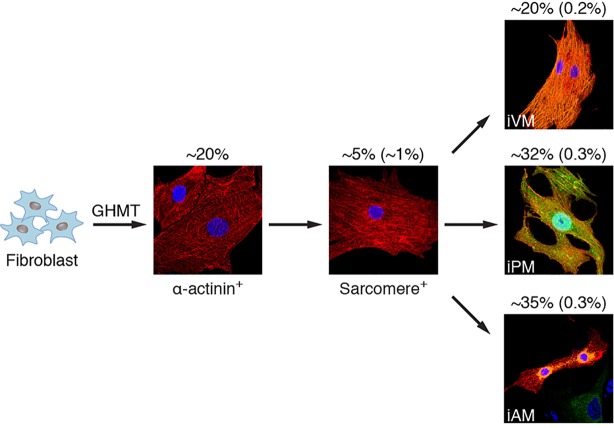
Previous population-based gene expression analyses of GHMT-reprogrammed cells were most consistent with an atrial-like fate (Song et al., 2012). Based on our results demonstrating a sizeable population of putative Hcn4-GFP+/cTnT+ iPM cells induced by GHMT (Fig. 4B), however, we re-evaluated the potential of GHMT to induce diverse cardiac phenotypes. Using the immunostaining criteria for CM subtypes established above (Fig. 3G), we investigated the subtype identity of CMs induced by GHMT-mediated reprogramming. To assess whether MEFs can be reprogrammed into atrial and PM CMs, we evaluated the expression of α-actinin, Nppa and Hcn4-GFP in reprogrammed cells derived from Hcn4-GFP reporter mice. Consistent with the flow cytometry data (Fig. 4B), we identified a subset of reprogrammed cells that co-expressed Hcn4-GFP and α-actinin, suggesting that these cells resemble PM CMs (Fig. 6A). Importantly, the atrial marker Nppa was excluded from presumptive PM-like myocytes (iPMs) (Fig. 6A). By contrast, we observed atrial-like myocytes (iAMs) that co-expressed Nppa and α-actinin in the absence of Hcn4-GFP (Fig. 6B). Furthermore, presumptive iAMs co-stained for Nppa and Myl7 but not for Hcn4-GFP, further substantiating that GHMT-mediated reprogramming generates atrial-like cells (supplementary material Fig. S4). Similarly, we performed multiplex immunostaining on GHMT-reprogrammed MEFs for Hcn4-GFP, α-actinin and the ventricular marker Myl2 to determine whether ventricular CMs can be induced by GHMT (Fig. 6C). We found that a subset of reprogrammed cells were α-actinin+/Hcn4-GFP−/Myl2+, similar to endogenous ventricular CMs. Taken together, these findings suggest that GHMT-mediated reprogramming is able to generate diverse cardiac phenotypes that mimic atrial, PM and ventricular CMs as defined by our multiplex immunostaining approach.
Fig. 6.
Diversity of cardiac phenotypes induced by reprogramming fibroblasts with GHMT. (A,B) Immunofluorescence staining of GHMT-transduced Hcn4-GFP MEFs for Hcn4-GFP (green), α-actinin (red) and Nppa (orange). Hcn4-GFP+/α-actinin+/Nppa− cells represent induced PM-like myocytes (iPMs) (A). Hcn4-GFP−/α-actinin+/Nppa+ cells represent induced atrial-like myocytes (iAMs) (B). (C) Immunofluorescence staining of GHMT-transduced Hcn4-GFP MEFs for Hcn4-GFP (green), α-actinin (red) and Myl2 (orange). Hcn4-GFP−/α-actinin+/Myl2+ cells represent induced ventricular-like myocytes (iVMs). (D-F) Quantification of iPMs, iAMs and iVMs using sarcomere+ cells. Two weeks after Hcn4-GFP MEFs were transduced by GHMT, immunofluorescence staining for α-actinin and for Hcn4-GFP, Nppa or Myl2 was performed. First, the number of α-actinin+ cells that developed organized sarcomere structures was counted on a 20× field of a confocal microscope. Then, the number of subtype/sarcomere double-positive cells were counted and divided by the total number of sarcomere+ cells [Hcn4-GFP+/ sarcomere+ cells (%), Nppa/sarcomere+ cells (%) and Myl2+/sarcomere+ cells (%), respectively]. Hcn4-GFP+/sarcomere+ cells: total 1010 cells in 16 independent experiments; Nppa+/sarcomere+ cells: total 132 cells in five independent experiments; Myl2+/sarcomere+ cells: total 59 cells in four independent experiments. Data are presented as mean±s.d. (G) Schematic diagram of cardiac subtype reprogramming by GHMT. Based on the criterion of organized sarcomere structure, ∼1% of reprogrammed cells become iCLMs that go on to diversify into various phenotypes defined by marker immunostaining and electrophysiology. (H) Representative multiplex immunostaining demonstrating three distinctive cardiac phenotypes along with specific criteria to define each subtype. Scale bars: 20 µm.
To quantitate the potential of GHMT to induce multiple cardiac subtypes, we assessed the percentage of sarcomere+ cells that constitute each individual subtype determined by multiplex immunostaining. Using this stringent approach, ∼32% of reprogrammed cells displaying organized sarcomeres also expressed Hcn4-GFP, indicating that nearly a third of directly reprogrammed fibroblasts adopt a PM-like or immature non-working CM identity (Fig. 6D). Among the remaining sarcomere+ reprogrammed cells, ∼35% expressed the atrial marker Nppa (Fig. 6E), whereas ∼22% were Myl2+ ventricular-like cells (Fig. 6F). Collectively, these experiments demonstrate that GHMT-mediated reprogramming generates unexpected CM diversity (summarized in Fig. 6G and H) based on our rigorous immunostaining criteria.
To test whether the acquisition of cell-type characteristics during cardiac reprogramming is time dependent, we performed two-color (cTnT and Hcn4-GFP) flow cytometry analysis on GHMT-reprogrammed fibroblasts at various time points (supplementary material Fig. S5A). These experiments demonstrated that the percentage of reprogrammed cells expressing either cTnT alone or both cTnT and Hcn4-GFP increased in a time-dependent fashion (supplementary material Fig. S5B), although the ratio of cTnT+Hcn4-GFP+ cells relative to the total population of cTnT+ cells remained surprisingly constant (supplementary material Fig. S5C). To evaluate more rigorously the time dependence of individual CM subtypes at the single-cell level, we applied our multiplex immunostaining criteria to GHMT-reprogrammed cells at various time points. Consistent with the flow cytometry data, we found that formation of iAMs, iVMs and iPMs increased in a time-dependent manner (supplementary material Fig. S6A), whereas their relative proportions remained constant throughout the time course (supplementary material Fig. S6B).
Our immunostaining experiments on directly reprogrammed MEFs suggest that multiple potential CM subtypes are formed: iAMs, iVMs (induced ventricular-like myocytes) and iPMs (induced PM-like myocytes). To confirm these observations, we performed patch clamping on individual, spontaneously contracting iCLMs (induced cardiac-like myocytes) (supplementary material Movies 7-13). The action potential of a typical Hcn4-GFP+ iPM (supplementary material Movie 7) resembled the spontaneous PM action potential of an endogenous Hcn4-GFP+ SAN cell, including prominent diastolic depolarization, smaller amplitude and slower upstroke velocity compared with working CMs (Fig. 7A, top). However, the spontaneous firing rate of the Hcn4-GFP+ iPM was slower and the action potential duration was longer than endogenous PM cells, indicating that the phenotypic conversion of fibroblasts to PM CMs by GHMT remains incomplete and might require additional factor(s).
Fig. 7.
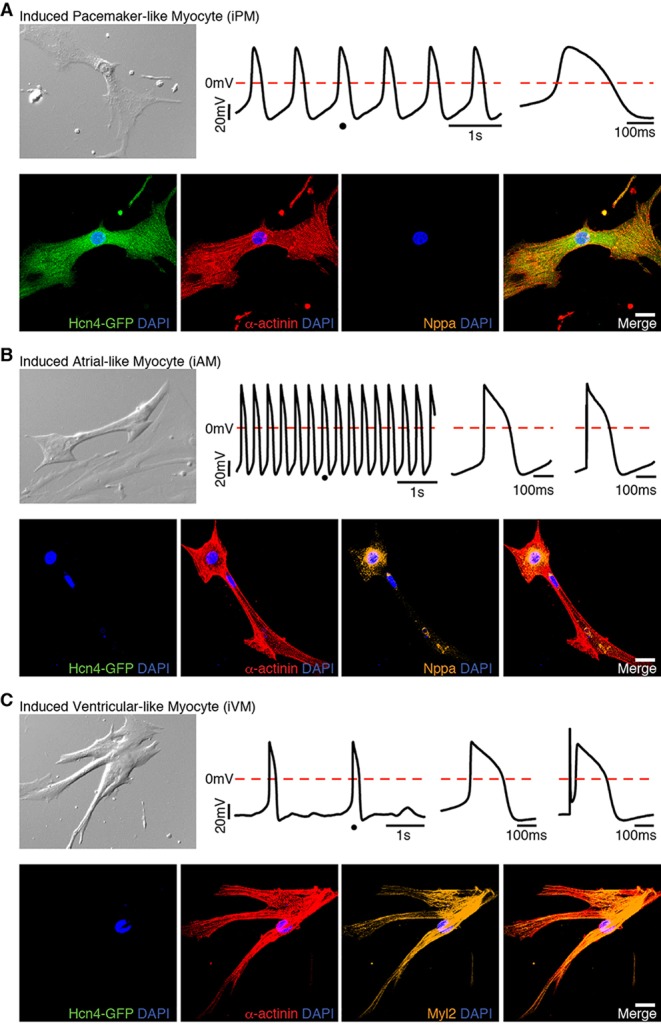
Primitive phenotype of reprogrammed cells correlates with retrospective immunostaining. Spontaneously beating cells were identified 2-3 weeks after GHMT transduction into Hcn4-GFP MEFs (supplementary material Movies 7-9). DIC images (top left in each panel) and movies were taken immediately prior to patch clamping. Action potentials of a single beating cell were recorded (top middle in each panel). The dot on the action potential tracing indicates the location in which the single action potential was zoomed. The enlarged view of a single action potential is shown to the right of the tracing. An electrically stimulated action potential is shown at the upper panel of the right ends in B and C. Multiplex immunostaining on the same cell following action potential recording indicates iPM (A), iAM (B) and iVM (C). Scale bars: 20 μm.
Immediately after patch clamping, we performed multiplex immunostaining on the same cell whose action potential was recorded to confirm expression of the expected panel of markers (Hcn4-GFP+/α-actinin+/Nppa−) (Fig. 7A, bottom). Beating Hcn4-GFP− iCLMs (supplementary material Movies 8,9) were similarly classified into atrial- and ventricular-like, based on their action potential characteristics. The action potential of a typical iAM (Fig. 7B, top) had a shorter duration and faster spontaneous rate than iVMs (Fig. 7C, top). Although the action potential of this iAM was wider than that of neonatal atrial myocytes (compare Fig. 7B with bottom of Fig. 1C), other iAMs displayed action potentials with a triangular shape (supplementary material Fig. S7, top), similar to endogenous atrial CMs. By contrast, iVMs showed a more prominent plateau phase compared with iAMs (Fig. 7C; supplementary material Figs S8-S10, top). Moreover, GHMT-induced iVMs displayed action potentials with a prolonged duration and slow spontaneous rate, similar to endogenous ventricular CMs (supplementary material Fig. S11). Collectively, our patch-clamping studies independently confirm that GHMT-mediated reprogramming generates diverse CM subtypes, and retrospective immunostaining of patch-clamped cells verified their subtype identity (atrial: Hcn4-GFP−/α-actinin+/Nppa+, Fig. 7B and supplementary material Fig. S7, bottom; ventricular: Hcn4-GFP−/α-actinin+/Myl2+, Fig. 7C and supplementary material Figs. S8-S10, bottom). However, it is important to emphasize that the electrophysiological properties of iAMs, iVMs and iPMs reprogrammed by GHMT are far from identical to those demonstrated in native CMs, as summarized in the supplementary material Table S1. Taken together, these studies show that GHMT reprograms fibroblasts into heterogeneous cell types that resemble specific CM subtypes, although these diverse iCLMs have not yet acquired all the functional properties of endogenous cells.
GHMT-reprogrammed PM-like cells do not arise from Nkx2.5+ cardiac progenitor cells
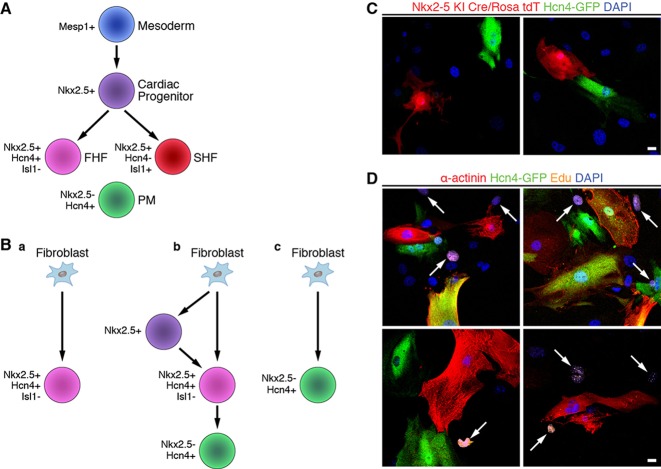
Recent studies have demonstrated that Hcn4 is expressed in the first heart field (FHF) during early embryogenesis (E7.5–E12.5) and becomes restricted to the cardiac conduction system by E16.5 (Liang et al., 2013; Spater et al., 2013). As expected, Hcn4-GFP reporter mice follow a similar pattern of developmental expression (Fig. 1A; supplementary material Fig. S12). Given the broad Hcn4 expression pattern during early heart development, we analyzed whether Hcn4-GFP+ reprogrammed cells represented genuine PM cells or merely FHF cells with embryonic action potentials. Prior studies have shown that GMT-mediated reprogramming does not involve a Mesp1+ mesodermal or Isl1+ second heart field (SHF) progenitor cell (Ieda et al., 2010) (Fig. 8A), although these experiments do not exclude the possibility of an FHF progenitor intermediate. Nkx2-5 is co-expressed with Hcn4 in FHF cells, but the Nkx2-5+ lineage does not give rise to Hcn4+ PM cells (Fig. 8A) (Liang et al., 2013). Although the exact origin of SAN PM cells remains uncertain (Bressan et al., 2013), Nkx2.5 is a reliable negative marker of these cells (Fig. 8A) (Mommersteeg et al., 2007). Therefore, if reprogrammed cells are merely embryonic FHF cells (Fig. 8Ba), then they will co-express Nkx2.5 and Hcn4. Even if reprogrammed cells are not embryonic FHF cells, they might have derived from such progenitor cells at some point during the reprogramming process (Fig. 8Bb). Lineage tracing with Nkx2.5Cre/+ mice can address both of these possibilities and simultaneously demonstrate that Hcn4-GFP+ cells are directly reprogrammed iPMs (Fig. 8Bc).
Fig. 8.
Reprogrammed Hcn4-GFP+ cells are not derived from Nkx2-5+ progenitors or actively dividing intermediates. (A) Schematic diagram of the progenitor cells involved in cardiomyocyte differentiation. A Mesp1+ mesodermal progenitor cell gives rise to an Nkx2.5+ committed cardiac progenitor cell. These cardiac progenitors diversify into the first heart field (FHF) and the Nkx2.5+/Hcn4−/Isl1+ second heart field (SHF). Nkx2.5−/Hcn4+ PM cells are derived from an unclear progenitor population. (B) Possible pathways for the generation of Hcn4-GFP+ iPMs: (a) fibroblasts convert into Nkx2.5+/Hcn4+/Isl1− FHF progenitor cells; (b) fibroblasts convert into Nkx2.5+ cardiac progenitors or Nkx2.5+/Hcn4+/Isl1− FHF progenitors prior to becoming iPM cells; or (c) fibroblasts convert directly into iPM cells without transition through a progenitor cell intermediate. (C) Hcn4-GFP+ cells do not originate from Nkx2-5+ cells. TTFs isolated from Nkx2.5Cre/+; Rosa26tdT/+; Hcn4-GFP+ triple-heterozygous mice were transduced with GHMT and then immunostained for Hcn4-GFP. None of the Hcn4-GFP+ cells co-stained with tdTomato in three independent experiments, each of which started with ∼12×103 TTFs. (D) Cells reprogrammed by GHMT do not proliferate. Forty-eight hours after Hcn4-GFP MEFs were transduced with GHMT, they were cultured with 5-ethynyl-2-deoxyuridine (EdU) to label cells that synthesize DNA. Ten days after transduction, GHMT-transduced cells were stained for Hcn4-GFP (green), a-actinin (red) and EdU (orange). EdU+ cells are indicated by white arrows. Almost no Hcn4-GFP+, α-actinin+ or double-positive cells (Hcn4-GFP+/α-actinin+) were labeled by EdU in three independent experiments, each of which started with ∼12×103 MEFs isolated from Hcn4-GFP mice. Scale bars: 20 µm.
To definitively evaluate whether GHMT-reprogrammed Hcn4-GFP+ cells are embryonic FHF cells or form via an FHF progenitor cell intermediate, we reprogrammed fibroblasts derived from Hcn4-GFP+; Nkx2.5Cre/+; Rosa26tdT/+ triple-heterozygous mice. We found that Hcn4-GFP does not co-localize with Tomato expression (Fig. 8C), strongly supporting the notion that Hcn4-GFP+ cells represent directly reprogrammed PM cells rather than FHF cells. Moreover, this experiment demonstrates that Hcn4-GFP+ cells do not derive from Nkx2.5+ progenitor cells. Interestingly, we counted a total of 213 Tomato+ cells without encountering a single cell that co-stained with α-actinin, suggesting that iAMs and iVMs also do not pass through an Nkx2.5+ progenitor cell. Further supporting the notion that Hcn4-GFP+ reprogrammed cells are not progenitor cells, we observed that GHMT-reprogrammed cells exit the cell cycle immediately following retroviral transduction (Fig. 8D). As progenitor cells by definition must exhibit some degree of proliferation and self-renewal, we believe that Hcn4-GFP+ cells do not possess typical progenitor cell characteristics. Taken together, our experimental evidence suggests that GHMT reprograms fibroblasts directly into Hcn4-GFP+ iPMs rather than FHF progenitor cells.
DISCUSSION
Various combinations of transcription factors have been shown to activate aspects of the cardiac phenotype when expressed in fibroblasts (Addis et al., 2013; Christoforou et al., 2013; Fu et al., 2013; Ieda, 2013; Nam et al., 2013; Song et al., 2012; Wada et al., 2013), but the potential diversity of cardiac cell types generated by this reprogramming approach has not been rigorously explored. Here, we established a multiplex immunostaining assay in combination with a PM-specific reporter mouse to distinguish individual endogenous cell types – atrial, ventricular and PM. Using this approach, we show that activation of Hcn4-GFP reporter expression alone is insufficient to identify functional PM CMs reprogrammed by selected transcription factors, due in part to a lack of contractile protein expression and sarcomere organization. Unexpectedly, we uncovered a remarkable degree of CM diversity generated by reprogramming fibroblasts with GHMT. Finally, we demonstrate that GHMT-mediated reprogramming of Hcn4-GFP+ iPMs is direct and does not involve an Nkx2.5+ progenitor cell intermediate. Taken together, our studies establish direct CM reprogramming as a tool for understanding cardiac subtype specification and as a potential vehicle for generating individual CM subtypes.
Bottlenecks for efficient cardiomyocyte generation by direct reprogramming
Collectively, our study highlights several bottlenecks during the CM reprogramming process that require further investigation (Fig. 9). Initial estimates of reprogramming efficiency determined by single-label flow cytometry suggested that cardiac reprogramming occurred in ∼20% of the initial fibroblasts (Ieda et al., 2010; Song et al., 2012) (Fig. 4B). Although this figure is likely to overestimate the efficiency of functional conversion, it remains unclear why only ∼20% of fibroblasts are reprogrammed. This inefficiency might be due to suboptimal viral delivery of transcription factors or might reflect the requirement of a precise stoichiometry of transcription factors, which is only achieved in a limited number of fibroblasts. Among the 20% of fibroblasts that express sarcomeric markers, only 5% of those (1% overall) form well-organized sarcomeric structures. This raises the interesting question of why sarcomere assembly represents a rate-limiting step during CM reprogramming. Aside from the technical issues detailed above, inadequate sarcomere formation might result from the absence of particular transcription factors necessary for optimal expression of specific structural proteins. Alternatively, external cues (e.g. signaling molecules or stretch) might not be present in vitro to facilitate sarcomere assembly, which is consistent with the observation that the efficiency of direct reprogramming in vivo far exceeds the efficiency expected from in vitro results (Qian et al., 2012; Song et al., 2012). Addressing these issues will not only answer important questions regarding the efficiency of cardiac reprogramming but might shed light on some of the fundamental mechanisms that regulate sarcomere formation.
Fig. 9.
Re-calculation of GHMT reprogramming efficiency based on criteria proposed in this study. Schematic diagram showing the various stages of fibroblast reprogramming with GHMT and the associated efficiency of each step (overall efficiency). The major bottlenecks for cardiac reprogramming are the activation of sarcomeric protein expression, organization of the contractile apparatus and CM diversification.
As the reprogramming potential of fibroblasts from various sources can differ substantially, we tested whether GHMT-mediated reprogramming of, for example, adult cardiac fibroblasts also displayed phenotypic diversity. Whereas atrial-like (Hcn4-GFP−/α-actinin+/Nppa+ or Hcn4-GFP−/α-actinin+/Myl7+) and PM-like (Hcn4-GFP+/α-actinin+/Nppa− or Hcn4-GFP+/α-actinin+/Myl2−) cells could be induced with GHMT (supplementary material Figs S13,S14), we were unable to identify ventricular-like Myl2+ cells in these experiments (supplementary material Fig. S15), indicating that GHMT is unable to overcome epigenetic barriers that prevent adult fibroblasts from adopting a ventricular phenotype. Based on previous studies suggesting induction of the ventricular phenotype by direct reprogramming in vivo (Qian et al., 2012; Song et al., 2012), we speculate that the ventricular environment might allow fibroblasts to clear specific epigenetic hurdles required for iVM reprogramming. In addition, it is likely that each combination of reprogramming factors used for direct cardiac reprogramming (Addis et al., 2013; Chen et al., 2012; Christoforou et al., 2013; Ieda et al., 2010; Jayawardena et al., 2012; Protze et al., 2012; Song et al., 2012) has its distinctive potential for subtype specification. Interestingly, our observation that embryonic and adult cardiac fibroblasts differ in their reprogramming potential confirms the notion that the cell source plays an important role in the acquisition of cardiac cell-type identity.
Among the 1% of original fibroblasts that ultimately assemble sarcomeres, GHMT induces immature forms of all three cardiac subtypes – atrial, ventricular and PM – at roughly equal proportions. This observation suggests that a remarkable degree of plasticity is hard-wired into iCLMs generated by GHMT. Furthermore, we surmise that a ‘ground-state’ GHMT-reprogrammed iCLM can be manipulated to favor formation of a specific subtype. The details of how cardiac subtype-specific reprogramming can be accomplished, however, remain to be elucidated. Nevertheless, several potential manipulations can be envisioned, including additional transcription factors, key small molecules or other, as yet uncharacterized features of the in vivo milieu.
Comparisons between direct reprogramming and heart development
Cardiac development proceeds through stereotypical stages guided by specific contextual cues, leading to anatomical separation of individual CM subtypes (Olson, 2006; Srivastava, 2006). By contrast, direct reprogramming occurs in a two-dimensional cell culture dish without passing through a cardiac progenitor state (Ieda et al., 2010). Although the core gene regulatory networks are likely to be shared between heart development and direct reprogramming, our observations strongly suggest that each process is distinct. As opposed to the strict compartmentalization of each CM subtype in vivo (atrial CMs: atria; ventricular CMs: ventricles; PM CMs: SAN and atrioventricular node), we often find iPMs directly adjacent to iAMs (supplementary material Fig. S16A, top) and iVMs (supplementary material Fig. S16A, bottom). Similarly, iAMs were found adjacent to iVMs (supplementary material Fig. S16B). Furthermore, the observation that GHMT-reprogrammed fibroblasts can adopt several cell fates suggests that the reprogramming process transitions through an intermediate endowed with sufficient plasticity to adopt one of multiple cell fates, including atrial-, ventricular- and PM-like myocytes (Fig. 9). Interestingly, the remarkable degree of plasticity inherent to GHMT-reprogrammed cells is distinct from the plasticity observed in progenitor cells, given that iCLMs do not arise from the Nkx2.5+ lineage. Whether a plastic iCLM intermediate exists and how subtype specification is accomplished are important questions deserving of further investigation.
Our results clearly show that iPMs do not originate from an Nkx2.5+ progenitor cell intermediate (Fig. 8C). Overall, these results are consistent with a direct lineage transformation as shown for GMT (Ieda et al., 2010); however, this contrasts with partial reprogramming approaches for generating CMs that clearly pass through a dividing progenitor cell intermediate (Efe et al., 2011). Interestingly, the fact that Hcn4-GFP+ reprogrammed cells completely exclude the Nkx2.5 lineage strongly argues that iPMs represent lineage-committed cells, as the absence of Nkx2.5 correlates with SAN differentiation. Therefore, the immaturity observed in our reprogrammed iPM cells is probably due to incomplete reprogramming rather than reprogramming of an immature PM progenitor cell. As iPMs are likely to be lineage-committed yet incompletely reprogrammed, we are confident that subtle manipulation of the reprogramming process will eventually yield fully functional PM cells. As discussed above, the nature of such manipulations remains to be identified.
The heterogeneous fate of GHMT-reprogrammed fibroblasts might reflect the particular combination of transcription factors expressed in a given cell. For example, fibroblasts transduced with a certain combination (e.g. GMT) might preferentially form iVMs or iAMs, whereas other cells receiving a different combination of factors might be more likely to form iPMs. Alternatively, a cell that becomes an iPM might express subtly different amounts of a particular transcription factor. Stochastic effects could also contribute to the heterogeneity observed during GHMT-mediated reprogramming. Interestingly, iPS reprogramming has been postulated to occur by distinct stochastic and deterministic phases (Buganim et al., 2012; Polo et al., 2012). By analogy, we speculate that the initial phase of GHMT-mediated reprogramming is a stochastic process; however, once a particular lineage – iAM, iVM or iPM – has been selected, reprogrammed cells follow a deterministic path towards their eventual fate. Identifying ways to bias this path rationally toward a specific lineage will provide a powerful method for creating individual CM subtypes. Furthermore, future studies exploring the mechanisms that underlie iCLM plasticity might clarify how subtype specificity is accomplished during fibroblast reprogramming, stem cell differentiation and normal heart development.
MATERIALS AND METHODS
Mice
All experimental procedures with animals were approved by the Institutional Animal Care and Use Committee at UT Southwestern Medical Center. Hcn4-GFP reporter mice were generated by artificial insemination with sperm derived from a transgenic line created for the GENSAT Project (Gong et al., 2003). The original Hcn4-GFP line was created by bacterial artificial chromosome (BAC) transgenesis using a ∼237 kb BAC clone (RP23-281H22) that encompasses the Hcn4 gene and flanking regulatory sequences. Nkx2-5Cre/+; Rosa26tdT/+ mice were generated by cross-breeding Nkx2-5Cre/+ mice (Moses et al., 2001) with Rosa26 tdTomato mice (Jackson Laboratory). Hcn4-GFP+; Nkx2.5Cre/+; Rosa26tdT/+ triple-heterozygous mice were generated by cross-breeding Nkx2-5Cre/+; Rosa26tdT/+ mice with Hcn4-GFP reporter mice.
Isolation of mouse fibroblasts
Mouse embryonic fibroblasts (MEFs) were isolated from E13.5 mouse embryos of Hcn4-GFP reporter or wild-type mice. The embryos were separated from the placenta and surrounding membranes. The head and the internal organs of the chest and abdominal cavities were removed from the embryos. The remaining tissues were minced and digested with trypsin (0.25%) for 15 min at 37°C to obtain single-cell suspensions. The isolated cells were cultured in fibroblast medium containing 10% FBS and 1% penicillin/streptomycin. These cells were trypsinized and replated the following day. Adult tail-tip fibroblasts (TTFs) were isolated using explant culture as described previously (Song et al., 2012). Adult tail tips were skinned and cut into small pieces. These small pieces of tail tips were cultured in fibroblast medium which was changed every 2-3 days. Adult TTFs migrated out from the explants and were harvested around day 7. Adult cardiac fibroblasts were isolated using explant culture as described previously (Song et al., 2012). Hearts from adult Hcn4-GFP mice (older than 4 weeks of age) were minced into small pieces, which were cultured in fibroblast medium that was changed every 2-3 days. After ∼10 days in culture, adult cardiac fibroblasts were harvested.
Isolation of CMs
Neonatal mouse ventricular CMs were isolated using the Neomyt kit (Cellutron) as per manufacturer's protocol. Neonatal atrial and PM CMs were isolated using methods modified from Sreejit et al. (2008). P0-P1 hearts were dissected, washed in ice-cold PBS and placed in cold Balanced Solution (20 mM HEPES pH 7.6, 130 mM NaCl, 1 mM NaH2PO4, 4 mM glucose and 3 mM KCl). The right atrium was manually dissected and minced extensively in a minimal volume of 0.05% trypsin. Atrial tissue was incubated with 0.25% trypsin and agitated for 4 min in a 37°C shaking water bath before allowing tissue to settle for 1 min without agitation. The first fraction was collected by removing the supernatant into a fresh tube containing culture medium (DMEM, 20% FBS, 2 mM L-glutamine and 3 mM sodium pyruvate). This cycle was repeated three times to collect a total of four fractions that were pooled, passed through a 100-μm filter and combined with additional culture medium. Cells were pelleted at 800 g for 4 min at 4°C and resuspended in warm culture medium before plating on glass coverslips that had been previously coated with 2% gelatin for at least 10 min. After initial plating, the medium was changed after 72 h and every 48 h thereafter.
Generation of retroviruses
Generation of retroviral constructs of mouse Gata4, Hand2, Mef2c and Tbx5 has been previously described (Song et al., 2012). Retroviral constructs were transfected using FuGENE 6 (Promega) into Platinum-E cells (Cell Biolabs). Twenty-four hours after transfection, the viral medium (the medium cultured with Platinum-E cells) was collected and hexadimethrine bromide (polybrene) was added to viral medium that had been filtered through a 0.45 µm filter at a concentration of 6 µg/µl. The mixture replaced the growth medium in the cell culture plate with mouse fibroblasts. Platinum-E cells were replenished with the growth medium (DMEM with 10% FBS). Twenty-four hours later, mouse fibroblasts were re-infected with the second viral medium from Platinum-E cell plate as described above for the first infection. Another 24 h later, viral medium on the plate with mouse fibroblasts was replaced with induction medium, composed of DMEM/199 (4:1), 10% FBS, 5% horse serum, 1% penicillin/streptomycin, 1% non-essential amino acids, 1% essential amino acids, 1% B-27, 1% insulin-selenium-transferrin, 1% vitamin mixture, 1% sodium pyruvate (Invitrogen) and 10% conditioned medium obtained from rat neonatal cardiomyocyte culture in DMEM/199 (4:1) with 5% FBS as described previously (Song et al., 2012). Conditioned medium was filtered through a 0.22 µm filter. This medium was changed every 2-3 days until cells were harvested.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with permeabilization buffer (0.05% Triton X-100 in PBS) three times for 5 min each at room temperature. Cells were blocked with blocking buffer (universal blocking buffer, BioGenex) for 30 min and then incubated with primary antibodies against cTnT (mouse monoclonal, Thermo Scientific, MS-295-P; 1:400), α-actinin (mouse monoclonal, Sigma, A7811; 1:400), GFP (chicken IgY fraction, Invitrogen, A10262; 1:400), Nppa (rabbit polyclonal, Abgent, AP8534a; 1:200), Myl2 (rabbit polyclonal, Protein Technologies, 10906-1-AP; 1:200), Myl7 (rabbit polyclonal, Protein Technologies, 17283-1-AP; 1:200), Myl7 (mouse monoclonal, Synaptic Systems, 311011; 1:200), Hcn4 (mouse monoclonal, NeuroMab, 75-150; 1:200) for 1 h at room temperature or overnight at 4°C (for anti-Myl2, anti-Myl7 and anti-Hcn4 antibodies). Following washing three times for 5 min each with permeabilization buffer, cells were incubated with appropriate Alexa fluorogenic secondary antibodies (Invitrogen or Abcam) to detect the signal at room temperature for 1 h. After another set of washing (three times for 5 min each with permeabilization buffer), cells were mounted with Vectashield with DAPI and images were captured with Zeiss LSM 500 confocal microscope. High-resolution confocal images can be found in the online version of this article.
Real-time PCR
Left atrium (LA), left ventricular apex, atrioventricular node (AVN) and SAN were dissected from wild-type C57BL/6 mice at P0. Total RNA was extracted from a pool of three individual heart tissues representing the specific heart regions listed above, 4F-transduced TTFs and uninfected TTFs, using TRIzol (Invitrogen) as per manufacturer's protocol. Complementary DNA (cDNA) was synthesized by reverse transcription for real-time (quantitative) PCR (RT-qPCR) using the SuperScript III First-Strand Synthesis System (Invitrogen). RT-qPCR was performed with Power SYBR Green 2× Master Mix in a Light Cycler and analyzed with SDS2.4 software (Applied Biosystems). Relative gene expression was normalized to GAPDH. Primer sequences are shown below. Hcn4 F: 5′-TTGACTCGGAGGTCTACAAAAC-3′; Hcn4 R: 5′-CA-GGTCATAGGTCATGTGGAAG-3′; Col1a1 F: 5′-TAGGCCATTGTGT-ATGCAGC-3′; Col1a1 R: 5′-ACATGTTCAGCTTTGTGGACC-3′; Col1a2 F: 5′-AGCAGGTCCTTGGAAACCTT-3′; Col1a2 R: 5′-AAGGAGTTTC-ATCTGGCCCT-3′; Col3a1 F: 5′-TAGGACTGACCAAGGTGGCT-3′; Col3a1 R: 5′-GGAACCTGGTTTCTTCTCACC-3′; GAPDH F: 5′-TTGA-TGGCAACAATCTCCAC-3′; GAPDH R: 5′-CGTCCCGTAGACAAAA-TGGT-3′.
Electrophysiology
Endogenous or induced CMs were cultured at low density to ensure identification of single cells. Beating cells were found under visual guidance using infrared differential interference contrast (IR-DIC) and GFP-fluorescence to identify GFP+ cells. Action potentials of neonatal endogenous CMs were recorded at 4-6 days in culture after isolation. Induced CMs were recorded at 2-3 weeks post infection. Recordings were made at 30°C in a submersion chamber perfused at 3 ml/min with oxygenated Tyrode’s solution containing: 150 mM NaCl, 4 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 10 mM glucose, 10 mM HEPES, pH 7.4. Whole-cell recordings were performed on cultured endogenous or induced CMs using recording pipettes (∼5 MΩ) filled with an intracellular solution containing: 135 mM KCl, 5 mM EGTA, 0.5 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 4 mM ATP-Mg, 0.4 mM GTP-Na, 14 mM phosphocreatine-di(Tris) at pH 7.2 and 285 mOsm. Series and input resistance were measured in voltage clamp mode with a 400 ms, 5 mV step from -60 mV holding potential (filtered at 30 kHz, sampled at 50 kHz). Cells were only used for analysis if the series resistance was less than 30 MΩ and was stable throughout the experiment. All recordings were obtained with a MultiClamp 700B amplifier (Molecular Devices). Currents were filtered at 2 kHz, acquired and digitized at 10 kHz using Clampex10.3 (Molecular Devices). Action potentials were recorded in current clamp mode and elicited by a brief pulse of depolarizing current, 4000 pA for 0.1-2 ms. Spontaneous action potentials were recorded in current clamp mode without any stimulation. In all current-clamp recordings, recordings were made at resting membrane potential or without any current injection. Data analysis was performed in Clampfit10.3 (Molecular Devices). Electrical recordings were successfully obtained from 23 reprogrammed cells. Based on action potential characteristics, a specific CM subtype could be clearly assigned for 15 of these 23 cells. In 12 out of 15 cells assigned in this manner, retrospective multiplex immunostaining was technically feasible; in all 12 cells, the subtype identity based on immunostaining matched the assignment based on electrophysiology.
Statistical analysis
All data are expressed as mean±s.d. except where indicated. P values were generated using Student's two-tailed t-test or one-way ANOVA, and statistical significance was considered for P<0.05.
Supplementary Material
Acknowledgements
We thank members of the Olson and Munshi labs for valuable intellectual input and experimental advice, and we gratefully acknowledge Jose Cabrera for excellent graphical assistance. We thank Robert Schwartz for kindly providing Nkx2.5Cre/+ mice. The mouse strain STOCK Tg(Hcn4-EGFP)JK158Gsat/Mmucd, identification number 030842-UCD, was obtained from the Mutant Mouse Regional Resource Center (MMRRC), an NCRR-NIH-funded strain repository, and was donated to the MMRRC by the NINDS-funded GENSAT BAC transgenic project.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
Y.-J.N. and N.V.M. conceived the study and experimental design. Y.-J.N., C.L., M.B. and A.F.-P. conducted experiments, and T.Z. performed electrophysiology studies. J.M. generated transgenic mice for this study. R.B.-D. and E.N.O provided intellectual input. Y.-J.N., E.N.O. and N.V.M. wrote the manuscript.
Funding
Y.-J.N. was funded by a K08 Award from the National Institutes of Health (NIH) [HL111420]. E.N.O. was supported by funds from the NIH [DK099653, HL077439, HL111665 and HL100401] and the American Heart Association (AHA). N.V.M. was funded by a K08 Award from the NIH [HL094699], the Robert A. Welch Foundation and a Career Award for Medical Scientists from the Burroughs Wellcome Fund [#1009838]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.114025/-/DC1
References
- Addis R. C., Ifkovits J. L., Pinto F., Kellam L. D., Esteso P., Rentschler S., Christoforou N., Epstein J. A. and Gearhart J. D. (2013). Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell. Cardiol. 60, 97-106 10.1016/j.yjmcc.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker M. L., Boink G. J. J., Boukens B. J., Verkerk A. O., van den Boogaard M., den Haan A. D., Hoogaars W. M. H., Buermans H. P., de Bakker J. M. T., Seppen J. et al. (2012). T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc. Res. 94, 439-449 10.1093/cvr/cvs120 [DOI] [PubMed] [Google Scholar]
- Bleeker W. K., Mackaay A. J., Masson-Pevet M., Bouman L. N. and Becker A. E. (1980). Functional and morphological organization of the rabbit sinus node. Circ. Res. 46, 11-22 10.1161/01.RES.46.1.11 [DOI] [PubMed] [Google Scholar]
- Bressan M., Liu G. and Mikawa T. (2013). Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science 340, 744-748 10.1126/science.1232877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Faddah D. A., Cheng A. W., Itskovich E., Markoulaki S., Ganz K., Klemm S. L., van Oudenaarden A. and Jaenisch R. (2012). Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150, 1209-1222 10.1016/j.cell.2012.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforou N., Chellappan M., Adler A. F., Kirkton R. D., Wu T., Addis R. C., Bursac N. and Leong K. W. (2013). Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS ONE 8, e63577 10.1371/journal.pone.0063577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe J. A., Hilcove S., Kim J., Zhou H., Ouyang K., Wang G., Chen J. and Ding S. (2011). Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 13, 215-222 10.1038/ncb2164 [DOI] [PubMed] [Google Scholar]
- Fu J. D., Stone N. R., Liu L., Spencer C. I., Qian L., Hayashi Y., Delgado-Olguin P., Ding S., Bruneau B. G. and Srivastava D. (2013). Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Reports 1, 235-247 10.1016/j.stemcr.2013.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S. C., Zheng C., Doughty M. L., Losos K., Didkovsky N., Schambra U. B., Nowak N. J., Joyner A., Leblanc G., Hatten M. E. et al. (2003). A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917-925 10.1038/nature02033 [DOI] [PubMed] [Google Scholar]
- He J.-Q., Ma Y., Lee Y., Thomson J. A. and Kamp T. J. (2003). Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ. Res. 93, 32-39 10.1161/01.RES.0000080317.92718.99 [DOI] [PubMed] [Google Scholar]
- Ieda M. (2013). Heart regeneration using reprogramming technology. Proc. Jpn. Acad. B Phys. 89, 118-128 10.2183/pjab.89.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M., Fu J.-D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G. and Srivastava D. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375-386 10.1016/j.cell.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagawa K., Miyamoto K., Yamakawa H., Muraoka N., Sadahiro T., Umei T., Wada R., Katsumata Y., Kaneda R., Nakade K. et al. (2012). Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ. Res. 111, 1147-1156 10.1161/CIRCRESAHA.112.271148 [DOI] [PubMed] [Google Scholar]
- Jayawardena T. M., Egemnazarov B., Finch E. A., Zhang L., Payne J. A., Pandya K., Zhang Z., Rosenberg P., Mirotsou M. and Dzau V. J. (2012). MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 110, 1465-1473 10.1161/CIRCRESAHA.112.269035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N., Liang W., Marbán E. and Cho H. C. (2013). Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol. 31, 54-62 10.1038/nbt.2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Wang G., Lin L., Lowe J., Zhang Q., Bu L., Chen Y., Chen J., Sun Y. and Evans S. M. (2013). HCN4 dynamically marks the first heart field and conduction system precursors. Circ. Res. 113, 399-407 10.1161/CIRCRESAHA.113.301588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathison M., Gersch R. P., Nasser A., Lilo S., Korman M., Fourman M., Hackett N., Shroyer K., Yang J., Ma Y. et al. (2012). In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. J. Am. Heart Assoc. 1, e005652 10.1161/JAHA.112.005652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommersteeg M. T., Hoogaars W. M. H., Prall O. W. J., de Gier-de Vries C., Wiese C., Clout D. E. W., Papaioannou V. E., Brown N. A., Harvey R. P., Moorman A. F. M. et al. (2007). Molecular pathway for the localized formation of the sinoatrial node. Circ. Res. 100, 354-362 10.1161/01.RES.0000258019.74591.b3 [DOI] [PubMed] [Google Scholar]
- Moses K. A., DeMayo F., Braun R. M., Reecy J. L. and Schwartz R. J. (2001). Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis 31, 176-180 10.1002/gene.10022 [DOI] [PubMed] [Google Scholar]
- Munshi N. V. (2012). Gene regulatory networks in cardiac conduction system development. Circ. Res. 110, 1525-1537 10.1161/CIRCRESAHA.111.260026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y.-J., Song K., Luo X., Daniel E., Lambeth K., West K., Hill J. A., DiMaio J. M., Baker L. A., Bassel-Duby R. et al. (2013). Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA 110, 5588-5593 10.1073/pnas.1301019110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E. N. (2006). Gene regulatory networks in the evolution and development of the heart. Science 313, 1922-1927 10.1126/science.1132292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J. M., Anderssen E., Walsh R. M., Schwarz B. A., Nefzger C. M., Lim S. M., Borkent M., Apostolou E., Alaei S., Cloutier J. et al. (2012). A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151, 1617-1632 10.1016/j.cell.2012.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze S., Khattak S., Poulet C., Lindemann D., Tanaka E. M. and Ravens U. (2012). A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J. Mol. Cell. Cardiol. 53, 323-332 10.1016/j.yjmcc.2012.04.010 [DOI] [PubMed] [Google Scholar]
- Qian L., Huang Y., Spencer C. I., Foley A., Vedantham V., Liu L., Conway S. J., Fu J.-d. and Srivastava D. (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593-598 10.1038/nature11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Nam Y.-J., Luo X., Qi X., Tan W., Huang G. N., Acharya A., Smith C. L., Tallquist M. D., Neilson E. G. et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599-604 10.1038/nature11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Später D., Abramczuk M. K., Buac K., Zangi L., Stachel M. W., Clarke J., Sahara M., Ludwig A. and Chien K. R. (2013). A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat. Cell Biol. 15, 1098-1106 10.1038/ncb2824 [DOI] [PubMed] [Google Scholar]
- Sreejit P., Kumar S. and Verma R. S. (2008). An improved protocol for primary culture of cardiomyocyte from neonatal mice. In Vitro Cell. Dev. Biol. Anim. 44, 45-50 10.1007/s11626-007-9079-4 [DOI] [PubMed] [Google Scholar]
- Srivastava D. (2006). Making or breaking the heart: from lineage determination to morphogenesis. Cell 126, 1037-1048 10.1016/j.cell.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Wada R., Muraoka N., Inagawa K., Yamakawa H., Miyamoto K., Sadahiro T., Umei T., Kaneda R., Suzuki T., Kamiya K. et al. (2013). Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA 110, 12667-12672 10.1073/pnas.1304053110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.