Abstract
An appropriate balance between self-renewal and differentiation is crucial for stem cell function during both early development and tissue homeostasis throughout life. Recent evidence from both pluripotent embryonic and adult stem cell studies suggests that this balance is partly regulated by reactive oxygen species (ROS), which, in synchrony with metabolism, mediate the cellular redox state. In this Primer, we summarize what ROS are and how they are generated in the cell, as well as their downstream molecular targets. We then review recent findings that provide molecular insights into how ROS signaling can influence stem cell homeostasis and lineage commitment, and discuss the implications of this for reprogramming and stem cell ageing. We conclude that ROS signaling is an emerging key regulator of multiple stem cell populations.
Keywords: Hematopoietic stem cells, ROS, Embryonic stem cells, Metabolism, Mitochondria
Introduction
Reactive oxygen species (ROS) have been increasingly implicated in the physiological regulation of crucial developmental processes, e.g. the emergence of embryonic blood stem cells or differentiation of embryonic cardiomyocytes (Harris et al., 2013; Hernández-García et al., 2010; Hom et al., 2011). There is also increasing evidence that ROS are implicated at many distinct levels of biological processes, from gene expression and protein translation to protein-protein interactions, etc. (Holmström and Finkel, 2014). They function in cellular signaling, propagating signals from one tissue to the next, and in translating environmental cues into cellular responses in order to balance cellular input, e.g. nutrients and cytokines, with the appropriate cellular response. ROS may function as a rheostat to coordinate various cellular processes and adjust cellular activity to the available bioenergetic sources (Liang and Ghaffari, 2014). With the advances in genomics and proteomics, there is also increasing information about various ways in which ROS are balanced and control cellular processes. Particularly in stem cells, changes in oxidation state, otherwise known as redox regulation, might be responsible for the communication between mitochondria and the nucleus (Gomes et al., 2013; Mouchiroud et al., 2013; Rimmelé et al., 2014). Redox-mediated mitochondria-nucleus crosstalk could explain the coordination of cellular metabolism with chromatin remodeling, gene expression, cell cycling, DNA repair and cell differentiation. ROS have also been implicated in the ageing process but less is known about whether and how ROS might be involved in the ageing of stem cells (Beckman and Ames, 1998; Harman, 1972). As slight variations in ROS content may have profound effects on stem cell fate (Ito et al., 2004, 2006; Jang and Sharkis, 2007), elucidating mechanisms whereby ROS metabolism influences stem cell fate could reveal how stem cell ageing relates to age-associated diseases. In this Primer, we review what ROS are, how they function and what is known about their role in different stem cell populations, both embryonic and adult. We describe the various sources of ROS in stem cells, as well as what is known about oxygen metabolism in stem cells and how this might influence stem cell fate. The role of ROS in regulating stem cell dynamics has implications for various diseases, including cancer and age-related illnesses. We conclude by considering the potential role of oxygen metabolism in stem cell ageing and discuss how the properties of ROS signaling can be exploited to manipulate stem cell fate.
What are ROS and how are they generated?
ROS arise from the one-electron reduction of molecular oxygen (Fig. 1). Intracellular ROS exist primarily in three forms: superoxide anions (O2−), hydrogen peroxide (H2O2) and hydroxyl radicals (OH−). The superoxide anion contains an unpaired electron that imparts high reactivity and necessitates a rapid reduction to H2O2 by the antioxidant enzyme superoxide dismutase (SOD) (Dröge, 2002). H2O2 can be further reduced to H2O and O2 by various cellular antioxidants (Fig. 1A). ROS can be detected intracellularly using a range of techniques; however, most assays for ROS do not discriminate between different ROS species (Box 1). Although ROS were originally thought to be merely a harmful byproduct of metabolism, accumulating evidence demonstrates a role for ROS in cell fate signaling, as discussed below (Finkel, 2003; Janssen-Heininger et al., 2008). H2O2 is thought to be the main ROS species involved with intracellular signaling, and in specific contexts can act directly as a second messenger, integrating environmental cues and passing them to downstream signal transduction cascades. This is due mostly to the longer half-life of H2O2 and its ability to diffuse easily through membranes relative to other types of ROS (Holmström and Finkel, 2014).
Fig. 1.
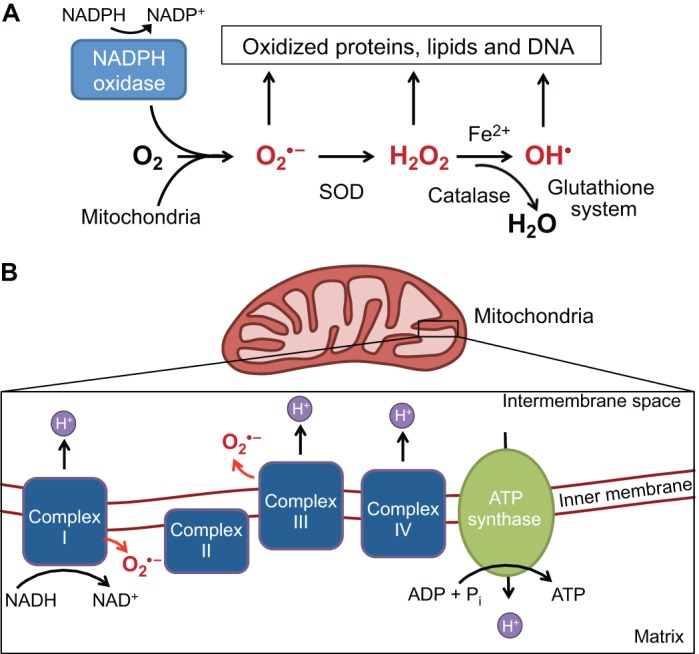
ROS generation and scavenging. (A) Reactive oxygen species (ROS) include superoxide (O2.−), hydrogen peroxide (H2O2) and the highly reactive hydroxyl radical (OH.) (shown in red). O2.− can be generated from complexes I and III (shown in B) or through the oxidation of NADPH by NADPH oxidases. Subsequent reduction to H2O2 is catalyzed by superoxide dismutase (SOD). H2O2 can be further reduced to water (H2O) by catalase or can spontaneously oxidize iron (Fe2+) to form the highly reactive OH.. Under conditions of oxidative stress, when ROS generation outpaces the ROS scavenging system, accumulating levels of ROS oxidize and damage various cellular components. (B) The electron transport chain complexes I-IV harness electrons from NADH in a series of redox reactions, which are coupled to pumping protons (H+) into the mitochondrial intermembrane space. The proton motive force, a combination of the membrane potential (charge) and the concentration gradient (pH), powers ATP synthase (complex V). Normally, O2 acts as the final electron acceptor at complex IV, but aberrant reduction of O2 can occur at complexes I and III (red arrows), leading to the generation of O2.− (red).
Box 1. Tools for ROS detection and their limitations
There are a great variety of reactive oxygen species (ROS) probes that allow analysis by flow cytometry or microscopy (Murphy et al., 2011; Winterbourn, 2014). However, most of these are not specific to any specific ROS species, are unstable and can be affected by other factors distinct from the oxidants. Therefore, data generated from the use of these probes should be carefully interpreted. Below is a brief description of the types of probes that are currently used.
Oxidized fluorescent probes
These are the most widely used probes. They diffuse through the cell membrane as non-fluorescent esterified compounds and fluoresce upon oxidation in the cytoplasm. The most common are dihydrodichlorofluorescein (DCFH2) and dihydrorhodamine, widely used to measure hydrogen peroxide levels. These probes, however, are not directly oxidized by H2O2, but require a peroxidase or a metal catalyst for the reaction to occur. As such, any change in fluorescence might simply indicate a change in catalyst levels.
Non-oxidized fluorescent probes
These are composed of fluorophores protected by a blocking group that is released upon oxidation, allowing them to fluoresce. The most commonly used form are the boronate-conjugated probes, which display a higher sensitivity than oxidized probes, although they are still not specific to any particular ROS species. Some other conjugates, such as benzene sulfonyl-ester or benzyl groups, have shown some specificity for H2O2. Future manipulation and protocol establishment for the use of these non-redox probes seem very promising.
Redox-sensitive green fluorescent proteins (GFP)
These are GFP protein variants in which redox-sensitive cysteines are incorporated in the beta-barrel of GFP (e.g. roGFP and HyPER). These probes constitute the most promising probes for in vivo analysis as they can be used, when combined with tissue-specific promoters, to generate transgenic animals. The disadvantage of these probes is that in freshly isolated primary cells, including stem cells, their use might be limited because of the need to introduce the reporter plasmids into the cells (Guzman et al., 2010).
Under normal physiological conditions, the generation of ROS is tightly regulated by the ROS scavenging system. ROS scavengers are antioxidant enzymes that can neutralize ROS by directly reacting with and accepting electrons from ROS. When ROS production outpaces ROS scavenging, an excessive accumulation of ROS occurs, leading to oxidative stress and producing adverse effects on multiple cellular components, including proteins, lipids and nucleotides. To counteract this, the cell contains multiple types of antioxidants that are specific to different species of ROS, which helps to prevent pathological levels of ROS and to repair oxidative damage to cellular components. These include superoxide dismutase (SOD), catalase, peroxiredoxins (PRX), thioredoxin (TRX), glutathione peroxidase (GPX) and glutathione reductase (GR). Glutathione, a tripeptide, is one of the most abundant antioxidants synthesized by the cell. Oxidized proteins and H2O2 are reduced by glutathione through the glutaredoxin and thioredoxin system. Other key antioxidants include SOD and catalase, which reduce O2− and H2O2, respectively. The subcellular localization of antioxidants at areas of high ROS generation, such as within the mitochondria, may further enhance the efficiency of ROS scavenging.
Sources of ROS
The electron transport chain, a component of mitochondria that is responsible for mitochondrial respiration, is the main source of ROS within the cell. The primary role of the electron transport chain is to generate the proton motive force, which leads to ATP production through ATP synthase in a process known as oxidative phosphorylation (Fig. 1B). However, ∼0.1-0.2% of O2 consumed by mitochondria is thought to form ROS through the premature electron flow to O2, mainly through electron transport chain complexes I and III (Tahara et al., 2009). The precise proportion of ROS generated from mitochondrial respiration can differ greatly depending on the cell type, environment and, ultimately, the activity of mitochondria (Murphy, 2009). Thus, another method of cellular regulation of ROS levels is through control of mitochondrial function and the regulation of metabolic pathways. Specifically, reduced ROS levels can be achieved by diverting substrates away from oxidative phosphorylation to decrease the rate of mitochondrial respiration. In addition, ROS levels can also be minimized by diverting metabolic substrates through processes that regenerate oxidized glutathione, such as the pentose phosphate pathway. Another major source of ROS is the membrane-bound protein NADPH oxidase (NOX) (Fig. 1), which consumes NADPH to generate O2− and, subsequently, H2O2. ROS produced by NOX have been shown to act as anti-microbial molecules and also to enhance growth factor signaling (Nathan and Cunningham-Bussel, 2013).
ROS signaling: molecular targets and downstream pathways
ROS were originally shown to have signaling properties when they were found to act as secondary messengers in growth factor and oncogenic signaling (Chandel et al., 1998; Irani et al., 1997; Lee, 1998; Salmeen et al., 2003; Sundaresan et al., 1995; Toledano and Leonard, 1991). However, not all ROS can be employed in signaling events. Only ROS with a substrate specificity that generates reversible oxidation, such as H2O2, are likely to trigger signaling cascade in in vivo physiological settings (Janssen-Heininger et al., 2008).
ROS can signal directly to proteins via amino acid oxidation (Box 2), the most common reaction being oxidation of cysteine residues. ROS signaling to amino acids can cause functional changes in range of different proteins (Table 1) and thus these types of modifications have established ROS as crucial regulators of cellular signaling. Such proteins are known as redox sensors, meaning that they are directly modified by ROS, undergoing a conformational change as a result of the oxidative modification (Box 2); this change influences their function, stability, subcellular localization, interactions with other proteins and other crucial processes (summarized in Table 1).
Box 2. Types of oxidative modification
Oxidation of the cysteine thiol group is the most extensively characterized type of protein modification that transduces reactive oxygen species (ROS) signaling. This results in sulfur-containing products, including disulfide bridges. In addition, a growing list of amino acids modifications by reactive oxygen and nitrogen species are described as knowledge on free radicals extends (Finkel, 2011).
Cysteine oxidation
Cysteines possess a reactive sulfur atom, the oxidation of which accounts for 1.9% of all protein modifications by ROS. Reactive cysteines can be easily converted to a sulphenic form (SOH) and re-converted to their reduced form, modulating protein activity. Oxidation of the cysteine thiol group is the most extensively characterized type of protein modification that transduces ROS signaling.
Cysteine nitrosylation
This is the reversible modification by nitric oxide (NO) that is substrate specific.
Cysteine glutathionylation
This process involves converting the reactive cysteines in proteins to the intermediate molecule SOH. These can be conjugated to glutathione to form a S-glutathionylated intermediate that subsequently modulates protein activity.
Protein carbonylation
This occurs through direct oxidation of side chains of lysines, arginines, prolines and threonines, or by covalent attachment of products from lipid peroxidation (e.g. unsaturated aldehydes). Carbonylation is an irreversible protein modification that leads to protein inactivation that is unlikely to mediate physiological cellular signaling.
Methionine oxidation
Methionines, like cysteines, posses reactive sulfur atoms and are therefore modified in a similar way to cysteines. The rate constant for methionine oxidation is much slower than for Cys oxidation.
Protein hydroxylation
Hydroxylation occurs on valines, leucines and lysines residues. The elevated oxidant property of the hydroxyl radical can even lead to the modification of these amino acids such that they have fewer reactive side chains. Hydroxyvalines and hydroxyleucines are common markers in advanced stages of human diseases, such as atherosclerosis.
Table 1.
Crucial regulators of ROS and redox sensor molecules

Although ROS can modify protein function, the opposite is also true: a growing network of proteins have been shown to modulate ROS levels (Fig. 2). Interestingly, many of these redox sensor proteins (Table 1) that are directly modulated by ROS in response to oxidative stress are also found to be crucial regulators of stem cell fate (Fig. 2). Among these proteins are transcription factors that have been connected to the regulation of cellular antioxidant machinery. These include members of the forkhead box O (FOXO) family, nuclear factor erythroid 2 (NRF2), PR domain containing 16 (PRDM16) and the p53 (TRP53) tumor suppressor (Chuikov et al., 2010; Miyamoto et al., 2007; Sablina et al., 2005; Tomko et al., 2006; Tothova et al., 2007; Yalcin et al., 2008). Modulations of ROS and p53 activity by thioredoxin-interacting protein (TXNIP) may be implicated in hematopoietic stem cell (HSC) function specifically during ageing (Jung et al., 2013) (Fig. 2). Other transcription factors, such as nuclear factor κB (NFκB), mediate the transactivation by ROS of hypoxia inducible factor 1α (HIF1α) (Bonello et al., 2007). Furthermore, other protein types, such as ATM (ataxia telangiectasia mutated kinase), p38 mitogen-activated protein kinase (MAPK), mammalian target of rapamycin (mTOR) and protein kinase B (AKT) protein kinases, as well as the multifunctional apurinic/apyrimidinic (AP) endonuclease1/redox factor 1 (APE/REF1) protein, PTEN (phosphate and tensin homolog) and sirtuins (specifically SIRT1 and SIRT3) are also considered to be redox sensors. The polycomb group member BMI1 is another protein that regulates stem cell function, modulates ROS levels and is crucial for mitochondrial function (Lessard and Sauvageau, 2003; Liu et al., 2009b; Molofsky et al., 2003). However, whether BMI1 is also directly modulated by ROS or whether BMI1 regulates mitochondria in HSCs remains to be determined. It is noteworthy that the redox-sensing property of many, if not all, of the proteins discussed above was established in somatic cells and often in cultured lines; whether these reactions also occur in primary stem cells or have a similar outcome remains to be established.
Fig. 2.
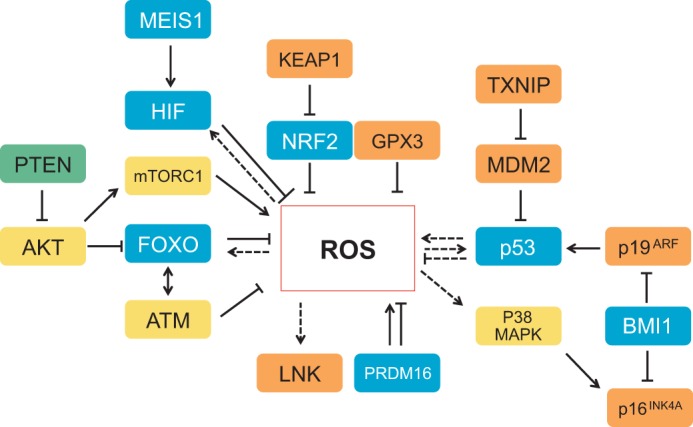
Redox sensor molecules. Intricate control of reactive oxygen species (ROS) can be either directly or indirectly mediated by several transcription factors (blue), as well as by kinases (yellow) and phosphatases (green). Other regulators, such as the cytokine signaling inhibitor LNK, the modulator KEAP1, the E3 ubiquitin ligase MDM2, the cell cycle inhibitors p16INK4A and p19ARF (which are negatively modulated by the polycomb group member BMI1), the complex mTORC1, TXNIP, and the antioxidant enzyme GPX3 (all shown in orange) can also control ROS levels. Dashed arrows and lines indicate regulations that have not been explicitly shown to occur in stem cells; unbroken lines represent interactions that have been shown in stem cells. p53 has both antioxidant and pro-oxidant functions (shown in somatic cells). AKT, protein kinase B; FOXO, forkhead box O protein; GPX3, glutathione peroxidase 3; HIF, hypoxia-inducible factor; KEAP1, kelch-like ECH-associated protein 1; MDM2, transformed mouse 3T3 cell double minute 2; MEIS1, Meis homeobox 1; mTORC1, mammalian target of rapamycin complex 1; NRF2, nuclear factor erythroid 2-related factor 2; PTEN, phosphate and tensin homolog; TXNIP, thioredoxin-interacting protein;
As well as a role in redox regulation, ROS might also function to alter the epigenetic landscape, which plays a particularly pertinent role in regulating stem cell fate (Challen et al., 2012; Mishra et al., 2014; Rimmelé et al., 2014; Singh et al., 2013; Trowbridge et al., 2009; Will et al., 2013). Many metabolic intermediates are necessary substrates for the post-translational modifications of histones that together establish the epigenetic landscape of stem cells. As the activity of glycolysis and oxidative phosphorylation can directly influence ROS, leading to changes in the concentrations of various metabolic intermediates, this might represent a potential mechanism of ROS-mediated epigenetic regulation, albeit indirect (Gut and Verdin, 2013; Sutendra et al., 2014; Xiao et al., 2012). For example, acetylation of the lysine tails of histones cannot occur without acetyl CoA, the TCA cycle metabolite, while deacetylation by sirtuin proteins (SIRTs) requires activation by nicotinamide adenine dinucleotide (NAD). Similarly, methylation of CpG islands in DNA requires the substrate SAM (s-adenosyl methionine), which is generated through threonine metabolism, a highly upregulated pathway in embryonic stem cells (ESCs) (Wang et al., 2009). Demethylation occurs through a series of hydroxylations of the methyl group catalyzed by TET (ten eleven translocase) enzymes and requires αKG (α ketogluterate) and O2 as substrates (Kohli and Zhang, 2013; Tahiliani et al., 2009). Both SIRT1 and TET enzymes are crucial factors in regulating hematopoietic stem cells (Moran-Crusio et al., 2011; Rimmelé et al., 2014; Singh et al., 2013). However, how flux through various metabolic pathways and nutrient availability control the concentrations of substrates required by histone-modifying enzymes has not yet been fully explored in stem cells.
ROS in stem cell metabolism
Cellular metabolism is the sum of catabolic and anabolic processes that involve the chemical conversion of carbon substrates to generate energy in the form of ATP and reducing co-factors (catabolic), or to produce macromolecular precursors in the form of nucleotides, lipids and amino acids (anabolic). The balance of catabolic and anabolic processes can shift depending on the cellular process being executed. Processes such as cellular growth and proliferation require mostly anabolic processes to generate building blocks for DNA, protein and membranes.
Manipulating metabolic pathways used by stem cells with either genetic approaches or drugs can directly affect whether stem cells remain quiescent, self-renew or differentiate (Takubo et al., 2013; Yu et al., 2013; Zhang et al., 2011b). One of the major ways in which metabolism can affect signaling pathways is through alterations of ROS levels. In turn, ROS can directly react with various proteins, such as kinases, phosphatases or transcription factors, to alter processes that regulate cell cycle progression, apoptosis, quiescence or differentiation (Dansen et al., 2009; Guo et al., 2010b; Velu et al., 2007). Furthermore, ROS can also directly modify metabolic enzymes or proteins that participate in nutrient-sensing pathways to direct the metabolic flux (Anastasiou et al., 2011; Brunelle et al., 2005; Sarbassov and Sabatini, 2005). In these contexts, ROS can be considered as signaling molecules that take part in the crosstalk between metabolism and stem cell fate decisions. Importantly though, metabolism can affect cell fate through multiple ROS-independent mechanisms or via mechanisms where the influence of ROS on metabolism is less obvious. Such mechanisms include changes in the epigenetic landscape brought about by metabolite abundances, as well as the ‘moonlighting’ functions of metabolic enzymes beyond their role in catalyzing metabolic reactions (De Bock et al., 2013; Gut and Verdin, 2013; Ritterson Lew and Tolan, 2013; Sutendra et al., 2014; Yang et al., 2011). However, compared with the effects of ROS, these other methods of crosstalk between metabolism and cell fate have not been as well characterized in stem cells.
Embryonic stem cells
Embryonic stem cells (ESCs) originate from the inner cell mass of the mammalian blastocyst and possess the ability to differentiate into all three germ layers of the embryo under defined in vitro conditions (Murry and Keller, 2008). Increased ROS levels in vitro induce only a transient G2/M cell cycle arrest in ESCs, suggesting that ESCs are highly resistant to oxidative stress (Guo et al., 2010a). However, like many other differentiated cells, continuous ROS exposure induces apoptosis in ESCs (Guo et al., 2010a). Consistent with these results, ESCs cultured under physiological oxygen levels (2%) maintain their genomic integrity and clonal recovery (Forsyth et al., 2006); however, the prolonged hypoxic environment leads to increased levels of ROS and to apoptosis (Urao and Ushio-Fukai, 2013).
ESCs self-renew rapidly due to a shortened G1 cell cycle phase. This process relies primarily on glycolysis and the pentose phosphate pathway, with a deliberate suppression of oxidative phosphorylation (Folmes et al., 2012; Prigione et al., 2010; Tohyama et al., 2012; Zhang et al., 2011b, 2012) (Fig. 3). Glycolysis allows for the quick generation of ATP, while the pentose phosphate pathway generates the precursors for nucleotide biosynthesis. Both ATP and nucleotides are required to power the rapid proliferation and DNA replication of ESCs (Ward and Thompson, 2012). The glycolytic requirement became apparent after studies that compared multiple metabolic parameters between ESCs and differentiated cells revealed increased lactate production and an uncoupling of electron transport chain flux from ATP production in ESCs, as well as immature mitochondrial morphology and a more-reduced redox environment (Yanes et al., 2010; Zhang et al., 2011b) (Fig. 3). Furthermore, forced activation of oxidative phosphorylation led to loss of stem cell properties and increased differentiation or apoptosis (Zhang et al., 2011b). This was shown by knock down of the uncoupling protein 2 (UCP2), a gatekeeper of pyruvate entry into the mitochondrial oxidative phosphorylation pathway (Fig. 3), as well as by delivery of metabolites that stimulate this pathway. Conversely, enhancing glycolysis through hypoxia-mediated HIF activation and inhibition of oxidative phosphorylation improved proliferation and maintenance of ESCs, while repressing differentiation (Mandal et al., 2011; Zhou et al., 2012). Both results also lead to concomitant decrease in ROS levels with improved stem cell maintenance. In mouse ESCs, endogenous ROS are elevated by a SIRT1-mediated inhibition of p53 antioxidant function (Han et al., 2008). In addition, SIRT1-mediated regulation of ROS in ESCs is central in coordinating p53 activity with the expression of pluripotentcy factor NANOG (Han et al., 2008). Evidence also suggests that SIRT1 is an important player in the regulation of ESC mitochondria (Ou et al., 2014). Together, these findings support the idea that stem cell fate may be directly modified by metabolism through ROS. They also support the notion that, in ESCs, the need for glycolysis meets the biosynthetic demands of highly proliferative cells, similar to the Warburg effect in cancer cells (Ward and Thompson, 2012). Interestingly, glucose metabolism increases the generation of hematopoietic stem cells via ROS-mediated HIF stabilization, a mechanism that might be implicated in leukemia in children exposed to high glucose in general (Harris et al., 2013; Hjalgrim et al., 2004).
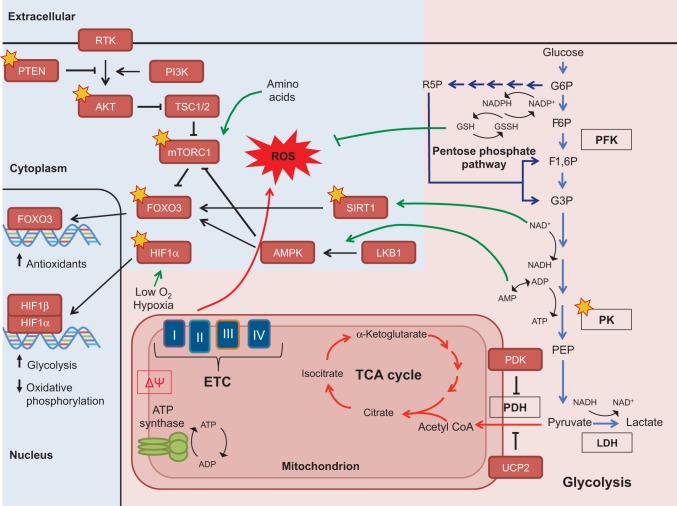
Fig. 3.
Metabolic crosstalk between key signaling pathways in stem cells via ROS and other metabolic co-factors. Glycolysis (depicted by light-blue arrows) is a catabolic process, creating energy via the conversion of glucose to pyruvate. The glycolytic intermediate glucose 6-phosphate (G6P) can also be shunted into the pentose phosphate pathway (dark-blue arrows) to produce precursors for nucleotide biosynthesis and also to regenerate NADPH, a co-factor that replenishes the reduced glutathione pools. In turn, the antioxidant glutathione (GSH) mitigates oxidative stress. The pentose phosphate pathway is especially important for the anabolic (energy consuming) demands of stem cells and for the regeneration of glutathione. Pyruvate can be catalyzed to lactate to regenerate NAD+, a required co-factor for continued flux through glycolysis and is the preferred path for stem cells. Pyruvate can also be further metabolized inside the mitochondria, beginning with the conversion to acetyl CoA, which feeds into the tricarboxylic acid cycle (TCA) cycle. The TCA cycle generates reducing co-factors that power the electron transport chain (ETC) and subsequent production of ATP, a process known as oxidative phosphorylation. Some key metabolic enzymes that are described in the text are outlined in black. The metabolic processes described can affect concentrations of mitochondrial membrane potential (ΔΨ), metabolic intermediates (acetyl CoA, α-ketoglutarate), co-factors (NADPH, NAD+, AMP/ADP) and ROS, which in turn affect the function of nutrient-sensing (depicted by green arrows) and redox-sensitive proteins (depicted by gold stars). Many of these proteins can then alter cellular processes and ultimately regulate stem cell fate. Additionally, some proteins, such as the transcription factors FOXO3 and HIF1α, can modulate the expression of metabolic genes to fit the needs of stem cells (pink background depicts the metabolic reactions and blue background indicates signaling pathways. AKT, protein kinase B; AMPK, 5′ adenosine monophosphate-activated protein kinase; BMI1, Bmi1 polycomb ring finger oncogene; F6P, fructose 6-phosphate; F1,6P, fructose 1,6-bisphosphate; FOXO, forkhead box O protein; G3P, glyceraldehyde 3-phosphate; G6P, glucose 6-phosphate; GSSH, oxidized glutathione; HIF1α, hypoxia-inducible factor 1α; LDH, lactate dehydrogenase; LKB1 (STK11), serine/threonine kinase 11; LNK, SH2B adaptor protein 3 (SH2B3; mTORC1, mammalian target of rapamycin complex 1; p16INK4A, cyclin-dependent kinase inhibitor 2A (CDKN2A); p19ARF, cyclin-dependent kinase inhibitor 2A (CDKN2A); p38 MAPK, p38 mitogen-activated protein kinase; p53, transformation related protein 53 (TRP53); PDH, pyruvate dehydrogenase complex; PDK, pyruvate dehydrogenase kinase; PEP, phosphoenolpyruvate; PFK, phosphofructokinase; PI3K, phosphatidylinositol 3-kinase; PK, pyruvate kinase; PRDM16, PR domain containing 16; PTEN, phosphate and tensin homolog; R5P, ribulose 5-phosphate; RTK, receptor tyrosine kinase; SIRT1, sirtuin 1; TSC1/2, tuberous sclerosis 1/2; UCP2, uncoupling protein 2.
Adult stem cells
During foetal life and later after birth, adult stem cells continue to replenish damaged and lost tissue. The potency of adult stem cells is limited to a subset of lineages, which necessitates a specialized stem cell that is specific to different tissue types, as well as a specialized niche in which the stem cell resides. Unlike ESCs, adult stem cells are mainly highly quiescent, a property that is crucial for their self-renewal capacity (Foudi et al., 2009; Saito et al., 2010; Wilson et al., 2008; reviewed by Liang and Ghaffari, 2014). Despite their quiescence, adult stem cells are empowered by an intrinsic potential to proliferate quickly in order to regenerate tissue within a limited time in response to damage or loss. This requires metabolic plasticity in order to adapt to either quiescence or to the highly proliferative state. Thus, adult stem cells such as HSCs require a delicate balance between the maintenance that prevents exhaustion of the stem cell pool and the differentiation that continually replenishes downstream lineages.
The maintenance of HSCs is also highly dependent on glycolysis, similar to ESCs (Unwin et al., 2006). Examination of metabolic parameters of HSCs showed that mitochondrial respiration and abundance are decreased relative to downstream progenitors (Norddahl et al., 2011; Simsek et al., 2010). In addition, HSCs show low levels of ROS and are enriched for glycolytic metabolites (Norddahl et al., 2011; Simsek et al., 2010). Similar analyses in neural stem cells (NSCs) and mesenchymal stem cells (MSCs) also revealed a preference for aerobic glycolysis and repression of oxidative phosphorylation (Funes et al., 2007; Paik et al., 2009; Yeo et al., 2013). The dependence on glycolysis and the pentose phosphate pathway of adult stem cells, and more specifically of HSCs, may be due to multiple factors, such as their location within a hypoxic niche, the low energy requirements of quiescence and the need to minimize oxidative stress from mitochondrial ROS (Jang and Sharkis, 2007; Kunisaki et al., 2013). Evidence of this comes from genetic ablation of HIFs, which causes activation of oxidative phosphorylation and an increase in ROS, resulting in the subsequent loss of quiescence and the self-renewal properties of HSCs (Rouault-Pierre et al., 2013; Takubo et al., 2010). In HSCs, MEIS1 regulates both HIF1α and HIF2α (Simsek et al., 2010; Kocabas et al., 2012). Loss of MEIS1 results in a phenotype almost identical to Hif−/− HSCs that is entirely reversible by ROS scavenging (Kocabas et al., 2012), suggesting that MEIS1 is an important regulator of HSC metabolism upstream of HIF. More recently, conditional deletion of the M2 isoform of pyruvate kinase 2 (PKM2) or lactate dehydrogenase A (LDHA), both crucial enzymes of glycolysis, further emphasizes the key function of glycolytic metabolism for normal HSCs and leukemic stem cells (Wang et al., 2014). Interestingly, the increase in ROS that results from loss of LDHA and not of PKM2 partially mediates Ldha−/− blood stem and progenitor cell defects (Wang et al., 2014).
When HSCs are activated to replenish downstream blood lineages, there is a shift from glycolysis to oxidative phosphorylation as they exit quiescence and begin to proliferate. This metabolic requirement is well exemplified by recent studies of proteins regulating key entry points of pyruvate oxidation by mitochondria, such as pyruvate dehydrogenase kinase (PDK) and PTEN-like mitochondrial phosphatase (PTPMT1) (Takubo et al., 2013; Yu et al., 2013) (Fig. 3). In a recent study, genetic ablation of PDK in mice increased oxidative phosphorylation, and led to loss of quiescence, to increased ROS and to exhaustion of the HSC pool (Takubo et al., 2013). By contrast, deletion of PTPMT1 in mice, which favors glycolysis, led to a large expansion of the HSC pool but prevented differentiation into downstream lineages (Yu et al., 2013).
Mitochondrial ROS in stem cells
The role of mitochondria in regulating stem cell fate appears more complex than the role of aerobic glycolysis. Mitochondria are highly dynamic organelles at the center of major signaling pathways. They control cellular processes such as apoptosis, Ca2+ signaling, oxidative phosphorylation and ROS production, to name a few. As such, mitochondria can manifest in multiple different morphologies and subcellular localizations, depending on their activity. Normally, actively respiring mitochondria exist as a filamentous network, with elongated shapes, and are densely packed with cristae (Fig. 4). Cristae are the folds made up by the inner mitochondrial membrane and allow for greater amounts of surface area to house the electron-transport chain complexes (Youle and van der Bliek, 2012). In ESCs, the mitochondrial network is punctate, with individual mitochondrion that are small and round with low numbers of swollen cristae (Prigione et al., 2010; Zhang et al., 2011b). These features of mitochondria are indicative of an immature and inactive mitochondrial network. When compared with fibroblasts, ESC mitochondria have a lower respiratory capacity but a higher mitochondrial membrane potential, an important component of the proton motive force (Chung et al., 2007; Folmes et al., 2011; Zhang et al., 2011b). High mitochondrial membrane potential can be an indicator of increased electron transport chain activity, whereas low mitochondrial membrane potential is associated with lower amounts of respiration; complete loss of mitochondrial membrane potential can trigger apoptosis (Vander Heiden et al., 1997). Similar to ESCs, HSCs also contain relatively immature mitochondria, suggesting that HSCs contain mitochondria with low levels of activity. This is supported by a lower respiratory rate and a low mitochondrial membrane potential when compared with downstream progenitors (Du et al., 2014; Simsek et al., 2010). The difference in mitochondrial membrane potential between ESCs and HSCs may represent the proliferative and ‘primed to differentiate’ nature of ESCs, in contrast to HSCs (which are mostly quiescent). It is therefore conceivable that it is the mitochondrial membrane potential and not the type of metabolism that is indicative of the degree to which stem cells are primed to differentiate; however, this requires further investigations (Schieke et al., 2008).
Fig. 4.
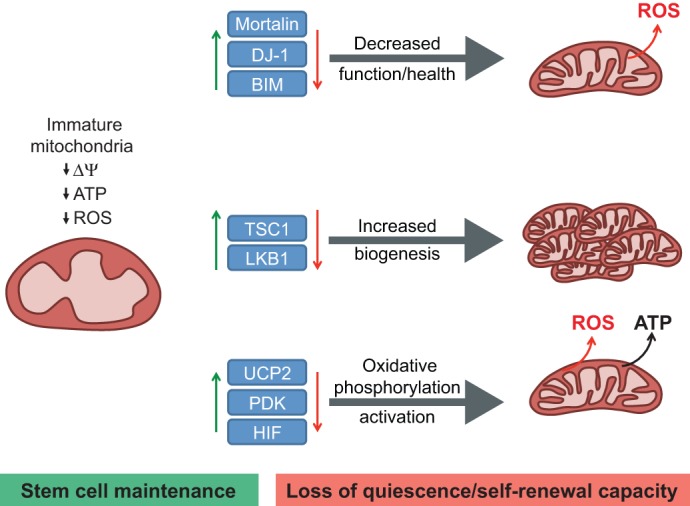
Effects of mitochondrial function on HSC maintenance and ROS production. Mitochondria in hematopoietic stem cells (HSCs) are characterized as having an immature morphology with lower metabolic activity, as determined by lower ATP output, O2 consumption, total mass and membrane potential (ΔΨ) when compared with more differentiated cells. These attributes appear to be required for stem cell and especially for HSC maintenance. The balance between stem cell maintenance (green) and the loss of quiescence and self-renewal capacity (red) can be influenced by the abundance and activity of certain proteins (blue boxes). Knock out of the corresponding genes leads to disruption of normal mitochondrial status in stem cells and changes in ROS either by decreasing the function and health of the mitochondria (top), increasing mitochondria biogenesis (middle) or activating the mitochondria oxidative phosphorylation pathway (bottom). BIM, BCL2-like 11; DJ-1, Parkinson disease (autosomal recessive, early onset) 7 (PARK7); HIF, hypoxia-inducible factor; LKB1 (STK11), serine/threonine kinase 11; PDK, pyruvate dehydrogenase kinase; TSC1, tuberous sclerosis 1; UCP2, uncoupling protein 2.
Compared with more differentiated cells, the mitochondria of stem cells are relatively metabolically inactive in terms of ATP production. Nonetheless, functional mitochondria are still required for proper maintenance of adult stem cells. In mice, deficiencies or mutations in important genes for mitochondrial function, such as Lkb1 (Stk11 – Mouse Genome Informatics), Bid, mortalin (Hspa9 – Mouse Genome Informatics), Dj-1 (Park7 – Mouse Genome Informatics) and Tsc1 (tuberous sclerosis 1), have been associated with loss of HSC quiescence and transplantation capacity (Chen et al., 2008; Gan et al., 2010; Gurumurthy et al., 2010; Maryanovich et al., 2012; Nakada et al., 2010; Tai-Nagara et al., 2014) (Fig. 4). Although all the models in these studies showed increased ROS levels, albeit to varying degrees, only the Lkb1−/− HSC phenotype was not rescued with N-acetyl-cysteine (NAC), a glutathione precursor able to reduce levels of oxidative stress. Together, these results point towards ROS as a major marker through which stem cells can sense mitochondrial health and activity, although this is not the only mechanism. The need to survey and maintain constantly the health and numbers of mitochondria within stem cells is emerging as a key aspect of stem cell biology (Joshi and Kundu, 2013). Based on this hypothesis, mitophagy machinery that regulate clearance of damaged mitochondria, and transcription factors such as PGC1α, a regulator of the mitochondrial biogenesis, may have important functions in regulating stem cells.
Given the complexity of the biochemical pathways and reactions that occur within mitochondria, it is likely that there are multiple metabolic checkpoints that regulate cell fate. Recently, mitochondrial fatty acid oxidation mediated by the promyelocytic leukaemia protein (PML)-peroxisome proliferator activator receptor δ (PPARδ) axis was shown to be necessary for the self-renewal of HSCs by promoting asymmetrical cell division (Ito et al., 2012). Given that mitochondrial ATP production is reduced in HSCs compared with committed progenitors, it has been proposed that the fatty acid oxidation in HSCs supports acetyl CoA generation (Ito et al., 2012). Acetyl CoA is fed into the TCA cycle to generate downstream substrates that are subsequently shuttled into the cytosol as citrate to reduce NADP to NADPH, a co-factor in replenishing reduced glutathione pools. Overall, the mechanism by which fatty acid oxidation promotes HSC self-renewal remains unknown and requires further investigation.
ROS as a mediator of stem cell fate and reprogramming
One of the eventual applications of stem cell biology is the generation of healthy differentiated cells to repair damaged or deteriorated tissues and organs. Given that ROS may influence a vast array of biological processes, and that we are limited in our knowledge of which species of ROS are implicated in any given physiological setting, it seems an immense challenge to explore how ROS metabolism can be manipulated to generate stem cells and influence stem cell fate. However, the study of metabolism and ROS mediated mechanisms of stem cell fate regulation has led to improved differentiation and reprogramming protocols. Differentiation of ESCs towards the cardiac lineage has been shown to rely on H2O2 signaling induced by NOX4 upregulation (Wang et al., 2007; Xiao et al., 2009). In the case of the cardiac lineage, not only are ROS important for differentiation, but the exclusive use of oxidative phosphorylation in cardiomyocytes compared with pluripotent stem cells (PSCs) can be taken advantage of to improve purification and differentiation efficiency (Chung et al., 2007; Tohyama et al., 2012). Interestingly, the degree of activation of mitochondrial metabolism is related to mouse ESC fate determination (Schieke et al., 2008). Finally, a recent study in human HSCs demonstrated that glutamine metabolism and pentose phosphate pathway-mediated generation of nucleotides is required for erythroid lineage commitment (Oburoglu et al., 2014). Chemical inhibition of these metabolic pathways led to commitment towards myeloid and granulocytic fates. Notably, as in ESCs, differentiation of MSCs towards adipocytes or neuron-like cells has also been shown to employ NOX4-mediated H2O2 signaling, as well as mitochondrial ROS (Kanda et al., 2011; Tormos et al., 2011). Further studies are required to reveal whether manipulation of ROS through metabolic pathways or directly can direct differentiation of other types of stem cells to various lineages (Box 3).
Box 3. Manipulation of ROS levels for eliminating the cancer stem cell
Cancer stem cells (CSCs) are defined as cells within a tumor that have acquired stem cell properties enabling them to reconstitute the whole tumor months or years after therapy (Baccelli and Trumpp, 2012). These cells are found in solid tumors such as prostate and breast cancers, as well as in leukemias. CSCs, in opposition to the bulk of cancer cells and similar to normal stem cells, display very low levels of reactive oxygen species (ROS), mainly due to increased activity of the antioxidant machinery and to their metabolic properties, which rely mainly on aerobic glycolysis. Leukemic stem cells (LSCs) are highly vulnerable to increases in ROS levels (Diehn et al., 2009; Kim et al., 2013). Subtle differences between normal cells and CSCs in their sensitivity to ROS can be exploited to target CSCs in therapy. Below, we summarize studies in which targeting ROS resulted in the efficient elimination of CSCs.
Targeting glutathione metabolism
Glutathione metabolism is central for ROS scavenging and glutathione peroxidase 3 (GPX3) expression correlates positively with the severity of acute myeloid leukemia. Knockdown of GPX3 or the use of the pharmacological inhibitor parthenolide, which depletes GPX1, efficiently induces apoptosis in LSCs and breast CSCs (Herault et al., 2012; Pei et al., 2013).
Increasing ROS by targeting mitochondrial energy production
Recently, BCL2 inhibition was shown to disrupt mitochondrial energy production, which increased ROS and induced apoptosis in LSCs, with little or no impact on normal stem cells (Lagadinou et al., 2013).
In contrast to differentiation, induced pluripotency occurs when a cell is reprogrammed to revert to a pluripotent state and becomes what is called an induced pluripotent stem cell (iPSC) (Takahashi and Yamanaka, 2006). The generation of iPSCs from differentiated cells has also benefited from careful regulation of ROS levels and metabolic flux. Metabolic rewiring from oxidative phosphorylation to glycolysis may even precede the activation of other key steps in the process of reprogramming (Folmes et al., 2011). Consistent with this, the key reprogramming factor OCT4 transcriptionally targets multiple metabolic genes (Kang et al., 2009). Moreover, new methods of small molecule-mediated iPSC generation have been shown to modulate the transition to aerobic glycolysis (Zhu et al., 2010). Although the precise effect of ROS on signaling pathways during the reprogramming process has not been evaluated, levels of ROS appear to increase during reprogramming and to cause damage to DNA, which can be minimized by the addition of NAC (Ji et al., 2014). The efficiency of reprogramming and continued maintenance of iPSCs can also be improved under low O2 conditions (Ezashi et al., 2005). Based on the fact that mitochondrial consumption of O2 is suppressed under hypoxia, leading to diminished levels of ROS, and that in iPSCs many ROS scavenging pathways are enhanced, it is logical to assume that increased levels of ROS might be detrimental to reprogramming efficiency (Armstrong et al., 2010). In light of these findings, it will also be interesting to evaluate the effects of FOXO factors, which are essential for the maintenance of pluripotency in ESCs, have been implicated in iPSC reprogramming, and are crucial for the regulation of ROS and cellular metabolism (Yeo et al., 2013; Zhang et al., 2011a, 2014).
ROS dynamics in stem cell homeostasis
In Drosophila, multipotent hematopoietic progenitors that are similar to mammalian myeloid progenitors display high ROS levels that decline upon differentiation (Owusu-Ansah and Banerjee, 2009). Modulation of ROS levels has been shown to direct the differentiation of Drosophila multipotent hematopoietic progenitor cells, supporting a signaling role for ROS in regulating hematopoietic cell fate (Owusu-Ansah and Banerjee, 2009). An increase in ROS is associated with mammalian blood stem cell differentiation and with increased production of their immediate progenitors, in which ROS mediate cell cycle progression (Jang and Sharkis, 2007). Consistent with this, increased ROS mediate myeloproliferation in Foxo3 mutant mice and in a mouse model of human myeloproliferative disorder (Marty et al., 2013; Yalcin et al., 2010).
In contrast to myeloid progenitors, HSCs with a long-term competitive repopulation capacity (LT-HSC) found within bone marrow compartments have been shown to have low levels of ROS. Indeed, a decrease in blood stem cell activity occurs within regions of bone marrow that have increased levels of ROS (Jang and Sharkis, 2007). Consistent with this, genetic ablation in mice of ataxia telangiectasia mutated kinase (Atm), Foxo1/3/4 (forkhead box O 1/3/4) transcription factors or just Foxo3, resulted in an accumulation of ROS in HSCs, which compromised their activity (Ito et al., 2004; Miyamoto et al., 2007; Tothova et al., 2007; Yalcin et al., 2008). The defects in Foxo−/− or Foxo3−/− HSC activity were suggested to be due to elevated ROS levels that resulted from the decreased expression of antioxidant enzymes, including catalase and superoxide dismutase 2 (SOD2); however, the source of increased ROS in Atm mutant HSC remains unclear (Ito et al., 2004; Miyamoto et al., 2007; Tothova et al., 2007; Yalcin et al., 2008; Zhang et al., 2011c). Nonetheless, mice with a Sod2 mutation do not exhibit any blood stem cell defects, which might indicate some potential functional redundancy between antioxidant enzymes of the ROS scavenging system (Friedman et al., 2004). The defective Atm−/− HSC activity is attributed to the ROS-mediated activation of p16Ink4a and of the retinoblastoma pathway (Ito et al., 2004). By contrast, the Foxo3 mutant HSC defects are likely to be mediated by ROS-induced activation of p53 tumor suppressor (Yalcin et al., 2008) (Fig. 5) or the activation of p38-MAPK (Miyamoto et al., 2007). In addition, the activation of p38-MAPK by elevated ROS compromises HSC self-renewal potential and impairs their engraftment (Ito et al., 2004, 2006; Yahata et al., 2011). ATM enzymatic activity and expression are regulated by FOXO3, but the extent to which ATM might contribute to the Foxo3 mutant HSC phenotype is unknown (Yalcin et al., 2008; Tsai et al., 2008). FOXO3 redox balance and transcriptional control of metabolic genes is also implicated in the maintenance of neural stem cells (NSCs) (Renault et al., 2009; Yeo et al., 2013). Nonetheless, highly proliferative NSCs require high ROS to maintain their self-renewal and neurogenesis properties (Le Belle et al., 2011). ROS generated by NADPH oxidases are also important for the self-renewal of spermatogonial stem cells (SSCs). However, elevated ROS in MSCs reduce their engraftment potential and induce apoptosis after transplantation (Morimoto et al., 2013; Rodrigues et al., 2012).
Fig. 5.
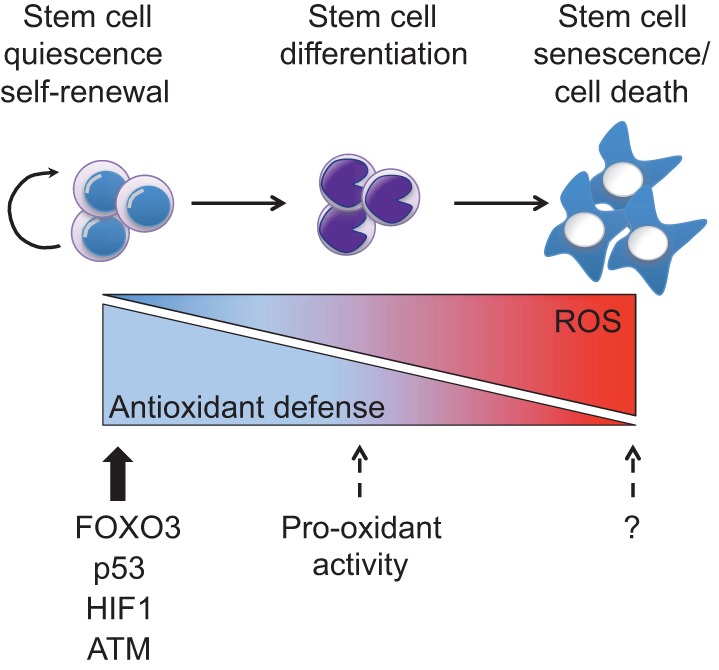
ROS effects on stem cells. Quiescent and/or self-renewing stem cells display low reactive oxygen species (ROS) levels due to their strong antioxidant machinery, which is maintained by proteins such as FOXO3 (forkhead box O3), p53 [transformation related protein 53 (TRP53)], HIF1 (hypoxia-inducible factor 1) and ATM (ataxia telangiectasia mutated kinase). Intermediate ROS levels prime stem cells for differentiation and under this context some proteins (such as p53) might act as pro-oxidant factors (dashed arrows). High ROS levels cause stem cell senescence and death. Question marks indicate unidentified proteins responsible for senescence and cell death under high ROS conditions in stem cells.
Redox modulation and ageing of stem cells
Stem cell function is compromised with increasing age (Liu et al., 2011; Signer and Morrison, 2013). The free radical theory of ageing proposes that it is caused by ROS-mediated damage to macromolecules (Harman, 1972). Although this has been challenged recently (Lapointe and Hekimi, 2010), increasing evidence implicates mitochondria in the ageing process of the whole organism (Gomes et al., 2013; Mouchiroud et al., 2013). However, the possible role of mitochondria in ageing is not necessarily due to generation of free radicals. Indeed, there is little evidence to suggest free radicals are involved in the ageing of adult stem cells and, furthermore, mitochondrial DNA mutations are not involved in the declining homeostasis of blood stem cells with age (Norddahl et al., 2011).
Although mitochondria have been implicated in whole organism ageing, it remains unclear whether mitochondrial metabolism is directly implicated in stem cell ageing. The NAD that serves as a redox regulator of oxido-reduction reactions in the cell has been recently implicated in the organismal ageing process, and thus could potentially be involved in stem cell ageing (Gomes et al., 2013; Mouchiroud et al., 2013). The NAD/NADH ratio is a measure of cellular redox status and is important for the maintenance of the glycolytic flux. Importantly, NAD serves also as an activator of several enzymes, including SIRT family deacetylases that regulate histones and non-histone proteins (Haigis and Sinclair, 2010). This activation is crucial for mitochondrial homeostasis, as SIRT1 regulates the expression of oxidative phosphorylation enzymes and PGC1, which are crucial for mitochondrial gene expression (Gomes et al., 2013). The NAD/SIRT pathway is also involved in the regulation of worm lifespan, although the underlying mechanism may be distinct and mediated by FOXO (Mouchiroud et al., 2013). These results show that communication between the nucleus and mitochondria mediated by NAD is crucial for decelerating the ageing process. In this context, recent findings implicating SIRT proteins in the regulation of blood stem cells and their ageing are notable (Brown et al., 2013; Rimmelé et al., 2014; Singh et al., 2013). Although SIRT3 is critical for the maintenance of the blood stem cell pool in old mice or under stress, SIRT1 is key to the maintenance of blood stem cells in young adult mice during both steady-state and stress conditions (Brown et al., 2013; Rimmelé et al., 2014; Singh et al., 2013). Loss of SIRT1 results in an ageing-like phenotype associated with defective lineage specification, as well as other hallmarks of stem cell ageing, including the accumulation of ROS and DNA damage in young adult mice, some of which is mediated by relative loss of FOXO3 activity in the Sirt1 mutant HSC (Rimmelé et al., 2014). Together, these findings raise the possibility that modulations of NAD might be important for the stem cell ageing process. In addition, they point to a potential function for SIRT/FOXO in the regulation of adult stem cell ageing. As well as regulating FOXO3 in blood stem cells, SIRT1 has many targets, including p53, PGC1α and HIF1, which suggests that SIRT1 may regulate stem cells through a panel of critical stem cell proteins (Lim et al., 2010; Rodgers et al., 2005; Vaziri et al., 2001). It will be necessary to devise reliable methods for the measurement of NAD levels in cell populations that contain few adult stem cells and, in this context, to clarify whether and how SIRT1 and/or SIRT3 regulation of mitochondria contribute to the correct lineage specification and/or ageing of stem cells.
Concluding remarks
Work in the past decade has uncovered the importance of redox signaling to the biology of stem cells. ROS signal the metabolic state of stem cells and, in doing this, can impact stem cell fate. Whether ROS globally impact the stem cell epigenome is not known; however, given the ability of metabolic intermediates to modify epigenetic machinery, it certainly appears possible. As is the case in cancer cells, the exact underlying mechanisms of metabolic regulation of stem cell epigenetics remains unknown, representing an exciting avenue for future exploration.
Acknowledgements
We apologize to authors whose papers we could not cite due to space limitations.
Footnotes
Competing interests
The authors declare no competing financial interests.
Funding
The authors’ research was supported by the National Institutes of Health, by a Myeloproliferative Neoplasm Foundation (MPN) award and by an Irma Hirschl/Weill-Caulier Trust Research award (S.G.); by a Roche TCRC Young Investigator award (C.L.B.); and by the National Institutes of Health (R.L.). Deposited in PMC for release after 12 months.
References
- Abbas H. A., MacCio D. R., Coskun S., Jackson J. G., Hazen A. L., Sills T. M., You M. J., Hirschi K. K. and Lozano G. (2010). Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell 7, 606-617 10.1016/j.stem.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D., Poulogiannis G., Asara J. M., Boxer M. B., Jiang J.-k., Shen M., Bellinger G., Sasaki A. T., Locasale J. W., Auld D. S. et al. (2011). Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334, 1278-1283 10.1126/science.1211485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angkeow P., Deshpande S. S., Qi B., Liu Y.-X., Park Y. C., Jeon B. H., Ozaki M. and Irani K. (2002). Redox factor-1: an extra-nuclear role in the regulation of endothelial oxidative stress and apoptosis. Cell Death Differ. 9, 717-725 10.1038/sj.cdd.4401025 [DOI] [PubMed] [Google Scholar]
- Armstrong L., Tilgner K., Saretzki G., Atkinson S. P., Stojkovic M., Moreno R., Przyborski S. and Lako M. (2010). Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells 28, 661-673 10.1002/stem.307 [DOI] [PubMed] [Google Scholar]
- Baccelli I. and Trumpp A. (2012). The evolving concept of cancer and metastasis stem cells. J. Cell Biol. 198, 281-293 10.1083/jcb.201202014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman K. B. and Ames B. N. (1998). The free radical theory of aging matures. Physiol. Rev. 78, 547-581. [DOI] [PubMed] [Google Scholar]
- Bhandari D. R., Seo K.-W., Roh K.-H., Jung J.-W., Kang S.-K. and Kang K.-S. (2010). REX-1 expression and p38 MAPK activation status can determine proliferation/differentiation fates in human mesenchymal stem cells. PLoS ONE 5, e10493 10.1371/journal.pone.0010493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonello S., Zähringer C., BelAiba R. S., Djordjevic T., Hess J., Michiels C., Kietzmann T. and Görlach A. (2007). Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler. Thromb. Vasc. Biol. 27, 755-761 10.1161/01.ATV.0000258979.92828.bc [DOI] [PubMed] [Google Scholar]
- Brown K., Xie S., Qiu X., Mohrin M., Shin J., Liu Y., Zhang D., Scadden D. T. and Chen D. (2013). SIRT3 reverses aging-associated degeneration. Cell Rep. 3, 319-327 10.1016/j.celrep.2013.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle J. K., Bell E. L., Quesada N. M., Vercauteren K., Tiranti V., Zeviani M., Scarpulla R. C. and Chandel N. S. (2005). Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metabol. 1, 409-414 10.1016/j.cmet.2005.05.002 [DOI] [PubMed] [Google Scholar]
- Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y. et al. (2004). Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011-2015 10.1126/science.1094637 [DOI] [PubMed] [Google Scholar]
- Challen G. A., Sun D., Jeong M., Luo M., Jelinek J., Berg J. S., Bock C., Vasanthakumar A., Gu H., Xi Y. et al. (2012). Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 44, 23-31 10.1038/ng.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel N. S., Maltepe E., Goldwasser E., Mathieu C. E., Simon M. C. and Schumacker P. T. (1998). Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. USA 95, 11715-11720 10.1073/pnas.95.20.11715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Liu Y., Liu R., Ikenoue T., Guan K.-L., Liu Y. and Zheng P. (2008). TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J. Exp. Med. 205, 2397-2408 10.1084/jem.20081297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S., Levi B. P., Smith M. L. and Morrison S. J. (2010). Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat. Cell Biol. 12, 999-1006 10.1038/ncb2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Dzeja P. P., Faustino R. S., Perez-Terzic C., Behfar A. and Terzic A (2007). Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat. Clin. Pract. Cardiovasc. Med. 4Suppl. 1, S60-S67 10.1038/ncpcardio0766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H., Hatta M., Matsuzaki H., Aratani S., Ohshima T., Miyagishi M., Nakajima T. and Fukamizu A. (2004). Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. U. S. A. 101, 10042-10047 10.1073/pnas.0400593101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansen T. B., Smits L. M. M., van Triest M. H., de Keizer P. L. J., van Leenen D., Koerkamp M. G., Szypowska A., Meppelink A., Brenkman A. B., Yodoi J. et al. (2009). Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat. Chem. Biol. 5, 664-672 10.1038/nchembio.194 [DOI] [PubMed] [Google Scholar]
- De Bock K., Georgiadou M., Schoors S., Kuchnio A., Wong B. W., Cantelmo A. R., Quaegebeur A., Ghesquière B., Cauwenberghs S., Eelen G. et al. (2013). Role of PFKFB3-driven glycolysis in vessel sprouting. Cell 154, 651-663 10.1016/j.cell.2013.06.037 [DOI] [PubMed] [Google Scholar]
- Diehn M., Cho R. W., Lobo N. A., Kalisky T., Dorie M. J., Kulp A. N., Qian D., Lam J. S., Ailles L. E., Wong M., et al. (2009). Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458, 780-783 10.1038/nature07733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Liang X.-G., Hu Y., Zhu D.-Y. and Lou Y.-J. (2008). Involvement of p38MAPK and reactive oxygen species in icariin-induced cardiomyocyte differentiation of murine embryonic stem cells in vitro. Stem Cells Dev. 17, 751-760 10.1089/scd.2007.0206 [DOI] [PubMed] [Google Scholar]
- Dröge W. (2002). Aging-related changes in the thiol/disulfide redox state: implications for the use of thiol antioxidants. Exp. Gerontol. 37, 1333-1345 10.1016/S0531-5565(02)00175-4 [DOI] [PubMed] [Google Scholar]
- Du J., Li Q., Tang F., Puchowitz M. A., Fujioka H., Dunwoodie S. L., Danielpour D. and Yang Y.-C. (2014). Cited2 is required for the maintenance of glycolytic metabolism in adult hematopoietic stem cells. Stem Cells Dev. 23, 83-94 10.1089/scd.2013.0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein A. C. R., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A. et al. (2001). C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43-54 10.1016/S0092-8674(01)00507-4 [DOI] [PubMed] [Google Scholar]
- Ezashi T., Das P. and Roberts R. M. (2005). Low O2 tensions and the prevention of differentiation of hES cells. Proc. Natl. Acad. Sci. USA 102, 4783-4788 10.1073/pnas.0501283102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. (2003). Oxidant signals and oxidative stress. Curr. Opin. Cell Biol. 15, 247-254 10.1016/S0955-0674(03)00002-4 [DOI] [PubMed] [Google Scholar]
- Finkel T. (2011). Signal transduction by reactive oxygen species. J. Cell Biol. 194, 7-15 10.1083/jcb.201102095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes C. D. L., Nelson T. J., Martinez-Fernandez A., Arrell D. K., Lindor J. Z., Dzeja P. P., Ikeda Y., Perez-Terzic C. and Terzic A. (2011). Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metabol. 14, 264-271 10.1016/j.cmet.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes C. D. L., Dzeja P. P., Nelson T. J. and Terzic A. (2012). Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 11, 596-606 10.1016/j.stem.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth N. R., Musio A., Vezzoni P., Simpson A. H. R. W., Noble B. S. and McWhir J. (2006). Physiologic oxygen enhances human embryonic stem cell clonal recovery and reduces chromosomal abnormalities. Cloning Stem Cells 8, 16-23 10.1089/clo.2006.8.16 [DOI] [PubMed] [Google Scholar]
- Foudi A., Hochedlinger K., Van Buren D., Schindler J. W., Jaenisch R., Carey V. and Hock H. (2009). Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27, 84-90 10.1038/nbt.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. S., Lopez M. F., Fleming M. D., Rivera A., Martin F. M., Welsh M. L., Boyd A., Doctrow S. R. and Burakoff S. J. (2004). SOD2-deficiency anemia: protein oxidation and altered protein expression reveal targets of damage, stress response, and antioxidant responsiveness. Blood 104, 2565-2573 10.1182/blood-2003-11-3858 [DOI] [PubMed] [Google Scholar]
- Funes J. M., Quintero M., Henderson S., Martinez D., Qureshi U., Westwood C., Clements M. O., Bourboulia D., Pedley R. B., Moncada S. et al. (2007). Transformation of human mesenchymal stem cells increases their dependency on oxidative phosphorylation for energy production. Proc. Natl. Acad. Sci. USA 104, 6223-6228 10.1073/pnas.0700690104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B., Hu J., Jiang S., Liu Y., Sahin E., Zhuang L., Fletcher-Sananikone E., Colla S., Wang Y. A., Chin L. et al. (2010). Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 468, 701-704 10.1038/nature09595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A. P., Price N. L., Ling A. J. Y., Moslehi J. J., Montgomery M. K., Rajman L., White J. P., Teodoro J. S., Wrann C. D., Hubbard B. P. et al. (2013). Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 155, 1624-1638 10.1016/j.cell.2013.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Einhorn L., Kelley M., Hirota K., Yodoi J., Reinbold R., Scholer H., Ramsey H. and Hromas R. (2004). Redox regulation of the embryonic stem cell transcription factor oct-4 by thioredoxin. Stem Cells 22, 259-264 10.1634/stemcells.22-3-259 [DOI] [PubMed] [Google Scholar]
- Guo Y.-L., Chakraborty S., Rajan S. S., Wang R. and Huang F. (2010a). Effects of oxidative stress on mouse embryonic stem cell proliferation, apoptosis, senescence, and self-renewal. Stem Cells Dev. 19, 1321-1331 10.1089/scd.2009.0313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Kozlov S., Lavin M. F., Person M. D. and Paull T. T. (2010b). ATM activation by oxidative stress. Science 330, 517-521 10.1126/science.1192912 [DOI] [PubMed] [Google Scholar]
- Gurumurthy S., Xie S. Z., Alagesan B., Kim J., Yusuf R. Z., Saez B., Tzatsos A., Ozsolak F., Milos P., Ferrari F. et al. (2010). The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 468, 659-663 10.1038/nature09572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut P. and Verdin E. (2013). The nexus of chromatin regulation and intermediary metabolism. Nature 502, 489-498 10.1038/nature12752 [DOI] [PubMed] [Google Scholar]
- Guzman J. N., Sanchez-Padilla J., Wokosin D., Kondapalli J., Ilijic E., Schumacker P. T. and Surmeier D. J. (2010). Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature 468, 696-700 10.1038/nature09536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis M. C. and Sinclair D. A. (2010). Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253-295 10.1146/annurev.pathol.4.110807.092250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.-K., Song E.-K., Guo Y., Ou X., Mantel C. and Broxmeyer H. E. (2008). SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell 2, 241-251 10.1016/j.stem.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. (1972). Free radical theory of aging: dietary implications. Am. J. Clin. Nutr. 25, 839-843. [DOI] [PubMed] [Google Scholar]
- Harris J. M., Esain V., Frechette G. M., Harris L. J., Cox A. G., Cortes M., Garnaas M. K., Carroll K. J., Cutting C. C., Khan T. et al. (2013). Glucose metabolism impacts the spatiotemporal onset and magnitude of HSC induction in vivo. Blood 121, 2483-2493 10.1182/blood-2012-12-471201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herault O., Hope K. J., Deneault E., Mayotte N., Chagraoui J., Wilhelm B. T., Cellot S., Sauvageau M., Andrade-Navarro M. A., Hébert J. et al. (2012). A role for GPx3 in activity of normal and leukemia stem cells. J. Exp. Med. 209, 895-901 10.1084/jem.20102386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-García D., Wood C. D., Castro-Obregón S. and Covarrubias L. (2010). Reactive oxygen species: a radical role in development? Free Radic. Biol. Med. 49, 130-143 10.1016/j.freeradbiomed.2010.03.020 [DOI] [PubMed] [Google Scholar]
- Hjalgrim L. L., Rostgaard K., Hjalgrim H., Westergaard T., Thomassen H., Forestier E., Gustafsson G., Kristinsson J., Melbye M. and Schmiegelow K. (2004). Birth weight and risk for childhood leukemia in Denmark, Sweden, Norway, and Iceland. J. Natl. Cancer Inst. 96, 1549-1556 10.1093/jnci/djh287 [DOI] [PubMed] [Google Scholar]
- Hochmuth C. E., Biteau B., Bohmann D. and Jasper H. (2011). Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell 8, 188-199 10.1016/j.stem.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmström K. M. and Finkel T. (2014). Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 15, 411-421 10.1038/nrm3801 [DOI] [PubMed] [Google Scholar]
- Hom J. R., Quintanilla R. A., Hoffman D. L., de Mesy Bentley K. L., Molkentin J. D., Sheu S.-S. and Porter G. A. (2011). The permeability transition pore controls cardiac mitochondrial maturation and myocyte differentiation. Dev. Cell 21, 469-478 10.1016/j.devcel.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani K., Xia Y., Zweier J. L., Sollott S. J., Der C. J., Fearon E. R., Sundaresan M., Finkel T. and Goldschmidt-Clermont P. J. (1997). Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science 275, 1649-1652 10.1126/science.275.5306.1649 [DOI] [PubMed] [Google Scholar]
- Ito K., Hirao A., Arai F., Matsuoka S., Takubo K., Hamaguchi I., Nomiyama K., Hosokawa K., Sakurada K., Nakagata N. et al. (2004). Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 431, 997-1002 10.1038/nature02989 [DOI] [PubMed] [Google Scholar]
- Ito K., Hirao A., Arai F., Takubo K., Matsuoka S., Miyamoto K., Ohmura M., Naka K., Hosokawa K., Ikeda Y. et al. (2006). Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat. Med. 12, 446-451 10.1038/nm1388 [DOI] [PubMed] [Google Scholar]
- Ito K., Carracedo A., Weiss D., Arai F., Ala U., Avigan D. E., Schafer Z. T., Evans R. M., Suda T., Lee C.-H. et al. (2012). A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 18, 1350-1358 10.1038/nm.2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D. and Yamamoto M. (1999). Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13, 76-86 10.1101/gad.13.1.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y.-Y. and Sharkis S. J. (2007). A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood 110, 3056-3063 10.1182/blood-2007-05-087759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen-Heininger Y. M. W., Mossman B. T., Heintz N. H., Forman H. J., Kalyanaraman B., Finkel T., Stamler J. S., Rhee S. G. and van der Vliet A. (2008). Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic. Biol. Med. 45, 1-17 10.1016/j.freeradbiomed.2008.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Sharma V., Qi S., Guarch M. E., Zhao P., Luo Z., Fan W., Wang Y., Mbabaali F., Neculai D. et al. (2014). Antioxidant supplementation reduces genomic aberrations in human induced pluripotent stem cells. Stem Cell Rep. 2, 44-51 10.1016/j.stemcr.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A. and Kundu M. (2013). Mitophagy in hematopoietic stem cells: the case for exploration. Autophagy 9, 1737-1749 10.4161/auto.26681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Kim M. J., Kim D. O., Kim W. S., Yoon S.-J., Park Y.-J., Yoon S. R., Kim T.-D., Suh H.-W., Yun S. et al. (2013). TXNIP maintains the hematopoietic cell pool by switching the function of p53 under oxidative stress. Cell Metabol. 18, 75-85 10.1016/j.cmet.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Juntilla M. M., Patil V. D., Calamito M., Joshi R. P., Birnbaum M. J. and Koretzky G. A. (2010). AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood 115, 4030-4038 10.1182/blood-2009-09-241000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda Y., Hinata T., Kang S. W. and Watanabe Y. (2011). Reactive oxygen species mediate adipocyte differentiation in mesenchymal stem cells. Life Sci. 89, 250-258 10.1016/j.lfs.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Kang J., Gemberling M., Nakamura M., Whitby F. G., Handa H., Fairbrother W. G. and Tantin D. (2009). A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress. Genes Dev. 23, 208-222 10.1101/gad.1750709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. and Wong P. K. Y. (2009). Loss of ATM impairs proliferation of neural stem cells through oxidative stress-mediated p38 MAPK signaling. Stem Cells 27, 1987-1998 10.1002/stem.125 [DOI] [PubMed] [Google Scholar]
- Kim Y. S., Kang M. J. and Cho Y. M. (2013). Low production of reactive oxygen species and high DNA repair: mechanism of radioresistance of prostate cancer stem cells. Anticancer Res. 33, 4469-4474. [PubMed] [Google Scholar]
- Kobayashi Y., Furukawa-Hibi Y., Chen C., Horio Y., Isobe K., Ikeda K. and Motoyama N. (2005). SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int. J. Mol. Med. 16, 237-243. [PubMed] [Google Scholar]
- Kocabas F., Zheng J., Thet S., Copeland N. G., Jenkins N. A., DeBerardinis R. J., Zhang C. and Sadek A. H. (2012). Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood 120, 4963-4972 10.1182/blood-2012-05-432260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli R. M. and Zhang Y. (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472-479 10.1038/nature12750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki Y., Bruns I., Scheiermann C., Ahmed J., Pinho S., Zhang D., Mizoguchi T., Wei Q., Lucas D., Ito K. et al. (2013). Arteriolar niches maintain haematopoietic stem cell quiescence. Nature 502, 637-643 10.1038/nature12612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagadinou E. D., Sach A., Callahan K., Rossi R. M., Neering S. J., Minhajuddin M., Ashton J. M., Pei S., Grose V., O'Dwyer K. M. et al. (2013). BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 12, 329-341 10.1016/j.stem.2012.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J. and Hekimi S. (2010). When a theory of aging ages badly. Cell. Mol. Life Sci. 67, 1-8 10.1007/s00018-009-0138-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Belle J. E., Orozco N. M., Paucar A. A., Saxe J. P., Mottahedeh J., Pyle A. D., Wu H. and Kornblum H. I. (2011). Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell 8, 59-71 10.1016/j.stem.2010.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M. (1998). Inhibition of p53-dependent apoptosis by the KIT tyrosine kinase: regulation of mitochondrial permeability transition and reactive oxygen species generation. Oncogene 17, 1653-1662 10.1038/sj.onc.1202102 [DOI] [PubMed] [Google Scholar]
- Lee S.-R., Yang K.-S., Kwon J., Lee C., Jeong W. and Rhee S. G. (2002). Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 277, 20336-20342 10.1074/jbc.M111899200 [DOI] [PubMed] [Google Scholar]
- Lessard J. and Sauvageau G. (2003). Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423, 255-260 10.1038/nature01572 [DOI] [PubMed] [Google Scholar]
- Liang R. and Ghaffari S. (2014). Stem cells, redox signaling, and stem cell aging. Antioxid. Redox Signal. 20, 1902-1916 10.1089/ars.2013.5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J.-H., Lee Y.-M., Chun Y.-S., Chen J., Kim J.-E. and Park J.-W. (2010). Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol. Cell 38, 864-878 10.1016/j.molcel.2010.05.023 [DOI] [PubMed] [Google Scholar]
- Liu H., Colavitti R., Rovira I. I. and Finkel T. (2005). Redox-dependent transcriptional regulation. Circ. Res. 97, 967-974 10.1161/01.RES.0000188210.72062.10 [DOI] [PubMed] [Google Scholar]
- Liu Y., Elf S. E., Miyata Y., Sashida G., Liu Y., Huang G., Di Giandomenico S., Lee J. M., Deblasio A., Menendez S. et al. (2009a). p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4, 37-48 10.1016/j.stem.2008.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao L., Chen J., Song S., Lee I. H., Quijano C., Liu H., Keyvanfar K., Chen H., Cao L.-Y. et al. (2009b). Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature 459, 387-392 10.1038/nature08040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao L. and Finkel T. (2011). Oxidants, metabolism, and stem cell biology. Free Radic. Biol. Med. 51, 2158-2162 10.1016/j.freeradbiomed.2011.10.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S., Lindgren A. G., Srivastava A. S., Clark A. T. and Banerjee U. (2011). Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells 29, 486-495 10.1002/stem.590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty C., Lacout C., Droin N., Le Couédic J.-P., Ribrag V., Solary E., Vainchenker W., Villeval J.-L. and Plo I. (2013). A role for reactive oxygen species in JAK2V617F myeloproliferative neoplasm progression. Leukemia 27, 2187-2195 10.1038/leu.2013.102 [DOI] [PubMed] [Google Scholar]
- Maryanovich M., Oberkovitz G., Niv H., Vorobiyov L., Zaltsman Y., Brenner O., Lapidot T., Jung S. and Gross A. (2012). The ATM-BID pathway regulates quiescence and survival of haematopoietic stem cells. Nat. Cell Biol. 14, 535-541 10.1038/ncb2468 [DOI] [PubMed] [Google Scholar]
- Mishra B. P., Zaffuto K. M., Artinger E. L., Org T., Mikkola H. K. A., Cheng C., Djabali M. and Ernst P. (2014). The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell Rep. 7, 1239-1247 10.1016/j.celrep.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K., Araki K. Y., Naka K., Arai F., Takubo K., Yamazaki S., Matsuoka S., Miyamoto T., Ito K., Ohmura M. et al. (2007). Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1, 101-112 10.1016/j.stem.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Molofsky A. V., Pardal R., Iwashita T., Park I.-K., Clarke M. F. and Morrison S. J. (2003). Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425, 962-967 10.1038/nature02060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K., Reavie L., Shih A., Abdel-Wahab O., Ndiaye-Lobry D., Lobry C., Figueroa M. E., Vasanthakumar A., Patel J., Zhao X. et al. (2011). Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20, 11-24 10.1016/j.ccr.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto H., Iwata K., Ogonuki N., Inoue K., Atsuo O., Kanatsu-Shinohara M., Morimoto T., Yabe-Nishimura C. and Shinohara T. (2013). ROS are required for mouse spermatogonial stem cell self-renewal. Cell Stem Cell 12, 774-786 10.1016/j.stem.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Motohashi H. and Yamamoto M. (2004). Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 10, 549-557 10.1016/j.molmed.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M. and Guarente L. (2004). Mammalian SIRT1 represses forkhead transcription factors. Cell 116, 551-563 10.1016/S0092-8674(04)00126-6 [DOI] [PubMed] [Google Scholar]
- Mouchiroud L., Houtkooper R. H., Moullan N., Katsyuba E., Ryu D., Cantó C., Mottis A., Jo Y.-S., Viswanathan M., Schoonjans K. et al. (2013). The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154, 430-441 10.1016/j.cell.2013.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata H., Ihara Y., Nakamura H., Yodoi J., Sumikawa K. and Kondo T. (2003). Glutaredoxin exerts an antiapoptotic effect by regulating the redox state of Akt. J. Biol. Chem. 278, 50226-50233 10.1074/jbc.M310171200 [DOI] [PubMed] [Google Scholar]
- Murphy M. P. (2009). How mitochondria produce reactive oxygen species. Biochem. J. 417, 1-13 10.1042/BJ20081386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. P., Holmgren A., Larsson N.-G., Halliwell B., Chang C. J., Kalyanaraman B., Rhee S. G., Thornalley P. J., Partridge L., Gems D. et al. (2011). Unraveling the biological roles of reactive oxygen species. Cell Metabol. 13, 361-366 10.1016/j.cmet.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry C. E. and Keller G. (2008). Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661-680 10.1016/j.cell.2008.02.008 [DOI] [PubMed] [Google Scholar]
- Nakada D., Saunders T. L. and Morrison S. J. (2010). Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 468, 653-658 10.1038/nature09571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C. and Cunningham-Bussel A. (2013). Beyond oxidative stress: an immunologist's guide to reactive oxygen species. Nat. Rev. Immunol. 13, 349-361 10.1038/nri3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norddahl G. L., Pronk C. J., Wahlestedt M., Sten G., Nygren J. M., Ugale A., Sigvardsson M. and Bryder D. (2011). Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell 8, 499-510 10.1016/j.stem.2011.03.009 [DOI] [PubMed] [Google Scholar]
- Oburoglu L., Tardito S., Fritz V., de Barros S. C., Merida P., Craveiro M., Mamede J., Cretenet G., Mongellaz C., An X. et al. (2014). Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell p15, 169-184. 10.1016/j.stem.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Ou X., Lee M. R., Huang X., Messina-Graham S. and Broxmeyer H. E. (2014). SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells 32, 1183-1194 10.1002/stem.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E. and Banerjee U. (2009). Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 461, 537-541 10.1038/nature08313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik J.-h., Ding Z., Narurkar R., Ramkissoon S., Muller F., Kamoun W. S., Chae S.-S., Zheng H., Ying H., Mahoney J. et al. (2009). FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5, 540-553 10.1016/j.stem.2009.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D., Bolinger R. and Mann K. (1997). Redox state regulates binding of p53 to sequence-specific DNA, but not to non-specific or mismatched DNA. Nucleic Acids Res. 25, 1289-1295 10.1093/nar/25.6.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei S., Minhajuddin M., Callahan K. P., Balys M., Ashton J. M., Neering S. J., Lagadinou E. D., Corbett C., Ye H., Liesveld J. L. et al. (2013). Targeting aberrant glutathione metabolism to eradicate human acute myelogenous leukemia cells. J. Biol. Chem. 288, 33542-33558 10.1074/jbc.M113.511170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., Xia Y., Zweier J. L., Kinzler K. W. and Vogelstein B. (1997). A model for p53-induced apoptosis. Nature 389, 300-305 10.1038/38525 [DOI] [PubMed] [Google Scholar]
- Prigione A., Fauler B., Lurz R., Lehrach H. and Adjaye J. (2010). The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells 28, 721-733 10.1002/stem.404 [DOI] [PubMed] [Google Scholar]
- Renault V. M., Rafalski V. A., Morgan A. A., Salih D. A. M., Brett J. O., Webb A. E., Villeda S. A., Thekkat P. U., Guillerey C., Denko N. C. et al. (2009). FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5, 527-539 10.1016/j.stem.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelé P., Bigarella C. L., Liang R., Izac B., Dieguez-Gonzalez R., Barbet G., Donovan M., Brugnara C., Blander J. M., Sinclair D. A. et al. . 2014). Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem Cell Rep. 3, 44-59 10.1016/j.stemcr.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritterson Lew C. and Tolan D. R. (2013). Aldolase sequesters WASP and affects WASP/Arp2/3-stimulated actin dynamics. J. Cell. Biochem. 114, 1928-1939 10.1002/jcb.24538 [DOI] [PubMed] [Google Scholar]
- Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M. and Puigserver P. (2005). Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434, 113-118 10.1038/nature03354 [DOI] [PubMed] [Google Scholar]
- Rodrigues M., Turner O., Stolz D., Griffith L. G. and Wells A. (2012). Production of reactive oxygen species by multipotent stromal cells/mesenchymal stem cells upon exposure to fas ligand. Cell Transplant. 21, 2171-2187 10.3727/096368912X639035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault-Pierre K., Lopez-Onieva L., Foster K., Anjos-Afonso F., Lamrissi-Garcia I., Serrano-Sanchez M., Mitter R., Ivanovic Z., de Verneuil H., Gribben J. et al. (2013). HIF-2α protects human hematopoietic stem/progenitors and acute myeloid leukemic cells from apoptosis induced by endoplasmic reticulum stress. Cell Stem Cell 13, 549-563 10.1016/j.stem.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Sablina A. A., Budanov A. V., Ilyinskaya G. V., Agapova L. S., Kravchenko J. E. and Chumakov P. M. (2005). The antioxidant function of the p53 tumor suppressor. Nat. Med. 11, 1306-1313 10.1038/nm1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Uchida N., Tanaka S., Suzuki N., Tomizawa-Murasawa M., Sone A., Najima Y., Takagi S., Aoki Y., Wake A. et al. (2010). Induction of cell cycle entry eliminates human leukemia stem cells in a mouse model of AML. Nat. Biotechnol. 28, 275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y., Kawabata M., Miyazono K. and Ichijo H. (1998). Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 17, 2596-2606 10.1093/emboj/17.9.2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeen A., Andersen J. N., Myers M. P., Meng T.-C., Hinks J. A., Tonks N. K. and Barford D. (2003). Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 423, 769-773 10.1038/nature01680 [DOI] [PubMed] [Google Scholar]
- Sarbassov D. D. and Sabatini D. M. (2005). Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J. Biol. Chem. 280, 39505-39509 10.1074/jbc.M506096200 [DOI] [PubMed] [Google Scholar]
- Schieke S. M., Ma M., Cao L., McCoy J. P., Liu C., Hensel N. F., Barrett A. J., Boehm M. and Finkel T. (2008). Mitochondrial metabolism modulates differentiation and teratoma formation capacity in mouse embryonic stem cells. J. Biol. Chem. 283, 28506-28512 10.1074/jbc.M802763200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer R. A. J. and Morrison S. J. (2013). Mechanisms that regulate stem cell aging and life span. Cell Stem Cell 12, 152-165 10.1016/j.stem.2013.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek T., Kocabas F., Zheng J., Deberardinis R. J., Mahmoud A. I., Olson E. N., Schneider J. W., Zhang C. C. and Sadek H. A. (2010). The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380-390 10.1016/j.stem.2010.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K., Williams C. A., Klarmann K., Burkett S. S., Keller J. R. and Oberdoerffer P. (2013). Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. J. Exp. Med. 210, 987-1001 10.1084/jem.20121608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. S., Cho H. H., Lee B.-J., Bae Y. C. and Jung J. S. (2011). Role of thioredoxin 1 and thioredoxin 2 on proliferation of human adipose tissue-derived mesenchymal stem cells. Stem Cells Dev. 20, 1529-1537 10.1089/scd.2010.0364 [DOI] [PubMed] [Google Scholar]
- Sundaresan M., Yu Z.-X., Ferrans V. J., Irani K. and Finkel T. (1995). Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 270, 296-299 10.1126/science.270.5234.296 [DOI] [PubMed] [Google Scholar]
- Sutendra G., Kinnaird A., Dromparis P., Paulin R., Stenson T. H., Haromy A., Hashimoto K., Zhang N., Flaim E. and Michelakis E. D. (2014). A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 158, 84-97 10.1016/j.cell.2014.04.046 [DOI] [PubMed] [Google Scholar]
- Tahara E. B., Navarete F. D. T. and Kowaltowski A. J. (2009). Tissue-, substrate-, and site-specific characteristics of mitochondrial reactive oxygen species generation. Free Radic. Biol. Med. 46, 1283-1297 10.1016/j.freeradbiomed.2009.02.008 [DOI] [PubMed] [Google Scholar]
- Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L. et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930-935 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai-Nagara I., Matsuoka S., Ariga H. and Suda T. (2014). Mortalin and DJ-1 coordinately regulate hematopoietic stem cell function through the control of oxidative stress. Blood 123, 41-50 10.1182/blood-2013-06-508333 [DOI] [PubMed] [Google Scholar]
- Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takubo K., Goda N., Yamada W., Iriuchishima H., Ikeda E., Kubota Y., Shima H., Johnson R. S., Hirao A., Suematsu M. et al. (2010). Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 7, 391-402 10.1016/j.stem.2010.06.020 [DOI] [PubMed] [Google Scholar]
- Takubo K., Nagamatsu G., Kobayashi C. I., Nakamura-Ishizu A., Kobayashi H., Ikeda E., Goda N., Rahimi Y., Johnson R. S., Soga T. et al. (2013). Regulation of glycolysis by pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12, 49-61 10.1016/j.stem.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TeKippe M., Harrison D. E. and Chen J (2003). Expansion of hematopoietic stem cell phenotype and activity in Trp53-null mice. Exp. Hematol. 31, 521-527. 10.1016/S0301-472X(03)00072-9 [DOI] [PubMed] [Google Scholar]
- Tohyama S., Hattori F., Sano M., Hishiki T., Nagahata Y., Matsuura T., Hashimoto H., Suzuki T., Yamashita H., Satoh Y. et al. (2012). Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 12, 127-137 10.1016/j.stem.2012.09.013 [DOI] [PubMed] [Google Scholar]
- Toledano M. B. and Leonard W. J. (1991). Modulation of transcription factor NF-kappa B binding activity by oxidation-reduction in vitro. Proc. Natl. Acad. Sci. USA 88, 4328-4332 10.1073/pnas.88.10.4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko R. J., Bansal P. and Lazo J. S. (2006). Airing out an antioxidant role for the tumor suppressor p53. Mol. Interv. 6, 23-25, 2 10.1124/mi.6.1.5 [DOI] [PubMed] [Google Scholar]
- Tormos K. V., Anso E., Hamanaka R. B., Eisenbart J., Joseph J., Kalyanaraman B. and Chandel N. S. (2011). Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metabol. 14, 537-544 10.1016/j.cmet.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B. J., Lee B. H., Castrillon D. H., Cullen D. E., McDowell E. P., Lazo-Kallanian S., Williams I. R., Sears C. et al. (2007). FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128, 325-339 10.1016/j.cell.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Trowbridge J. J., Snow J. W., Kim J. and Orkin S. H. (2009). DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell 5, 442-449 10.1016/j.stem.2009.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W. B., Chung Y. M., Takahashi Y., Xu Z., Hu, M. C., (2008). Functional interaction between FOXO3a and ATM regulates DNA damage response. Nat. Cell Biol. 10, 460-467 10.1038/ncb1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J. J., Dudakov J. A., Takahashi K., Shieh J.-H., Velardi E., Holland A. M., Singer N. V., West M. L., Smith O. M., Young L. F. et al. (2013). Nrf2 regulates haematopoietic stem cell function. Nat. Cell Biol. 15, 309-316 10.1038/ncb2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin R. D., Smith D. L., Blinco D., Wilson C. L., Miller C. J., Evans C. A., Jaworska E., Baldwin S. A., Barnes K., Pierce A., et al. (2006). Quantitative proteomics reveals posttranslational control as a regulatory factor in primary hematopoietic stem cells. Blood 107, 4687-4694 10.1182/blood-2005-12-4995 [DOI] [PubMed] [Google Scholar]
- Urao N. and Ushio-Fukai M. (2013). Redox regulation of stem/progenitor cells and bone marrow niche. Free Radic. Biol. Med. 54, 26-39 10.1016/j.freeradbiomed.2012.10.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M. G., Chandel N. S., Williamson E. K., Schumacker P. T. and Thompson C. B. (1997). Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell 91, 627-637 10.1016/S0092-8674(00)80450-X [DOI] [PubMed] [Google Scholar]
- Van der Horst A., Tertoolen L. G. J., de Vries-Smits L. M. M., Frye R. A., Medema R. H. and Burgering B. M. T. (2004). FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1). J. Biol. Chem. 279, 28873-28879 10.1074/jbc.M401138200 [DOI] [PubMed] [Google Scholar]
- Vaziri H., Dessain S. K., Ng Eaton E., Imai S.-I., Frye R. A., Pandita T. K., Guarente L. and Weinberg R. A. (2001). hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149-159 10.1016/S0092-8674(01)00527-X [DOI] [PubMed] [Google Scholar]
- Velu C. S., Niture S. K., Doneanu C. E., Pattabiraman N. and Srivenugopal K. S. (2007). Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry 46, 7765-7780 10.1021/bi700425y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal R. and Jaiswal A. K. (1996). Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. USA 93, 14960-14965 10.1073/pnas.93.25.14960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. J., Robson C. N., Black E., Gillespie D. and Hickson I. D. (1993). Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol. Cell. Biol. 13, 5370-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Xie K., Huo S., Zhao J., Zhang S. and Miao J. (2007). Suppressing phosphatidylcholine-specific phospholipase C and elevating ROS level, NADPH oxidase activity and Rb level induced neuronal differentiation in mesenchymal stem cells. J. Cell. Biochem. 100, 1548-1557 10.1002/jcb.21139 [DOI] [PubMed] [Google Scholar]
- Wang J., Alexander P., Wu L., Hammer R., Cleaver O. and McKnight S. L. (2009). Dependence of mouse embryonic stem cells on threonine catabolism. Science 325, 435-439 10.1126/science.1173288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-H., Israelsen W. J., Lee D., Yu V. W. C., Jeanson N. T., Clish C. B., Cantley L. C., Vander Heiden M. G. and Scadden D. T. (2014). Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell 158, 1309-1323 10.1016/j.cell.2014.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P. S. and Thompson C. B. (2012). Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21, 297-308 10.1016/j.ccr.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will B., Vogler T. O., Bartholdy B., Garrett-Bakelman F., Mayer J., Barreyro L., Pandolfi A., Todorova T. I., Okoye-Okafor U. C., Stanley R. F. et al. (2013). Satb1 regulates the self-renewal of hematopoietic stem cells by promoting quiescence and repressing differentiation commitment. Nat. Immunol. 14, 437-445 10.1038/ni.2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R. C., Blanco-Bose W., Jaworski M., Offner S., Dunant C. F., Eshkind L., Bockamp E. et al. (2008). Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118-1129 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Winterbourn C. (2014). Current methods to study reactive oxygen species--pros and cons. Preface. Biochim. Biophys. Acta 1840, 707 10.1016/j.bbagen.2013.09.021 [DOI] [PubMed] [Google Scholar]
- Xiao Q., Luo Z., Pepe A. E., Margariti A., Zeng L. and Xu Q. (2009). Embryonic stem cell differentiation into smooth muscle cells is mediated by Nox4-produced H2O2. Am. J. Physiol. Cell Physiol. 296, C711-C723 10.1152/ajpcell.00442.2008 [DOI] [PubMed] [Google Scholar]
- Xiao M., Yang H., Xu W., Ma S., Lin H., Zhu H., Liu L., Liu Y., Yang C., Xu Y. et al. (2012). Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26, 1326-1338 10.1101/gad.191056.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahata T., Takanashi T., Muguruma Y., Ibrahim A. A., Matsuzawa H., Uno T., Sheng Y., Onizuka M., Ito M., Kato S. et al. (2011). Accumulation of oxidative DNA damage restricts the self-renewal capacity of human hematopoietic stem cells. Blood 118, 2941-2950 10.1182/blood-2011-01-330050 [DOI] [PubMed] [Google Scholar]
- Yalcin S., Zhang X., Luciano J. P., Mungamuri S. K., Marinkovic D., Vercherat C., Sarkar A., Grisotto M., Taneja R. and Ghaffari S. (2008). Foxo3 is essential for the regulation of ataxia telangiectasia mutated and oxidative stress-mediated homeostasis of hematopoietic stem cells. J. Biol. Chem. 283, 25692-25705 10.1074/jbc.M800517200 [DOI] [PubMed] [Google Scholar]
- Yalcin S., Marinkovic D., Mungamuri S. K., Zhang X., Tong W., Sellers R. and Ghaffari S. (2010). ROS-mediated amplification of AKT/mTOR signalling pathway leads to myeloproliferative syndrome in Foxo3(-/-) mice. EMBO J. 29, 4118-4131 10.1038/emboj.2010.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes O., Clark J., Wong D. M., Patti G. J., Sánchez-Ruiz A., Benton H. P., Trauger S. A., Desponts C., Ding S. and Siuzdak G. (2010). Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 6, 411-417 10.1038/nchembio.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Xia Y., Ji H., Zheng Y., Liang J., Huang W., Gao X., Aldape K. and Lu Z. (2011). Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 478, 118-122 10.1038/nature10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo H., Lyssiotis C. A., Zhang Y., Ying H., Asara J. M., Cantley L. C. and Paik J.-H. (2013). FoxO3 coordinates metabolic pathways to maintain redox balance in neural stem cells. EMBO J. 32, 2531-2656 10.1038/emboj.2013.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Hong S., Suzuki T., Nada S., Mannan A. M., Wang J., Okada M., Guan K.-L. and Inoki K. (2011). Redox regulates mammalian target of rapamycin complex 1 (mTORC1) activity by modulating the TSC1/TSC2-Rheb GTPase pathway. J. Biol. Chem. 286, 32651-32660 10.1074/jbc.M111.238014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R. J. and van der Bliek A. M. (2012). Mitochondrial fission, fusion, and stress. Science 337, 1062-1065 10.1126/science.1219855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W.-M., Liu X., Shen J., Jovanovic O., Pohl E. E., Gerson S. L., Finkel T., Broxmeyer H. E. and Qu C.-K. (2013). Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell 12, 62-74 10.1016/j.stem.2012.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee R. S., Yoo C. B., Pimentel D. R., Perlman D. H., Burgoyne J. R., Hou X., McComb M. E., Costello C. E., Cohen R. A. and Bachschmid M. M. (2010). Redox regulation of sirtuin-1 by S-glutathiolation. Antioxid. Redox Signal. 13, 1023-1032 10.1089/ars.2010.3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yalcin S., Lee D.-F., Yeh T.-Y. J., Lee S.-M., Su J., Mungamuri S. K., Rimmelé P., Kennedy M., Sellers R. et al. (2011a). FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat. Cell Biol. 13, 1092-1099 10.1038/ncb2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Khvorostov I., Hong J. S., Oktay Y., Vergnes L., Nuebel E., Wahjudi P. N., Setoguchi K., Wang G., Do A. et al. (2011b). UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 30, 4860-4873 10.1038/emboj.2011.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Rielland M., Yalcin S. and Ghaffari S. (2011c). Regulation and function of FoxO transcription factors in normal and cancer stem cells: what have we learned? Curr. Drug Targets 12, 1267-1283 10.2174/138945011796150325 [DOI] [PubMed] [Google Scholar]
- Zhang J., Nuebel E., Daley G. Q., Koehler C. M. and Teitell M. A. (2012). Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 11, 589-595 10.1016/j.stem.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Campreciós G., Rimmelé P., Liang R., Yalcin S., Mungamuri S. K., Barminko J., D'Escamard V., Baron M. H., Brugnara C. et al. (2014). FOXO3-mTOR metabolic cooperation in the regulation of erythroid cell maturation and homeostasis. Am. J. Hematol. 89, 954-963 10.1002/ajh.23786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C. A., Horwitz M. S., Hockenbery D., Ware C. et al. (2012). HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 31, 2103-2116 10.1038/emboj.2012.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K. and Ding S. (2010). Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7, 651-655 10.1016/j.stem.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou G.-M., Luo M.-H., Reed A., Kelley M. R. and Yoder M. C. (2007). Ape1 regulates hematopoietic differentiation of embryonic stem cells through its redox functional domain. Blood 109, 1917-1922 10.1182/blood-2006-08-044172 [DOI] [PubMed] [Google Scholar]