Abstract
Large-scale genomic rearrangements are a major force of evolutionary change and the ascertainment of such events between the human and great ape genomes is fundamental to a complete understanding of the genetic history and evolution of our species. Here, we present the results of an evolutionary analysis utilizing array comparative genomic hybridization (array CGH), measuring copy-number gains and losses among these species. Using an array of 2460 human bacterial artificial chromosomes (BACs) (12% of the genome), we identified a total of 63 sites of putative DNA copy-number variation between humans and the great apes (chimpanzee, bonobo, gorilla, and orangutan). Detailed molecular characterization of a subset of these sites confirmed rearrangements ranging from 40 to at least 175 kb in size. Surprisingly, the majority of variant sites differentiating great ape and human genomes were found within interstitial euchromatin. These data suggest that such large-scale events are not restricted solely to subtelomeric or pericentromeric regions, but also occur within genic regions. In addition, 5/9 of the verified variant sites localized to areas of intrachromosomal segmental duplication within the human genome. On the basis of the frequency of duplication in humans, this represents a 14-fold positional bias. In contrast to previous cytogenetic and comparative mapping studies, these results indicate extensive local repatterning of hominoid chromosomes in euchromatic regions through a duplication-driven mechanism of genome evolution.
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: O. Ryder, L. Faust, and E. Adams.]
The evolution of human and nonhuman primate genomes has been studied at two levels, karyotypic differences and single-base-pair nucleotide differences (Yunis et al. 1980; Yunis and Prakash 1982; Muller et al. 1989; Kaessmann et al. 2001; Muller and Weinberg 2001). To date, no ascertainment has been made of the frequency, extent, or distribution of large-scale sequence gain or loss events, defined here as copy-number altering events involving >10 kb of sequence, yet events that are undetectable at the cytogenetic level. Events of this size have the potential to significantly impact the gene complement and genome structure of closely related species; however, genome-wide experimental comparative analyses at this scale have been impossible due to the lack of efficient methods of comparing genomes with accuracy and a high level of resolution. With the advent of the human genome reference sequence, however, new approaches have emerged that facilitate the simultaneous detection and high-resolution mapping of large-scale DNA variation across the entire genome (Snijders et al. 2001). One such method, array comparative genomic hybridization (array CGH), has demonstrated the ability to reliably detect DNA copy-number changes between genomic DNA samples with the resolution of a single BAC clone (Pinkel et al. 1998; Albertson et al. 2000; Snijders et al. 2001).
Our array CGH procedure involves the hybridization of differentially labeled specimen and reference genomic DNA to an array of large-insert genomic clones. The hybridization intensity ratio at each array locus is proportional to the copy-number ratio between genomic DNA samples, which is used as a measure of putative regional gains and losses. The microarray used in this study consisted of 2460 large-insert human BAC clones that had been mapped previously by STS content and validated by FISH (Snijders et al. 2001). The entire clone set encompassed ∼12% (370 Mb) of the entire human genome, providing a resolution of 1 BAC every 1.4 Mb of DNA. To date, the array CGH technique has been used primarily to assess within-species DNA copy-number variation associated with tumor progression or recurrent structural rearrangements of the human genome (Pinkel et al. 1998; Albertson et al. 2000; Bruder et al. 2001; Hodgson et al. 2001). In this study, we applied array CGH to detect fixed interspecific copy-number differences between the genomes of humans and great apes.
Large-scale sequence duplication and deletion events have the potential to significantly impact the structure and evolution of the primate genome. At the simplest level, sequence gain and loss events can alter the gene complement of an organism, which may result in phenotypic variation susceptible to selection pressures. Gene loss has been proposed as an important force in driving the evolution of eukaryotic genomes in response to selective pressures such as altered growth conditions in yeast, or infectious disease in the human population (Olson 1999). This “less is more” hypothesis can only be addressed with respect to primate genomes through the application of high-throughput sequence variation detection that allows for direct and efficient correlation with genome sequence information (or cDNA expression analysis). In addition, segmental duplications characterized from analysis of the human genome have proven to be considerably variable during the course of primate evolution, with alterations seen in copy number and location in the genomes of the great apes, particularly within the pericentromeric regions of the genome (Guy et al. 2000; Horvath et al. 2000b). Very little is known in regard to the role of segmental duplications in restructuring euchromatin, although they have been hypothesized to provide the molecular substrate for chromosomal rearrangement among humans and great ape chromosomes (Stankiewicz et al. 2001; Samonte and Eichler 2002).
Using normal human genomic DNA as the reference, we compared relative hybridization intensities among four great ape species, chimpanzee (Pan troglodytes), bonobo (Pan paniscus), gorilla (Gorilla gorilla), and orangutan (Pongo pygmaeus), in pairwise comparisons with human. Multiple individuals of each species were examined in independent experiments, and array loci were scored as potential variants only if a consistent increase or decrease in hybridization intensity ratio was observed across all trials and all individuals of each species (see Methods). This approach ensured the detection of fixed differences between the species, but eliminated potential large-scale polymorphisms within species for further analysis. The application of such conservative criteria, however, was essential to identify sites of interspecific copy number differences as opposed to spurious artefacts. We examined the distribution and the sequence context of these sites to provide some insight into the significance of such variation as a potential force underlying chromosomal change among humans and nonhuman primates. This study, therefore, presents the first genome-wide comparison of great ape species, with the level of resolution afforded by array CGH.
RESULTS
Genomic DNA from multiple unrelated individuals of each primate species were analyzed using array CGH, and only sites displaying copy-number variation in all individuals were scored as positive (Fig. 1). Four great ape species were examined in this study, common chimpanzee (n = 2), pygmy chimpanzee (n = 4), gorilla (n = 5), and orangutan (n = 4). In addition, several hybridizations with individual primate genomic DNA samples were repeated and the results compared with previous hybridizations to test the consistency of the hybridization results. On the basis of these criteria, 63 sites of variant intensity ratios (38 reductions and 25 increases) were consistently identified in primate–human comparisons and mapped to the human reference genome sequence (Fig. 1; Supplemental Table 1). Both lineage-specific and shared-ratio differences were observed among the great apes. Lineage-specific differences, however, predominated (Fig. 2B). As expected, the quantity of variant sites detected in each great ape species was in proportion to the estimated divergence times of each species (Goodman 1999), as orangutan showed the greatest number of ratio differences and the chimpanzee species demonstrated the fewest ratio differences.
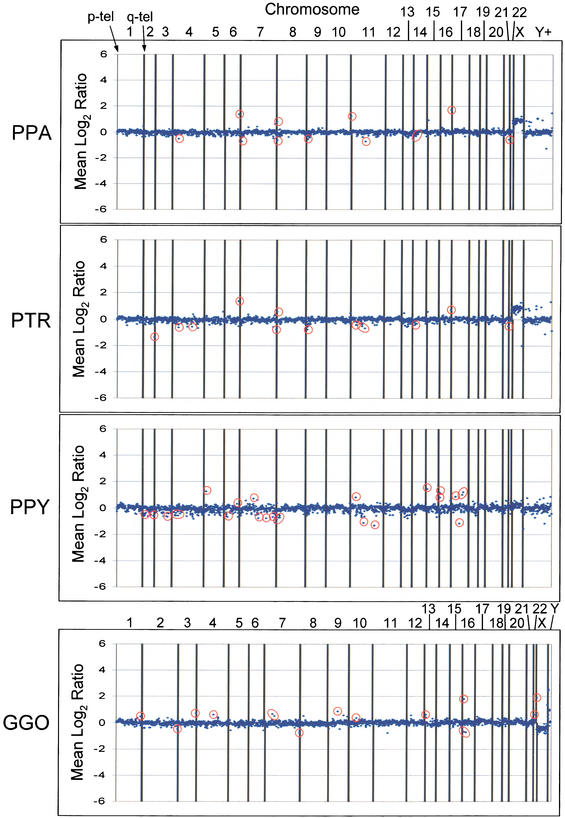
Figure 1.
Examples of the array CGH data. Graphs depicting data for 2460 array loci are presented for a single individual of each species. (PPA) Pan paniscus, (PTR) Pan troglodytes, (GGO) Gorilla gorilla, (PPY) Pongo pygmaeus. Putative variant sites are circled in red. Vertical lines separate the loci from each chromosome, with the p-arm telomere oriented toward the left of each interval and the q-arm telomere toward the right. The Y+ interval contains loci from the Y chromosome in addition to potentially duplicated clones, determined by FISH characterization, and variants in this interval were disregarded for this analysis. Not all variant sites detected in a particular species are represented by the individuals shown. Hybridization of human male reference DNA with female primate DNA or vice versa will yield a constitutive gain or loss for the X chromosome, as seen for the PTR, PPA, and GGO hybridizations. The GGO hybridization depicted here presents data from version 2.0 of the human BAC array, whereas the PTR, PPA, and PPY hybridizations depicted are from version 1.14 of the human BAC array.
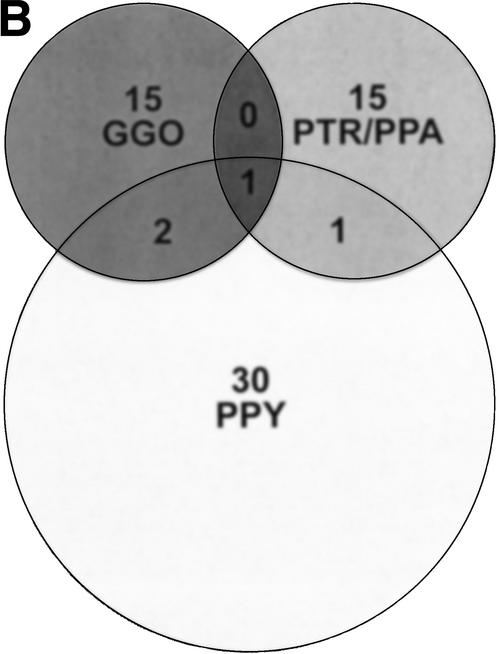
Figure 2.
Sites of great ape/human variation detected by array CGH. (A) A histogram shows the chromosomal distribution of loci detected by array CGH with variant signal intensities consistent with gains and losses of genomic DNA in each great ape species when compared with human. The number of variant sites per 100 Mb is shown for each chromosome. Approximately 12% of the human genome (2460 BAC and PAC clones) was screened by this assay, identifying a total of 63 loci for all species comparisons. (B) A Venn diagram demonstrates that the majority of sites detected by array CGH are specific to each great ape lineage. The number of variant sites per species, PTR and PPA are grouped as they share a majority of variant sites, is shown above the species abbreviation and shared sites are indicated in the respective overlapping regions. The Venn diagram describes 64 rearrangements, yet 63 sites of variation were detected in our analysis. This is due to the fact that one rearrangement site (RP11-88L18) demonstrated a duplication in orangutan and a deletion in gorilla, which are considered separate events. A complete description of each site including Log2 ratio intensity statistics is available (Supplemental Table 1).
We chose a subset of the 63 putative variant sites for detailed experimental validation and verification of the array CGH approach. Seven sites with increased primate signal intensity ratios, therefore potential duplications with respect to the human genome, were assessed by interphase and metaphase FISH. In all cases, a genomic duplication was detected among the great apes when probing with the human single-copy arrayed BAC. Among these variants, we observed both intra- and interchromosomal duplications, duplications within homologous chromosomes and between nonhomologous chromosomes, respectively, which were readily resolved at the metaphase and interphase levels (Fig. 3A–C). Similarly, seven sites with reduced primate signal intensity ratios, potential interspecific deletions, were examined by FISH. We encountered one instance in which there was a complete absence of signal, suggesting that a deletion of at least the size of the arrayed BAC (175 kb by PFGE, data not shown) had occurred (Table 1; Fig. 3A). The extent of this deletion was subsequently confirmed by STS-content mapping and Southern analysis (data not shown). Reduced FISH signal intensity was observed for several of the remaining six sites tested, yet deletions could not be demonstrated convincingly by comparative FISH. Considering that the Log2 ratios for these unverified sites were decreased to a lesser extent than the complete deletion demonstrated in orangutan by RP11-171I8, we believe these sites were partial deletions of the arrayed BAC sequence. Thus, we sought an alternative approach to validate these potential partial deletions.
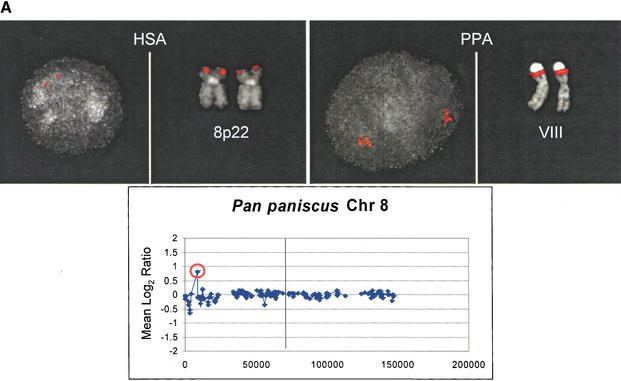
Figure 3.
Comparative FISH Validation of duplications. Interphase and metaphase images are presented for three array CGH duplications verified by FISH; all signals are shown in red. Array CGH results for an individual of the respective primate species are shown below each FISH comparison, with the variant BAC used as a probe in each respective FISH experiment circled in red. The x-axis represents the map position in megabases along the chromosome and the y-axis represents the Log2 ratio. The p-telomere is oriented to the left in each plot, and the q-telomere to the right. The centromere is indicated as a vertical line. An intrachromosomal duplication of RP11-218N24 is confirmed in bonobo when compared with human (A). Similarly, a gorilla intrachromosomal duplication is illustrated by interphase FISH using probe RP11-64I2 in contrast to the single signal on human 7p14 (B). Extensive interchromosomal duplication of BAC RP11-88L18 is shown for orangutan acrocentric chromosomes when compared with the unique 5p15 human locus (C). See Table 1 for more details.
Table 1.
Summary of Verified Variant Sites of Genomic Rearrangement
| Clone ID | Event | Chrom | Fish | STS | Dup. contenta | Gene contenta | Avg. Log2 ratio | |||
| PPA | PTR | GGO | PPY | |||||||
| RP11-88L18 | Duplication | 5 | 5p15 | AFM105XG1 | Intra, ∼50 kb @ 99% | None | −0.25 | −0.10 | −1.13 | 0.65 |
| RP11-64I02 | Duplication | 7 | 7p14 | SHGC-33366 | None | RFP-1 | 0.15 | 0.15 | 0.71 | −0.18 |
| RP11-218N24 | Duplication | 8 | 8p22 | SHGC-11994 | None | CTSB, FDFT1, GATA4 | 0.67 | 0.51 | 0.08 | 0.32 |
| RP11-209M09 | Duplication | 11 | 11p15 | SHGC-14668 | Intra, ∼50 kb @ 98% | RRM1 | 0.81 | 0.29 | −0.09 | 0.27 |
| RP11-56J22 | Duplication | 11 | 11p15.1 | AFM210yb8 | None | None | −0.09 | −0.14 | −0.11 | 0.69 |
| RP11-194H07 | Duplication | 15 | 15q14 | SHGC-24145 | Intra, ∼40 kb @ 99% | SLC12A6, NOLA3, KIAA0855 | 0.47 | 0.36 | −0.06 | 1.23 |
| RP11-49M06 | Duplication | 16 | 16p13.2 | SHGC-11261 | None | PIG7, SNN, LOC51061 | 0.04 | 0.10 | 0.13 | 1.20 |
| RP11-171I08 | Deletion | 11 | 11p11.12 | WI-636 | Intra, ∼30 kb @ 95% | None | −0.01 | −0.06 | −0.49 | −0.96 |
| RP11-75H24 | Deletion | 11 | 11q12.3 | G29694 | Intra, ∼50 kb @ 95% | None | −0.61 | −0.64 | 0.00 | −0.46 |
Defined as within 100 kb proximal or distal of the BAC-linked STS according to the UCSC Genome Browser, August 2001 assembly.
Average Log2 ratios exceeding the +/−0.5 threshold are indicated in bold type.
Experimentally verified gains and losses among great ape species, with respect to the human genome, were mapped and characterized for local duplication and genic content. The species average Log2 ratios for each variant site are shown for all species examined. Duplication content is indicated with the type, approximate size, and percent similarity of the duplications involved.
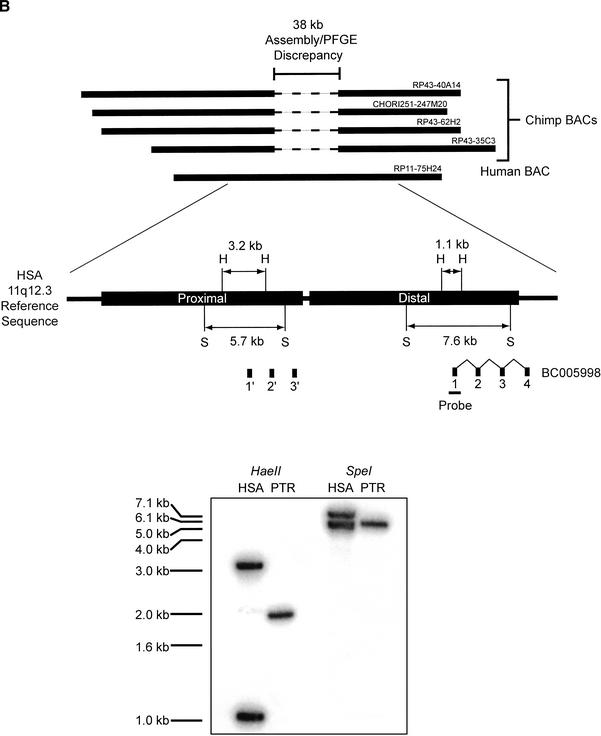
We developed a BAC end sequence-based strategy to verify the potential partial deletions that were not confirmed by comparative FISH. This strategy compared the insert size of primate BACs linked to a variant site with the estimated BAC insert size according to the human genome reference sequence (Methods; Fig. 4B). A disparity between the experimentally determined primate BAC insert size, and the estimated insert size on the basis of sequence similarity searches against the human genome sequence, was indicative that a partial deletion had occurred. For a putative deletion in chimpanzee 11q12, this approach identified a ∼38-kb disparity between chimpanzee BAC insert size and the human equivalent insert size (Fig. 3B). Examination of the underlying human sequence revealed the presence of a ∼40-kb tandem intrachromosomal duplication event that contained a partial duplication of the BC005998 gene. Subsequent Southern analysis confirmed the deletion of the proximal BC005998 duplicon in the chimpanzee genome (Fig. 3B). Thus, we had successfully identified a deletion in the chimpanzee genome using array CGH that was below the level of cytogenetic detection and involved the partial deletion of a BAC clone on the array. We believe that the tandem duplication was present prior to the divergence of the great ape lineages, as the gorilla and orangutan did not show reduced intensity ratios at this locus. Two other predicted sites were investigated for possible deletions by this method; however, no size discrepancies were detected by this BAC end-mapping approach.
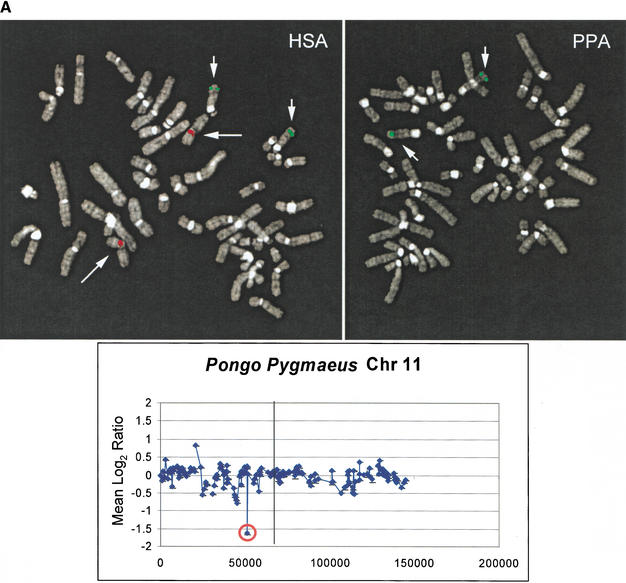
Figure 4.
Validation of array CGH-detected deletions. (A) Validation of a deletion by comparative FISH. FISH images are shown for orangutan variant array BAC RP11-171I8. To control for variability in FISH conditions, human and orangutan cells were mixed on a slide prior to fixation. Hybridization with BAC RP11-171I8 (red) and an unrelated control probe, RP11-233C13 (green) is shown for human (HSA) and orangutan (PPY). The complete absence of signal was observed for RP11-171I8 against the orangutan metaphase chromosomes, but not RP11-233C13. A genomic deletion at least the size of the BAC (175 kb by PFGE; see text) was confirmed by STS content mapping and Southern Analysis (data not shown). (B) Deletion validation by primate BAC analysis and Southern blot. Human BAC clone RP11-75H24 showed a reduced relative fluorescent signal intensity ratio of chimpanzee compared with human (Table 1). Four chimpanzee BAC clones from libraries RP-43 and CHORI-251 orthologous to this site were obtained, end-sequenced, and restriction mapped. A ∼38-kb discrepancy in size was determined by comparing BAC PFGE size and BAC end sequence placement of the chimpanzee loci against both public and private assembly versions of the human genome. Analysis of the human sequence identified ∼40 kb-tandem duplications (black boxes labeled Proximal and Distal), both containing exons 1–3 of gene BC005998. Using a probe spanning exon 1 of BC005998, Southern analysis of HaeII and SpeI-digested human (RP11-75H24) and chimpanzee (RP43-35C3) BAC DNA show a loss of a duplication within the chimpanzee genome (HaeII and SpeI restriction sites are indicated by the H and S, respectively).
Combined, the experimental validation results suggest that array CGH offers excellent sensitivity in detecting genomic duplications, as 100% of array CGH-detected variant sites were verified experimentally by comparative FISH; however, deletions were not detected with similar efficiency, as only two deletions have been verified experimentally. Unlike intensity ratio increases, the intensity ratio decreases reported by array CGH may be due to a number of other factors, such as extensive sequence divergence or dramatic restructuring of loci as a result of repeat-content variation. Therefore, sites of reduced intensity ratios, without significant size differentials between species by our BAC end-sequence-based approach, remain important targets for further comparative sequence analysis. As large-insert genomic libraries become available from a number of the great ape species (Eichler and DeJong 2002), high-quality BAC sequencing of such regions will provide the most efficient method for assessing the molecular nature of these differences.
DISCUSSION
Previous studies have suggested that the majority of fixed large-scale variation between the genomes of humans and great apes localize to gene-poor heterchromatic regions (Verma and Luke 1991; Archidiacono et al. 1995; Trask et al. 1998). In light of these predictions, we examined the genomic landscape around the nine experimentally verified duplications (n = 7) and deletions (n = 2). Gene content within the vicinity of a bonafide genomic rearrangement site was assessed by considering the proximal and distal 100 kb flanking the array BAC-linked STS, or by placing the BAC end sequences for the variant clone against the human genome assembly by sequence similarity searches (Methods). Overall, we found that six of nine experimentally verified sites of rearrangement lie within or in close proximity to actively transcribed euchromatic regions (Table 1). These results suggest that gene-rich regions are susceptible to copy-number changes between humans and great apes, and that array CGH can effectively detect such rearrangements. Such regional differences in copy number could, in theory, underlie gene-expression differences that have been predicted and observed between humans and great apes (King and Wilson 1975; Enard et al. 2002).
The distribution of sites along the interstitial euchromatic regions of human chromosomes, determined by viewing the BAC-linked STS on the human assembly, shows that these events show no bias toward the subtelomeric and pericentromeric regions of human chromosomes. A total of 9 of the 63 variant sites occurred within 2 Mb of a subtelomeric region or pericentromeric region, and the remaining 86% of sites (54/63) mapped to euchromatic regions of the human genome. This result demonstrates that the large genomic insertion and deletion events occurred in euchromatic, and potentially gene-rich regions, and were not limited to the heterochromatic expansions observed in previous karyotypic analyses (Yunis et al. 1980; Yunis and Prakash 1982).
During our analysis of potential large-scale variation, we noticed that certain chromosomes showed a disproportionately large number of variant sites (Fig. 2A). Interestingly, many of the same chromosomes enriched for rearrangements (chromosomes 4, 7, 8, 16, 17, and 22) have also been shown to be enriched for segmental duplications (Bailey et al. 2002). A positive correlation (r2 = 0.50) is observed when segmental duplication content and the number of variant sites per chromosome, adjusted for array clone coverage per chromosome, are compared. Several studies have shown that highly homologous sequences within genomes (also known as segmental duplications) may predispose to homologous unequal recombination, leading to large-scale deletions and duplications (Lupski 1998; Mazzarella and Schlessinger 1998; Ji et al. 2000; Emanuel and Shaikh 2001). More recently, nonallelic homologous recombination between such sequences has been postulated to underlie evolutionary chromosome rearrangements (Tunnacliffe et al. 1993; Nickerson et al. 1999; Valero et al. 2000; Dehal et al. 2001; Stankiewicz et al. 2001).
To more specifically test this association, we examined the sequence context for the nine verified structural rearrangements detected in this study. Segmental duplications comprise an estimated 5% of the total human genome sequence, yet we found 5/9 (56%) of our validated rearrangements were within close proximity to segmental duplications, an 11-fold bias (Bailey et al. 2002). Considering the majority of duplications associated with the variant sites were intrachromosomal duplications (2.8% of total genome sequence), this bias increases to 14-fold (Bailey et al. 2002). These results indicate a highly significant (G = 19.63, P < 0.0001) nonrandom association of large-scale structural variation and segmental duplication. Although the sample size of verified duplications and deletions is small, we believe this trend suggests the importance segmental duplications may play in evolutionary rearrangements. Furthermore, the sites chosen for the array used in our great ape comparisons were selected due to their seemingly single-copy nature as determined by extensive testing on human material. This inherent bias against sites of duplication may thus provide a fairly conservative estimate of the role segmental duplications play in interstitial euchromatic evolutionary rearrangements. A microarray targeted to regions of recent segmental duplication should provide a more accurate view of the dynamics of these regions and their relative importance in contributing to great ape and human evolution.
In summary, our results show that array CGH technology is a powerful approach for interrogating large-scale differences among the genomes of closely related species. It can successfully identify large-scale deletions and duplications too small to be detected by standard karyotype analysis, yet too large to be readily resolved by whole-genome shotgun sequencing approaches. Our analysis of the orangutan genome using human BAC microarrays suggests that 3% sequence divergence is sufficient for cross-species comparisons by this method. Array CGH is therefore a valuable first step to target regions for specialized study—and is therefore amenable to many cross-species comparisons in which entire genomes are unlikely to be sequenced. Second, our analysis of primate genomes indicates that large-scale events are not uncommon within genic regions. Whereas large-scale differences among great apes have been documented within heterochromatic regions such as pericentromeric DNA, this is among the first demonstrations of large-scale differences within euchromatic DNA. We have characterized deletions and duplications ranging in size from 40–175 kb and have provided a first approximation the frequency of such events at a genome-wide level. Rate estimation of such events will require more uniform representation of the genome as well as resolution of regions enriched for segmental duplications. Perhaps the most striking finding is that the events are nonrandomly distributed. Regions within or in the vicinity of segmental duplications show a proclivity to delete and duplicate. Whereas such genomic variation has commonly been reported in association with de novo rearrangements associated with disease, these data implicate a homology-driven mechanism for the generation of fixed differences that distinguish closely related species. The biological significance of these events and their relationship to gene expression variation among primates await further characterization.
METHODS
Human BAC Arrays
Arrays were prepared as described by Snijders et al. (2001). Briefly, ligation-mediated PCR was used to prepare DNA representations of ∼2460 human BAC and P1 artificial chromosome (PAC) clones. The DNA was suspended in 20% DMSO and spotted in triplicate onto chromium-coated microscope slides using a custom-built printer with capillary printing pins. The entire array of ∼7500 spots filled a 12-mm × 12-mm square. Each clone on the array contained at least one STS mapped to the human reference assembly sequence, allowing the underlying genomic sequence to be assessed. All clones on the array were cytogenetically mapped by FISH, confirming 93.4% as single copy. In addition, extensive analyses of cell lines containing known single-copy aberrations were performed to allow recognition of clones that contained significant amounts of sequence that mapped to multiple sites in the human genome. Data from such clones were excluded from the analysis presented here. On average, the array provides a resolution of one genomic clone for every 1.4 Mb of human genomic sequence. It should be noted that by performing this analysis with a human BAC array, deletions that have occurred specifically within the human lineage with respect to other primate genomes cannot be readily detected. Deletions in the human lineage with respect to the chimpanzee may become apparent as the chimpanzee genome project progresses, and sites detected via this method will provide landmarks for where the chimpanzee and human sequence maps may vary significantly. For the other primate species for which no dedicated genome project is anticipated, it may be necessary to develop reciprocal primate BAC arrays to detect large-scale deletions that have occurred within the human genome.
Primate DNA Samples
Four great ape species were examined in this study, common chimpanzee (n = 2), pygmy chimpanzee (n = 4), gorilla (n = 5), and orangutan (n = 4). For each species, all individuals were unrelated with the exception of two gorilla samples that were related as half-sisters. Chimpanzee samples (PTR BC449, BC450, and PPA OR833, KB8763, BB501, BB502) were derived from lymphoblastoid and primary fibroblast cell lines. Of the four orangutan DNAs (O100, O101, GM04272, Segundo), three were isolated from EBV-transformed lymphoblastoid cell lines and were compared with results from an unrelated blood sample. The gorilla cohort (Kwan, KB6278, 9247, 324, 465) consisted of four DNA samples extracted from peripheral blood lymphocytes and one primary fibroblast cell line. The use of DNA isolated from both transformed cell line material and peripheral blood served as a valuable control to assess the quality of the cell line material. For microarray analysis, only sites in which consistent increases and decreases in Log2 ratios were observed in all individuals were considered. Although this requirement eliminated potential false-positive signals due to small-scale rearrangements within cell lines, it also removed potential structural polymorphic variation from further analysis.
Array Comparative Genomic Hybridization
Genomic DNA samples were prepared from blood using the PureGene Genomic DNA Isolation Kit (Gentra Systems). Great ape and human DNAs were labeled directly with Cy3 and Cy5 fluorochromes, respectively, by random primer labeling. Arrays were simultaneously hybridized with a primate and a human genomic DNA probe for at least 48 h, using unlabeled human Cot-1 DNA to block repetitive sequences. After post-hybridization washing, the arrays were imaged with a custom-built CCD camera system, and quantitative measurements of the fluorescence intensity ratios were obtained using the software package UCSF SPOT (Pinkel et al. 1998; Jain et al. 2002). Ratios for the triplicate spots were averaged. For each hybridization, the primate-to-human fluorescence intensity ratio (Log2 ratio) at every array locus was assessed for variation. Arrays fabricated with two different sets of print stocks and printed in several different batches were used to eliminate false positives due to inconsistencies in array production. If a particular site appeared to be variant in a primate species, the Log2 ratios for all hybridizations of DNAs from individuals of that species were averaged. Sites with average Log2 ratios >0.5 and <−0.5 were selected as putative variants.
The ratio variation among clones at constant copy number (most of the clones on the array) was significantly higher for great ape/human comparisons than for human/human comparisons. This ratio variation complicated recognition of interspecies copy-number differences that might be affecting only a portion of a BAC, and would therefore show a ratio change of reduced magnitude compared with the expected value for single- or multi-copy changes. Fixed copy-number differences between species should result in no copies in the genome for a deletion event, or four copies for a duplication event. Thus, if the change affected the entire BAC, one should see Log2 ratios of minus infinity or 1 for deletions and duplications, respectively. However, several factors modify this expectation, including the following: (1) changes that affect only part of the BAC, which are predicted to produce less dramatic ratio differences, (2) whether or not the arrayed BAC contains duplicated sequences, and (3) incomplete suppression of the repetitive sequences in the great ape genome by human Cot-1 DNA.
Fluorescence in Situ Hybridization
Lymphoblastoid cell lines derived from humans (Homo sapiens) and four great ape species (bonobo—Pan paniscus, chimpanzee—Pan troglodytes, lowland gorilla—Gorilla gorilla, and orangutan—Pongo pygmaeus) were used to prepare metaphase and interphase nuclei. In situ hybridizations with BAC probes corresponding to the arrayed clones, and control probes where appropriate, were conducted using standard techniques (Lichter et al. 1990). To prevent cross-hybridization due to the presence of repetitive sequence within BAC probes, Cot-1 DNA was used to block potential hybridization of high-copy repeat sequences. A minimum of 20 interphase and metaphase nuclei were examined in each hybridization experiment for the assessment of genomic duplications and deletions. For confirmation of the RP11-171I8 deletion in Pongo pygmaeus, cells from Homo sapiens and Pongo pygmaeus were mixed on a slide prior to fixation to ensure hybridization conditions were equivalent for cells of both species.
BAC Analysis
Large-insert genomic chimpanzee BAC clones corresponding to sites of putative deletion were isolated from the RPCI-43 and CHORI-251 BAC libraries. Amplicons generated by PCR from end-sequences of the variant human BAC were used as hybridization probes against the chimpanzee large-insert clone libraries as described previously (Horvath et al. 2000a). The resulting chimpanzee BAC clones were end-sequenced (T7 and SP6), and the end sequences were aligned to the human genome assembly using sequence similarity searches (BLAST). The human equivalent insert size of the chimpanzee BACs was then calculated. The insert size of chimpanzee BAC clones was determined by pulsed-field gel electrophoresis. A disparity between the insert size of the chimpanzee BAC clone and the human equivalent insert size indicated that a deletion in the chimpanzee genome occurred. Southern blot analysis was used to confirm the presence of the deletion in the chimpanzee BACs with respect to the human array BAC.
Acknowledgments
We thank Oliver Ryder, Lisa Faust, and Erin Adams for providing primate material for this study. This work was supported, in part, by NIH grants GM58815 and HD043569 and U.S. Department of Energy grant ER62862 to E.E.E., NCI grants CA83040 to D.P. and CA84118 to D.A., and the financial support of Telethon, CEGBA (Centro di Eccellenza Geni in campo Biosanitario e Agroalimentare, and MIUR (Ministero Italiano della Istruzione e della Ricerca) to N.A. The financial support of the W.M. Keck Foundation, Vysis Inc., and a grant from the Charles B. Wang Foundation to the Center for Computational Genomics, Case Western Reserve University are also gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL eee@cwru.edu; FAX (216) 368-3432.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1003303.
REFERENCES
- 1.Albertson D.G., Ylstra, B., Segraves, R., Collins, C., Dairkee, S.H., Kowbel, D., Kuo, W.L., Gray, J.W., and Pinkel, D. 2000. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat. Genet. 25: 144-146. [DOI] [PubMed] [Google Scholar]
- 2.Archidiacono N., Antonacci, R., Marzella, R., Finelli, P., Lonoce, A., and Rocchi, M. 1995. Comparative mapping of human alphoid sequences in great apes using fluorescence in situ hybridization. Genomics 25: 477-484. [DOI] [PubMed] [Google Scholar]
- 3.Bailey J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- 4.Bruder C.E., Hirvela, C., Tapia-Paez, I., Fransson, I., Segraves, R., Hamilton, G., Zhang, X.X., Evans, D.G., Wallace, A.J., Baser, M.E., et al. 2001. High resolution deletion analysis of constitutional DNA from neurofibromatosis type 2 (NF2) patients using microarray-CGH. Hum. Mol. Genet. 10: 271-282. [DOI] [PubMed] [Google Scholar]
- 5.Dehal P., Predki, P., Olsen, A.S., Kobayashi, A., Folta, P., Lucas, S., Land, M., Terry, A., Ecale Zhou, C.L., Rash, S., et al. 2001. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science 293: 104-111. [DOI] [PubMed] [Google Scholar]
- 6.Eichler E.E. and DeJong, P.J. 2002. Biomedical applications and studies of molecular evolution: A proposal for a primate genomic library resource. Genome Res. 12: 673-678. [DOI] [PubMed] [Google Scholar]
- 7.Emanuel B.S. and Shaikh, T.H. 2001. Segmental duplications: An ‘expanding’ role in genomic instability and disease. Nat. Rev. Genet. 2: 791-800. [DOI] [PubMed] [Google Scholar]
- 8.Enard W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296: 340-343. [DOI] [PubMed] [Google Scholar]
- 9.Goodman M. 1999. The genomic record of Humankind's evolutionary roots. Am. J. Hum. Genet. 64: 31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guy J., Spalluto, C., McMurray, A., Hearn, T., Crosier, M., Viggiano, L., Miolla, V., Archidiacono, N., Rocchi, M., Scott, C., et al. 2000. Genomic sequence and transcriptional profile of the boundary between pericentromeric satellites and genes on human chromosome arm 10q. Hum. Mol. Genet. 9: 2029-2042. [DOI] [PubMed] [Google Scholar]
- 11.Hodgson G., Hager, J.H., Volik, S., Hariono, S., Wernick, M., Moore, D., Albertson, D.G., Pinkel, D., Collins, C., Hanahan, D., et al. 2001. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat. Genet. 29: 459-464. [DOI] [PubMed] [Google Scholar]
- 12.Horvath J., Schwartz, S., and Eichler, E. 2000a. The mosaic structure of a 2p11 pericentromeric segment: A strategy for characterizing complex regions of the human genome. Genome Res. 10: 839-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horvath J., Viggiano, L., Loftus, B., Adams, M., Rocchi, M., and Eichler, E. 2000b. Molecular structure and evolution of an α/non-α satellite junction at 16p11. Hum. Mol. Genet. 9: 113-123. [DOI] [PubMed] [Google Scholar]
- 14.Jain A.N., Tokuyasu, T.A., Snijders, A.M., Segraves, R., Albertson, D.G., and Pinkel, D. 2002. Fully automatic quantification of microarray image data. Genome Res. 12: 325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji Y., Eichler, E.E., Schwartz, S., and Nicholls, R.D. 2000. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 10: 597-610. [DOI] [PubMed] [Google Scholar]
- 16.Kaessmann H., Wiebe, V., Weiss, G., and Paabo, S. 2001. Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nat. Genet. 27: 155-156. [DOI] [PubMed] [Google Scholar]
- 17.King M.C. and Wilson, A.C. 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107-116. [DOI] [PubMed] [Google Scholar]
- 18.Lichter P., Tang, C.J., Call, K., Hermanson, G., Evans, G.A., Housman, D., and Ward, D.C. 1990. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 247: 64-69. [DOI] [PubMed] [Google Scholar]
- 19.Lupski J.R. 1998. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 14: 417-422. [DOI] [PubMed] [Google Scholar]
- 20.Mazzarella R. and Schlessinger, D. 1998. Pathological consequences of sequence duplications in the human genome. Genome Res. 8: 1007-1021. [DOI] [PubMed] [Google Scholar]
- 21.Muller S. and Wienberg, J. 2001. “Bar-coding” primate chromosomes: Molecular cytogenetic screening for the ancestral hominoid karyotype. Hum. Genet. 109: 85-94. [DOI] [PubMed] [Google Scholar]
- 22.Muller S., Stanyon, R., O'Brien, P.C., Ferguson-Smith, M.A., Plesker, R., and Wienberg, J. 1999. Defining the ancestral karyotype of all primates by multidirectional chromosome painting between tree shrews, lemurs and humans. Chromosoma 108: 393-400. [DOI] [PubMed] [Google Scholar]
- 23.Nickerson E., Gibbs, R.A., and Nelson, D.L. 1999. Sequence analysis of the breakpoints of a pericentric inversion distinguishing the human and chimpanzee chromosomes 12. Am. J. Hum. Genet. 65: A56. [Google Scholar]
- 24.Olson M.V. 1999. When less is more: Gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 64: 18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinkel D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W.L., Chen, C., Zhai, Y., et al. 1998. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 20: 207-211. [DOI] [PubMed] [Google Scholar]
- 26.Samonte R.V. and Eichler, E.E. 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3: 65-72. [DOI] [PubMed] [Google Scholar]
- 27.Snijders A.M., Nowak, N., Segraves, R., Blackwood, S., Brown, N., Conroy, J., Hamilton, G., Hindle, A.K., Huey, B., and Kimura, K. 2001. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat. Genet. 29: 263-264. [DOI] [PubMed] [Google Scholar]
- 28.Stankiewicz P., Park, S.S., Inoue, K., and Lupski, J.R. 2001. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res. 11: 1205-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trask B.J., Massa, H., Brand-Arpon, V., Chan, K., Friedman, C., Nguyen, O.T., Eichler, E.E., van den Engh, G., Rouquier, S., Shizuya, H., et al. 1998. Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum. Mol. Genet. 7: 2007-2020. [DOI] [PubMed] [Google Scholar]
- 30.Tunnacliffe A., Liu, L., Moore, J.K., Leversha, M.A., Jackson, M.S., Papi, L., Ferguson-Smith, M.A., Thiesen, H.J., and Ponder, B.A.J. 1993. Duplicated KOX zinc finger gene clusters flank the centromere of human chromosome 10: Evidence for a pericentric inversion during primate evolution. Nucleic Acids Res. 21: 1409-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valero M.C., de Luis, O., Cruces, J., and Perez Jurado, L.A. 2000. Fine-scale comparative mapping of the human 7q11.23 region and the orthologous region on mouse chromosome 5G: The low-copy repeats that flank the Williams-Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s). Genomics 69: 1-13. [DOI] [PubMed] [Google Scholar]
- 32.Verma R.S. and Luke, S. 1991. Heteromorphisms of pericentromeric heterochromatin of chromosome 19. Genet. Anal. Tech. Appl. 8: 179-180. [DOI] [PubMed] [Google Scholar]
- 33.Yunis J.J. and Prakash, O. 1982. The origin of man: A chromosomal pictorial legacy. Science 215: 1525-1530. [DOI] [PubMed] [Google Scholar]
- 34.Yunis J.J., Sawyer, J.R., and Dunham, K. 1980. The striking resemblance of high-resolution G-banded chromosomes of man and chimpanzee. Science 208: 1145-1148. [DOI] [PubMed] [Google Scholar]