Abstract
Nodal/TGFβ signaling regulates diverse biological responses. By combining RNA-seq on Foxh1 and Nodal signaling loss-of-function embryos with ChIP-seq of Foxh1 and Smad2/3, we report a comprehensive genome-wide interaction between Foxh1 and Smad2/3 in mediating Nodal signaling during vertebrate mesendoderm development. This study significantly increases the total number of Nodal target genes regulated by Foxh1 and Smad2/3, and reinforces the notion that Foxh1-Smad2/3-mediated Nodal signaling directly coordinates the expression of a cohort of genes involved in the control of gene transcription, signaling pathway modulation and tissue morphogenesis during gastrulation. We also show that Foxh1 may function independently of Nodal signaling, in addition to its role as a transcription factor mediating Nodal signaling via Smad2/3. Finally, we propose an evolutionarily conserved interaction between Foxh1 and PouV, a mechanism observed in Pou5f1-mediated regulation of pluripotency in human embryonic stem and epiblast cells.
Keywords: TGFβ, Foxh1, Smad, Mesendoderm, Morphogenesis, Cell fate, Oct4, Pou5f1, Xenopus tropicalis, Signaling, Genomics, ChIP-seq, RNA-seq
INTRODUCTION
Cell signaling is essential for coordination of the dynamic spatiotemporal expression of genes. Systems level comprehension of the signaling inputs that control cell specification will facilitate decoding the fundamental regulatory mechanisms dictating animal development. Here, we examine Nodal signaling to uncover the network control system specifying mesendoderm development in Xenopus tropicalis. Nodal signaling is conserved during animal evolution and controls diverse biological activities, including left-right specification, stem cell pluripotency and mesendoderm development (Chea et al., 2005; Schier, 2003). Additionally, in regenerative biology, activation of the Nodal signaling pathway promotes the differentiation of embryonic stem cells (ESCs) toward mesendodermal lineages (Kubo et al., 2004). These diverse activities raise the issue of the underlying mechanism conferring specificity of gene regulation by Nodal in various developmental contexts.
Nodal signaling is triggered by ligand binding to type I and II receptors, which in turn phosphorylate Smad2 or Smad3. Phospho-Smads complex with Smad4, translocate to the nucleus and, together with other transcription factors (TFs) such as Foxh1, regulate target gene expression (Chen et al., 1996, 1997). Currently, the relative contribution of Foxh1 in mediating Nodal signaling has not been systematically examined in vivo. Nodal signaling is crucial for the proper induction of mesoderm and endoderm. The Nodal mutant mouse lacks the primitive streak and fails to form normal mesoderm, leading to developmental arrest shortly after gastrulation (Conlon et al., 1994; Varlet et al., 1997; Zhou et al., 1993). Zebrafish cyclops (cyc)/squint (sqt) double mutants fail to form the shield and gastrulation is disrupted (Feldman et al., 1998). In amphibians, these genes are zygotically expressed under the control of the maternal T-box transcription factor Vegt (Heasman, 2006). Inhibition of Nodal signaling in Xenopus via the overexpression of Nodal antagonists or by treatment with small-molecule Nodal receptor inhibitors leads to gastrulation defects (Agius et al., 2000; Ho et al., 2006; Osada and Wright, 1999; Sun et al., 1999). Three other Smad2/3-activating TGFβs function during mesendoderm formation: Gdf1/Vg1 is maternal, while Gdf3/Derriere and Inhbb/ActivinβB are zygotic. These are also important for normal early mesendodermal development (Birsoy et al., 2006; Piepenburg et al., 2004; Sun et al., 1999) and probably have overlapping functions with the Nodal proteins; we therefore refer to collective signaling by these ligands as Nodal signaling for simplicity. These results suggest that Nodal ligands are essential for vertebrate mesendoderm development.
Foxh1 (Fast1, forkhead activin signal transducer 1), is a winged-helix TF that is maternally expressed in Xenopus (Chen et al., 1996). It was first discovered as a mediator of Activin-like signaling via binding to the activin response element of the mix1 gene in conjunction with Smad2/3 (Chen et al., 1996). Subsequently, goosecoid (gsc) (Labbé et al., 1998), lhx1 (Watanabe et al., 2002), nodal1 (Osada et al., 2000) and pitx2 (Shiratori et al., 2001) were shown to be regulated by Foxh1. In mouse, Foxh1−/− embryos display a spectrum of phenotypes from severe gastrulation defects to milder anterior and midline deficiencies (Hoodless et al., 2001; Yamamoto et al., 2001). In zebrafish, schmalspur and midway mutants, both defective in the foxh1 gene, are deficient in prechordal plate, notochord and some axial mesoderm development (Pogoda et al., 2000; Sirotkin et al., 2000; Slagle et al., 2011). Loss of Foxh1 in Xenopus laevis results in reduced expression of mesendodermal markers, including gsc, nodal1 and mix1, but axial mesoderm was still present, albeit obviously abnormal (Howell et al., 2002; Kofron et al., 2004b; Watanabe and Whitman, 1999). The stronger phenotypes from loss of Nodal signaling, compared with foxh1 loss of function (LOF) in frog, fish and mouse, suggests that Nodal signals through both Foxh1-dependent and -independent pathways. Mixer, Foxh1.2, Gtf2ird1, Gtf2i, Tp53, Eomes and Tcf3 (also known as E2a) have been implicated in activation of Nodal targets in Xenopus (Cordenonsi et al., 2003; Germain et al., 2000; Howell et al., 2002; Ku et al., 2005; Ring et al., 2002; Slagle et al., 2011; Teo et al., 2011; Yoon et al., 2011), although the extent that any of these TFs play in the broader regulation of Nodal signaling gene batteries in developing embryos remains ill defined. Thus, despite advances made in dissecting the Nodal molecular cascade, significant gaps remain in our knowledge.
To comprehend how mutations in Nodal signaling affect transcriptional networks and cause developmental defects, we examined the cohort of genes regulated by Nodal in the context of early vertebrate embryonic development at a genome-wide level. We determined which of these genes are directly bound and regulated by Foxh1 and/or Smad2/3. Foxh1 and Smad2/3 interact with thousands of genomic sites and affect the transcriptional responses of hundreds of Nodal target genes. We uncovered a large set of new targets, and have begun understanding the complexity associated with the Nodal signaling network. Although Foxh1 and Smad2/3 directly bound and regulated some Nodal-activated genes, we found a Nodal signaling-independent function for Foxh1. Additionally, Foxh1- and Smad2/3-binding site analysis revealed the involvement of Pou family TFs in Nodal target gene expression. PouV family genes (pou5f3s) modulate the expression of some Nodal targets, perhaps functioning as regulators of differentiation.
RESULTS
Foxh1 is crucial for mesendoderm formation
In Xenopus, there are two foxh1 genes: foxh1 and foxh1.2 (Chen et al., 1996; Howell et al., 2002). foxh1 is expressed maternally and plays a central role in mediating Nodal signaling (Chen et al., 1996; Howell et al., 2002; Kofron et al., 2004b; Watanabe and Whitman, 1999; Yeo et al., 1999). foxh1.2 is expressed after the mid-blastula transition (MBT) (Howell et al., 2002), and its function is unclear. Since the initial activation of Nodal signaling is detected at MBT, before foxh1.2 expression, and foxh1 transcripts are more abundant than foxh1.2 in early gastrulae (Howell et al., 2002; Lee et al., 2001), we examined the contribution of Foxh1 in mediating Nodal signaling.
In situ hybridization analyses in Xenopus laevis have shown that foxh1 transcripts are preferentially localized to the animal and marginal zones, but are excluded from deep mesendodermal cells (Howell et al., 2002). However, these results are inconsistent with the widespread notion that Foxh1 is the key player in mediating Nodal signaling during mesendoderm formation (Chen et al., 1996, 1997; Watanabe and Whitman, 1999; Yeo et al., 1999). RT-qPCR analysis from dissected tissue fragments of X. tropicalis early gastrulae confirmed that foxh1 is ubiquitously expressed (Fig. 1A).
Fig. 1.
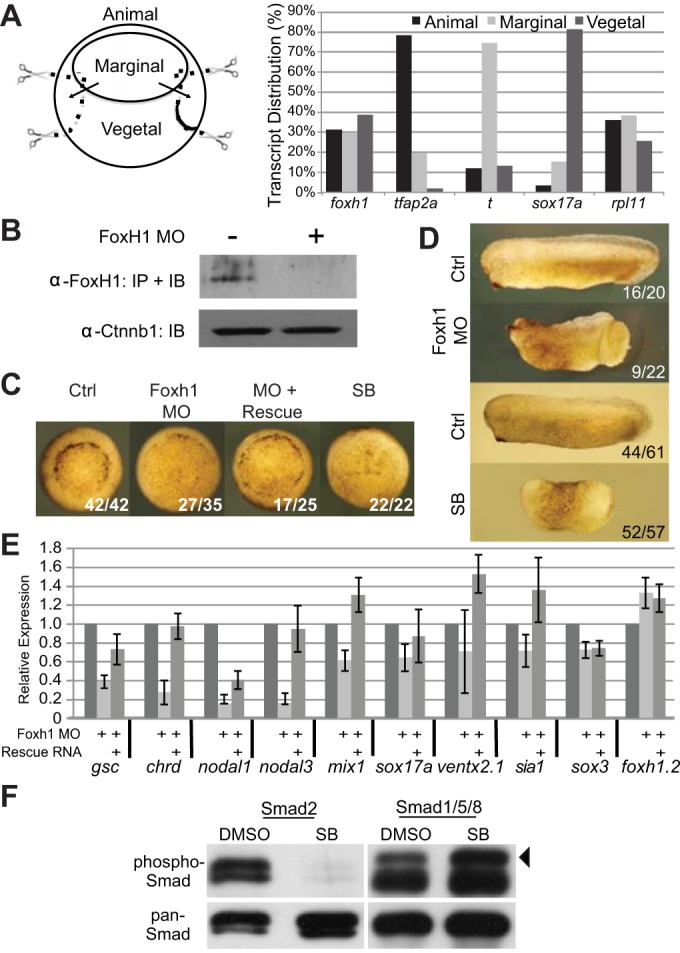
Foxh1 is crucial for mesendoderm formation. (A) Distribution of foxh1 transcripts in X. tropicalis early gastrula. Total RNAs from animal, marginal and vegetal fragments were subjected to RT-qPCR analyses. tfap2a, ectodermal marker; t/brachyury, mesodermal marker; sox17a, endodermal marker; rpl11, expressed throughout the embryo. (B) Embryonic lysates from control or foxh1-MO injected embryos were subjected to immunoprecipitation followed by western blot using anti-Foxh1 antibody. Ctnnb1 protein levels in crude embryo lysates are unaffected by the MO. (C) Both Foxh1 morphant and SB431542-treated embryos exhibit gastrulation delay (vegetal views). (D) Early tailbud stage Foxh1 morphants displaying anterior defects and incomplete blastopore closure; SB431542-treated embryos lack distinctive A-P or D-V features. (E) Examination of foxh1 MO effects on different germ-layer markers by RT-qPCR. gsc, chrd, nodal1, nodal3 and mix1 are mesoendodermal markers; sox17a is an endodermal marker; ventx2.1 is a BMP target gene; sia1 is a Wnt target gene. The ectodermally enriched markers sox3 and foxh1.2 are included as non-Foxh1 targets for comparison. (F) Early gastrula cleared lysates were immunoprecipitated using either pan anti-Smad2 or anti-Smad1 polyclonal antibodies covalently coupled to beads. Bound proteins were subjected to western immunoblotting using anti-P-Smad2 or anti-P-Smad1, respectively. After detection, membranes were re-probed with anti-Smad2 or anti-Smad1 to show efficiency of the immunoprecipitations. Arrowhead indicates the P-Smad1 band; the lower band in the P-Smad1 lanes is low level primary antibody release from the beads.
Expression of Foxh1 protein was inhibited by microinjecting translation-blocking foxh1 morpholino antisense oligonucleotides (MO). We generated an affinity-purified antibody against X. tropicalis Foxh1 protein (supplementary material Fig. S1). Immunoprecipitation (IP) of embryonic extracts followed by western immunoblotting (Fig. 1B) detected a single ∼55 kDa band that matches the predicted molecular weight (56.6 kDa) of Foxh1. MO injection strongly knocked down Foxh1 protein expression (Fig. 1B) and the morphant phenotypes were reminiscent of that in X. laevis (Howell et al., 2002; Kofron et al., 2004a). Dorsal lip formation was delayed (>75%, n=35), and was partially rescued by co-injecting MO-insensitive foxh1 mRNA (Fig. 1C). We also observed incomplete gastrulation and a shortened anteroposterior (A/P) axis in tailbud embryos, which were rescued by co-injecting MO-insensitive foxh1 mRNA (Fig. 1D; supplementary material Fig. S2).
Next, we examined the effective concentrations of the Nodal receptor (ALK4/5/7) inhibitor SB431542 (Inman et al., 2002) during X. tropicalis mesendoderm development. Embryos were treated with increasing concentrations of SB431542 and the expression of well-characterized Nodal target genes was effectively inhibited at 100 µM (supplementary material Fig. S3), which is also the effective concentration reported in X. laevis (Ho et al., 2006). SB431542-treated embryos displayed varying phenotypic severity. Typically, embryos fail to form any blastopore lip, arrest during gastrulation and die during neurulation. In some clutches, SB431542-treated embryos had a severe delay in blastopore lip formation, but survived to tailbud stages. Generally, the phenotypes of embryos treated with SB431542 (Fig. 1C,D) were more severe than those of Foxh1-depleted embryos. We also examined the specificity of SB431542 treatment on Smad2/3 activation (Fig. 1F). SB431542 inhibited the phosphorylation of Smad2/3, whereas the phosphorylation of Smad1/5/8 was not diminished. We conclude that 100 μM SB431542 specifically inhibits Nodal signaling in X. tropicalis embryos.
Foxh1 morphants showed significant reduction in expression of the dorsal mesendoderm-enriched genes gsc, chrd, nodal1 and nodal3 (Fig. 1E). Expression of the early endodermal markers mix1 and sox17a was also reduced, whereas expression of the ventral marker ventx2.1, the animally enriched marker sox3, and sia1 (a Wnt target) were minimally affected. Interestingly, foxh1 itself was consistently upregulated 8- to 15-fold in foxh1 morphants (supplementary material Fig. S4), whereas expression of foxh1.2 transcripts was only slightly increased (∼1.3-fold; Fig. 1E). Delivery of foxh1 rescue mRNA to morphants either fully or partially reversed foxh1 MO effects (Fig. 1E). We conclude that Foxh1 is required in X. tropicalis for normal mesendoderm development. Other findings are: (1) foxh1 expression is negatively autoregulated; (2) Foxh1 antibody specifically recognizes endogenous protein; and (3) foxh1 MO efficiently knocks down Foxh1 protein and downstream mesendodermal gene expression.
Significantly different sets of genes are responsive to Foxh1 and Nodal
To examine the contribution of Foxh1 to Nodal signaling, we compared RNA-seq transcriptome profiles between SB431542-treated embryos and foxh1 morphants. Sequence reads were mapped to the X. tropicalis v.7 genome assembly (supplementary material Table S2) and differential gene expression levels between control and perturbed embryos were calculated. SB431542 treatment and foxh1 MO injection each affected the mRNA expression of hundreds of genes, both positively and negatively (supplementary material Fig. S5). Affected genes include well-studied Nodal targets such as gsc, nodal1, cer1 and hhex (supplementary material Table S1). Fig. 2A-D show read-mapping densities (tracks 3-6 in each panel) across these four genes.
Fig. 2.
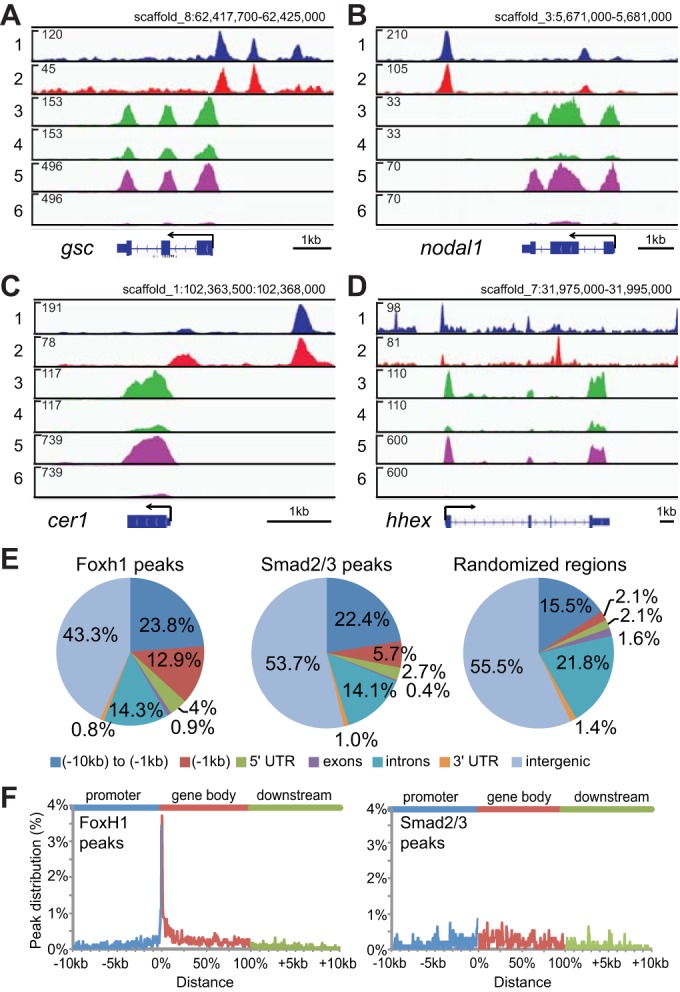
Genome-wide survey of Nodal and Foxh1 targets in early gastrulae. (A-D) IGV genome browser views of gsc (A), nodal1 (B), cer1 (C) and hhex (D) genes. Track contents: (1) Foxh1 ChIP-seq; (2) Smad2/3 ChIP-seq; (3) RNA-seq of uninjected control embryos; (4) RNA-seq of foxh1 MO-injected embryos; (5) RNA-seq of mock-treated control embryos; and (6) RNA-seq of SB431542-treated embryos. The numbers in the upper left of each track indicate track heights. (E) Genome-wide analyses of Foxh1 and Smad2/3 peaks. Pie chart distributions of Foxh1 and Smad2/3 peaks across seven defined genomic features are shown. Randomized regions were also analyzed for comparison (right). These were generated by randomly redistributing the intervals of Foxh1 peaks throughout the genome. In parallel, Smad2/3 peak intervals were similarly randomized and showed a nearly identical genomic distribution (data not shown). (F) Distribution of Foxh1 (left) and Smad2/3 (middle) peaks within the intervals of 10 kb upstream of gene 5′ ends, gene bodies and 10 kb downstream of gene 3′ ends. Supplementary material Fig. S7 contains a distribution of randomly selected regions. As individual gene bodies are highly variable in length, we normalized these segments on a 0-100% scale (Zhang et al., 2012).
Using a 1.5-fold cutoff (empirically derived from Kolmogorov-Smirnov analysis below), 259 genes were differentially affected by SB431542, 707 were affected by foxh1 MO and 87 were affected by both foxh1 MO injection and SB431542 treatment. These expression changes represent both direct and indirect target genes. That only 34% (87) of the 259 SB431542-sensitive genes were affected by foxh1 knockdown suggests the involvement of additional TFs in Nodal signaling. In addition to its role in mediating Nodal signaling, Foxh1 also regulates the expression of a set of non-Nodal targets. Expression differences between Nodal and Foxh1 loss of function may explain the phenotypic differences observed (Fig. 1).
Genome-wide binding patterns of Foxh1 and Smad2/3 identify many potential co-regulated target genes
Differential expression profiling alone is unable to distinguish between direct and indirect transcriptional regulation by Foxh1 and Smad2/3. Therefore, we correlated gene expression data with physical binding of Foxh1 and Smad2/3. The suitability of Foxh1 and Smad2/3 antibodies was validated by ChIP-qPCR (supplementary material Fig. S6), and ChIP-seq (Fig. 2A-D, tracks 1 and 2) was performed on stage 10.25-10.5 early gastrulae using two biologically independent replicates. MACS2 and SISSRs (Jothi et al., 2008; Zhang et al., 2008) identified bound regions (‘peaks’) and high-confidence peaks were those defined by the intersection of output of these programs (supplementary material Table S2). ChIP-seq analyses revealed 2266 Foxh1 and 939 Smad2/3 peaks. We assigned nearest genes to peaks located within 10 kb upstream of gene 5′ ends, within 10 kb downstream of 3′ ends and within gene transcription units (supplementary material Table S3). These analyses mapped 70.1% (1588) of Foxh1 peaks to 1321 genes, and 65.5% (615) of Smad2/3 peaks to 501 genes, with 297 genes being bound by both Foxh1 and Smad2/3. Most of the peaks and genes identified in Smad2/3 ChIP-seq by Yoon et al. (2011) are also included in our data analysis (see supplementary material Fig. S12).
We found that 12.9% of Foxh1 peak summits were located within 1 kb upstream of the 5′ ends of nearby genes (Fig. 2E), representing a 6-fold enrichment over the distribution of computationally randomized peaks (2.1%). Little to no enrichment of Foxh1 peaks was found for the other six genomic intervals. A similar analysis for Smad2/3 showed that 5.7% of peaks are within this 1 kb upstream interval, representing an ∼3-fold enrichment. The highest abundance of Foxh1 binding is located within a narrow window flanking the 5′ end of genes, whereas Smad2/3 did not reveal such localization (Fig. 2F).
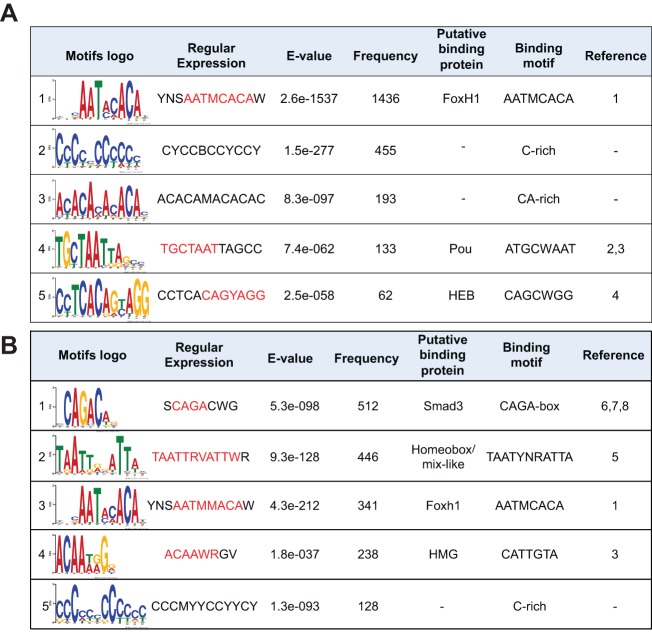
We performed a de novo motif search of the DNA sequences extending 75 bp to either side of the summits of the 2266 Foxh1 peaks. The Foxh1 motif was the most common sequence found (1436 peaks) (Fig. 3A). Although a similar analysis of Smad2/3 peaks revealed the Smad2/3 motif to be the most abundant, the Foxh1 motif was also frequently found (Fig. 3B, supplementary material Fig. S11), supporting the view that Foxh1 is a major transcriptional partner of Smad2/3.
Fig. 3.
Motif analyses of Foxh1 and Smad2/3 peaks. Foxh1-bound (A) and Smad2/3-bound (B) regions (151 bp centered on peak summits) were retrieved to perform de novo motif analysis. Binding motifs in the sixth column were matched manually (see citations in ‘Reference’ column). Base positions in red in the ‘Regular Expression’ column match these published motifs. References: 1, Zhou et al., 1998; 2, Mason et al., 2010; 3, Chen et al., 2008; 4, Yoon et al., 2011; 5, Wilson et al., 1993; 6, Dennler et al., 1998; 7, Shi et al., 1998; 8, Zawel et al., 1998. While the search by STAMP of TRANSFAC identifies motif 5 in A as HEB, the search by TOMTOM of JASPAR and UniPROBE assigns this to Zic-related proteins.
The primary role of Foxh1 in Nodal signaling is to activate target genes
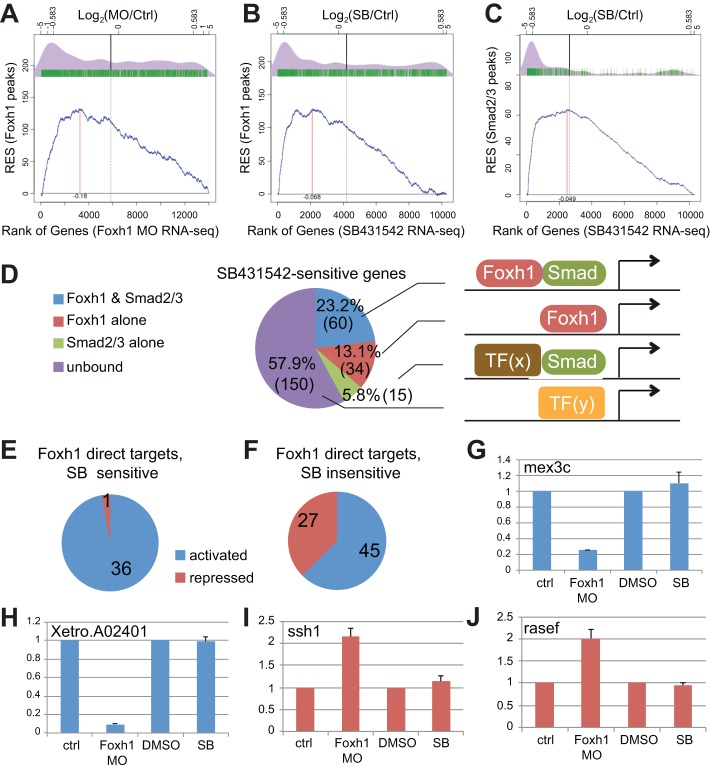
To examine whether Foxh1 functions as an activator or repressor, or both, we applied the Kolmogorov-Smirnov (KS) test to determine the statistical significance of the correlation between TF binding to genes and changes in gene expression after loss of function (Subramanian et al., 2005). The differential expression values of over 14,000 genes in foxh1 morphants versus those of control embryos were used to rank the genes in ascending order along the x-axis (Fig. 4A,B). This ranking was correlated with the Foxh1 ChIP-seq results by scoring for the presence/absence (green tick marks) of Foxh1 peaks over these genes. A running (cumulative) enrichment score (RES, y-axis) was calculated based on the presence/absence of a Foxh1 peak (blue line). This plot is significantly different from that expected for the null hypothesis (P<5.6e-16), i.e. that peaks are randomly distributed with respect to the foxh1 MO effects on differential expression. Therefore, there is a statistical correlation between Foxh1 binding and changes in gene expression. The density plot (purple) of the genes containing peaks (green tick marks) reveals a tight correlation between the presence of bound Foxh1 and genes downregulated upon Foxh1 depletion, but not with genes upregulated, suggesting that Foxh1 functions primarily as a transcriptional activator.
Fig. 4.
Functional relationships between Foxh1 and Smad2/3 binding, and differential gene expression. (A-C) Kolmogorov-Smirnov (KS) tests for comparing relatedness between TF binding (ChIP-seq data sets) and loss-of-function analyses (RNA-seq data sets). Comparisons between Foxh1 peaks and either Foxh1-regulated targets (A) or Nodal-regulated targets (B). The x-axis represents genes ranked by ascending fold change (bottom scale), depicted as log2 ratios (top scale) between either foxh1 MO or SB431542 and controls. Log2 of ±0.583 corresponds to ±1.5-fold change. The y-axis scale is the running enrichment score (RES). (C) Comparison between Smad2/3 peaks and Nodal-regulated targets. (D) A pie chart that depicts the distributions of Nodal regulated targets that are (1) co-bound by both Foxh1 and Smad2/3 (blue); (2) bound by Foxh1 alone (red); (3) bound by Smad2/3 alone (green); or (4) not bound by either Foxh1 or Smad2/3 (purple). (E) A pie chart that depicts the distributions of 37 genes that are both Foxh1 direct targets (Foxh1 bound and change expression in response to Foxh1 MO) and Nodal targets (SB sensitive): 36 are activated by Foxh1 (blue), whereas only 1 is repressed (red). (F) Among 72 Foxh1 direct targets that are independent of Nodal regulation (SB insensitive), 45 are activated by Foxh1 (blue) and 27 are repressed. (G-J) RT-qPCR validations of the Nodal-independent Foxh1 targets. (G,H) mex3c and Xetro.A02401, from the group of 45 downregulated targets, were further validated by showing downregulation in foxh1 MO-injected embryos, when compared with control, but are unaffected by SB431542. (I,J) ssh1 and rasef were validated by showing upregulation after foxh1 MO injection but are unaffected by SB431542.
A strong positive correlation between Foxh1 binding and downregulation of genes in response to SB431542 was also observed in the density plot in Fig. 4B, consistent with the activating role of Foxh1 in Nodal signaling. Finally, a strong correlation was observed between the binding of Smad2/3 and downregulation of genes in response to SB431542 (Fig. 4C). The RESs (blue plots) for both Foxh1-Nodal and Smad2/3-Nodal signaling are significantly different than those expected for their corresponding null hypotheses (P-values in each were <2.2e-16). We conclude that binding of Foxh1 and Smad2/3 are prerequisites for the activation of many Nodal target genes. Interestingly, a comparison between the density plots for Foxh1 and Smad2/3 binding on SB431542-sensitive genes (Fig. 4B,C) shows that Smad2/3 correlates 2.5-fold more closely with Nodal targets than does Foxh1 (see supplementary materials and methods). This is consistent with the RNA-seq results (Fig. 2) that also implicated Foxh1 in Nodal-independent gene regulation.
A large cohort of direct Nodal target genes
Among 259 genes regulated by Nodal signaling, 218 are positively regulated by Nodal and 41 genes are negatively regulated (supplementary material Table S1). Seventy-five genes are putative direct Nodal targets as they are bound by Smad2/3 (blue and green sectors in Fig. 4D).
Among these 75 genes, 60 are co-bound by both Foxh1 and Smad2/3 (Fig. 4D). Fifty-nine of these are activated by Nodal signaling (downregulated by SB431542) (supplementary material Table S4), and thus are potential Foxh1-mediated Nodal direct targets. This list includes the previously identified Nodal targets gsc, mix1, rnd1, flrt3, pitx2, nodal1 and nodal2. Some new direct Nodal targets are wnt8a, frzb, dkk1, prickle2 and cxcr4. Only one gene is repressed by Nodal: bambi, a transmembrane decoy receptor that inhibits both Nodal and BMP signaling (Karaulanov et al., 2004).
Thirty-four SB431542-sensitive genes had Foxh1 peaks without Smad2/3 peaks (Fig. 4D, supplementary material Table S5). Additionally, 15 genes show the presence of Smad2/3 peaks without Foxh1 binding (Fig. 4D, supplementary material Table S6). This latter group contains some known mesendodermal genes, including dkk1, hnf1b and gata6, and genes with poorly characterized roles in the early embryo. The remaining 150 SB431542-sensitive genes are not bound by either Foxh1 or Smad2/3, and thus likely represent a group of indirect Nodal targets.
GO (gene ontology) term analysis (supplementary material Table S7) of the 59 Foxh1-Smad2/3-bound and Nodal-activated genes includes cell surface receptor-linked signal transduction, pattern specification/cellular morphogenesis and negative regulation of signal transduction. These 59 genes were preferentially expressed in mesendoderm [W.T.C., I.L.B. and K.W.Y.C., unpublished; see Xenbase (James-Zorn et al., 2013)] where Nodal signaling is most active. These findings reinforce the notion that Foxh1-Smad2/3-mediated Nodal signaling coordinates the expression of a large cohort of genes regulating cell fate specification, morphogenesis and cell signaling.
Foxh1 is a dual regulator and functions independent of Nodal signaling
We investigated the role of Foxh1 in early embryogenesis by comparing foxh1 MO knockdown and SB431542 RNA-seq together with ChIP-seq. Among the 1321 Foxh1-bound genes (Fig. 2D), the expression of 109 is affected (>1.5-fold) following foxh1 MO injection. We consider these to be Foxh1 direct targets. Thirty-seven of these genes are SB431542 sensitive (Fig. 4E), suggesting that they are either regulated by Nodal directly or indirectly. The Smad2/3 ChIP-seq data show that 26 (70%) of these are bound (supplementary material Table S8) and therefore likely to be direct Nodal targets.
Among the 109 Foxh1 direct target genes, 37 are Nodal-dependent targets and all except one (fos) are positively regulated by Foxh1 (Fig. 4E). Conversely, 72 lack Smad2/3 binding and are insensitive to SB431542 (Fig. 4F), suggesting that Foxh1 regulates these genes in a Nodal-independent fashion. Of these 72 genes, 45 are downregulated in the absence of Foxh1, whereas 27 are upregulated, suggesting that Foxh1 functions as a dual-acting TF to activate or repress target genes in a Nodal-independent manner. RT-qPCR analysis independently confirmed both activator and repressor behavior of Foxh1 (Fig. 4G-J and supplementary material Fig. S8). The apparent Nodal-independent Foxh1 gene regulation is indeed a specific effect of foxh1 MO knockdown as normal expression of these genes was rescued via Foxh1 expression using a MO-resistant foxh1 mRNA (supplementary material Fig. S9). All 72 Nodal-independent Foxh1 regulated genes appear to be ubiquitously expressed in the early gastrulae, based on public expression data (Xenbase) and RNA-seq results from dissected early gastrulae (W.T.C., I.L.B. and K.W.Y.C., unpublished), in contrast to Nodal-dependent gene expression, which is confined to the mesendoderm. GO term analysis failed to reveal any term enrichment, and the developmental significance of these genes will require further investigation.
Xenopus PouV class transcription factors interact with Foxh1 to regulate Nodal targets
To identify additional TFs that interact with Foxh1 to regulate target genes, we searched for over-represented sequence motifs in the vicinity of Foxh1 peak summits. The five most statistically significant motifs (based on E-values) ranked by abundance are shown in Fig. 3A, with the Foxh1 motif being the most frequent. Although searches of various motif databases (UniPROBE, JASPAR and TRANSFAC) failed to identify candidate factors that matched the second and third motifs, the fourth and fifth resemble Pou and Heb/Zic motifs. Because Heb, also known as Tcf12, was previously implicated as a Foxh1/Smad partner (Yoon et al., 2011), we investigated the role of Pou TFs that participate in Nodal signaling regulation.
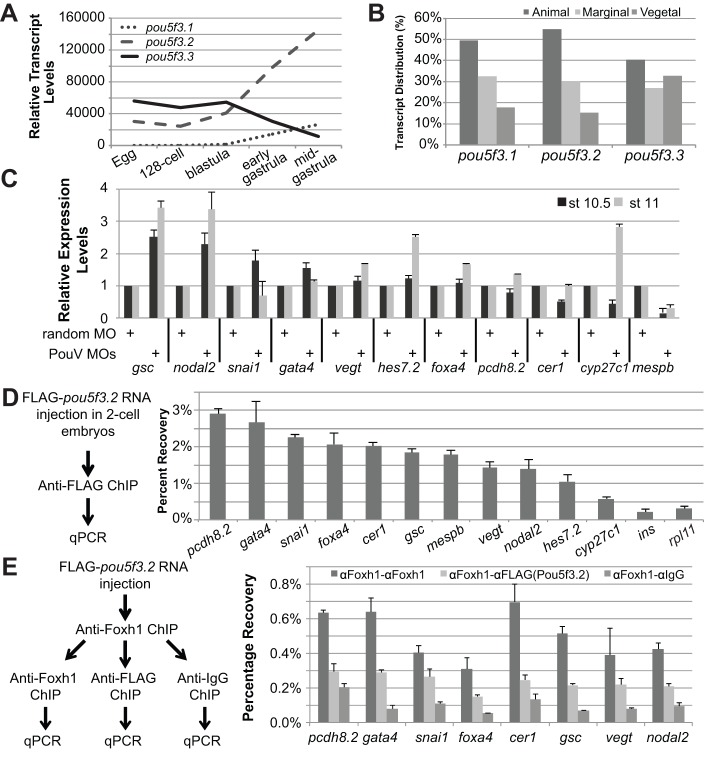
Xenopus and zebrafish PouV family TFs have been associated with the regulation of early mesendodermal development (Burgess et al., 2002; Cao et al., 2007, 2006, 2008; Lunde et al., 2004; Morrison and Brickman, 2006; Reim and Brand, 2006; Reim et al., 2004), and are evolutionarily closely related to mammalian Pou5f1/Oct4 (Frankenberg et al., 2010; Frankenberg and Renfree, 2013). Xenopus PouV genes (pou5f3.1, pou5f3.2 and pou5f3.3; previously oct91, oct25 and oct60, respectively) are locally triplicated in the genome (Hellsten et al., 2010) and differentially expressed in the early embryo. pou5f3.2 and pou5f3.3 are expressed maternally, whereas pou5f3.1 is zygotic (Fig. 5A) (Hinkley et al., 1992). The three PouV genes are expressed in all three primary germ layers (Fig. 5B).
Fig. 5.
Functional analysis of PouV genes in regulating Nodal targets. (A) RT-qPCR analysis of pou5f3.1, pou5f3.2 and pou5f3.3 transcript levels at egg, 128-cell, blastula (stage 9), early (stage 10) and mid-gastrula (stage 10.5) stages. Transcript levels were normalized to the pou5f3.1 level in egg RNA. (B) RT-qPCR analysis of pou5f3.1, pou5f3.2 and pou5f3.3 in animal, marginal and vegetal fragments of the gastrula (stage 10-10.5) stage embryo. (C) RT-qPCR of mesendodermal targets in PouV-depleted embryos at early gastrula (stage 10.5) and mid-gastrula (stage 11). (D) ChIP-qPCR strategy to show FLAG-Pou5f3.2 binding to Pou motif-containing regions within Foxh1 peaks. (E) Sequential ChIP-qPCR analyses for Foxh1 and PouV co-binding on Nodal targets. Chromatin from embryos expressing FLAG-Pou5f3.2 was immunoprecipitated using anti-Foxh1 antibody, followed by a second immunoprecipitation using anti-FLAG antibody or anti-IgG antibody (negative control).
We examined whether PouV proteins directly regulate Foxh1-Smad2/3-mediated Nodal target genes as a general mechanism. We bioinformatically mined the Foxh1 and Smad2/3 peak sequences on the 60 direct Nodal targets (SB-sensitive) bound by these TFs (Fig. 4D, supplementary material Table S4) for the presence of Pou motifs. Among these, 22 contain Pou motifs within these regions (supplementary material Table S4), but only 11 are affected in Foxh1 MO-injected embryos (based on RNA-seq). After PouV knockdown, three out of these 11 genes (gsc, nodal2 and mespb), by conservative criteria, were consistently affected at both stages examined (>2-fold, Fig. 5C), and expression of these genes was rescued after co-injecting a cocktail containing MO-insensitive versions of all three PouV mRNAs (supplementary material Fig. S10). Expression of many of the other genes was affected to varying degrees in a stage-dependent manner that makes both molecular phenotypes and rescue rather difficult to assess (see also Livigni et al., 2013). gsc and nodal2 are upregulated in response to the PouV MO, suggesting that these genes are negatively regulated by PouV proteins, whereas mespb expression is reduced in MO-injected embryos suggesting it may instead be positively regulated by PouV. Such a dual role for mammalian Pou5f1/Oct4 has been well documented (Pan et al., 2002; Hammachi et al., 2012). These results suggest that PouV proteins regulate the expression of a subset of Nodal target genes that are crucial to early mesendodermal development.
We also examined whether PouV proteins directly interact with the Pou motif regions found in these genes, which would imply direct PouV regulation. Because antibodies against Xenopus PouVs are not available, we microinjected mRNA encoding FLAG-tagged pou5f3.2 into embryos and performed ChIP-qPCR to interrogate binding to the identified regions containing the Pou motif. FLAG-Pou5f3.2 protein preferentially bound to the cis-regulatory regions of all genes (Fig. 5D) that respond to PouV knockdown (Fig. 5C), but not to two genes (ins and rpl11) that were used as negative controls. We next investigated whether both PouV and Foxh1 proteins can co-occupy the cis-regulatory regions of a subset of these genes. A sequential ChIP-qPCR analysis (Geisberg and Struhl, 2004) showed that six of the eight genes tested were co-bound by Foxh1 and PouV proteins (>2-fold compared with an IgG control; Fig. 5E). These results suggest that the combinatorial action of inputs both from Foxh1-Smad2/3(Nodal) signaling and PouV proteins is required for the establishment of proper expression levels of some of these genes during mesendodermal specification and patterning. Interestingly, Mullen et al. (2011) reported that Pou5f1 and Smad3 co-occupy target genes in mouse ESCs, suggesting that PouV proteins may have a more general role in modulating Activin/Nodal signaling.
DISCUSSION
We report the genome-wide examination of a Smad ‘partner’ in a developing vertebrate embryo. This study greatly expands the number of bona fide Nodal direct target genes operating in early gastrulation. Our analyses strongly support the notion that the coordinated action of Foxh1-Smad2/3 is a major in vivo driver of the mesendoderm transcriptional program. We also uncovered a new dual function for Foxh1 that is Nodal independent. PouV TFs were found to interact with Foxh1 to regulate Nodal target genes. And, finally, a cohort of Nodal target genes was identified that likely function in morphogenesis. Therefore, this study significantly broadens our view of the mechanisms that underlie TGFβ signaling control over the coordination of cell fate and behavior in early embryos.
Mesendodermal specification and patterning
Xenopus mesendodermal GRNs containing numerous connections between genes have been published (Koide et al., 2005; Loose and Patient, 2004). Furthermore, an endodermal core GRN based on studies from a variety of vertebrate systems suggests the involvement of Mix, Gata, Foxa, Hnf1b and Sox17 family members (Zorn and Wells, 2007). The molecular details of the interactions between Nodal signaling and these TFs are only partially understood. Here, we have confirmed six out of 11 suggested connections for Foxh1 direct regulation (gsc, chrd, otx2, mix1, gata4, bix1) and nine out of 16 connections for direct Nodal regulation (nodal1, nodal2, pitx2, bix1, cer1, mix1, gata4, gata6 and lefty). Additionally, numerous new direct connections were found for Foxh1 and Nodal regulation, respectively. Thus, this study has not only validated many interactions that were previously inferred, but also increased the total number of direct Nodal targets, providing a more sophisticated blueprint of the vertebrate mesendoderm GRN.
We found a number of genes encoding secreted signaling ligands and extracellular modifiers of signaling are direct Nodal targets. nodal1, nodal2 and wnt8a are directly upregulated by Nodal, whereas fgf16 and wnt11b are indirectly regulated. These Nodal genes and wnt8a are also direct Foxh1 targets. Secreted antagonists of Nodal, BMP and Wnt signaling, including cer1, lefty, frzb and dkk1, are direct targets of Nodal, implying that cells responding to Nodal signaling secrete these antagonists to coordinate the integration of multiple signaling activities. Others have attempted to identify direct Foxh1 and Smad2/3 targets. Microarray experiments by Pei et al. (2007) using zebrafish Foxh1 morphants identified cytokeratins 1 and 2, and keratins 4, 8 and 18 to be possible Foxh1 targets. These genes are not found in our list of direct Foxh1 targets and may be indirect targets of Foxh1 depletion in zebrafish. Silvestri et al. (2008) searched for human, mouse and rat genes with conserved Foxh1-Smad2/3 DNA motifs to identify potential targets of these factors. Among 21 candidates, excluding five well-known Nodal target genes (gsc, mix1, lefty, pitx2 and nodal), the remaining 16 genes are not found in our list of Nodal or Foxh1 target genes. Perhaps these genes are under Nodal regulation at later stages in development, but not gastrulation.
TGFβ signaling and Smad partner proteins
Foxh1 is a major Smad2/3 co-factor acting in early development, and several other Smad2/3 partners (e.g. Mixer, Gtf2i, Eomes) have also been implicated. The broader roles of these additional TFs in Nodal target regulation are unknown. We performed a motif analysis on Smad2/3-bound regions to determine the relative contribution of putative Smad2/3 partners. We confirmed that a large percentage of Smad2/3-bound Nodal-responsive genes contain Smad motifs. This analysis identified 5′-CAGAC-3′ in only 26 out of 75 direct Nodal targets. However, when we searched for the minimal Smad2/3-binding motif (5′-AGAC-3′) (Shi et al., 1998), this was found on 88% (66/75) of direct Nodal target genes. Sequences matching the Foxh1 motif 5′-AATMCACA-3′ were found on 66% (50/75) of direct Nodal targets, consistent with the view that Foxh1 is a major regulator of this signaling pathway.
The motif 5′-TAATYNRATTA-3′ is also enriched under Smad peaks and contains an inverted repeat (underlined) recognized by paired-family homeodomains (Wilson et al., 1993). This motif was found on 37% (28/75) of direct Nodal targets. The paired homeodomain proteins Mixer and Bix2 were shown to physically interact with Smad2 to regulate gsc expression (Germain et al., 2000) and finding this element on over one-third of direct Nodal target genes suggests that these proteins play an important role in Nodal signaling.
Another enriched motif we identified is the Heb (bHLH family) motif (supplementary material Fig. S11), also previously reported by Yoon et al. (2011) as being over-represented under Foxh1 and Smad peaks in human ESCs. The Heb sequence was also associated with 18 out of 75 (24%) Foxh1-bound regions in our ChIP-seq data, consistent with evidence from Yoon et al. that Heb/Tcf12 and E2a/Tcf3 may also regulate Nodal targets in Xenopus.
A T-box motif is enriched under Smad2/3 peaks on 20% (15/75) of direct Nodal target genes (supplementary material Fig. S11). Eomes, a T-box protein, binds Smad2 and has been implicated in Nodal-mediated mesendoderm induction in Xenopus, zebrafish and mammalian epiblast stem cells (Arnold et al., 2008; Picozzi et al., 2009; Slagle et al., 2011; Teo et al., 2011). Recently, ChIP-seq analysis of Eomes has been reported in Xenopus (Gentsch et al., 2013), allowing for a direct test of whether both Smad2/3 and Eomes are bound to the aforementioned 15 genes. Our analysis confirms that 12 out of 15 (80%) genes indeed have Eomes bound to the T-box motifs found under Smad2/3 peaks.
We found an HMG/Sox motif under Smad2/3 peaks on 27% (21/78) of direct Nodal target genes. Although there has been no demonstration that HMG/Sox family TFs interact with Smads, two SoxF genes, Sox7 and Sox17, are both expressed vegetally and have been implicated in mesendoderm specification. We hypothesize that these SoxF proteins might cooperate with Smad2/3. Finally, the Pou motif is enriched in both our Foxh1 and Smad2/3 ChIP-seq datasets (see below). We did not find enrichment for Gtf2ird1, Gtf2i or Tp53 motifs.
PouV in mesendoderm and pluripotency
Among 60 SB431542-sensitive Foxh1-Smad2/3 bound genes, 31 genes (Fig. 4D blue sector, e.g. cxcr4 and hes7.2) were unaffected by Foxh1 MO injection. We hypothesize that these genes are Foxh1-Smad2/3 targets that receive strong inputs from other TFs that may act together with Foxh1, or in parallel. TFs predicted by motif analyses of Foxh1 peaks may be involved in this regulation.
Motif enrichment analysis found 47% (37/75) of direct Nodal targets contain a Pou motif under Smad2/3 peaks (supplementary material Tables S4 and S6). The involvement of PouV TFs in antagonizing expression of two Nodal target genes, gsc and mix2 (currently known as mix1), has previously been suggested (Cao et al., 2008; Levigni et al., 2013). Our analysis of PouV MO-injected embryos further extends these observations by identifying two other Nodal targets (nodal2, mespb) that are subjected to regulation by PouV (no Pou motif is present under Smad2/3 peaks on the X. tropicalis mix1 gene). Interestingly, 37% (13/35) of the direct Nodal targets containing Pou motifs are not co-bound by Foxh1, suggesting that the Foxh1-PouV interaction may not be the only mechanism for regulation of these genes.
Human ESCs (hESCs) are likely of epiblast origin, which is comparable to late blastula in Xenopus. Frog embryonic cells at this stage are pluripotent, can be directed into all three germ layers by appropriate signals and thus have characteristics that mirror ESCs. Furthermore, although hESCs require Activin/Nodal signaling for maintenance of pluripotency, high doses of Activin drive these cells into mesendodermal lineages. Perhaps PouV-mediated regulation of Nodal targets in Xenopus is analogous to Pou5f1 regulation of pluripotency in epiblast and hESCs. Experiments differentiating hESCs into definitive endoderm have indeed shown that Pou5f1 represses a number of Nodal-induced endodermal markers, including gsc (Teo et al., 2011), which we showed are similarly repressed in Xenopus. These observations support a model for evolutionarily conserved PouV-mediated repression of Nodal-dependent mesendodermal differentiation.
Nodal-independent Foxh1 functions and repression
Our KS tests confirm the view that FoxH1 generally functions as a transcriptional activator. The nodal5, nodal6 and flk1 genes were previously implicated to be repressed by Foxh1 in either X. laevis or zebrafish (Choi et al., 2007; Kofron et al., 2004b), but in X. tropicalis, neither Foxh1 nor Smad2/3 binding to these genes was detected. Thus, the observed repression by Foxh1 may be indirect or occurs at stages other than the early gastrula. Our analysis also suggests that foxh1 is negatively autoregulated as foxh1 MO injection increases the expression of foxh1 itself. However, ChIP-seq fails to find Foxh1 binding to the foxh1 gene, implying that negative autoregulation is indirect.
Our analysis revealed 72 Foxh1 direct targets that are regulated independently of Nodal signaling (supplementary material Table S8). Knockdown of foxh1 reveals that it acts as both an activator and repressor, independent of Nodal-Smad2/3. The spatial expression patterns of Nodal-independent Foxh1 target genes were variable in gastrula stage embryos (W.T.C., I.L.B. and K.W.Y.C., unpublished), unlike the Foxh1-Smad2/3 targets, which are either mesendoderm-specific or significantly enriched in this region. Although GO term analysis for the 71 Nodal-independent Foxh1 targets did not reveal useful functional information, the list includes a noteworthy mix of secreted growth factors, TFs, GTPase modifiers and adhesion molecules.
Regulation of morphogenesis
Nodal signaling regulates mesodermal morphogenesis by affecting cellular activities involved in tissue dynamics. Among the 60 direct Nodal targets, many were previously shown to affect cell behaviors. One Nodal-regulated TF, snai1, is involved in epithelial-mesenchymal transitions (EMTs) in numerous contexts, and has been implicated in the morphogenesis of gastrulation (Spring et al., 2002).
The direct Nodal induction of flrt3, a transmembrane protein that regulates cell adhesion, is necessary for proper morphogenesis of the organizer cells (Ogata et al., 2007). We found rnd1, pdgfra and pcdh8.2, confirming previous reports that they were either responsive to Nodal signaling or implicated as direct targets (Luxardi et al., 2010; Ogata et al., 2007). We also identified plekhg5, prickle2, cmtm8, efnb2, cxcr4, ntn3 and cass4 as direct Nodal targets that likely participate in morphogenetic movements during Xenopus gastrulation. Plekhg5 regulates Rnd1 protein activity (Goh and Manser, 2012) and thus might function in an adhesion pathway with rnd1 and flrt3. prickle2 regulates cell polarity in response to non-canonical Wnt signaling (Antic et al., 2010; Tao et al., 2012, 2011). cxcr4 regulates endodermal cell migration during zebrafish gastrulation (Nair and Schilling, 2008) but its function in Xenopus gastrulation is unknown. cmtm8, efnb2, ntn3 and cass4 have all been implicated in EMT, cell migration and/or differential cell adhesion in various cell types. Here, we have identified a gene battery that is directly regulated by Nodal and likely functions in coordinating complex cellular behaviors that affect morphogenesis during gastrulation. Our future goal is to further examine the mesendoderm network of Xenopus tropicalis and better elucidate the conserved GRN architecture involved in vertebrate gastrulation.
MATERIALS AND METHODS
Embryo handling
X. tropicalis embryos were obtained by in vitro fertilization. Four-cell stage embryos were immersed in 100 μM SB431542 (Tocris Bioscience) in 1/9×MMR and cultured at 25°C until mock (solvent)-treated siblings reached gastrula stage. MO injections were performed at the 2-cell stage.
Polyclonal antibody generation
A GST fusion protein containing amino acids 14-113 of X. tropicalis Foxh1 was produced in BL21 cells and purified using glutathione-agarose chromatography. Rabbit polyclonal antisera were produced by Covance and affinity purified.
MO knockdowns and rescue constructs
The foxh1 MO sequence is 5′-TCATCCTGAGGCTCCGCCCTCTCTA-3′. A 5′UTR deletion was created by BstBI/Bsu36I digestion of X. tropicalis foxh1 cDNA TGas103n06 (Gilchrist et al., 2004) followed by self-ligation to generate a MO-resistant foxh1. PouV MO sequences are: pou5f3.1, 5′-CCTGTTGGTTATACATGGTCGGCTC-3′; pou5f3.2, 5′-GCTGTTG-GCTGTACATGGTGTC-3′; pou5f3.3, 5′-GTACAGAACGGGTTGGTCC-ATGTTC-3′. Full details of the construction of PouV rescue mRNA plasmid templates can be found in the methods in the supplementary material.
Immunoprecipitation and western blot
Embryonic extracts from either control or Foxh1 MO-injected mid-gastrula embryos were immunoprecipitated followed by western blot analysis. Anti-Foxh1 antibody coupled to CNBr-activated sepharose (GE Healthcare) was used for immunoprecipitation. Beads were washed, boiled in SDS sample buffer, and eluent was subjected to western blot analysis. The blot was probed with anti-Foxh1 and then HRP-coupled anti-protein A antibodies. Anti-β-catenin antibody was used to control for morpholino specificity. For Smad IP-westerns, extracts were incubated with either Smad2/3 or Smad1/5/8 antibodies coupled to CNBr-activated beads. After washing, eluted Smad protein was subjected to western blotting and probed with either anti-pSmad2 or anti-pSmad1/5/8. Bands were visualized using HRP-coupled secondary antibody using an ECL Prime kit. Membranes were re-probed with anti-Smad2/3 or anti-Smad1/5/8. Details of antibodies are given in supplementary material Table S11.
Quantitative RT-PCR analysis
RNA samples from morphant or control were reverse transcribed using MMLV reverse transcriptase (Invitrogen). Quantitative RT-PCR of cDNA samples was performed using a Roche LightCycler 480. Details of primers are given in supplementary material Table S9.
RNA-seq analyses
Total RNAs were extracted from control and experimental early gastrulae as described previously (Chomczynski and Sacchi, 1987). Total RNA was oligo(dT) selected and fragmented, and libraries were prepared for single-end sequencing using a TruSeq RNA-seq sample preparation kit (Illumina). More details can be found in the methods in the supplementary material.
ChIP
Anti-Smad2/3 (1.5 μg) and custom anti-Foxh1 (4 µg) antibodies were used per 100-embryo-equivalents of chromatin for ChIP. Two thousand early gastrulae were subjected to ChIP for each antibody. Formaldehyde fixation, sonication, immunoprecipitation and washing steps were performed as previously described (Lee et al., 2006, Stewart et al., 2006) with modifications. Immunoprecipitated chromatin was purified and resuspended in Qiagen EB solution for subsequent library generation using NEXTflex ChIP-Seq Kit (Bioo Scientific). More details can be found in the methods in the supplementary material. Details of primers are given in supplementary material Table S10.
For sequential ChIP, embryos microinjected with FLAG-tagged Pou5f3.2 mRNA were fixed at late blastula stage 9. Crosslinked chromatin was first immunoprecipitated using anti-Foxh1 antibody, followed by the regular ChIP washes. Immunocomplexes were eluted and subjected to a second immunoprecipitation with mouse anti-FLAG M2, anti-Foxh1 or rabbit anti-IgG antibodies, then washed and eluted. DNAs were extracted, purified and subjected to qPCR.
Bioinformatics
Differential gene expression analysis of RNA-seq data was performed using TopHat (v. 1.3.3) (Trapnell et al., 2009) and Cuffdiff (v. 1.3.0) (Roberts et al., 2011). ChIP-seq reads were mapped using Bowtie (v.0.12.9) (Langmead et al., 2009); peaks were called using MACS (v.2.0.10) (Zhang et al., 2008) and SISSRs (v.1.4) (Jothi et al., 2008). De novo motif analyses used MEME (v.4.9.0) (Bailey and Elkan, 1994). STAMP/TOMTOM were used to find candidate proteins in public databases: TRANSFAC (v.11.3), UniPROBE and JASPAR CORE 2009. FIMO (v.4.9.0) (Grant et al., 2011) was used to search for motifs within genomic regions. GO analyses were performed using DAVID Bioinformatic Resources 6.7 (Huang et al., 2009). More details can be found in the methods in the supplementary material. The Gene Expression Omnibus accession number for high-throughput sequencing data reported in this paper is GSE53654.
Kolmogorov-Smirnov analysis
ChIP peak calls were compared with RNA-seq differential expression data (Foxh1 MO or SB-431542) using a Kolmogorov-Smirnov (KS) plot, similar to Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005). More details can be found in the methods in the supplementary material.
Supplementary Material
Acknowledgements
We thank the UCI Genomics High-Throughput Facility, which is supported by NIH P30CA062203, and the UCR Institute for Integrative Genome Biology for sequencing support. We thank Xenbase for Xenopus resources.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
W.T.C., R.C.L., M.B.F. and I.L.B. performed wet bench experiments. W.T.C., Y.L., J.B. and X.X. performed bioinformatics analyses. W.T.C., I.L.B., R.C.L., M.B.F. and K.W.Y.C. drafted the manuscript. K.W.Y.C. conceived and coordinated the study. All authors read and approved the final manuscript.
Funding
Research was supported by the National Institutes of Health and National Science Foundation [NSF IOS-1147270 and NIH HD073179 to K.W.Y.C.; NIH R01HG006870 and NSF DBI-0846218 to X.X.; NIH/NICHD T32-HB60555 to W.T.C.; and NSF IGERT DGE 0549479 to R.L.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.107227/-/DC1
References
- Agius E., Oelgeschläger M., Wessely O., Kemp C. and De Robertis E. M. (2000). Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development 127, 1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antic D., Stubbs J. L., Suyama K., Kintner C., Scott M. P. and Axelrod J. D. (2010). Planar cell polarity enables posterior localization of nodal cilia and left-right axis determination during mouse and xenopus embryogenesis. PLoS ONE 5, e8999 10.1371/journal.pone.0008999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S. J., Hofmann U. K., Bikoff E. K. and Robertson E. J. (2008). Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development 135, 501-511 10.1242/dev.014357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L. and Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28-36. [PubMed] [Google Scholar]
- Birsoy B., Kofron M., Schaible K., Wylie C. and Heasman J. (2006). Vg1 is an essential signaling molecule in Xenopus development. Development 133, 15-20 10.1242/dev.02144 [DOI] [PubMed] [Google Scholar]
- Burgess S., Reim G., Chen W., Hopkins N. and Brand M. (2002). The zebrafish spiel-ohne-grenzen (spg) gene encodes the POU domain protein Pou2 related to mammalian Oct4 and is essential for formation of the midbrain and hindbrain, and for pre-gastrula morphogenesis. Development 129, 905-916. [DOI] [PubMed] [Google Scholar]
- Cao Y., Siegel D. and Knöchel W. (2006). Xenopus POU factors of subclass V inhibit activin/nodal signaling during gastrulation. Mech. Dev. 123, 614-625 10.1016/j.mod.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Cao Y., Siegel D., Donow C., Knöchel S., Yuan L. and Knöchel W. (2007). POU-V factors antagonize maternal VegT activity and beta-Catenin signaling in Xenopus embryos. EMBO J. 26, 2942-2954 10.1038/sj.emboj.7601736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Siegel D., Oswald F. and Knöchel W. (2008). Oct25 represses transcription of nodal/activin target genes by interaction with signal transducers during Xenopus gastrulation. J. Biol. Chem. 283, 34168-34177 10.1074/jbc.M803532200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chea H. K., Wright C. V. and Swalla B. J. (2005). Nodal signaling and the evolution of deuterostome gastrulation. Dev. Dyn. 234, 269-278 10.1002/dvdy.20549 [DOI] [PubMed] [Google Scholar]
- Chen X., Rubock M. and Whitman M. (1996). A transcriptional partner for MAD proteins in TGF-β signalling. Nature 383, 691-696 10.1038/383691a0 [DOI] [PubMed] [Google Scholar]
- Chen X., Weisberg E., Fridmacher V., Watanabe M., Naco G. and Whitman M. (1997). Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 389, 85-89 10.1038/38008 [DOI] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J. et al. (2008). Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106-1117 10.1016/j.cell.2008.04.043 [DOI] [PubMed] [Google Scholar]
- Choi J., Dong L., Ahn J., Dao D., Hammerschmidt M. and Chen J.-N. (2007). FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev. Biol. 304, 735-744 10.1016/j.ydbio.2007.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P. and Sacchi N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156-159 10.1016/0003-2697(87)90021-2 [DOI] [PubMed] [Google Scholar]
- Conlon F. L., Lyons K. M., Takaesu N., Barth K. S., Kispert A., Herrmann B. and Robertson E. J. (1994). A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development 120, 1919-1928. [DOI] [PubMed] [Google Scholar]
- Cordenonsi M., Dupont S., Maretto S., Insinga A., Imbriano C. and Piccolo S. (2003). Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell 113, 301-314 10.1016/S0092-8674(03)00308-8 [DOI] [PubMed] [Google Scholar]
- Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S. and Gauthier J.-M. (1998). Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17, 3091-3100 10.1093/emboj/17.11.3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman B., Gates M. A., Egan E. S., Dougan S. T., Rennebeck G., Sirotkin H. I., Schier A. F. and Talbot W. S. (1998). Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature 395, 181-185 10.1038/26013 [DOI] [PubMed] [Google Scholar]
- Frankenberg S. and Renfree M. B. (2013). On the origin of POU5F1. BMC Biol. 11, 56 10.1186/1741-7007-11-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg S., Pask A. and Renfree M. B. (2010). The evolution of class V POU domain transcription factors in vertebrates and their characterisation in a marsupial. Dev. Biol. 337, 162-170 10.1016/j.ydbio.2009.10.017 [DOI] [PubMed] [Google Scholar]
- Geisberg J. V. and Struhl K. (2004). Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 32, e151 10.1093/nar/gnh148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch G. E., Owens N. D. L., Martin S. R., Piccinelli P., Faial T., Trotter M. W. B., Gilchrist M. J. and Smith J. C. (2013). In vivo T-box transcription factor profiling reveals joint regulation of embryonic neuromesodermal bipotency. Cell Rep. 4, 1185-1196 10.1016/j.celrep.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain S., Howell M., Esslemont G. M. and Hill C. S. (2000). Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 14, 435-451. [PMC free article] [PubMed] [Google Scholar]
- Gilchrist M. J., Zorn A. M., Voigt J., Smith J. C., Papalopulu N. and Amaya E. (2004). Defining a large set of full-length clones from a Xenopus tropicalis EST project. Dev. Biol. 271, 498-516 10.1016/j.ydbio.2004.04.023 [DOI] [PubMed] [Google Scholar]
- Goh L. L. and Manser E. (2012). The GTPase-deficient Rnd proteins are stabilized by their effectors. J. Biol. Chem. 287, 31311-31320 10.1074/jbc.M111.327056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C. E., Bailey T. L. and Noble W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017-1018 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammachi F., Morrison G. M., Sharov A. A., Livigni A., Narayan S., Papapetrou E. P., O'Malley J., Kaji K., Ko M. S. H., Ptashne M. et al. (2012). Transcriptional activation by Oct4 is sufficient for the maintenance and induction of pluripotency. Cell Rep. 1, 99-109 10.1016/j.celrep.2011.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J. (2006). Patterning the early Xenopus embryo. Development 133, 1205-1217 10.1242/dev.02304 [DOI] [PubMed] [Google Scholar]
- Hellsten U., Harland R. M., Gilchrist M. J., Hendrix D., Jurka J., Kapitonov V., Ovcharenko I., Putnam N. H., Shu S., Taher L. et al. (2010). The genome of the Western clawed frog Xenopus tropicalis. Science 328, 633-636 10.1126/science.1183670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkley C. S., Martin J. F., Leibham D. and Perry M. (1992). Sequential expression of multiple POU proteins during amphibian early development. Mol. Cell. Biol. 12, 638-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho D. M., Chan J., Bayliss P. and Whitman M. (2006). Inhibitor-resistant type I receptors reveal specific requirements for TGF-beta signaling in vivo. Dev. Biol. 295, 730-742 10.1016/j.ydbio.2006.03.050 [DOI] [PubMed] [Google Scholar]
- Hoodless P. A., Pye M., Chazaud C., Labbé E., Attisano L., Rossant J. and Wrana J. L. (2001). FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev. 15, 1257-1271 10.1101/gad.881501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M., Inman G. J. and Hill C. S. (2002). A novel Xenopus Smad-interacting forkhead transcription factor (XFast-3) cooperates with XFast-1 in regulating gastrulation movements. Development 129, 2823-2834. [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. and Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Inman G. J., Nicolás F. J., Callahan J. F., Harling J. D., Gaster L. M., Reith A. D., Laping N. J. and Hill C. S. (2002). SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I Activin receptor-Like Kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 62, 65-74 10.1124/mol.62.1.65 [DOI] [PubMed] [Google Scholar]
- James-Zorn C., Ponferrada V. G., Jarabek C. J., Burns K. A., Segerdell E. J., Lee J., Snyder K., Bhattacharyya B., Karpinka J. B., Fortriede J. et al. (2013). Xenbase: expansion and updates of the Xenopus model organism database. Nucleic Acids Res. 41, D865-D870 10.1093/nar/gks1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R., Cuddapah S., Barski A., Cui K. and Zhao K. (2008). Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 36, 5221-5231 10.1093/nar/gkn488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaulanov E., Knöchel W. and Niehrs C. (2004). Transcriptional regulation of BMP4 synexpression in transgenic Xenopus. EMBO J. 23, 844-856 10.1038/sj.emboj.7600101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofron M., Puck H., Standley H. and Wylie C. (2004a). New roles for FoxH1 in patterning the early embryo. Development131, 5065-5078. [DOI] [PubMed] [Google Scholar]
- Kofron M., Puck H., Standley H., Wylie C., Old R., Whitman M. and Heasman J. (2004b). New roles for FoxH1 in patterning the early embryo. Development 131, 5065-5078 10.1242/dev.01396 [DOI] [PubMed] [Google Scholar]
- Koide T., Hayata T. and Cho K. W. Y. (2005). Xenopus as a model system to study transcriptional regulatory networks. Proc. Natl. Acad. Sci. USA 102, 4943-4948 10.1073/pnas.0408125102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M., Sokol S. Y., Wu J., Tussie-Luna M. I., Roy A. L. and Hata A. (2005). Positive and negative regulation of the transforming growth factor beta/activin target gene goosecoid by the TFII-I family of transcription factors. Mol. Cell. Biol. 25, 7144-7157 10.1128/MCB.25.16.7144-7157.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Shinozaki K., Shannon J. M., Kouskoff V., Kennedy M., Woo S., Fehling H. J. and Keller G. (2004). Development of definitive endoderm from embryonic stem cells in culture. Development 131, 1651-1662 10.1242/dev.01044 [DOI] [PubMed] [Google Scholar]
- Labbé E., Silvestri C., Hoodless P. A., Wrana J. L. and Attisano L. (1998). Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol. Cell 2, 109-120 10.1016/S1097-2765(00)80119-7 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. and Salzberg S. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. A., Heasman J. and Whitman M. (2001). Timing of endogenous activin-like signals and regional specification of the Xenopus embryo. Development 128, 2939-2952. [DOI] [PubMed] [Google Scholar]
- Lee T. I., Johnstone S. E. and Young R. A. (2006). Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1, 729-748 10.1038/nprot.2006.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livigni A., Peradziryi H., Sharov A. A., Chia G., Hammachi F., Migueles R. P., Sukparangsi W., Pernagallo S., Bradley M., Nichols J. et al. (2103). A conserved Oct4/POUV-dependent network links adhesion and migration to progenitor maintenance. Curr. Biol. 23, 2233-2244 10.1016/j.cub.2013.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M. and Patient R. (2004). A genetic regulatory network for Xenopus mesendoderm formation. Dev. Biol. 271, 467-478 10.1016/j.ydbio.2004.04.014 [DOI] [PubMed] [Google Scholar]
- Lunde K., Belting H.-G. and Driever W. (2004). Zebrafish pou5f1/pou2, homolog of mammalian Oct4, functions in the endoderm specification cascade. Curr. Biol. 14, 48-55 10.1016/j.cub.2003.11.022 [DOI] [PubMed] [Google Scholar]
- Luxardi G., Marchal L., Thomé V. and Kodjabachian L. (2010). Distinct Xenopus Nodal ligands sequentially induce mesendoderm and control gastrulation movements in parallel to the Wnt/PCP pathway. Development 137, 417-426 10.1242/dev.039735 [DOI] [PubMed] [Google Scholar]
- Mason M. J., Plath K. and Zhou Q. (2010). Identification of context-dependent motifs by contrasting ChIP binding data. Bioinformatics 26, 2826-2832 10.1093/bioinformatics/btq546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison G. M. and Brickman J. M. (2006). Conserved roles for Oct4 homologues in maintaining multipotency during early vertebrate development. Development 133, 2011–2022 10.1242/dev.02362 [DOI] [PubMed] [Google Scholar]
- Mullen A. C., Orlando D. A., Newman J. J., Lovén J., Kumar R. M., Bilodeau S., Reddy J., Guenther M. G., DeKoter R. P. and Young R. A. (2011). Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147, 565-576 10.1016/j.cell.2011.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S. and Schilling T. F. (2008). Chemokine signaling controls endodermal migration during zebrafish gastrulation. Science 322, 89-92 10.1126/science.1160038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata S., Morokuma J., Hayata T., Kolle G., Niehrs C., Ueno N. and Cho K. W. Y. (2007). TGF-beta signaling-mediated morphogenesis: modulation of cell adhesion via cadherin endocytosis. Genes Dev. 21, 1817-1831 10.1101/gad.1541807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada S. I. and Wright C. V. (1999). Xenopus nodal-related signaling is essential for mesendodermal patterning during early embryogenesis. Development 126, 3229-3240. [DOI] [PubMed] [Google Scholar]
- Osada S. I., Saijoh Y., Frisch A., Yeo C. Y., Adachi H., Watanabe M., Whitman M., Hamada H. and Wright C. V. (2000). Activin/nodal responsiveness and asymmetric expression of a Xenopus nodal-related gene converge on a FAST-regulated module in intron 1. Development 127, 2503-2514. [DOI] [PubMed] [Google Scholar]
- Pan G. J., Chang Z. Y., Schöler H. R. and Pei D. (2002). Stem cell pluripotency and transcription factor Oct4. Cell Res. 12, 321-329 10.1038/sj.cr.7290134 [DOI] [PubMed] [Google Scholar]
- Pei W., Noushmehr H., Costa J., Ouspenskaia M. V., Elkahloun A. G. and Feldman B. (2007). An early requirement for maternal FoxH1 during zebrafish gastrulation. Dev. Biol. 310, 10-22 10.1016/j.ydbio.2007.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picozzi P., Wang F., Cronk K. and Ryan K. (2009). Eomesodermin requires transforming growth factor-beta/activin signaling and binds Smad2 to activate mesodermal genes. J. Biol. Chem. 284, 2397-2408 10.1074/jbc.M808704200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O., Grimmer D., Williams P. H. and Smith J. C. (2004). Activin redux: specification of mesodermal pattern in Xenopus by graded concentrations of endogenous activin B. Development 131, 4977-4986 10.1242/dev.01323 [DOI] [PubMed] [Google Scholar]
- Pogoda H.-M., Solnica-Krezel L., Driever W. and Meyer D. (2000). The zebrafish forkhead transcription factor FoxH1/Fast1 is a modulator of Nodal signaling required for organizer formation. Curr. Biol. 10, 1041-1049 10.1016/S0960-9822(00)00669-2 [DOI] [PubMed] [Google Scholar]
- Randall R. A., Howell M., Page C. S., Daly A., Bates P. A. and Hill C. S. (2004). Recognition of phosphorylated-Smad2-containing complexes by a novel Smad interaction motif. Mol. Cell. Biol. 24, 1106-1121 10.1128/MCB.24.3.1106-1121.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim G. and Brand M. (2006). Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development 133, 2757-2770 10.1242/dev.02391 [DOI] [PubMed] [Google Scholar]
- Reim G., Mizoguchi T., Stainier D. Y., Kikuchi Y. and Brand M. (2004). The POU domain protein Spg (Pou2/Oct4) is essential for endoderm formation in cooperation with the HMG domain protein casanova. Dev. Cell 6, 91-101 10.1016/S1534-5807(03)00396-4 [DOI] [PubMed] [Google Scholar]
- Ring C., Ogata S., Meek L., Song J., Ohta T., Miyazono K. and Cho K. W. Y. (2002). The role of a Williams-Beuren syndrome-associated helix-loop-helix domain-containing transcription factor in activin/nodal signaling. Genes Dev. 16, 820-835 10.1101/gad.963802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pimentel H., Trapnell C. and Pachter L. (2011). Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325-2329 10.1093/bioinformatics/btr355 [DOI] [PubMed] [Google Scholar]
- Schier A. F. (2003). Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 19, 589-621 10.1146/annurev.cellbio.19.041603.094522 [DOI] [PubMed] [Google Scholar]
- Shi Y., Wang Y.-F., Jayaraman L., Yang H., Massagué J. and Pavletich N. P. (1998). Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell 94, 585-594 10.1016/S0092-8674(00)81600-1 [DOI] [PubMed] [Google Scholar]
- Shiratori H., Sakuma R., Watanabe M., Hashiguchi H., Mochida K., Sakai Y., Nishino J., Saijoh Y., Whitman M. and Hamada H. (2001). Two-step regulation of left-right asymmetric expression of Pitx2: initiation by nodal signaling and maintenance by Nkx2. Mol. Cell 7, 137-149 10.1016/S1097-2765(01)00162-9 [DOI] [PubMed] [Google Scholar]
- Silvestri C., Narimatsu M., von Both I., Liu Y., Tan N. B. J., Izzi L., McCaffery P., Wrana J. L. and Attisano L. (2008). Genome-wide identification of Smad/Foxh1 targets reveals a role for Foxh1 in retinoic acid regulation and forebrain development. Dev. Cell 14, 411-423 10.1016/j.devcel.2008.01.004 [DOI] [PubMed] [Google Scholar]
- Sirotkin H. I., Gates M. A., Kelly P. D., Schier A. F. and Talbot W. S. (2000). fast1 is required for the development of dorsal axial structures in zebrafish. Curr. Biol. 10, 1051-1054 10.1016/S0960-9822(00)00679-5 [DOI] [PubMed] [Google Scholar]
- Slagle C. E., Aoki T. and Burdine R. D. (2011). Nodal-dependent mesendoderm specification requires the combinatorial activities of FoxH1 and Eomesodermin. PLoS Genet. 7, e1002072 10.1371/journal.pgen.1002072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring J., Yanze N., Jösch C., Middel A. M., Winninger B. and Schmid V. (2002). Conservation of Brachyury, Mef2, and Snail in the myogenic lineage of jellyfish: a connection to the mesoderm of bilateria. Dev. Biol. 244, 372-384 10.1006/dbio.2002.0616 [DOI] [PubMed] [Google Scholar]
- Stewart D., Tomita A., Shi Y.-B. and Wong J. (2006). Chromatin immunoprecipitation for studying transcriptional regulation in Xenopus oocytes and tadpoles. Methods Mol. Biol. 322, 165-181 10.1007/978-1-59745-000-3_12 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S. et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B. I., Bush S. M., Collins-Racie L. A., LaVallie E. R., DiBlasio-Smith E. A., Wolfman N. M., McCoy J. M. and Sive H. L. (1999). derriere: a TGF-beta family member required for posterior development in Xenopus. Development 126, 1467-1482. [DOI] [PubMed] [Google Scholar]
- Tao H., Manak J. R., Sowers L., Mei X., Kiyonari H., Abe T., Dandaleh N. S., Yang T. A., Wu S., Chen S. et al. (2011). Mutations in prickle orthologs cause seizures in flies, mice, and humans. Am. J. Hum. Genet. 88, 138-149 10.1016/j.ajhg.2010.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H., Inoue K.-i., Kiyonari H., Bassuk A. G., Axelrod J. D., Sasaki H., Aizawa S. and Ueno N. (2012). Nuclear localization of Prickle2 is required to establish cell polarity during early mouse embryogenesis. Dev. Biol. 364, 138-148 10.1016/j.ydbio.2012.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A. K. K., Arnold S. J., Trotter M. W. B., Brown S., Ang L. T., Chng Z., Robertson E. J., Dunn N. R. and Vallier L. (2011). Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 25, 238-250 10.1101/gad.607311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. and Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105-1111 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlet I., Collignon J. and Robertson E. J. (1997). nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development 124, 1033-1044. [DOI] [PubMed] [Google Scholar]
- Watanabe M. and Whitman M. (1999). FAST-1 is a key maternal effector of mesoderm inducers in the early Xenopus embryo. Development 126, 5621-5634. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Rebbert M. L., Andreazzoli M., Takahashi N., Toyama R., Zimmerman S., Whitman M. and Dawid I. B. (2002). Regulation of the Lim-1 gene is mediated through conserved FAST-1/FoxH1 sites in the first intron. Dev. Dyn. 225, 448-456 10.1002/dvdy.10176 [DOI] [PubMed] [Google Scholar]
- Wilson D., Sheng G., Lecuit T., Dostatni N. and Desplan C. (1993). Cooperative dimerization of paired class homeo domains on DNA. Genes Dev. 7, 2120-2134 10.1101/gad.7.11.2120 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Meno C., Sakai Y., Shiratori H., Mochida K., Ikawa Y., Saijoh Y. and Hamada H. (2001). The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes Dev. 15, 1242-1256 10.1101/gad.883901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C.-Y., Chen X. and Whitman M. (1999). The role of FAST-1 and Smads in transcriptional regulation by activin during early Xenopus embryogenesis. J. Biol. Chem. 274, 26584-26590 10.1074/jbc.274.37.26584 [DOI] [PubMed] [Google Scholar]
- Yoon S.-J., Wills A. E., Chuong E., Gupta R. and Baker J. C. (2011). HEB and E2A function as SMAD/FOXH1 cofactors. Genes Dev. 25, 1654-1661 10.1101/gad.16800511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L., Dai J. L., Buckhaults P., Zhou S., Kinzler K. W., Vogelstein B. and Kern S. E. (1998). Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1, 611-617 10.1016/S1097-2765(00)80061-1 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W. et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wu Y., Schnable J. C., Zeng Z., Freeling M., Crawford G. E. and Jiang J. (2012). High-resolution mapping of open chromatin in the rice genome. Genome Res. 22, 151-162 10.1101/gr.131342.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Sasaki H., Lowe L., Hogan B. L. M. and Kuehn M. R. (1993). Nodal is a novel TGF-[beta]-like gene expressed in the mouse node during gastrulation. Nature 361, 543-547 10.1038/361543a0 [DOI] [PubMed] [Google Scholar]
- Zhou S., Zawel L., Lengauer C., Kinzler K. W. and Vogelstein B. (1998). Characterization of human FAST-1, a TGFβ and activin signal transducer. Mol. Cell 2, 121-127 10.1016/S1097-2765(00)80120-3 [DOI] [PubMed] [Google Scholar]
- Zorn A. M. and Wells J. M. (2007). Molecular basis of vertebrate endoderm development. Int. Rev. Cytol. 259, 49-111 10.1016/S0074-7696(06)59002-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.