Abstract
Identifying coronary artery progenitors and their developmental pathways could inspire novel regenerative treatments for heart disease. Multiple sources of coronary vessels have been proposed, including the sinus venosus (SV), endocardium and proepicardium, but their relative contributions to the coronary circulation and the molecular mechanisms regulating their development are poorly understood. We created an ApjCreER mouse line as a lineage-tracing tool to map SV-derived vessels onto the heart and compared the resulting lineage pattern with endocardial and proepicardial contributions to the coronary circulation. The data showed a striking compartmentalization to coronary development. ApjCreER-traced vessels contributed to a large number of arteries, capillaries and veins on the dorsal and lateral sides of the heart. By contrast, untraced vessels predominated in the midline of the ventral aspect and ventricular septum, which are vessel populations primarily derived from the endocardium. The proepicardium gave rise to a smaller fraction of vessels spaced relatively uniformly throughout the ventricular walls. Dorsal (SV-derived) and ventral (endocardial-derived) coronary vessels developed in response to different growth signals. The absence of VEGFC, which is expressed in the epicardium, dramatically inhibited dorsal and lateral coronary growth but left vessels on the ventral side unaffected. We propose that complementary SV-derived and endocardial-derived migratory routes unite to form the coronary vasculature and that the former requires VEGFC, revealing its role as a tissue-specific mediator of blood endothelial development.
Keywords: VEGF-C, Angiogenesis, Apelin receptor (APJ, APLNR), Coronary vessel development, Endothelium, Sinus venosus
INTRODUCTION
Given the devastating effects of coronary heart disease, there is an important need to fully understand coronary artery development and regeneration. Mature coronary arteries are composed of a blood-filled endothelial lumen wrapped in a layer of smooth muscle and then by an adventitial exterior. These three elements develop sequentially during embryogenesis, with the endothelial layer being established first through the assembly of an immature vascular plexus. Thus, identifying the progenitors of the early coronary plexus and their mechanisms of development is the first step in understanding how coronary arteries are constructed.
Several studies have characterized the pattern by which the coronary endothelial plexus populates the heart (Fig. 1A) (Kattan et al., 2004; Lavine et al., 2006; Red-Horse et al., 2010; Zeini et al., 2009). It first appears on the dorsal aspect at the atrioventricular groove near the sinus venosus (SV) (inflow tract) (Lavine et al., 2006; Red-Horse et al., 2010). From here, the plexus expands toward the apex and laterally around the sides toward the ventral aspect of the heart, eventually covering the entire left and right ventricles. These vessels are first seen just beneath the epicardium (subepicardial) and later appear to migrate from this location deeper into the myocardium. By contrast, on the ventral side of the heart, coronary vessels first emerge deep within the myocardium at the interventricular groove. From here, they appear to migrate laterally, populating the ventral aspect as they meet the vessels that traveled around from the dorsal side (Red-Horse et al., 2010). As this process proceeds, so-called ‘blood islands’, or endothelial spheres that are connected to the endocardium and contain erythrocytes, also appear (Hiruma and Hirakow, 1989; Hutchins et al., 1988; Red-Horse et al., 2010; Tian et al., 2013). Blood islands are most dense along the ventral interventricular groove (Tian et al., 2013) and eventually join with developing coronary vessels (Red-Horse et al., 2010). These endothelial migratory paths proceed to fill the entire heart muscle with a primitive vascular plexus, which eventually connects with the aorta to initiate blood flow and remodels into mature arteries, capillaries and veins.
Fig. 1.
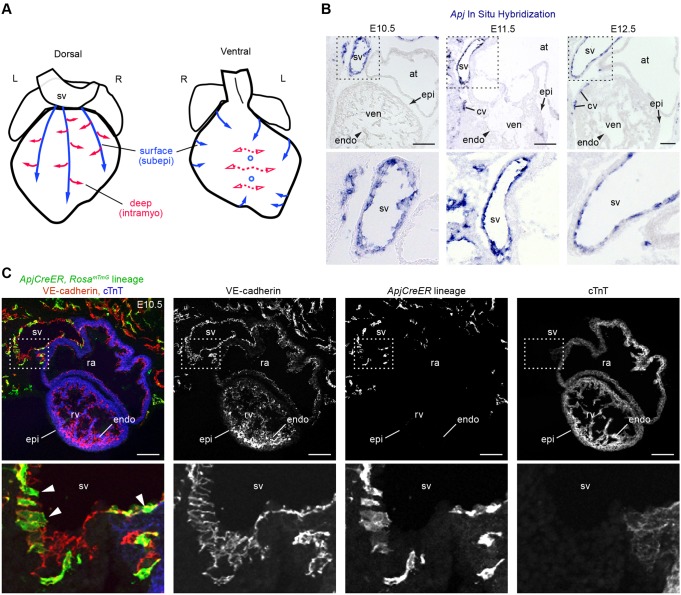
Apj and ApjCreER are expressed in the SV. (A) Schematics showing the direction of vessel migration (arrows) during coronary vascular plexus formation. Vessels take subepicardial (surface, subepi) and intramyocardial (deep, intramyo) routes as they populate the entire heart muscle. Solid and dashed lines represent proposed SV and endocardial migratory paths, respectively. (B) Apj mRNA is highly expressed in the SV endothelium but not at appreciable levels in the endocardium (arrowhead) or epicardium (arrow) from E10.5 to E12.5 as shown by in situ hybridization (ISH). Lower panels are magnifications of the boxed regions. (C) A tissue section from ApjCreER, RosamTmG embryos dosed with tamoxifen at E9.5 and analyzed at E10.5. Cre recombination is marked with GFP (green), endothelial cells with VE-cadherin (red) and myocardium with cTnT (blue). Recombination occurs in the SV (arrowheads), but not in the endocardium or epicardium. Lower panels are magnifications of the boxed regions. at, atrium; cv, coronary vessel; endo, endocardium; epi, epicardium; L, left; R, right; ra, right atrium; rv, right ventricle; sv, sinus venosus; ven, ventricle. Scale bars: 100 µm.
Recent reports have proposed that, in mammals, the nascent coronary endothelial plexus enters the heart by sprouting from the SV (Red-Horse et al., 2010; Tian et al., 2013) and endocardium (Red-Horse et al., 2010; Wu et al., 2012). Subsets of the proepicardium also contribute, and may do so by differentiating first into SV or endocardial cells (Katz et al., 2012). Clonal analysis detected a lineage relationship between coronary vessels and both the SV and endocardium (Red-Horse et al., 2010). A Cre line expressed in the endocardium lineage (Nfatc1Cre) labels coronary vessels (Wu et al., 2012), and this pathway is mediated by VEGFA produced by cardiomyocytes. A subset of the proepicardium expresses Sema3d and scleraxis (Scx), and Cre lines driven by their enhancer/promoter regions trace portions of the SV, endocardium and a subset of coronary endothelial cells (Katz et al., 2012). These data support a model whereby SV, endocardial and proepicardial cells all converge to produce the mature coronary tree. To fully understand how these cells develop into coronary vessels, it is necessary to define the exact extent of contribution by the different sources and whether similar or distinct signals guide their development.
Here, we define the spatial contributions of the different coronary progenitors within the entire heart. Our data show that the SV and endocardium give rise to complementary regions of the coronary vasculature in an inversely proportional manner along the dorsal to ventral axis. The proepicardium forms a smaller portion of the vasculature uniformly around the heart. We also show that VEGFC is important for the SV-derived pathway, activating vessel migration along the surface of the ventricles. Delineating the spatial contributions of the different coronary sources and identifying a mechanism that supports SV-derived vessel growth are major steps toward a comprehensive understanding of coronary development.
RESULTS
ApjCreER can be used to lineage trace SV-derived coronary vessels
To specifically follow SV-derived sprouts in the developing heart, we created a mouse line that expresses CreER under the control of the Apj enhancer/promoter. Apj is highly expressed in the SV endothelium, but not at appreciable levels in the endocardium or epicardium (Fig. 1B). Recombineering (Sharan et al., 2009; Warming et al., 2005) was used to insert CreERT2 at the Apj start site of a bacterial artificial chromosome (BAC). ApjCreER animals were crossed with the RosamTmG Cre reporter, a widely expressed allele that switches from membrane tomato to membrane GFP upon Cre-mediated recombination (Muzumdar et al., 2007).
Animals were dosed with tamoxifen to activate Cre activity on embryonic day (E) 9.5 and dissected at E10.5. Assessing recombination within the entire embryo revealed that ApjCreER was expressed in a pattern that appeared identical to that of Apj mRNA (supplementary material Fig. S1A-D). With the RosamTmG reporter line and the described dosing schedule, the endothelium of the SV was labeled whereas the epicardium was negative and recombination within the endocardium was very rare (Fig. 1C). Although epicardial labeling was not seen following E9.5 dosing, rare WT1+ proepicardial cells were occasionally observed when tamoxifen was given at E8.5 (data not shown). Thus, using the above-described dosing strategy (E9.5) with the RosamTmG reporter line, ApjCreER could be used to lineage trace SV-derived coronary vessels.
ApjCreER-marked lineages contribute extensively to the developing coronary vasculature
We next analyzed coronary vessel development in ApjCreER, RosamTmG animals to define the regions populated by SV-derived sprouts. A single tamoxifen dose at E9.5 was used to pulse-label ApjCreER-expressing cells after the SV had developed but before coronary vessels had formed. Hearts immunostained for VE-cadherin (also known as cadherin 5) to label endothelium and cTnT (also known as TNNT2) to label myocardium were analyzed using whole-mount confocal microscopy. Visualizing recombination in the whole heart revealed a robust labeling of the SV: hundreds of cells were marked in this compartment (Fig. 2A-F). Quantification showed that, on average, 68% of cells within the SV were marked using our dosing strategy (Fig. 2G). Apj expression within the SV has been reported to be heterogeneous (Arita et al., 2014) and could explain the observed labeling density. However, if increased amounts of tamoxifen were given over multiple days (E8.5, E9.5 and E10.5), lineage labeling of the SV was almost 100% (Fig. 2H). We did not proceed with this dose for most experiments, except to confirm final lineage patterns, to ensure that labeling occurred only in the SV and not in nascent coronary vessels, which express Apj as they first migrate onto the heart (Fig. 1B). In addition to SV labeling, rare recombination events did occur in endocardial cells, but this was minimal, ranging from 2-15 randomly distributed cells per heart (average=6).
Fig. 2.
The ApjCreER lineage traces early SV sprouts. (A-F,H,I) Whole-mount confocal images immunostained for VE-cadherin (endothelium/endocardium) and cTnT (myocardium). Direct GFP fluorescence indicates lineage labeling. (A-C) Robust SV labeling at E10.5 (A) progressively spreads onto the heart at subsequent developmental stages (B,C). Little or no ventral labeling is detected at these stages. (D-F) High magnifications of the SV, endocardium (endo) and coronary vessels (CV) from the white-boxed regions in A-C. (G) Quantification of SV and coronary vessel labeling with a single E9.5 dose of tamoxifen. Error bars indicate one s.d. above and below the mean. Each dot represents the quantification for one heart. (H) Near complete labeling of the SV is achieved when tamoxifen is given once daily at E8.5, E9.5 and E10.5. (I) Depth-coded confocal images of red boxed regions in B and C showing that lineage-labeled cells first migrate subepicardially (red) at E11.5 and then into the myocardium (green) at E12.5. bi, blood island; cv, coronary vessels; la, left atrium; lv, left ventricle; ot, outflow tract; ra, right atrium; rv, right ventricle. Scale bars: 100 µm.
From E10.5 through E12.5, the regions populated by lineage-labeled vessels expanded progressively over the dorsal side of the heart from the SV (Fig. 2A-C), first subepicardially then spreading into the myocardium (Fig. 2I). Dorsal vessels were labeled at a frequency similar to that of the SV (Fig. 2G), and marked cells were present at the leading front of the migrating vascular plexus (Fig. 2E,F). During this period, little labeling was seen on the ventral side of the heart, and blood islands were usually GFP negative (Fig. 2A-C). Importantly, no recombination was detected in control hearts that did not receive tamoxifen (supplementary material Fig. S1E). We conclude that this lineage trace accurately represents cellular contribution from the SV based on the following observations: (1) labeled cells progress in a contiguous fashion from the heavily labeled SV as development proceeds; and (2) there are negligible numbers of labeled cells in the endocardium and in regions derived from the endocardium (Tian et al., 2014, 2013) (see below). These observations support a model whereby a significant portion of the subepicardial and intramyocardial coronary sprouts on the dorsal side of the early heart arise by SV sprouting angiogenesis.
The precise origin of coronary endothelial cells has been a matter of controversy in recent years. Seemingly disparate findings could be reconciled by presenting lineage-tracing data as they appear on the whole heart. In order to do so, our experiments required a molecular marker to specifically label coronary vessels, but not the vast number of endocardial cells, for whole-mount confocal microscopy. We found that DACH1 could fulfill this purpose. This homolog to the Drosophila protein Dachshund was initially identified in a screen that used Cre-inducible TU-tagging of RNA to discover endothelial-specific genes from whole hearts (Gay et al., 2013). Commercially available antibodies to DACH1 are specific, as evidenced by the absence of immunostaining in hearts from knockout animals (supplementary material Fig. S2A). In wild-type hearts, DACH1 distinguishes coronary endothelium from endocardium, marking the former but not the latter (supplementary material Fig. S2A-C). Although expressed in endothelial cells of other organs, this protein, because of its specificity in the heart, was used in subsequent lineage-tracing experiments to more clearly visualize coronary endothelial cells.
We next investigated which coronary vessels were traced in ApjCreER, RosamTmG embryos at stages when arteries, capillaries and veins were apparent. Whole-mount immunohistochemistry on E13.75-17.5 hearts revealed a large number of lineage-labeled coronary vessels on most of the dorsal aspect of the heart (Fig. 3A). By contrast, the coronary endothelial population on the ventral side was highly compartmentalized, with the mid-portion of the ventral side consisting primarily of DACH1-expressing coronary vessels that did not contain the ApjCreER lineage label (Fig. 3B). Likewise, the ventricular septum contained almost no lineage-labeled coronary vessels compared with the ventricular walls (Fig. 3C). ApjCreER-traced coronary vessels were not segregated among the arterial, capillary and venous layers, but were in all vessel types at surface and intramyocardial locations (Fig. 3C-E) and always contributed to large arteries (Fig. 3E). Strikingly, there was sometimes a distinct border between ApjCreER-traced and non-ApjCreER-traced coronary vessels within the compact myocardium (Fig. 3E), which was observed in the ventral region of the heart. Immunostaining for connexin 40 (also known as gap junction protein alpha 5) and COUP-TFII (also known as NR2F2) showed that traced vessels were positive for both arterial and venous markers, respectively (Fig. 3F). The reduced ApjCreER lineage contribution on the ventral side and septum was not a consequence of Apj-negative and/or unlabeled cells within the SV since a similar lineage pattern was obtained with a triple dosing schedule (4 mg of tamoxifen daily on E8.5, E9.5 and E10.5), which almost completely labeled the SV (Fig. 2H and Fig. 4A). These data indicate that SV-derived coronary arteries, capillaries and veins vascularize large areas of the embryonic heart and that another source contributes to distinct regions, particularly the ventricular septum and mid-portion of the ventral side.
Fig. 3.
ApjCreER lineage-traced vessels give rise to coronary artery, capillaries and veins of the ventricular walls. Whole-mount confocal images (A,B,D,E) or tissue sections (C,F) of hearts from ApjCreER, RosamTmG crosses dosed with tamoxifen at E9.5 and isolated at the indicated ages. The lineage label is shown in green. (A,B) Many coronary vessels (DACH1+, red) on the dorsal and lateral sides of the heart are ApjCreER lineage traced (green), whereas parts of the apex and central region of the ventral side are not (asterisks). (C) Tissue sections through the left and right ventricular walls (lw and rw, respectively) and septum (sep, outlined by dotted line) show the paucity of ApjCreER lineage-traced vessels in the latter structure. (D) A series of optical sections through the ventricles (dorsal to ventral from left to right) shows lineage-traced cells on the surface (subepicardial) and within the myocardium (intramyocardial). (E) The boxed region from D showing a partially labeled coronary artery (ca) at the border between lineage-labeled (sv-derived) and non-labeled (non-sv-derived) vessels. (F) Immunostaining tissue sections for connexin 40 (Cx40) and COUP-TFII shows ApjCreER lineage contribution to arteries and veins, respectively. lv, left ventricle; rv, right ventricle. Scale bars: 100 µm.
Fig. 4.
ApjCreER-traced and Nfatc1Cre-traced coronary lineages populate complementary and overlapping regions of the heart. (A) Whole-mount confocal images of ApjCreER (left) and Nfatc1Cre (right) lineage traces immunostained with anti-DACH1 antibodies (red). Nfatc1Cre-labeled ventral vessels are dense on the ventral face, where ApjCreER-labeled vessels are sparse. High magnification of the boxed region is shown beneath. Arrowheads point to examples of GFP-positive ventral coronary vessels. (B) Representative tissue sections through the right lateral ventricular wall, left lateral ventricular wall, and septum of hearts from the indicated Cre lines highlight complementary contributions. Endothelial/endocardial cells are immunostained for VE-cadherin (top row) or PECAM1 (bottom row). (C) Quantification of ApjCreER-traced, Nfatc1Cre-traced and Sema3dCre-traced vessels shows that the latter population is at fairly consistent, low levels throughout the heart. ApjCreER and Nfatc1Cre lineages tend to contribute inversely proportional numbers to the cardiac compartments when considered from the dorsal to ventral aspect. Error bars represent one s.d. above and below the mean. cv, coronary vessels; endo, endocardium. Scale bars: 100 µm in A; 50 µm in B.
The SV and endocardium contribute to complementary and overlapping regions of the coronary vasculature
ApjCreER lineage tracing suggests that the SV contributes to the coronary vasculature in a region-specific manner. Since other sources contribute to the coronary vascular bed, we hypothesized that they might preferentially give rise to regions not populated by ApjCreER-labeled vessels. We tested this hypothesis by quantifying the number of lineage-labeled cells induced by three Cre mouse lines – ApjCreER (SV), Nfatc1Cre (endocardium) and Sema3dCre (proepicardium) – at five different regions of the heart: dorsal, left lateral, right lateral, ventral and septum. The pattern and quantification of each lineage trace were compared to determine if the contributions were complementary.
SV and endocardial lineage tracing were compared first, since these give rise to the majority of coronary vessels (Red-Horse et al., 2010; Wu et al., 2012). Initially, whole-mount images of the ventral and dorsal sides were compared between ApjCreER and Nfatc1Cre lines crossed to the RosamTmG reporter mouse. For this comparison, ApjCreER was dosed once daily on E8.5, E9.5 and E10.5 to exclude the possibility that incomplete or timing-specific labeling might account for non-traced areas. The differences were most striking on the ventral side at the mid-portion. In this region, Nfatc1Cre labeling was high, whereas ApjCreER-derived vessels were rare (Fig. 4A). Tissue sections through deeper regions of the heart showed a similar complementarity. ApjCreER traces were more numerous on the right lateral side and absent in the septum; Nfatc1Cre labeled more coronary cells on the left lateral wall and labeling was heavy in the septum (Fig. 4B).
The percentage of vessels in each of the five analyzed regions was then quantified (Fig. 4C). ApjCreER (single E9.5 dose) contributed to an average of 78% of coronary vessels on the dorsal and right lateral side. The left lateral side contained 40% ApjCreER-labeled cells, whereas only 4% of the ventral and septal regions were traced. The Nfatc1Cre line exhibited a corresponding pattern. The dorsal side was marked at 47%, the right lateral at 41%, and the left lateral at 65%. The ventral side and septum were traced at 71% and 83%, respectively. Dorsal labeling was highest by ApjCreER, but Nfatc1Cre did contribute to vessels on this side. Since Nfatc1Cre labels a small portion of the SV (Wu et al., 2012), it is difficult to determine which vessels were labeled as a result of SV or endocardial labeling, but it is likely that endocardial sprouting contributes at least in part, given that there are non-SV-derived dorsal vessels, particularly at the heart apex (Fig. 3A,B, Fig. 4A). This overlap in labeling at the SV could also explain why the dorsal and right lateral percentages (regions where the SV contributes most) sum to greater than 100%. These data indicate that endocardium-derived and SV-derived vessels coalesce to produce the coronary circulation, with the former producing vessels in areas not invested by the latter (ventral mid-portion and septum).
The distribution of Sema3dCre-derived coronary vessels was subsequently analyzed. Sema3dCre mice were crossed with the RosatdTomato reporter line, and tissues from E16.5 and postnatal day (P) 0 hearts were quantified (values from both time points were identical) (Fig. 4C). Using the RosatdTomato reporter, there were similar percentages of cells (9-11%) labeled at the left lateral, right lateral and ventral regions (Fig. 4C). Dorsal was slightly higher at 17%, which could be due to additional labeling of the SV by this Cre recombinase (Katz et al., 2012). Only 4% of the DACH1+ coronary endothelial cells in the septum were labeled, which is consistent with the low number of epicardial-derived cells in this region. Therefore, Sema3dCre labeling does not follow a spatial pattern that is similar to the SV or endocardial traces, supporting the notion that the epicardium contributes to the coronary vasculature through a direct pathway (i.e. not by first differentiating into SV or endocardial endothelium).
VEGFC is required for coronary vessel development
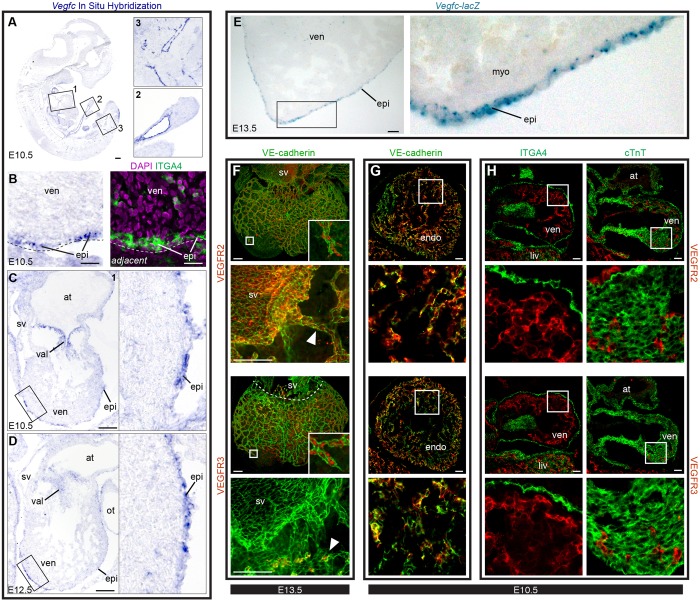
We next aimed to identify factors that mediate SV-derived coronary growth. It has been reported that VEGFA stimulates endocardial cells to migrate into the myocardium but that subepicardial coronary vessels are less dependent on this factor (Wu et al., 2012). Since SV sprouting could be completely blocked by injection of the VEGFR inhibitor Axitinib (data not shown), we looked for other VEGF family members that could trigger this process. Whereas VEGFA is heavily expressed in the myocardium (data not shown) (Miquerol et al., 1999), VEGFC was found to be most highly expressed in the epicardium and valves at E10.5-13.5 (Fig. 5A-E). There was also high expression within the outflow tract (data not shown). All developing coronary vessels expressed the VEGFC receptors VEGFR2 (also known as KDR) and VEGFR3 (also known as FLT4), but the SV was only positive for VEGFR2 (Fig. 5F; supplementary material Fig. S3A,B). However, coronary sprouts induced VEGFR3 shortly after exiting the SV (Fig. 5F; supplementary material Fig. S3B). Endocardial cells also expressed both receptors (Fig. 5G; supplementary material Fig. S3A,B). No expression of the receptors was detected in non-endothelial cell types present at the time when coronary vessel development is initiated (myocardium and epicardium; Fig. 5H; supplementary material Fig. S3C,D), suggesting that VEGFC signals primarily to cardiac endothelial and endocardial cells.
Fig. 5.
VEGFC is expressed in the epicardium, and its receptors are expressed in cardiac endothelial cells. (A-D) ISH for Vegfc mRNA. (A) Whole embryo and corresponding insets highlight probe specificity. (B) Adjacent sections subjected to either ISH (left) or immunostaining (right). Vegfc-positive cells (purple) overlap with α4-integrin (ITGA4)+ epicardial cells (epi). Dashed lines delineate the surface of the heart. (C) E10.5 heart (box 1 in A) showing high Vegfc expression in the epicardium and valves (val). (D) Epicardial expression at E12.5. (E) Tissue sections from Vegfc-lacZ mice in which X-Gal staining (blue) shows high Vegfc expression in the epicardium. (C-E) Boxed regions are shown at high magnification to the right. (F) Whole-mount confocal images of E13.5 wild-type hearts immunostained for VE-cadherin and either VEGFR2 or VEGFR3. Images with arrowheads show close-up views of receptor expression in the SV and in nearby coronary vessels. Both receptors are present in all coronary vessels, but VEGFR3 is absent from the SV. Insets are magnifications of the boxed regions showing receptor staining at the leading front of the growing coronary plexus. Arrowheads indicate coronary sprouts near the SV. (G,H) Tissue sections through E10.5 wild-type hearts showing colocalization of VEGFRs with VE-cadherin (G) but not ITGA4 (epicardium) or cTnT (myocardium) (H). Boxed regions (G,H) are magnified beneath. at, atrium; endo, endocardium; myo, myocardium; ot, outflow tract; ven, ventricle. Scale bars: 100 µm, except 20 µm in B.
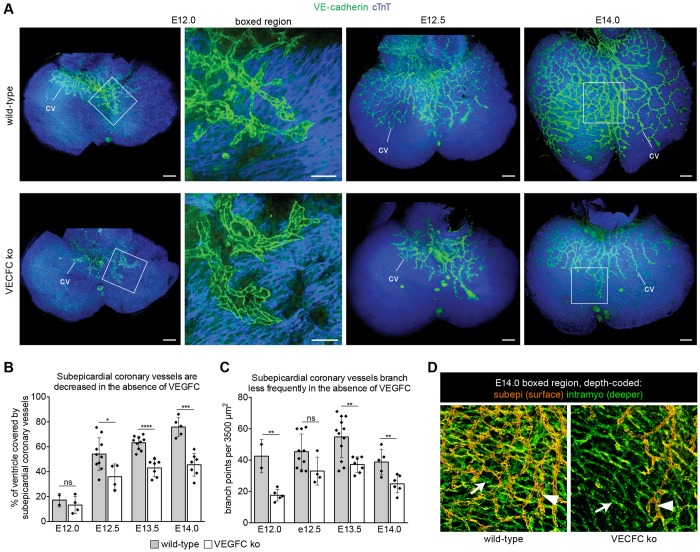
To probe the function of VEGFC, we observed coronary development in VEGFC-deficient mice. In E12.5 confocal images projected to highlight subepicardial vessels (transparent projection), dorsal coronary sprouts were severely stunted, having migrated over an average of 36±5% of the ventricles, whereas 54±4% were covered in wild-type hearts (Fig. 6A,B). Additionally, vessels that did migrate in mutants were structurally different from those in wild type. Vegfc knockout coronary vessels displayed fewer branch points (Fig. 6C) and they were sometimes wider and flatter (Fig. 6A, boxed region). This phenotype persisted through at least E14.0 (Fig. 6A-C). Intramyocardial vessels on the dorsal side of knockouts were not as extensive or mature as in wild type, but were much less affected by Vegfc deletion. Usually, subepicardial vessels on the dorsal side precede those within the myocardium (Fig. 2I). However, in Vegfc mutant hearts, intramyocardial vessels extended beyond subepicardial vessels (Fig. 6D). These data show that dorsal subepicardial vessels rely on VEGFC for normal progression over the heart and suggest that intramyocardial vessels might not be as dependent on this growth factor, an observation that is consistent with epicardial expression of VEGFC.
Fig. 6.
VEGFC is required for SV-derived coronary vessel development. (A) Confocal images (transparent projections) showing that coronary vessels (cv, green) on the dorsal surface of the heart are stunted in VEGFC-deficient hearts. Myocardium is blue (cTnT). High magnification of E12.0 boxed regions shows less elaborate vessel branching in knockout (ko) hearts. (B,C) Ventricle coverage by subepicardial coronary vessels (B) and vessel branching (C) are decreased in Vegfc mutant hearts. Bar heights show mean coronary coverage and number of vessel branch points, respectively. Error bars represent one s.d. above and below the mean. Each dot represents quantification for one heart. ****P<0.0001; ***P=0.0001 to 0.001; **P=0.001 to 0.01; *P=0.01 to 0.05; ns, P≥0.05. (D) Depth-coded confocal images from boxed regions on E14.0 hearts in A, in which subepicardial (subepi) vessels appear orange (arrowheads) and intramyocardial vessels (intramyo) appear green (arrows). In mutants, intramyocardial vessels extend farther beyond subepicardial vessels than in wild type. Scale bars: 100 µm.
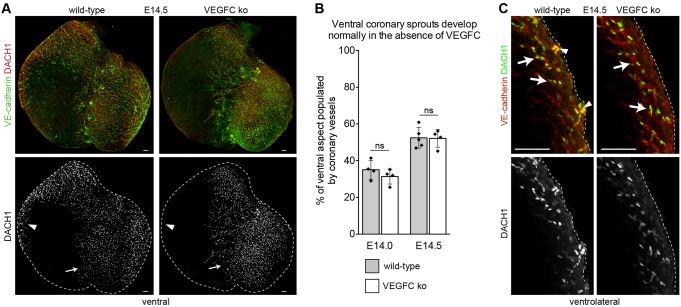
Coronary vessels on the ventral mid-portion of the heart migrate first into the intramyocardial space (Fig. 1A) and are derived mostly from the endocardium and not from the SV (Fig. 4A,C). This provides a location where the role of VEGFC in endocardium-derived intramyocardial migration could be investigated without possible secondary effects from defective subepicardial SV sprouting. In contrast to subepicardial cells on the dorsal side of the heart, ventral vessels were not dramatically altered by Vegfc deletion (Fig. 7A). Quantifying the percentage of the ventral mid-portion populated by coronary vessels at E14.0 and E14.5 showed that there was not a significant difference between wild-type and knockout animals (Fig. 7B). In optical sections showing lateral positions, VEGFC-independent vessels were in an intramyocardial location, while subepicardial vessels were missing in mutant hearts (Fig. 7C). Thus, ventral coronary vessels that were shown to arise mostly from the endocardium (Fig. 4A,C) migrate into the myocardium in response to molecular signals distinct from VEGFC.
Fig. 7.
The migration of endocardial-derived ventral coronary vessels does not require VEGFC. (A) Ventral coronary vessels (DACH1+, red) corresponding to endocardial-derived regions of the vasculature are normal in Vegfc knockout (ko) hearts (arrows), as compared with wild type. By contrast, few coronary vessels are in areas that arise from the SV (arrowheads). (B) The area occupied by ventral intramyocardial coronary vessels is not significantly different (ns) between wild-type and Vegfc knockout embryos. (C) Subepicardial vessels (arrowheads) are present in wild-type but not in Vegfc knockout myocardium as shown in optical sections through the ventrolateral region of the heart. Intramyocardial vessels (arrows) are present in both genotypes. Scale bars: 100 µm in A; 25 µm in C.
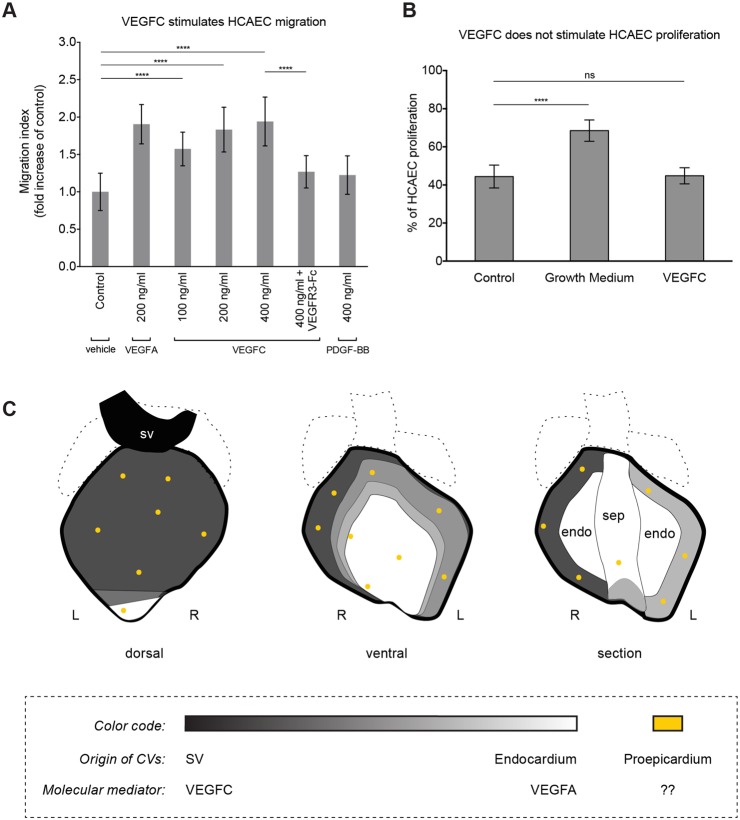
To obtain evidence that VEGFC directly signals to coronary endothelial cells, we performed migration and proliferation assays on human coronary artery endothelial cells (HCAECs). VEGFC stimulated HCAEC migration in a dose-responsive manner and at levels similar to those induced by VEGFA (Fig. 8A). In addition, the effect of VEGFC was abrogated by the addition of a soluble VEGFR3-Fc protein (Fig. 8A). In proliferation assays, there was no induction of additional cell division by VEGFC compared with control conditions, whereas growth media robustly increased proliferation (Fig. 8B). Thus, VEGFC can directly activate migration, but does not induce proliferation, in coronary endothelial cells, suggesting that the reduced ability of subepicardial vessels to progress towards the heart apex in the absence of VEGFC is a result of dysfunctional migration.
Fig. 8.
VEGFC stimulates human coronary artery endothelial cell migration. (A) The addition of VEGFC stimulates HCAEC migration. (B) Proliferation is not increased by VEGFC. Error bars are one s.d. above and below the mean. ****P<0.0001; ns, P≥0.05. (C) Schematics showing the complementary, but overlapping, regions proposed to derive from the SV (black), endocardium (white) and proepicardium (yellow). endo, endocardium; L, left; R, right; sep, septum.
Coronary developmental defects are often secondary to epicardial or myocardial growth abnormalities (Smart et al., 2009), but this did not appear to be true in VEGFC-deficient embryos. Hearts were grossly normal at E12.0-13.5 (Fig. 6A) and myocardial thickness was not significantly different between wild type and mutant at any stage (supplementary material Fig. S4A). The epicardium and valves were not obviously affected (supplementary material Fig. S4B; data not shown). Although not statistically significant, E14.0 myocardium trended towards being thinner and the hearts were slightly misshapen (Fig. 6A; supplementary material Fig. S4A), which could be secondary to defective coronary growth. VEGFC is known for its role in lymphangiogenesis, but the effect on coronary sprouting is unlikely to be secondary to effects on lymphatics for two reasons. First, lymphatic vessels do not appear on the heart until E14.5, which is 2 days after the deficiency begins to affect coronary development (supplementary material Fig. S5A). Second, in VEGFC pathway mutants that lack cardiac lymphatics, coronary development is normal (supplementary material Fig. S5A-C). The phenotype was also unlikely to be due to embryonic degeneration and death. VEGFC-deficient embryos die between E15.5 and birth (Karkkainen et al., 2004), and control vascular beds (in tissues lacking VEGFC expression) developed normally (supplementary material Fig. S4C). Taken together, these data suggest a precise role for VEGFC in which this factor signals to endothelial cells to stimulate coronary vessel development specifically along the SV subepicardial sprouting pathway (Fig. 8C).
DISCUSSION
Here, we show that different sources of coronary vessels populate complementary regions of the heart as they converge to vascularize the myocardium, and we identify a crucial role for VEGFC in this process. We generated an ApjCreER mouse line that robustly expresses inducible Cre recombinase in the SV endothelium. Our data show that SV-derived coronary vessels contribute to a large number of arteries, capillaries and veins on the dorsal and lateral sides of the heart. However, they do not extensively populate the ventricular septum or the mid-portion of the ventral face. Instead, we found that coronary vessels in these locations primarily arise from the endocardium (as shown by Nfatc1Cre lineage tracing). We also probed the distribution of proepicardium-derived coronary vessels, which contributed to a minor subset of the coronary vasculature. Unlike the SV and endocardium, the proepicardium gave rise to vessels relatively uniformly throughout the ventricular walls of the heart. Intramyocardial, but not subepicardial, coronary vessel migration is reported to be highly dependent on VEGFA produced by cardiomyocytes (Wu et al., 2012). Our data indicate that subepicardial vessels respond instead to VEGFC, which is expressed in the epicardium. Deletion of Vegfc disrupted subepicardial vessel growth emanating from the SV, severely affecting coronary vascularization on the dorsal and lateral sides of the heart. This work furthers knowledge of coronary vessel development by mapping how disparate endothelial sources cooperate to vascularize the heart and by identifying a crucial factor that stimulates their growth.
Our understanding of mammalian coronary artery formation has evolved in recent years following technical advances in Cre-lox-driven lineage tracing. Based on seminal work in the chick embryo, it was previously believed that all the components of the coronary vasculature derived from the proepicardium (Mikawa and Fischman, 1992). However, lineage tracing the Wt1-, Tbx18- and Tcf21-expressing (pro)epicardium in mice showed that these cells were progenitors of smooth muscle but not the endothelium (Acharya et al., 2012; Cai et al., 2008; Wilm et al., 2005; Zhou et al., 2008). Subsequent studies indicated that sprouting and budding of the SV and endocardium give rise to coronary endothelium (Red-Horse et al., 2010; Wu et al., 2012). Interestingly, it was later shown that the proepicardium is a heterogeneous population (Katz et al., 2012). Lineage tracing the Sema3d- and Scx-expressing proepicardial subsets revealed that they contribute to some coronary endothelial cells. In postnatal studies, the endocardium was shown to give rise to coronary vessels in the inner half of the compact myocardium through trabeculae coalescence, whereas those in the outer half are derived from the embryonic vasculature (Tian et al., 2014). In total, these experiments identified three sources of coronary endothelium, namely the SV, endocardium and proepicardium, but it was still unclear how each source is spatially related to the others within the intact organ. We explored this ambiguity using ApjCreER (SV), Nfatc1Cre (endocardium) and Sema3dCre (proepicardium) to compare, in the context of the whole intact heart, the regions of coronary vasculature traced from each source.
Defining the regional localization of coronary lineage labeling helps to explain how multiple populations can function as progenitors and how different studies can reach seemingly disparate conclusions. Our study shows that the SV and endocardium migrate into complementary areas of the heart. This is logical since SV sprouting begins at the dorsal side of the heart, progressively expanding over and into the heart muscle, while endocardial cells emerge outward from the cardiac lumen, particularly on the ventral side farthest from the SV (Fig. 1A). In contrast to the SV and endocardial routes, Sema3dCre-derived vessels from the proepicardium were more evenly dispersed throughout the ventricular walls of the heart. This observation suggests that there is a direct contribution from the proepicardium to the coronary endothelium, without necessarily first transitioning through the SV or endocardium. The combination of all pathways might have evolved as the most efficient and robust way to vascularize the rapidly growing ventricular walls, which contain high levels of angiogenic proteins that are likely to be capable of affecting any nearby competent cell.
The regionalized lineage-tracing patterns highlight the importance of considering the entire heart during quantification. Wu et al. (2012) described the endocardium (using the RCSfsEGFP reporter) as contributing 72% of coronary arteries, 81% of intramyocardial vessels, and 37% of subepicardial vessels, but regional differences were not taken into account. When quantifying five different areas of the heart, we found that Nfatc1Cre and ApjCreER contributions were inversely proportional. Although it is difficult to determine absolute numbers with the currently available tools because of overlapping labeling of the SV, the overall trend is highly informative. Furthermore, the major findings are consistent with previous studies, i.e. the SV and endocardium give rise to the majority of coronary vessels. Taken together, the current data support the following model (Fig. 8C). Vascular growth from the SV initiates subepicardially down the dorsal side of the heart progressing towards the apex and ventral side and into the underlying myocardium. This growing plexus meets endocardial-derived sprouts emerging from the lumen of the heart, which are especially dense in the septum and nearby ventral regions. Concomitantly, epicardial cells sparsely differentiate into endothelial cells. Ultimately, some intermixing occurs to form an interconnected, functional embryonic coronary vasculature.
Little is known about the molecules directly stimulating coronary vascularization since most reported defects in coronary development are secondary to epicardial insufficiency and/or myocardial hypoplasia (Smart et al., 2009). VEGFA mediates vascular development in many different organs and is highly expressed in the embryonic myocardium (Miquerol et al., 1999). Inhibition of VEGF family members during chick coronary development decreases vascular density in the heart (Tomanek et al., 2006). Deletion of Vegfa in the murine myocardium has been reported to reduce coronary microvasculature with little effect on larger arteries (Giordano et al., 2001). A different study reported that cardiac Vegfa deletion dramatically inhibits intramyocardial arteries and capillaries while subepicardial veins are less affected (Wu et al., 2012). These data suggest that subepicardial migration may be reliant on factors other than VEGFA. Our data indicate that VEGFA and VEGFC might have complementary functions stimulating intramyocardial and subepicardial sprouting, respectively. Receptor expression suggests that both SV-derived and endocardial-derived coronary vessels are capable of responding to VEGFA and/or VEGFC. The different effects of Vegfa and Vegfc deletion are likely to reflect their different expression domains and/or their proximity to the migratory pathways usually taken by either SV or endocardial sprouts. Interestingly, a recent report has shown that myocardial-derived angiopoietin 1 is required for subepicardial coronary vessel development but dispensable for intramyocardial growth (Arita et al., 2014). Thus, there is growing evidence that subepicardial and intramyocardial pathways develop in response to distinct molecular signals.
VEGFC is well known for its role in lymphangiogenesis (Lohela et al., 2009), but is becoming increasingly appreciated for its effects on blood vasculature (Cao et al., 1998; Lohela et al., 2008; Tammela et al., 2011; Villefranc et al., 2013). VEGFC binds VEGFR3, which is expressed in lymphatic vessels and in embryonic blood endothelial cells (Tammela et al., 2008). In its highly processed form, VEGFC also binds VEGFR2 (Joukov et al., 1997), a receptor that is expressed by most blood endothelial cells and that strongly stimulates vascular growth upon binding VEGF family members. Notch signaling, which plays an important role in vascular development by inhibiting vessel growth, restricts endothelial sprouting by limiting VEGFR3 levels and signaling (Benedito et al., 2012; Lawson et al., 2001; Tammela et al., 2008). Although it is clear that the VEGFR3 pathway is important in angiogenesis models such as retinal vasculature and tumors (Benedito et al., 2012; Tammela et al., 2008, 2011), the role of VEGFC in these contexts is considerably less well studied. In addition, the role of this VEGF family member in organ-specific blood vessel development has not previously been explored.
Analyzing VEGFC-deficient hearts revealed that subepicardial sprouts emanating from the SV were dramatically delayed in the absence of any other obvious cardiac defects. Sprouts appeared to exit the SV, but did not efficiently branch or progress toward the apex of the heart once they entered the subepicardial space. The ligand and receptor expression data suggest a paracrine signaling pathway in which VEGFC is produced by epicardial cells and signals directly to growing coronary vessels. However, confirmation of the role of epicardial VEGFC awaits a tissue-specific knockout. In the heart, VEGFC may function to stimulate endothelial migration. We found that VEGFC can induce migration of HCAECs at levels similar to those induced by VEGFA. VEGFC deficiency did not affect all embryonic vasculature, as vessel distribution in other tissues, such as in the digits and whiskers, was similar to that in wild type. Thus, VEGFC has context-specific angiogenic roles with a particularly important function in the SV developmental path during cardiac vascularization. VEGFC might be a useful additive to therapeutic protocols aimed at stimulating coronary regrowth following injury or during disease.
MATERIALS AND METHODS
Mice
Mouse use followed Stanford IACUC guidelines. Strains used were wild type (CD1 and FVB, Charles River Laboratories), ApjCreER (see below), Nfatc1Cre (Wu et al., 2012), Sema3dCre (Katz et al., 2012), Vegfc-lacZ (Karkkainen et al., 2004), Vegfr3-lacZ (Dumont et al., 1998), Vegfcflox (Chen et al., 2014), Dach1 (Davis et al., 2001), RosamTmG Cre reporter (The Jackson Laboratory) and Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo, RosatdTomato Cre reporter (The Jackson Laboratory).
ApjCreER was created through recombineering (Sharan et al., 2009; Warming et al., 2005). CreERT2 was inserted at the Apj start site of a BAC procured from the Children's Hospital Oakland Research Institute (CHORI) (clone RP24-301A16, 153,553 bp in length). Pronuclear injection (Cyagen) resulted in two founder lines, which exhibited similar expression patterns.
ApjCreER was crossed to RosamTmG (Muzumdar et al., 2007). Pregnant females were dosed intraperitoneally (4 mg of tamoxifen in corn oil at E9.5 or once daily at E8.5, E9.5 and E10.5). For single dose, hearts were analyzed at E10.5 (n=25), E11.5 (n=7), E12.5 (n=4), E13.75 (n=6), E14.5 (n=14), E15.5 (n=15) and E17.5 (n=7). For triple dose, hearts were dissected and analyzed at E11.5 (n=2), E12.5 (n=4) and E15.5 (n=4).
Immunohistochemistry and imaging
Embryos from timed pregnancies (morning of plug designated E0.5) were fixed in 4% paraformaldehyde (PFA) for 1 h. Fixed tissues were left intact or sectioned. Immunofluorescence staining was performed in either 1.5 ml tubes with constant rotation (whole mount) or on microscope slides (tissue sections). Primary antibodies in blocking solution (5% goat or donkey serum, 0.5% Triton X-100 in PBS) were incubated overnight at 4°C followed by PBT (PBS with 0.5% Triton X-100) washes for 6 h. Secondary antibodies diluted in blocking solution were incubated overnight at 4°C and washed again. Samples were imaged in Vectashield (Vector Labs) using either a Zeiss LSM-700 or Axioimager A2 epifluorescence microscope. Image processing used ImageJ (NIH) and/or Photoshop (Adobe Systems).
Antibodies: VE-cadherin (BD Pharmingen, 550548; 1:100); cTnT (DSHB, CT3; 1:500); DACH1 (Proteintech, 10914-1-AP; 1:1000); VEGFR2 (R&D Systems, AF644; 1:100); VEGFR3 (R&D Systems, AF743; 1:100); PROX1 (R&D Systems, AF2727; 1:300); WT1 (Abcam, ab15249; 1:500); ITGA4 (BD Pharmingen, 553314; 1:100); PECAM1 (BD Pharmingen, 553370; 1:100). Secondary antibodies were Alexa Fluor conjugates (488, 555, 594, 633, 635, 647, Life Technologies; 1:250).
In situ hybridization
Embryos were fixed in 4% PFA (pH 7.0) for 1-2 h at 4°C, washed in PBS, and cryoprotected in 30% sucrose for 1 h before snap freezing and sectioning. Slides were dried, washed in PBS, and fixed in 4% PFA (pH 7.0) for 20 min. A proteinase K incubation (10 µg/ml, 15 min) and PBS wash preceded a second fixation in PFA (15 min) and water rinse. Slides were then acetylated with 0.25% acetic anhydride (Sigma) in 0.1 M triethanolamine (pH 8.0) (Sigma) for 10 min, washed in PBS, and blocked for 2-3 h at 60°C with pre-hybridization solution: 50% formamide, 5× saline sodium citrate (SSC), 0.1% Tween 20, 0.1% CHAPS detergent, 5 mM EDTA, and water. Denatured digoxigenin-labeled (Roche) antisense probes in hybridization solution [50% formamide, 5× SSC, 1 mg/ml baker's yeast RNA (Sigma), 100 µg/ml heparin, 1× Denhardt's solution (Invitrogen or Sigma), 0.1% Tween 20, 0.1% CHAPS detergent, 5 mM EDTA, and water] were incubated with tissue sections at 60°C overnight in an airtight container. Sections were then washed twice for 30 min in 2× SSC, 50% formamide at 60°C and twice for 10 min in 2× SSC at 37°C followed by a 30 min RNase A (0.2 µg/ml in 2× SSC) incubation. Next were two 15 min washes in 2× SSC at 37°C, two 15 min washes in a 1:1 mixture of 2× SSC and maleic acid buffer with 0.1% Tween 20 (MABT) at 60°C, two 10 min washes in MABT at 60°C, and two 10 min washes in MABT at room temperature. Sections were blocked with 2% bovine serum albumin in MABT for 2-3 h at room temperature before incubating overnight at 4°C with alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche) diluted in blocking solution. Signal was detected with NBT-BCIP (Roche).
Quantification of lineage labeling
ApjCreER,RosamTmG, Nfatc1Cre,RosamTmG and Sema3dCre,Rosatdtomato hearts were isolated at E16.5 (and P0 for Sema3dCre). ApjCreER (n=3) and Sema3dCre (n=6) hearts were cryosectioned and immunostained with anti-DACH1 and anti-VE-cadherin; recombination was detected by direct fluorescence. Paraffin sections of Nfatc1Cre (n=4) were subjected to antigen retrieval (1 mM EDTA pH 8, 0.1% Tween 20) in a pressure cooker for 10 min before immunostaining with anti-GFP (Aves Labs, GFP-1020; 1:500) and anti-DACH1. The percentage of DACH1-positive, lineage-labeled cells was quantified from at least three fields of view from five regions of the heart.
Analysis of Vegfc knockout hearts
To calculate the area occupied by coronary vessels, ImageJ was used to circumscribe the whole heart, and the region containing vessels was expressed as a percentage. Branch points were counted within 3500 µm2 at the leading edge of the dorsal plexus on the left ventricle. Myocardial thickness was measured in ZEN (Zeiss) from optical sections of the right lateral side of the heart. Two measurements were taken from the dorsal wall of the right ventricle and averaged. Results were reported using Prism 6 (GraphPad). Unpaired t-tests were used to determine the two-tailed P-value for each comparison.
In vitro assays
HCAEC migration was assessed using Falcon cell culture inserts (8 µm pores, Corning) coated with fibronectin (Sigma, F0895; 1:100). 5×104 HCAECs in EBM-2 medium (Lonza) were added to the top chamber while the bottom contained EBM-2 with either vehicle, recombinant human (rh) PDGF-BB (100-14B), recombinant mouse (rm) VEGFA (450-32A), rhVEGFC (100-20C) or rhVEGFC, all from Peprotech. rmVEGFR3-Fc was from R&D Systems (743-R3-100). Cells were cultured for 4 h, fixed, permeabilized with 100% methanol, and stained with 0.1% Crystal Violet acetate. Cells remaining on the top were removed and those that migrated were quantified from ten fields of view from duplicate wells. The experiment was repeated three times.
For the proliferation assay, 5×104 HCAECs were cultured in 24-well plates, starved for 2 h, and cultured in EBM-2 containing 20 µM Edu (Sigma) with or without VEGFC (400 ng/ml) or with rich EGM-2MV (Lonza). After 16 h, cells were fixed and Edu incorporation was detected using the Click-iT Edu Alexa Fluor 488 Imaging Kit (Invitrogen). Four fields per well were photographed and proliferation rates were calculated.
All in vitro assay results were reported using Prism 6 (GraphPad). Unpaired t-tests were used to determine the two-tailed P-value for each comparison.
Supplementary Material
Acknowledgements
We thank Dr Bin Zhou for the Nfatc1Cre line. Drs Caroline Hu and Scott Juntti provided invaluable technical assistance with the in situ hybridization protocol.
Footnotes
Competing interests
K.A. has consulted for Herantis Pharma.
Author contributions
H.I.C. performed VEGFC experiments, edited and compiled the manuscript and figures. B.S. performed ApjCreER lineage tracing and in vitro experiments, the latter with A.H.J. B.N.A. and K.S. conducted Nfatc1Cre lineage tracing. H.J.N., R.K. and K.A. provided Vegfc knockout embryos. H.A. and J.A.E. provided the Sema3dCre hearts. P.S. developed Vegfcflox mice. A.H.C. and K.S. characterized DACH1 as a coronary vessel marker. A.S.M. and P.E.B. assisted in ApjCreER generation. K.A. contributed intellectual input. K.R. designed the study, performed experiments and wrote the manuscript.
Funding
H.I.C. and A.H.J. were partially supported by a summer research grant administered by the Office of the Vice Provost for Undergraduate Education at Stanford University. H.J.N., R.K., P.S. and K.A. were supported by grants from the Academy of Finland [130446 to P.S.], the European Research Council [ERC-2010-AdG-268804] and the Leducq Foundation [11CVD03]. A.H.C. was partially supported by a Stanford Graduate Fellowship in Science & Engineering (Tom and Susan Ford Fellow) administered by the Office of the Vice Provost for Graduate Education at Stanford University. H.A. and J.A.E. were supported by the National Institutes of Health (NIH) [R01HL118768]. B.N.A. and K.S. were supported by the NIH [1R01HL115294]. H.I.C., B.S., A.S.M., A.H.C. and K.R. were supported by the NIH [4R00HL10579303] and the Searle Scholar Foundation. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.113639/-/DC1
References
- Acharya A., Baek S. T., Huang G., Eskiocak B., Goetsch S., Sung C. Y., Banfi S., Sauer M. F., Olsen G. S., Duffield J. S. et al. (2012). The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development 139, 2139-2149 10.1242/dev.079970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita Y., Nakaoka Y., Matsunaga T., Kidoya H., Yamamizu K., Arima Y., Kataoka-Hashimoto T., Ikeoka K., Yasui T., Masaki T. et al. (2014). Myocardium-derived angiopoietin-1 is essential for coronary vein formation in the developing heart. Nat. Commun. 5, 4552 10.1038/ncomms5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito R., Rocha S., Woeste M., Zamykal M., Radtke F., Casanovas O., Duarte A., Pytowski B. and Adams R. (2012). Notch-dependent VEGFR3 upregulation allows angiogenesis without VEGF-VEGFR2 signalling. Nature 484, 110-114 10.1038/nature10908 [DOI] [PubMed] [Google Scholar]
- Cai C.-L., Martin J. C., Sun Y., Cui L., Wang L., Ouyang K., Yang L., Bu L., Liang X., Zhang X. et al. (2008). A myocardial lineage derives from Tbx18 epicardial cells. Nature 454, 104-108 10.1038/nature06969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Linden P., Farnebo J., Cao R., Eriksson A., Kumar V., Qi J.-H., Claesson-Welsh L. and Alitalo K. (1998). Vascular endothelial growth factor C induces angiogenesis in vivo. Proc. Natl. Acad. Sci. USA 95, 14389-14394 10.1073/pnas.95.24.14389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. I., Poduri A., Numi H., Kivela R., Saharinen P., McKay A. S., Raftrey B., Churko J., Tian X., Zhou B. et al. (2014). VEGF-C and aortic cardiomyocytes guide coronary artery stem development. J. Clin. Invest. pii: 77483 10.1172/JCI77483(in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. J., Shen W., Sandler Y. I., Amoui M., Purcell P., Maas R., Ou C.-N., Vogel H., Beaudet A. L. and Mardon G. (2001). Dach1 mutant mice bear no gross abnormalities in eye, limb, and brain development and exhibit postnatal lethality. Mol. Cell. Biol. 21, 1484-1490 10.1128/MCB.21.5.1484-1490.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont D. J., Jussila L., Taipale J., Lymboussaki A., Mustonen T., Pajusola K., Breitman M. and Alitalo K. (1998). Cardiovascular failure in mouse embryos deficient in VEGF receptor-3. Science 282, 946-949 10.1126/science.282.5390.946 [DOI] [PubMed] [Google Scholar]
- Gay L., Miller M. R., Ventura P. B., Devasthali V., Vue Z., Thompson H. L., Temple S., Zong H., Cleary M. D., Stankunas K. et al. (2013). Mouse TU tagging: a chemical/genetic intersectional method for purifying cell type-specific nascent RNA. Genes Dev. 27, 98-115 10.1101/gad.205278.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F. J., Gerber H.-P., Williams S.-P., VanBruggen N., Bunting S., Ruiz-Lozano P., Gu Y., Nath A. K., Huang Y., Hickey R. et al. (2001). A cardiac myocyte vascular endothelial growth factor paracrine pathway is required to maintain cardiac function. Proc. Natl. Acad. Sci. USA 98, 5780-5785 10.1073/pnas.091415198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma T. and Hirakow R. (1989). Epicardial formation in embryonic chick heart: computer-aided reconstruction, scanning, and transmission electron microscopic studies. Am. J. Anat. 184, 129-138 10.1002/aja.1001840204 [DOI] [PubMed] [Google Scholar]
- Hutchins G. M., Kessler-Hanna A. and Moore G. W. (1988). Development of the coronary arteries in the embryonic human heart. Circulation 77, 1250-1257 10.1161/01.CIR.77.6.1250 [DOI] [PubMed] [Google Scholar]
- Joukov V., Sorsa T., Kumar V., Jeltsch M., Claesson-Welsh L., Cao Y., Saksela O., Kalkkinen N. and Alitalo K. (1997). Proteolytic processing regulates receptor specificity and activity of VEGF-C. EMBO J. 16, 3898-3911 10.1093/emboj/16.13.3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkkainen M. J., Haiko P., Sainio K., Partanen J., Taipale J., Petrova T. V., Jeltsch M., Jackson D. G., Talikka M., Rauvala H. et al. (2004). Vascular endothelial growth factor C is required for sprouting of the first lymphatic vessels from embryonic veins. Nat. Immunol. 5, 74-80 10.1038/ni1013 [DOI] [PubMed] [Google Scholar]
- Kattan J., Dettman R. W. and Bristow J. (2004). Formation and remodeling of the coronary vascular bed in the embryonic avian heart. Dev. Dyn. 230, 34-43 10.1002/dvdy.20022 [DOI] [PubMed] [Google Scholar]
- Katz T. C., Singh M. K., Degenhardt K., Rivera-Feliciano J., Johnson R. L., Epstein J. A. and Tabin C. J. (2012). Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev. Cell 22, 639-650 10.1016/j.devcel.2012.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine K. J., White A. C., Park C., Smith C. S., Choi K., Long F., Hui C.-C. and Ornitz D. M. (2006). Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 20, 1651-1666 10.1101/gad.1411406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N. D., Scheer N., Pham V. N., Kim C. H., Chitnis A. B., Campos-Ortega J. A. and Weinstein B. M. (2001). Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development 128, 3675-3683. [DOI] [PubMed] [Google Scholar]
- Lohela M., Heloterä H., Haiko P., Dumont D. J. and Alitalo K. (2008). Transgenic induction of vascular endothelial growth factor-C is strongly angiogenic in mouse embryos but leads to persistent lymphatic hyperplasia in adult tissues. Am. J. Pathol. 173, 1891-1901 10.2353/ajpath.2008.080378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohela M., Bry M., Tammela T. and Alitalo K. (2009). VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr. Opin. Cell Biol. 21, 154-165 10.1016/j.ceb.2008.12.012 [DOI] [PubMed] [Google Scholar]
- Mikawa T. and Fischman D. A. (1992). Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proc. Natl. Acad. Sci. USA 89, 9504-9508 10.1073/pnas.89.20.9504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquerol L., Gertsenstein M., Harpal K., Rossant J. and Nagy A. (1999). Multiple developmental roles of VEGF suggested by a LacZ-tagged allele. Dev. Biol. 212, 307-322 10.1006/dbio.1999.9355 [DOI] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L. and Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Red-Horse K., Ueno H., Weissman I. L. and Krasnow M. A. (2010). Coronary arteries form by developmental reprogramming of venous cells. Nature 464, 549-553 10.1038/nature08873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan S. K., Thomason L. C., Kuznetsov S. G. and Court D. L. (2009). Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206-223 10.1038/nprot.2008.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N., Dubé K. N. and Riley P. R. (2009). Coronary vessel development and insight towards neovascular therapy. Int. J. Exp. Pathol. 90, 262-283 10.1111/j.1365-2613.2009.00646.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammela T., Zarkada G., Wallgard E., Murtomäki A., Suchting S., Wirzenius M., Waltari M., Hellström M., Schomber T., Peltonen R. et al. (2008). Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature 454, 656-660 10.1038/nature07083 [DOI] [PubMed] [Google Scholar]
- Tammela T., Zarkada G., Nurmi H., Jakobsson L., Heinolainen K., Tvorogov D., Zheng W., Franco C. A., Murtomäki A., Aranda E. et al. (2011). VEGFR-3 controls tip to stalk conversion at vessel fusion sites by reinforcing Notch signalling. Nat. Cell Biol. 13, 1202-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Hu T., Zhang H., He L., Huang X., Liu Q., Yu W., He L., Yang Z., Zhang Z. et al. (2013). Subepicardial endothelial cells invade the embryonic ventricle wall to form coronary arteries. Cell Res. 23, 1075-1090 10.1038/cr.2013.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Hu T., Zhang H., He L., Huang X., Liu Q., Yu W., He L., Yang Z., Yan Y. et al. (2014). De novo formation of a distinct coronary vascular population in neonatal heart. Science 345, 90-94 10.1126/science.1251487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanek R. J., Ishii Y., Holifield J. S., Sjogren C. L., Hansen H. K. and Mikawa T. (2006). VEGF family members regulate myocardial tubulogenesis and coronary artery formation in the embryo. Circ. Res. 98, 947-953 10.1161/01.RES.0000216974.75994.da [DOI] [PubMed] [Google Scholar]
- Villefranc J. A., Nicoli S., Bentley K., Jeltsch M., Zarkada G., Moore J. C., Gerhardt H., Alitalo K. and Lawson N. D. (2013). A truncation allele in vascular endothelial growth factor c reveals distinct modes of signaling during lymphatic and vascular development. Development 140, 1497-1506 10.1242/dev.084152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D. L., Jenkins N. A. and Copeland N. G. (2005). Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33, e36 10.1093/nar/gni035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm B., Ipenberg A., Hastie N. D., Burch J. B. E. and Bader D. M. (2005). The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development 132, 5317-5328 10.1242/dev.02141 [DOI] [PubMed] [Google Scholar]
- Wu B., Zhang Z., Lui W., Chen X., Wang Y., Chamberlain A. A., Moreno-Rodriguez R. A., Markwald R. R., O'Rourke B. P., Sharp D. J. et al. (2012). Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell 151, 1083-1096 10.1016/j.cell.2012.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeini M., Hang C. T., Lehrer-Graiwer J., Dao T., Zhou B. and Chang C.-P. (2009). Spatial and temporal regulation of coronary vessel formation by calcineurin-NFAT signaling. Development 136, 3335-3345 10.1242/dev.037903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Ma Q., Rajagopal S., Wu S. M., Domian I., Rivera-Feliciano J., Jiang D., von Gise A., Ikeda S., Chien K. R. et al. (2008). Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109-113 10.1038/nature07060 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.