Abstract
Under osmotic stress conditions such as drought and high salinity, the plant hormone abscisic acid (ABA) plays important roles in stress-responsive gene expression mainly through three bZIP transcription factors, AREB1/ABF2, AREB2/ABF4 and ABF3, which are activated by SNF1-related kinase 2s (SnRK2s) such as SRK2D/SnRK2.2, SRK2E/SnRK2.6 and SRK2I/SnRK2.3 (SRK2D/E/I). However, since the three AREB/ABFs are crucial, but not exclusive, for the SnRK2-mediated gene expression, transcriptional pathways governed by SRK2D/E/I are not fully understood. Here, we show that a bZIP transcription factor, ABF1, is a functional homolog of AREB1, AREB2 and ABF3 in ABA-dependent gene expression in Arabidopsis. Despite lower expression levels of ABF1 than those of the three AREB/ABFs, the areb1 areb2 abf3 abf1 mutant plants displayed increased sensitivity to drought and decreased sensitivity to ABA in primary root growth compared with the areb1 areb2 abf3 mutant. Genome-wide transcriptome analyses revealed that expression of downstream genes of SRK2D/E/I, which include many genes functioning in osmotic stress responses and tolerance such as transcription factors and LEA proteins, was mostly impaired in the quadruple mutant. Thus, these results indicate that the four AREB/ABFs are the predominant transcription factors downstream of SRK2D/E/I in ABA signalling in response to osmotic stress during vegetative growth.
Abscisic acid (ABA) plays important roles in osmotic stress-responsive gene expression mainly through three bZIP transcription factors, AREB1, AREB2, and ABF3, which are activated by SnRK2s such as SRK2D, SRK2E, and SRK2I (SRK2D/E/I). However, transcription factors other than the three AREB/ABFs that function downstream of SRK2D/E/I remain obscure. Here, we report that ABF1 is a functional homolog of AREB1, AREB2, and ABF3 in ABA-dependent gene expression from a comparative analysis between the areb1 areb2 abf3 abf1 and areb1 areb2 abf3 mutants. Moreover, genome-wide transcriptome analyses revealed that expression of downstream genes of SRK2D/E/I were mostly impaired in the areb1 areb2 abf3 abf1 quadruple mutant, suggesting that the four AREB/ABFs are the predominant transcription factors downstream of SRK2D/E/I in ABA signaling in response to osmotic stress.
Keywords: ABF1, transcriptional regulation, transcriptome analysis.
Introduction
Higher plants, as sessile organisms, have evolved adaptive robustness to environmental variation at the molecular and cellular levels, as well as at the physiological level. Drought and high-salinity stress conditions, in which water availability is severely limited, have an adverse effect on plant growth, survival, distribution and productivity. Under stress conditions, a myriad of genes that function in the stress tolerance and response are induced in diverse plant species (Bartels & Sunkar 2005; Yamaguchi-Shinozaki & Shinozaki 2006). Abscisic acid (ABA) is a key phytohormone involved in a host of biological processes, including plant development and responses to biotic and abiotic stresses (Finkelstein et al. 2002; Raghavendra et al. 2010). Endogenous ABA levels in plant cells are increased in response to osmotic stresses such as drought and high salinity, leading to expression of stress-responsive genes. Indeed, exogenous application of ABA can induce many dehydration-responsive genes (Zhu 2002; Yamaguchi-Shinozaki & Shinozaki 2006). ABA-dependent gene expression plays an essential part of transcriptional regulatory networks under osmotic stress conditions as well as ABA-independent gene expression (Yamaguchi-Shinozaki & Shinozaki 2006).
Promoter analyses of ABA-inducible genes identified the ABA-responsive element (ABRE; PyACGTGG/TC) as a conserved cis-element (Guiltinan et al. 1990; Mundy et al. 1990; Busk & Pagès 1998). By using ABREs as bait in yeast one-hybrid screening, a group of bZIP transcription factors, designated as ABRE-binding (AREB) proteins or ABRE-binding factors (ABFs), were isolated (Choi et al. 2000; Uno et al. 2000). Among the nine members of AREB/ABFs in Arabidopsis (Yoshida et al. 2010), AREB1/ABF2, AREB2/ABF4 and ABF3 are highly induced by ABA and osmotic stress treatments in vegetative tissues (Choi et al. 2000; Uno et al. 2000; Fujita et al. 2005). Further overexpression analyses showed that AREB1/ABF2, AREB2/ABF4 and ABF3 are involved in ABA signalling in response to osmotic stresses (Kang et al. 2002; Kim et al. 2004; Fujita et al. 2005; Furihata et al. 2006). Moreover, the areb1 areb2 abf3 triple knockout mutant displayed impaired expression of ABA- and osmotic stress-responsive genes, resulting in increased sensitivity to drought and decreased sensitivity to ABA in primary root growth (Yoshida et al. 2010). Overall, AREB1, AREB2 and ABF3 are master transcription factors in ABA signalling involved in drought stress tolerance.
ABA-dependent phosphorylation is a crucial post-transcriptional regulation of AREB/ABF transcription factors. Multiple conserved RXXS/T sites in AREB/ABFs are phosphorylated by SNF1-related kinase 2 (SnRK2) protein kinases in an ABA-dependent manner, as shown by in-gel kinase assays (Uno et al. 2000; Furihata et al. 2006; Fujii et al. 2007). Among the 10 Arabidopsis SnRK2s, three subclass III SnRK2s – SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3 (SRK2D/E/I) – are strongly activated by ABA and osmotic stresses (Yoshida et al. 2002; Boudsocq et al. 2004), and these SnRK2s co-localize and interact with AREB/ABFs in plant cell nuclei (Fujita et al. 2009; Yoshida et al. 2010). The srk2d/e/i triple mutant displays greatly reduced tolerance to drought stress and extreme ABA-insensitive phenotypes throughout its life cycle (Fujii & Zhu 2009; Fujita et al. 2009; Nakashima et al. 2009). Genome-wide transcriptome analyses revealed that expression of a vast number of ABA- and osmotic stress-responsive genes is impaired in the srk2d/e/i triple mutant, and that the downstream genes of SRK2D/E/I are highly overlapped with those of AREB1, AREB2 and ABF3, which were determined based upon microarray data of the areb1 areb2 abf3 triple mutant (Fujita et al. 2009).
Recently, our understanding of the molecular mechanisms underlying ABA signalling has been improved by a series of studies inspired by identification of novel ABA receptors, namely pyrabactin resistance 1/PYR1-like/regulatory components of ABA receptors (PYR/PYL/RCARs) (Ma et al. 2009; Park et al. 2009). Based upon structural insights into the ternary complex of ABA, PYR/PYL/RCARs and protein phosphatase 2Cs (PP2Cs) (Melcher et al. 2009; Miyazono et al. 2009; Nishimura et al. 2009) and a reconstituent study in vitro (Fujii & Zhu 2009; Fujii et al. 2009), a core component in ABA signalling consisting of PYR/PYL/RCARs, PP2Cs and SRK2D/E/I has been elucidated. Under the control of PYR/PYL/RCAR and PP2Cs, the AREB/ABF-SnRK2 pathway has a pivotal role as a positive regulator in ABA-dependent gene expression (Fujita et al. 2013). Although downstream genes of the AREB/ABFs are highly overlapped with those of SRK2D/E/I, two-thirds of ABA-responsive genes down-regulated in the srk2d/e/i triple mutant were not down-regulated in the areb1 areb2 abf3 triple mutant (Fujita et al. 2009). This finding implies that AREB1, AREB2 and ABF3 are key, but not exclusive, transcription factors that regulate gene expression downstream of SRK2D/E/I in ABA signalling. However, transcription factors other than the AREB/ABFs that function downstream of SRK2D/E/I remain obscure.
Here, we report that ABF1 is a functional homolog of AREB1, AREB2 and ABF3 in ABA signalling downstream of SRK2D/E/I during vegetative stages from a comparative analysis between the areb1 areb2 abf3 abf1 and areb1 areb2 abf3 mutants. Moreover, a large-scale transcriptome analysis of the areb1 areb2 abf3 abf1 quadruple mutant revealed that a majority of downstream genes of SRK2D/E/I are directly and indirectly regulated by the four AREB/ABFs. On the basis of our results, we discuss the pivotal roles of AREB/ABFs downstream of SRK2D/E/I in ABA signalling.
Materials and Methods
Plant materials, growth conditions and generation of transgenic plants
Arabidopsis thaliana ecotype Columbia (Col-0) plants were grown, transformed and treated as described previously (Fujita et al. 2005). The T-DNA insertion lines of abf1-1 (SALK_043079) and abf1-2 (SALK_132819) were obtained from the Arabidopsis Biological Resource Center (ABRC). A series of multiple areb1, areb2, abf3 and abf1 mutants in the Col-0 ecotype were constructed by genetic crosses between a series of multiple areb1, areb2 and abf3 mutants (Yoshida et al. 2010) and abf1 mutants, and were screened using primers recommended by the ABRC. We confirmed that the AREB1, AREB2, ABF3 and ABF1 genes were not expressed in these single and multiple mutants using reverse transcription-PCR (RT-PCR) analysis as described previously (Fujita et al. 2005). The detailed procedures for construction of pGreenII-based plasmids for plant transformation and plasmids for transient expression in plant cells are described in Supporting Information Appendix S1 and Table S1. The plant transformation vectors were transformed into Arabidopsis by the vacuum infiltration method using Agrobacterium tumefaciens strain GV3101 as described previously (Fujita et al. 2005).
Green fluorescent protein (GFP) fluorescence analysis
GFP fluorescence was observed with a confocal laser scanning microscope (LSM5 PASCAL; Carl Zeiss, Oberkochen, Germany).
Transient expression assays with Arabidopsis protoplasts
Transient expression assays using protoplasts derived from Arabidopsis leaf mesophyll cells were performed as described by Yoshida et al. (2010). The RD29B-GUS plasmid (Uno et al. 2000) and pBI35S-ELUC plasmid (Mizoi et al. 2013) were co-transfected with each effector plasmid as a reporter and an internal control, respectively.
Bimolecular fluorescence complementation (BiFC) analysis
BiFC analysis in onion epidermal cells was performed as described by Yoshida et al. (2010). The yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) fluorescence was observed with a confocal laser scanning microscope (LSM5 PASCAL; Carl Zeiss).
Drought tolerance assays, analysis of plant water relations and ABA analysis
Analyses of drought tolerance of potted seedlings, plant water relations and ABA sensitivity at the seedling stage were performed as described previously (Yoshida et al. 2010). For drought tolerance assays within a single pot, 2- to 3-week-old seedlings were transferred from growth medium (GM) agar plates to soil. After acclimatization for 1 week, the seedlings were subjected to drought stress by withholding watering for 11 to 12 d. The plants were re-watered after significant differences in wilting were observed.
Microarray analysis
Total RNA was isolated from 12-day-old seedlings of the areb1 areb2 abf3 abf1 quadruple mutant and wild-type (WT) plants grown on GM agar plates with or without stress treatments of 50 μm ABA, dehydration, or 250 mm NaCl for 6 h, as described previously (Yoshida et al. 2010). Genome-wide transcriptome analysis using the Arabidopsis 3 Oligo Microarray Kit (Agilent Technologies, http://www.home.agilent.com/agilent/home.jspx) was conducted with total RNA as described previously (Fujita et al. 2005). For each biological replicate, material from five plants was pooled to form a single sample for RNA purification. Two independent RNA samples were used for each experiment. The reproducibility of the microarray analysis was verified by a dye swap in each experiment. Based upon the microarray data obtained, the genes comprising the 27 206 nuclear protein-coding genes defined by The Arabidopsis Information Resource 10 (TAIR10) release were further studied. To search for cis-acting elements in promoters, 1 kb upstream sequences was retrieved from TAIR10. Real-time quantitative reverse transcription-PCR (qRT-PCR) analyses were carried out with total RNA using an ABI 7500 real-time PCR system as described previously (Yoshida et al. 2010). The primers used in this work are listed in Supporting Information Table S1. The microarray design and data have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress; accession numbers E-MTAB-2115).
Results
ABF1 is a candidate bZIP transcription factor in ABA signalling in vegetative tissues
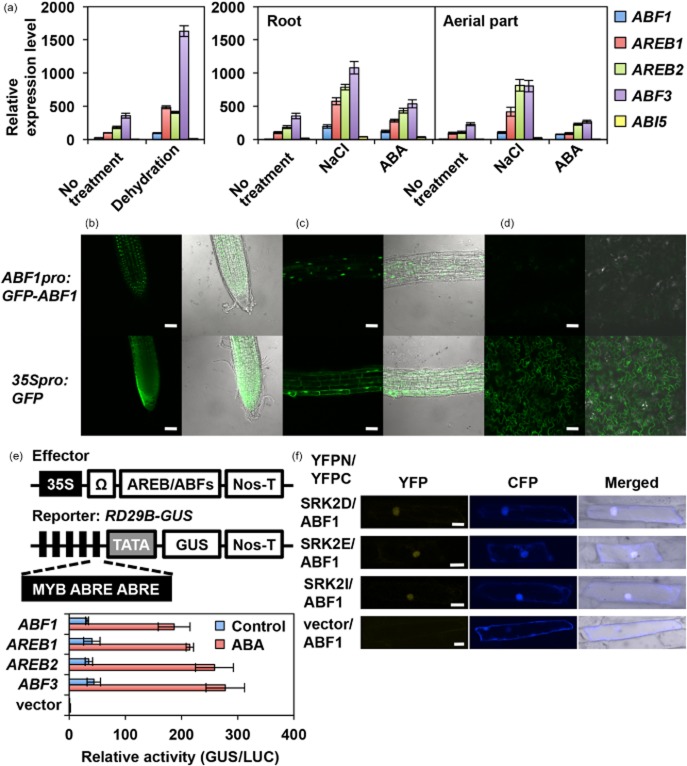
To identify transcription factors that regulate gene expression downstream of SRK2D/E/I other than AREB1, AREB2 and ABF3, we considered the AREB/ABF family genes. Among the nine AREB/ABF members in Arabidopsis, AREB1, AREB2 and ABF3 are highly induced by osmotic stress and ABA treatment in vegetative tissues (Fujita et al. 2005). Moreover, our microarray analysis indicated that ABF1 and ABI5 were also up-regulated by dehydration, high salinity and ABA treatment even though their expression intensities were markedly lower than those of AREB1, AREB2 and ABF3 (Supporting Information Table S2). We confirmed the expression levels of four AREB/ABFs and ABI5 under stress conditions using qRT-PCR (Fig. 1). In contrast to AREB1, AREB2 and ABF3, expression of ABF1 and ABI5 was barely detected in 12-day-old seedlings grown under non-stressed conditions. With dehydration, high salinity and ABA treatment, ABF1 was induced at lower levels compared with AREB1, AREB2 and ABF3. The expression profiles of the four AREB/ABF genes were similar in roots and aerial parts (Fig. 1; Yoshida et al. 2010). Whereas the role of ABI5 is well characterized in seed maturation and germination (Finkelstein & Lynch 2000; Lopez-Molina & Chua 2000; Lopez-Molina et al. 2002), ABF1 is thought to function in vegetative stages (Sharma et al. 2011). Consistent with the previous study, we observed GFP signal in root cell nuclei but not in leaf cells of transgenic Arabidopsis expressing GFP-ABF1 fusion proteins under the control of the ABF1 promoter (Fig. 1b–d). The localization pattern of GFP-ABF1 in roots was similar to those of AREB2 and ABF3 rather than that of AREB1 (Yoshida et al. 2010).
Figure 1.
ABF1 was induced by stress treatments in vegetative tissues at lower levels compared with AREB1, AREB2 and ABF3. (a) Gene expression profiles of AREB/ABFs and ABI5 analysed using real-time quantitative reverse transcription-PCR (qRT-PCR). cDNAs were synthesized from 1 μg total RNA prepared from 12-day-old whole seedlings with or without dehydration stress treatments, and from the root and aerial parts of 12-day-old seedlings with or without abscisic acid (ABA) and NaCl treatments. For each gene, the expression level was quantified from standard curves derived from each pre-quantified CDS fragment as template for qRT-PCR. The expression levels of AREB1 under non-stressed conditions or AREB1 in the root under non-stressed conditions were defined as 100. Data represent means and standard deviations of three replicate reactions. (b–d) Cellular localization of GFP-ABF1 proteins in the roots and leaf epidermis. Confocal images of GFP fluorescence in root tips (b), root elongation zone (c) and leaf epidermal tissues (d) of ABF1pro:GFP-ABF1 and 35Spro:GFP plants. The GFP fluorescence images and images merged with Nomarski images are shown. Bars = 50 μm. (e) Transient expression analysis of ABF1. Schemes of the effector and reporter constructs used in the transactivation analysis are shown on the left. The effector constructs contain the cauliflower mosaic virus (CaMV) 35S promoter and tobacco mosaic virus Ω sequence fused to ABF1, AREB1, AREB2 or ABF3 cDNA fragments. The reporter construct RD29B-GUS contains five tandem repeats of a 77 bp fragment of the RD29B promoter. The promoters were fused to the −51 RD29B minimal TATA promoter-GUS construct. Protoplasts were co-transfected with the RD29B-GUS reporter and the effector construct. To normalize for transfection efficiency, the pBI35SΩ-ELUC reporter was co-transfected as a control in each experiment. Transactivation experiments were performed three times, of which a representative result is shown. Bars indicate the SD; n = 3. ‘Relative activity’ indicates the multiples of expression compared with the value obtained with the vector control. Nos-T, nopaline synthase terminator. (f) Bimolecular fluorescence complementation (BiFC) visualization of interaction of ABF1 with SRK2D, SRK2E and SRK2I following transient expression in onion epidermal cells. Yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) fluorescence and merged images from the same field of transfected cells are shown for each transfection combination. A 35Spro:CFP (pGKX-CFP) control plasmid was always co-bombarded to identify transformed cells prior to the analysis of YFP fluorescence. In each transfected cell, YFP fluorescence was normalized against CFP fluorescence. Bars = 50 μm.
Our transient expression analysis using Arabidopsis leaf mesophyll protoplasts and a reporter plasmid, which was constructed by fusing five tandem copies of a 77 bp fragment of the Arabidopsis RD29B promoter containing two ABRE motifs (Uno et al. 2000) to the β-glucuronidase (GUS) gene, revealed that expression of ABF1 as well as that of AREB1, AREB2 and ABF3 in protoplasts produced dramatically enhanced activation of the reporter gene in the presence of ABA compared with that in the absence of ABA (Fig. 1e). Moreover, BiFC analysis in onion epidermal cells showed that ABF1 co-localizes and interacts with SRK2D/E/I in nuclei (Fig. 1f), as does AREB1 (Supporting Information Fig. S1; Fujita et al. 2009). Taken together with phylogenetic relationships among the four AREB/ABFs (Fujita et al. 2013), we hypothesized that ABF1 is a functional homolog of AREB1, AREB2 and ABF3.
Generation of an areb1 areb2 abf3 abf1 quadruple mutant
Our findings thus far indicate that ABF1 is a putative functional homolog of AREB1, AREB2 and ABF3 in ABA signalling, but it remains to be determined whether ABF1 plays roles in osmotic stress signalling mediated by ABA in planta. Considering functional redundancy among AREB/ABFs, knockout of ABF1 in the areb1 areb2 abf3 triple mutant is useful to study the role of ABF1 in planta. To investigate whether the phenotypes of the areb1 areb2 abf3 triple mutant, which have been described previously (Yoshida et al. 2010), are enhanced by lack of ABF1, we generated areb1 areb2 abf3 abf1 quadruple mutants by crossing the areb1 areb2 abf3 triple mutant and two alleles of abf1 mutants, in both of which the ABF1 transcript was shown to be absent (Supporting Information Fig. S2; Finkelstein et al. 2005; Sharma et al. 2011). Although the ABF1 locus is located 1.2 Mb from the AREB1 locus, we obtained an areb1 areb2 abf3 abf1-2 quadruple mutant but not an areb1 areb2 abf3 abf1-1 mutant. Considering artefacts due to two proximal T-DNA insertions, two quadruple mutant lines produced by independent crossing were used for further analyses. Together with the quadruple mutant, areb2 abf3 abf1-1 and areb2 abf3 abf1-2 triple mutants were generated.
All of the triple and quadruple mutants showed phenotypes similar to that of WT plants, except that the inflorescence height of multiple mutants was significantly shorter than that of WT plants (Supporting Information Fig. S3). This result is consistent with the previous observation that the areb2 abf3 and areb1 abf3 double and areb1 areb2 abf3 triple mutants had slightly shorter inflorescences compared with those of WT plants (Yoshida et al. 2010). To address the reason why the triple and quadruple mutants of AREB1, AREB2, ABF3 and/or ABF1 had a shorter inflorescence compared with WT plants, the bolting time of the mutants and WT plants was measured as number of days after germination (DAG) and number of rosette leaves at bolting (Supporting Information Fig. S3e,f). WT plants developed a 1-cm-high inflorescence at 21–24 DAG with 5–6 rosette leaves present, whereas all of the triple and quadruple mutants examined bolted at 28–30 DAG with 8–10 leaves present. Although it is still unclear whether the AREB/ABFs are indeed involved in the transition to flowering, a series of phenotypic analyses indicated that knockout of ABF1 has little effect on the growth of the areb1 areb2 abf3 triple mutant under normal growth conditions.
Lack of ABF1 reduces drought tolerance and ABA sensitivity in primary root growth of the areb1 areb2 abf3 triple mutant
The areb1 areb2 abf3 triple mutant grew similar to the WT plants, except for the shorter inflorescences, in normal growth conditions, whereas the mutant displayed higher sensitivity to drought compared with WT plants (Yoshida et al. 2010). To determine whether the T-DNA insertion in ABF1 enhances drought sensitivity of the triple mutant, we compared the difference in recovery after dehydration with plants grown in soil between the areb1 areb2 abf3 triple and areb1 areb2 abf3 abf1-2 quadruple mutants (Fig. 2a,b). Despite the slight difference in survival rates of WT plants, significant decrease in survival rates in the areb1 areb2 abf3 triple mutant compared with that of WT plants was consistently observed both in a previous study (Yoshida et al. 2010) and in the present work. Whereas the drought stress tolerance of the areb2 abf3 abf1-1 and areb2 abf3 abf1-2 triple mutants was slightly decreased compared with that of the WT plants, reduction of drought stress tolerance in the areb1 areb2 abf3 triple and areb1 areb2 abf3 abf1-2 quadruple mutants was remarkable (Fig. 2a). However, the difference between the triple and quadruple mutants was not as marked in the experiment. To assess the slight difference in survival rates between the areb1 areb2 abf3 abf1-2 quadruple and areb1 areb2 abf3 triple mutants, both mutants were grown in the same pot with WT plants and watering was withheld (Fig. 2b). After 11–12 d without watering, the survival rates of the areb1 areb2 abf3 triple and areb1 areb2 abf3 abf1-2 quadruple mutants decreased to 90 and 55%, respectively, which indicated that the quadruple mutant was more sensitive to drought than the triple mutant. These results suggest that ABF1 functions in drought stress tolerance and that its role is relatively minor compared with that of AREB1. To assess whether enhanced sensitivity to drought stress in the quadruple mutant compared with the triple mutant is due to altered transpiration rates, we measured water loss rates and standardized water contents of whole plants under dehydration conditions (Supporting Information Fig. S4). A slightly higher reduction was observed for both the water loss rates and the standardized water contents of the areb1 areb2 abf3 abf1-2 quadruple mutant in comparison with WT plants at 47% relative humidity (RH), but the reduction in the quadruple mutant was similar to that in the areb1 areb2 abf3 triple mutant, which was consistent with the previous observation at 70% RH (Yoshida et al. 2010). In contrast, water loss rates and standardized water content of the quadruple and triple mutants were comparable to those of the WT at lower RH (25%). ABF1 may have little effect on transpiration, unlike AREB1, AREB2 and ABF3, which may be partially associated with stomatal closure under conditions of drought stress (Yoshida et al. 2010), although no remarkable difference was observed in the stomatal aperture among the quadruple and triple mutants and WT plants (Supporting Information Fig. S4e).
Figure 2.
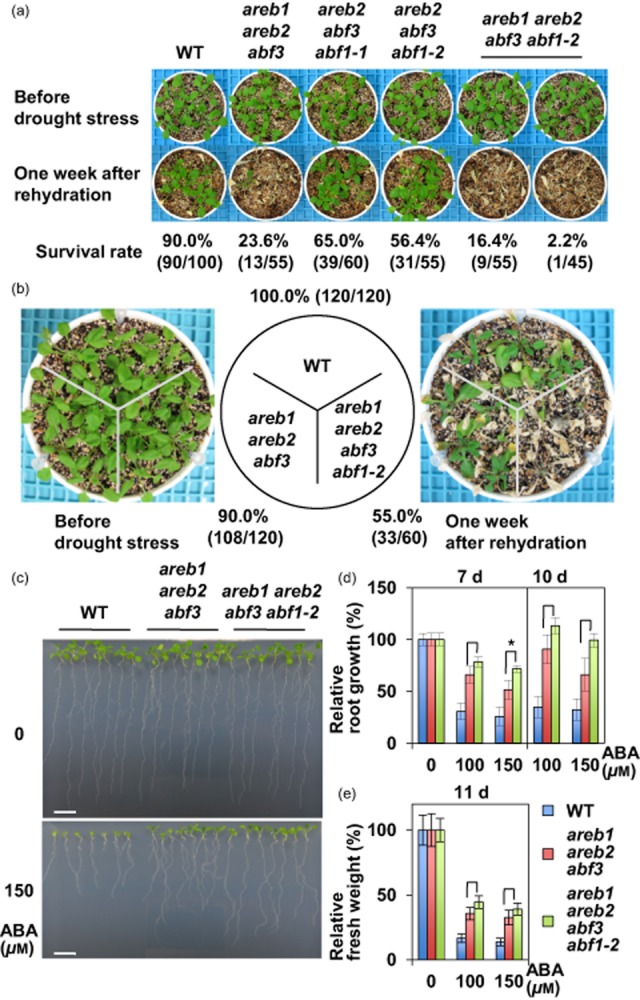
The areb1 areb2 abf3 abf1 quadruple mutant shows increased sensitivity to drought and reduced sensitivity to abscisic acid (ABA) in primary root growth compared with the areb1 areb2 abf3 triple mutant. (a, b) Plants before and after stress treatment. Watering was withheld from 4-week-old plants for 11–12 d, and then plants were re-watered for 1 week before the photograph was taken. Wild-type (WT) and the mutant plants (n = 5 each) were grown in soil in independent 6 cm pots (a) or a 9 cm pot (b). Survival rates were calculated from the results of four independent experiments (a; n ≥ 10 for each experiment) and three independent experiments (b; n ≥ 20 for each experiment). Similar results were obtained for two quadruple mutant lines in (b), thus representative results from one experiment are shown. (c) Seedlings at 7 and 10 d after transfer to control agar plates (GM with 1% sucrose) or plates containing 150 μm ABA. Seedlings were 4 d old at the time of transfer. (d, e) Relative root growth (d) and fresh weight (e) of seedlings, which were treated as described in (c) at the indicated number of days after transfer, were calculated from the results of three independent experiments (n ≥ 5 for each experiment). Similar results were obtained for two quadruple mutant lines, thus representative results from one experiment are shown. Bars indicate the SD; n ≥ 17. *P < 0.05 (t-test).
Exogenous ABA application induced ABF1 gene expression and activated the transactivation activity of ABF1 in vivo in a similar manner as AREB1, AREB2 and ABF3, which implied that ABF1 plays a role in ABA signalling. To reveal whether ABF1 is involved in ABA signalling, we compared ABA sensitivity between the areb1 areb2 abf3 triple and areb1 areb2 abf3 abf1-2 quadruple mutants during vegetative growth stages (Fig. 2c–e). Primary root growth of the areb1 areb2 abf3 triple and areb1 areb2 abf3 abf1-2 quadruple mutants was significantly less inhibited by ABA compared with that of WT plants. The areb1 areb2 abf3 abf1-2 quadruple mutant was more resistant to ABA than the areb1 areb2 abf3 triple mutant, which was significant on GM plates containing 150 μm ABA at 7 d after transfer. Consistent with this result, the areb1 areb2 abf3 abf1-2 quadruple mutant grew better on GM plates containing ABA than the areb1 areb2 abf3 mutant with regard to fresh weight (Fig. 2e). These results suggest that ABF1 plays a role in ABA signalling during vegetative growth.
SnRK2-mediated gene expression is more impaired in the areb1 areb2 abf3 abf1 quadruple mutant than in the areb1 areb2 abf3 triple mutant
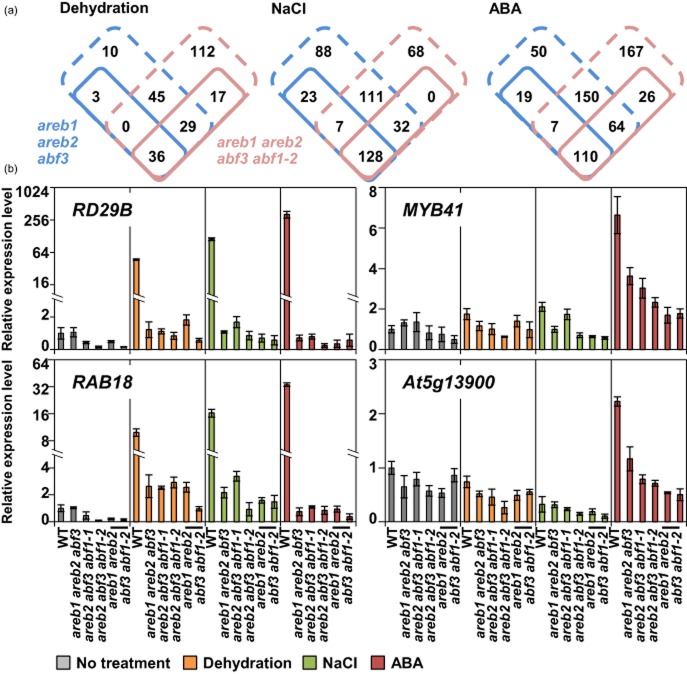
The areb1 areb2 abf3 abf1-2 quadruple mutant was slightly sensitive to drought stress and insensitive to ABA in terms of primary root growth compared with the areb1 areb2 abf3 triple mutant. To examine the role of ABF1 in the transcriptional network in response to osmotic stress, we compared the expression profiles of 12-day-old areb1 areb2 abf3 abf1-2 quadruple mutants with those of WT plants under both normal and stress conditions using an Agilent Arabidopsis 3 Oligo Microarray (44K feature format), and the data obtained were compared with those for the areb1 areb2 arbf3 triple and the srk2d/e/i triple mutants (Fujita et al. 2009; Yoshida et al. 2010). The results derived from the microarray analyses are summarized in Supporting Information Table S3. After 6 h of treatment with dehydration stress, 250 mm NaCl and 50 μm ABA, 95, 180 and 220 genes, respectively, showed reduced expression levels (greater than fourfold) in the quadruple mutant in comparison with expression levels in the WT (P < 0.05). To better understand the roles of AREB/ABFs in osmotic stress signalling, we hereafter focused upon genes induced by dehydration, high salinity or ABA in WT plants (Fujita et al. 2009). Comparison of microarray results from the quadruple and triple mutants showed that expression of more than 80% (dehydration, 36/39; NaCl, 128/158; ABA, 110/136) of down-regulated genes in the areb1 areb2 abf3 mutant was impaired in the areb1 areb2 abf3 abf1-2 mutant (Fig. 3a). Although dozens of the genes were down-regulated only in the areb1 areb2 abf3 mutant, these genes tended to show lower intensities in hybridization signals compared with the genes down-regulated only in the quadruple mutant (Supporting Information Fig. S5). Considering technical limitations in the reliable detection of low abundance genes in microarrays (Draghici et al. 2006), the data of the quadruple mutant were considered to be inclusive of those of the triple mutant.
Figure 3.
Number of down-regulated genes in the areb1 areb2 abf3 triple mutant was increased by lack of ABF1. (a) Microarray data from the areb1 areb2 abf3 triple mutant (Fujita et al. 2009; Yoshida et al. 2010) were compared with those from the areb1 areb2 abf3 abf1-2 quadruple mutant. Genes induced by dehydration, high salinity or abscisic acid (ABA) in wild-type (WT) plants (Fujita et al. 2009) were used for further analyses. Venn diagrams show the number of dehydration-, NaCl- or ABA-responsive genes showing reduced expression levels in each multiple mutant in comparison with the WT expression level after 6 h of treatment with dehydration, 250 mm NaCl or 50 μm ABA, respectively. Solid and dashed lines indicate the number of genes reduced more than fourfold and two- to fourfold, respectively. (b) Experimental validation of selected AREB/ABF downstream genes by real-time quantitative reverse transcription-PCR (qRT-PCR) analysis, where the expression level in the WT plants under non-stressed conditions was defined as 1.0. Data represent means and SDs of three replicate reactions.
Moreover, dehydration- and ABA-inducible genes were more impaired in the areb1 areb2 abf3 abf1-2 quadruple mutant than in the areb1 areb2 abf3 triple mutant (Fig. 3a). Notably, the expression of 17 dehydration-responsive and 26 ABA-responsive genes, which were not significantly down-regulated in the areb1 areb2 abf3 triple mutant, was markedly decreased in the areb1 areb2 abf3 abf1-2 quadruple mutant by dehydration stress and ABA treatment, respectively (Supporting Information Tables S4 & S5). Conversely, the number of NaCl-inducible genes down-regulated in the quadruple mutant was comparable to that in the triple mutant (Fig. 3a and Supporting Information Table S6). qRT-PCR was performed to confirm the difference in transcriptional profile between the areb1 areb2 abf3 abf1-2 quadruple and areb1 areb2 abf3 triple mutants (Fig. 3b and Supporting Information Fig. S6). Decreased expression of the typical stress-inducible genes, including RD29B and RAB18, in the areb1 areb2 abf3 abf1-2 quadruple mutant treated with osmotic stress and/or ABA was comparable to that in the areb1 areb2 abf3 triple mutant, whereas the expression of MYB41 and its downstream genes, including At5g13900, At3g22620 and ASFT/HHT/RWP1 (Cominelli et al. 2008; Lippold et al. 2009), was more impaired in the quadruple mutant after treatment with ABA than in the triple mutant. Consistent with the different induction patterns of MYB41 in response to ABA and abiotic stress treatments (Supporting Information Fig. S7; Cominelli et al. 2008), MYB41 and its downstream genes were not markedly induced in WT plants after 6 h of treatment with dehydration or NaCl in the present experiment. Taken together with our findings that the areb1 areb2 abf3 abf1-2 quadruple mutant displayed enhanced drought stress-sensitive and ABA-insensitive phenotypes compared with the areb1 areb2 abf3 triple mutant, these results suggest that ABF1 plays important roles in osmotic stress- and ABA-responsive gene expression in vegetative tissues similar to AREB1, AREB2 and ABF3.
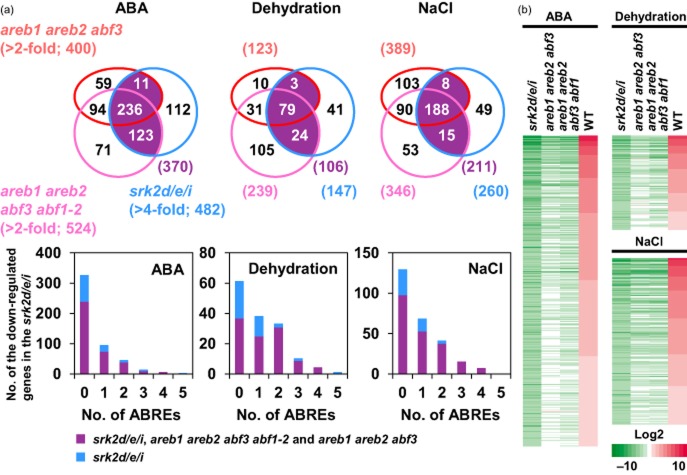
The relationship between AREB/ABF downstream genes and SRK2D/E/I downstream genes was further examined by comparing expression profiles of genes in the areb1 areb2 abf3 abf1-2 quadruple and the areb1 areb2 abf3 triple mutants with those in the srk2d/e/i triple mutants (Fujita et al. 2009; Yoshida et al. 2010). To determine the range of regulation by the AREB/ABFs, ABA-, dehydration- or NaCl-inducible genes down-regulated in the srk2d/e/i triple mutants were each divided into two groups based upon their expression changes in the areb1 areb2 abf3 abf1-2 quadruple and the areb1 areb2 abf3 triple mutants. Three hundred seventy out of 482, 106 out of 147, and 211 out of 260 genes were commonly down-regulated in the three mutants examined after ABA, dehydration and NaCl treatment, respectively (Fig. 4a). Expression of less than 56% (ABA, 247/482; dehydration, 82/147) of down-regulated genes in the srk2d/e/i triple mutant was impaired in the areb1 areb2 abf3 mutants, whereas more than 70% (ABA, 359/482; dehydration, 103/147) of genes were down-regulated both in the srk2d/e/i mutant and in the areb1 areb2 abf3 abf1-2 mutant in this analysis (Fig. 4a). In contrast, expression of more than 75% of NaCl-inducible genes down-regulated in the srk2d/e/i mutant was impaired both in the areb1 areb2 abf3 abf1-2 mutant and in the areb1 areb2 abf3 mutant. Remarkably, SRK2D/E/I downstream genes harbouring two or more ABREs were mostly down-regulated at least twofold either in the areb1 areb2 abf3 abf1-2 quadruple mutant or in the areb1 areb2 abf3 triple mutant in response to ABA, dehydration and high-salinity stress treatment (Fig. 4a). Consistent with Venn diagrams, among the genes down-regulated in the srk2d/e/i mutants, many genes not down-regulated in the areb1 areb2 abf3 triple mutants showed clearly decreased expression in the areb1 areb2 abf3 abf1-2 quadruple mutant (Fig. 4b). These results indicate that the four AREB/ABFs, including ABF1, are largely involved in gene expression downstream of SRK2D/E/I in ABA signalling.
Figure 4.
Most downstream genes of SRK2D/E/I were down-regulated in the areb1 areb2 abf3 abf1-2 quadruple mutant. (a) Venn diagrams show the number of ABA-, dehydration- or NaCl-responsive genes down-regulated in the areb1 areb2 abf3, the srk2d/e/i triple mutants (Fujita et al. 2009; Yoshida et al. 2010) and the areb1 areb2 abf3 abf1-2 quadruple mutant in comparison with the wild-type (WT) expression level after 6 h of treatment with abscisic acid (ABA), dehydration or NaCl, respectively. Among significantly (greater than fourfold) down-regulated genes in the srk2d/e/i triple mutant, the genes down-regulated by at least twofold either in the areb1 areb2 abf3 triple mutant or in the areb1 areb2 abf3 abf1-2 quadruple mutant are indicated as purple. Bar graphs show the numbers of ABRE sequences in each 1 kb upstream region of genes down-regulated by at least fourfold in the srk2d/e/i triple mutant. Each bar is colour-coded according to the Venn diagrams. (b) Gene expression profiles of the down-regulated genes in the srk2d/e/i triple mutant. The genes are listed from top down in the order of their expression changes in WT treated with ABA, dehydration or NaCl.
The four AREB/ABFs are predominant transcription factors downstream of SRK2D/E/I
To gain insight into the transcriptional regulation that occurred via SRK2D/E/I in ABA signalling, we further characterized downstream genes of SRK2D/E/I. A comprehensive gene ontology (GO) analysis revealed that many genes encoding transcription factors, LEA/Dehydrin proteins and signalling-related proteins were among the commonly down-regulated genes in the srk2d/e/i, areb1 areb2 abf3 abf1-2 and areb1 areb2 abf3 mutants, but few genes for LEA/Dehydrin proteins were among the down-regulated genes only observed in the srk2d/e/i mutant (Fig. 5). Considering that transcription factors were the major downstream genes of SRK2D/E/I (Fig. 5), we further examined their gene expression in the mutants (Supporting Information Fig. S8). Among 77 transcription factors significantly down-regulated in the srk2d/e/i triple mutant under either ABA, dehydration or NaCl stress, 61 transcription factor genes were down-regulated in the areb1 areb2 abf3 abf1-2 and/or areb1 areb2 abf3 mutants (Supporting Information Fig. S8a), and 16 transcription factor genes were down-regulated only in the srk2d/e/i triple mutant (Supporting Information Fig. S8b).
Figure 5.
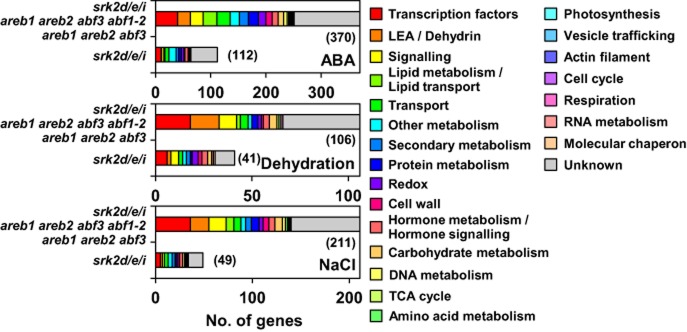
Gene ontology (GO) analysis of the down-regulated genes in the srk2d/e/i triple mutant. Bar graphs separately display the numbers of genes classified into each GO term among the genes down-regulated in the srk2d/e/i, areb1 areb2 abf3 abf1-2 and areb1 areb2 abf3 mutants, and the genes down-regulated only in the srk2d/e/i mutant. GO terms are based upon the PageMan profiling tool (Usadel et al. 2006) and the Arabidopsis Functional Modules Supporting Data (Heyndrickx & Vandepoele 2012). The numbers of genes in each group are indicated in parentheses.
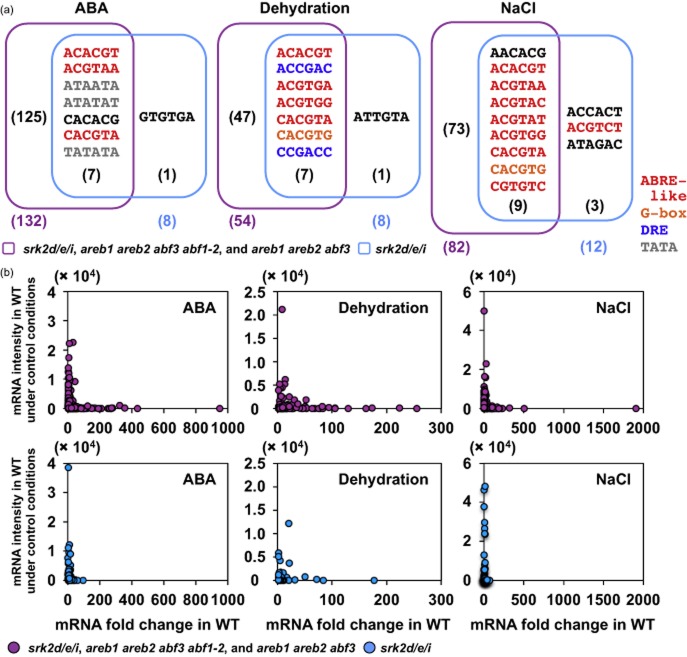
To investigate whether transcription factors other than the AREB/ABFs function downstream of SRK2D/E/I, promoter analysis was performed using the Element tool (Fig. 6a; Mockler et al. 2007). Although 8, 8 and 12 over-represented motifs were identified in the 1 kb promoter regions of the genes that were down-regulated only in the srk2d/e/i triple mutant under ABA, dehydration and high-salinity stress, respectively, most of the motifs were also observed in the promoters of the genes that were commonly down-regulated in the three mutants in each stress condition; thus, five motifs in all were specifically identified in the group. GTGTGA is similar to the TGTCACA motif, which is a cis-regulatory enhancer element involved in fruit-specific expression of the cucumisin gene in the fruit of melon, Cucumis melo L. (Yamagata et al. 2002); the other motifs were absent from the plant cis-acting regulatory DNA elements (PLACE) database (Higo et al. 1999), except an ABRE-like motif ACGTCT. Interestingly, expression of the commonly down-regulated genes in the srk2d/e/i, areb1 areb2 abf3 abf1-2 and areb1 areb2 abf3 mutants was more highly induced by ABA or osmotic stress treatments in the WT and less intensely expressed in WT plants under control conditions than those of the down-regulated genes observed only in the srk2d/e/i mutant (Fig. 6b). The trends in expression changes of the commonly down-regulated genes were significant compared to all ABA-, dehydration- and NaCl-inducible genes in Arabidopsis plants (Supporting Information Fig. S9). Taken together, these results suggest that active gene expression downstream of SRK2D/E/I is mainly mediated by AREB1, AREB2, ABF3 and ABF1 transcription factors and ABRE cis-elements in ABA signalling in response to osmotic stresses.
Figure 6.
Four AREB/ABFs are involved in most of active gene expression downstream of SRK2D/E/I. (a) The over-represented promoter motifs found in the two gene groups; the genes commonly down-regulated in the srk2d/e/i, the areb1 areb2 abf3 and the areb1 areb2 abf3 abf1-2 mutants, and the genes down-regulated only in the srk2d/e/i mutant. The 6-mer motifs were extracted using the Element web tool (Mockler et al. 2007). The numbers of over-represented motifs are indicated in parentheses. (b) Scatter plot of the downstream genes of SRK2D/E/I. The genes were divided into two groups as in (a). The x- and y-axes represent mRNA fold change in wild-type (WT) treated with abscisic acid (ABA), dehydration and NaCl and mRNA intensities in WT without the treatments, respectively, determined by microarray analyses (Fujita et al. 2009).
Discussion
Previously, we have shown that three bZIP transcription factors AREB1, AREB2 and ABF3 function in the regulation of ABA-dependent gene expression downstream of SRK2D/E/I (Yoshida et al. 2010). Given that the three AREB/ABFs are important, but not exclusive, for the SRK2D/E/I-mediated gene expression, we expected that other transcription factors function in the transcriptional regulation governed by SRK2D/E/I. Expression levels of AREB/ABF family genes other than AREB1, AREB2 and ABF3 were reported to be very low in the vegetative tissues even under stress conditions (Fujita et al. 2005). However, in the current study, we found that ABF1 was clearly induced by drought, high salinity and ABA treatments, although its expression levels were low even under stress conditions compared with those of AREB1, AREB2 and ABF3 (Fig. 1). Moreover, ABF1 behaved similarly to the other three AREB/ABFs in terms of subcellular localization in transgenic plants, transcriptional activity in protoplasts and co-localization and interaction with SRK2D/E/I in plant cell nuclei (Fig. 1; Fujita et al. 2009; Yoshida et al. 2010), suggesting that the role of ABF1 overlapped with those of the three AREB/ABFs in ABA signalling in response to drought stress. Among the nine AREB/ABF members in Arabidopsis, the other five transcription factors may not function in vegetative tissues because expression of these genes is rarely detected in such tissues (Fig. 1 and Supporting Information Table S2; Fujita et al. 2005). Therefore, it is likely that the quadruple mutant allows identification of most target genes of AREB/ABF-type transcription factors in vegetative tissues.
Using large-scale transcriptome data for the areb1 areb2 abf3 abf1-2, areb1 areb2 abf3 and srk2d/e/i mutants, we analysed the downstream transcription factors of SRK2D/E/I in vegetative tissues. In contrast to a previous study of the areb1 areb2 abf3 triple mutant (Fujita et al. 2009), expression of more than 70% of ABA-, dehydration- and NaCl-inducible genes down-regulated in the srk2d/e/i mutant was impaired in the areb1 areb2 abf3 abf1-2 quadruple mutant (Fig. 4). The numbers of ABA- and dehydration-inducible genes commonly down-regulated in the srk2d/e/i mutant were greatly increased in the areb1 areb2 abf3 abf1-2 quadruple mutant compared with the areb1 areb2 abf3 triple mutant, indicating that ABF1 functions in transcriptional regulation downstream of SRK2D/E/I similar to AREB1, AREB2 and ABF3. It is noteworthy that expression of the stress-responsive SRK2D/E/I downstream genes, which harbour two or more ABREs in their promoter regions, was almost completely impaired in the areb1 areb2 abf3 abf1-2 quadruple mutant (Fig. 4a). Considering that successful ABA-responsive gene expression requires multiple ABREs or the combination of an ABRE with one of several coupling elements in the promoter regions (Shen et al. 1996), which are often present as an ABRE motif in Arabidopsis (Zhang et al. 2005; Gómez-Porras et al. 2007), most ABRE-dependent gene expression downstream of SRK2D/E/I might be regulated by AREB1, AREB2, ABF3 and ABF1.
In contrast to the common downstream genes of the AREB/ABFs and SRK2D/E/I, the genes down-regulated only in the srk2d/e/i mutant tend to be slightly induced by ABA and osmotic stresses, and the known cis-elements potentially recognized by transcription factors other than AREB/ABFs were not observed to be specific over-represented motifs in their promoter regions (Fig. 6). Although a small number of ABRE-like motifs were over-represented in their promoter regions, some of them were not canonical ABRE sequences; thus, it is uncertain whether all of these ABRE-like motifs are involved in gene expression in ABA signalling. These data suggest the possibility that most of the genes down-regulated only in the srk2d/e/i mutant are not directly regulated by ABA or osmotic stress. SRK2D/E/I functions not only in ABA-responsive gene expression but also in ABA-responsive physiological changes, such as stomatal closure in vegetative tissues. We consider that these physiological changes probably affect the expression of some genes as a secondary effect. Such genes may be included among the genes down-regulated only in the srk2d/e/i mutant. Therefore, AREB1, AREB2, ABF3 and ABF1 are the predominant transcription factors downstream of SRK2D/E/I in ABA signalling in vegetative tissues under osmotic stress conditions.
Our data also revealed that ABF1, the expression of which is less abundant compared with AREB1, AREB2 and ABF3, has physiologically important roles in ABA signalling under drought stress conditions. The areb2 abf3 abf1 triple mutant displayed lower sensitivity to drought than the areb1 areb2 abf3 triple mutant (Fig. 2), indicating that ABF1 plays a less essential role in drought stress tolerance than AREB1. Nevertheless, lack of ABF1 in the areb1 areb2 abf3 triple mutant reduced the tolerance to drought and the sensitivity to ABA in primary root growth (Fig. 2), and more severely impaired the expression of dehydration- and ABA-inducible genes (Fig. 3). On the contrary, lack of ABF1 in the areb1 areb2 abf3 triple mutant did not alter transpiration rate and stomatal aperture (Supporting Information Fig. S4). The areb1 areb2 abf3 abf1-2 quadruple mutant is more sensitive to drought than the areb1 areb2 abf3 triple mutant probably because of reduced expression of the downstream genes of the AREB/ABFs, such as many genes encoding transcription factors, LEA/Dehydrin proteins and signalling-related proteins (Figs 3 & 5). Interestingly, the number of NaCl-inducible genes down-regulated in the areb1 areb2 abf3 abf1-2 mutant was comparable to that in the areb1 areb2 abf3 mutants (Fig. 3), despite ABF1 expression in response to high-salinity stress as well as dehydration and ABA (Fig. 1). These observations imply that transcript abundances of the AREB/ABFs are not directly associated with their physiological roles because AREB/ABFs are also regulated by post-transcriptional modifications such as phosphorylation. In addition to AREB1, AREB2 and ABF3, ABF1 is likely to be required for ABRE-dependent gene expression under conditions of drought but not high salinity.
Two recent phosphoproteomic studies using the knockout mutants of SRK2D/SnRK2.2, SRK2E/SnRK2.6 and SRK2I/SnRK2.3 revealed the possible substrates of SRK2D/E/I. Among the 32 phosphopeptides down-regulated in the srk2d/e/i mutant compared with the WT, except for two phosphopeptides corresponding to AREB1, AREB2, ABF3 or ABF1, the remaining 30 phosphopeptides for SRK2D/E/I candidate substrates did not include any transcription factors (Umezawa et al. 2013). On the contrary, among 84 phosphopeptides identified as possible substrates of SRK2D/E/I in ABA signalling, those corresponding to eight proteins, including AREB1/ABF2, AREB3, EEL, FLOWERING BHLH 3 (FBH3) and TATA binding protein-associated factor 5 (TAF5), were classified in the DNA binding and transcription category by GO analysis (Wang et al. 2013). AREB3/AtDPBF3 and EEL/AtDPBF4, which belong to the AREB/ABF subfamily of group-A bZIP transcription factors (Fujita et al. 2013), are predominantly expressed during early silique development (Bensmihen et al. 2002, 2005; Kim et al. 2002) and are rarely expressed in vegetative tissues even under stress conditions (Supporting Information Table S2; Fujita et al. 2005), consistent with the role of EEL in seed maturation (Bensmihen et al. 2002). Despite the experimental evidence that AREB3 was originally identified as an ABRE-binding protein using cDNA libraries prepared from rosette plants (Uno et al. 2000), and that phosphopeptides corresponding to AREB3 were altered in response to ABA during vegetative stages (Kline et al. 2010), the role of AREB3 in vegetative tissues has not yet been reported. Given that Arabidopsis plants used in these experiments were grown in liquid culture medium, expression of EEL and AREB3 might be induced non-specifically in vegetative tissues.
FBH3 and TAF5 are likely to be involved in developmental processes (Ito et al. 2012; Mougiou et al. 2012). FBH3 and its homologs are the transcriptional activators of CONSTANS (CO), which is the photoperiodic flowering regulator (Ito et al. 2012). Notably, expression of FBH3 and CO was impaired in the areb1 areb2 abf3 abf1-2, areb1 areb2 abf3 and srk2d/e/i mutants in comparison with the expression levels in WT plants treated with ABA and osmotic stresses (Supporting Information Fig. S8). Given that FBH3 and CO activate FT expression, resulting in early flowering (Ito et al. 2012), our expression data are consistent with the late-flowering phenotype of the areb1 areb2 abf3 abf1-2 and areb1 areb2 abf3 mutants (Supporting Information Fig. S3), but not with the early flowering phenotype of the srk2d/e/i triple mutant (Wang et al. 2013). Considering that expression of several genes involved in floral development, such as AP3, were up-regulated in the srk2d/e/i mutant after dehydration stress (Fujita et al. 2009), SRK2D/E/I may be involved in flowering in a complex manner. Conversely, in spite of the pivotal roles of SRK2D/E/I in ABA-induced stomatal closure (Fujii & Zhu 2009; Fujita et al. 2009), known substrates of SRK2D/E/I such as SLAC1 and KAT1, which play key roles in guard cells (Joshi-Saha et al. 2011), have not been detected in the two phosphoproteome analyses (Umezawa et al. 2013; Wang et al. 2013). Intriguingly, a recent study has shown that SRK2E/SnRK2.6 is involved in phosphorylation-dependent inactivation of AKS transcription factors, which have important roles in stomatal opening (Takahashi et al. 2013). Although it is also noteworthy that CO and FT are involved in stomatal opening in response to blue light (Kinoshita et al. 2011; Ando et al. 2013), further studies of the substrates of SnRK2s (Umezawa et al. 2013; Wang et al. 2013) are required to obtain a comprehensive understanding of the gene expression networks regulated by SnRK2s in various plant processes, including osmotic stress responses, stomatal movements and flowering. Collectively, these phosphoproteome analyses demonstrated that AREB1, AREB2, ABF3 and ABF1 are the main substrate transcription factors of SRK2D/E/I in gene expression in ABA signalling in response to osmotic stresses during vegetative growth, which supports our conclusion, derived from transcriptome analyses, that the AREB/ABFs function predominantly in stress-responsive gene expression downstream of SRK2D/E/I.
Acknowledgments
We thank Y. Tanaka, S. Murasaki, K. Amano, K. Murai and E. Kishi for the technical assistance; K. Yoshiwara for microarray analysis; and E. Toma and M. Toyoshima for skilful editorial assistance. This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas (No. 22119004 to K.Y.-S.) and Scientific Research (C) (No. 24510312 to Y. F.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Program for the Promotion of Basic Research Activities for Innovative Biosciences (BRAIN) of Japan, and the Science and Technology Research Partnership for Sustainable Development (SATREPS) of the Japan Science and Technology Agency/Japan International Cooperation Agency (to K.Y.-S.).
Supporting Information
Figure S1. BiFC visualization of interaction of AREB1 with SRK2D, SRK2E and SRK2I following transient expression in onion epidermal cells.
Figure S2. Generation of an areb1 areb2 abf3 abf1 quadruple mutant.
Figure S3. Growth phenotypes of areb1 areb2 abf3 abf1 quadruple mutants.
Figure S4. Analyses of plant water relations in the areb1 areb2 abf3 abf1 quadruple mutants.
Figure S5. Expression intensities of the genes down-regulated either in the areb1 areb2 abf3 abf1 quadruple mutant or in the areb1 areb2 abf3 triple mutant.
Figure S6. Experimental validation of selected AREB/ABF downstream genes by qRT-PCR analysis.
Figure S7. Expression patterns of MYB41 in response to osmotic stresses and ABA.
Figure S8. Transcription factor genes down-regulated in the srk2d/e/i triple mutant.
Figure S9. Fold changes in expression of the downstream genes of SRK2D/E/I in WT.
Table S1. Primer pairs used in the study.
Table S2. Expression intensities of AREB/ABF genes in microarray experiments.
Table S3. Summary of the microarray analyses.
Table S4. Genes significantly down-regulated either in the areb1 areb2 abf3 abf1-2 mutant or in the areb1 areb2 abf3 mutant under dehydration stress.
Table S5. Genes significantly down-regulated either in the areb1 areb2 abf3 abf1-2 mutant or in the areb1 areb2 abf3 mutant by ABA treatment.
Table S6. Genes significantly down-regulated either in the areb1 areb2 abf3 abf1-2 mutant or in the areb1 areb2 abf3 mutant by NaCl treatment.
Appendix S1. Materials and methods.
References
- Ando E, Ohnishi M, Wang Y, Matsushita T, Watanabe A, Hayashi Y. Kinoshita T. TWIN SISTER OF FT, GIGANTEA, and CONSTANS have a positive but indirect effect on blue light-induced stomatal opening in Arabidopsis thaliana. Plant Physiology. 2013;162:1529–1538. doi: 10.1104/pp.113.217984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D. Sunkar R. Drought and salt tolerance in plants. Critical Reviews in Plant Sciences. 2005;24:23–58. [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F. Parcy F. The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. The Plant Cell. 2002;14:1391–1403. doi: 10.1105/tpc.000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S, Giraudat J. Parcy F. Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. Journal of Experimental Botany. 2005;56:597–603. doi: 10.1093/jxb/eri050. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Barbier-Brygoo H. Laurière C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. The Journal of Biological Chemistry. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- Busk PK. Pagès M. Regulation of abscisic acid-induced transcription. Plant Molecular Biology. 1998;37:425–435. doi: 10.1023/a:1006058700720. [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J. Kim SY. ABFs, a family of ABA-responsive element binding factors. The Journal of Biological Chemistry. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Sala T, Calvi D, Gusmaroli G. Tonelli C. Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. The Plant Journal. 2008;53:53–64. doi: 10.1111/j.1365-313X.2007.03310.x. [DOI] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Eklund AC. Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends in Genetics. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Gampala SS, Lynch TJ, Thomas TL. Rock CD. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Molecular Biology. 2005;59:253–267. doi: 10.1007/s11103-005-8767-2. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR. Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS. Rock CD. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;14(Suppl):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H. Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Verslues PE. Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. The Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY. Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M. Yamaguchi-Shinozaki K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. The Plant Cell. 2005;17:3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N. Yamaguchi-Shinozaki K. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant and Cell Physiology. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Yoshida T. Yamaguchi-Shinozaki K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiologia Plantarum. 2013;147:15–27. doi: 10.1111/j.1399-3054.2012.01635.x. [DOI] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K. Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Porras JL, Riaño-Pachón DM, Dreyer I, Mayer JE. Mueller-Roeber B. Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics. 2007;8:260. doi: 10.1186/1471-2164-8-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WR. Quatrano RS. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;250:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- Heyndrickx KS. Vandepoele K. Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiology. 2012;159:884–901. doi: 10.1104/pp.112.196725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M. Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG. Imaizumi T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3582–3587. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Saha A, Valon C. Leung J. A brand new START: abscisic acid perception and transduction in the guard cell. Sci Signal. 2011;4:re4. doi: 10.1126/scisignal.2002164. [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY. Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. The Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH. Kim SY. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. The Plant Journal: For Cell and Molecular Biology. 2004;40:75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- Kim SY, Ma J, Perret P, Li Z. Thomas TL. Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiology. 2002;130:688–697. doi: 10.1104/pp.003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Ono N, Hayashi Y, Morimoto S, Nakamura S, Soda M. Shimazaki K. FLOWERING LOCUS T regulates stomatal opening. Current Biology. 2011;21:1232–1238. doi: 10.1016/j.cub.2011.06.025. [DOI] [PubMed] [Google Scholar]
- Kline KG, Barrett-Wilt GA. Sussman MR. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15986–15991. doi: 10.1073/pnas.1007879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible WR, Hincha DK. Udvardi MK. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiology. 2009;149:1761–1772. doi: 10.1104/pp.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L. Chua NH. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant and Cell Physiology. 2000;41:541–547. doi: 10.1093/pcp/41.5.541. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT. Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. The Plant Journal. 2002;32:317–328. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A. Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM. Xu HE. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A. Tanokura M. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- Mizoi J, Ohori T, Moriwaki T, Kidokoro S, Todaka D, Maruyama K. Yamaguchi-Shinozaki K. GmDREB2A;2, a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiology. 2013;161:346–361. doi: 10.1104/pp.112.204875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA. Chory J. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harbor Symposium on Quantitative Biology. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- Mougiou N, Poulios S, Kaldis A. Vlachonasios K. Arabidopsis thaliana TBP-associated factor 5 is essential for plant growth and development. Molecular Breeding. 2012;30:355–366. [Google Scholar]
- Mundy J, Yamaguchi-Shinozaki K. Chua NH. Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1406–1410. doi: 10.1073/pnas.87.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S. Yamaguchi-Shinozaki K. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant and Cell Physiology. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR. Getzoff ED. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y. Cutler SR. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A. Grill E. ABA perception and signalling. Trends in Plant Science. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Sharma PD, Singh N, Ahuja PS. Reddy TV. Abscisic acid response element binding factor 1 is required for establishment of Arabidopsis seedlings during winter. Molecular Biology Reports. 2011;38:5147–5159. doi: 10.1007/s11033-010-0664-3. [DOI] [PubMed] [Google Scholar]
- Shen Q, Zhang P. Ho TH. Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. The Plant Cell. 1996;8:1107–1119. doi: 10.1105/tpc.8.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Ebisu Y, Kinoshita T, Doi M, Okuma E, Murata Y. Shimazaki K. bHLH transcription factors that facilitate K+ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci Signal. 2013;6:ra48. doi: 10.1126/scisignal.2003760. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC. Shinozaki K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal. 2013;6:rs8. doi: 10.1126/scisignal.2003509. [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K. Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H. Stitt M. PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics. 2006;7:535. doi: 10.1186/1471-2105-7-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue L, Batelli G, Lee S, Hou YJ, Van Oosten MJ. Zhu JK. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11205–11210. doi: 10.1073/pnas.1308974110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Yonesu K, Hirata A. Aizono Y. TGTCACA motif is a novel cis-regulatory enhancer element involved in fruit-specific expression of the cucumisin gene. The Journal of Biological Chemistry. 2002;277:11582–11590. doi: 10.1074/jbc.M109946200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K. Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J. Shinozaki K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant and Cell Physiology. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J. Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. The Plant Journal. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- Zhang W, Ruan J, Ho TH, You Y, Yu T. Quatrano RS. Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics (Oxford, England) 2005;21:3074–3081. doi: 10.1093/bioinformatics/bti490. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. BiFC visualization of interaction of AREB1 with SRK2D, SRK2E and SRK2I following transient expression in onion epidermal cells.
Figure S2. Generation of an areb1 areb2 abf3 abf1 quadruple mutant.
Figure S3. Growth phenotypes of areb1 areb2 abf3 abf1 quadruple mutants.
Figure S4. Analyses of plant water relations in the areb1 areb2 abf3 abf1 quadruple mutants.
Figure S5. Expression intensities of the genes down-regulated either in the areb1 areb2 abf3 abf1 quadruple mutant or in the areb1 areb2 abf3 triple mutant.
Figure S6. Experimental validation of selected AREB/ABF downstream genes by qRT-PCR analysis.
Figure S7. Expression patterns of MYB41 in response to osmotic stresses and ABA.
Figure S8. Transcription factor genes down-regulated in the srk2d/e/i triple mutant.
Figure S9. Fold changes in expression of the downstream genes of SRK2D/E/I in WT.
Table S1. Primer pairs used in the study.
Table S2. Expression intensities of AREB/ABF genes in microarray experiments.
Table S3. Summary of the microarray analyses.
Table S4. Genes significantly down-regulated either in the areb1 areb2 abf3 abf1-2 mutant or in the areb1 areb2 abf3 mutant under dehydration stress.
Table S5. Genes significantly down-regulated either in the areb1 areb2 abf3 abf1-2 mutant or in the areb1 areb2 abf3 mutant by ABA treatment.
Table S6. Genes significantly down-regulated either in the areb1 areb2 abf3 abf1-2 mutant or in the areb1 areb2 abf3 mutant by NaCl treatment.
Appendix S1. Materials and methods.