Abstract
There is a growing body of evidence that small subpopulations of cells with stem cell-like characteristics within most solid tumors are responsible for the malignancy of aggressive cancer cells and that targeting these cells might be a good therapeutic strategy to reduce the risk of tumor relapse after therapy. Here, we examined the effects of emodin (1,3,8-trihydroxy-6-methylanthraquinone), an active component of the root and rhizome of Rheum palmatum that has several biological activities, including antitumor effects, on primary cultured glioma stem cells (GSCs). Emodin inhibited the self-renewal activity of GSCs in vitro as evidenced by neurosphere formation, limiting dilution, and soft agar clonogenic assays. Emodin inhibited the maintenance of stemness by suppressing the expression of Notch intracellular domain, nonphosphorylated β-catenin, and phosphorylated STAT3 proteins. In addition, treatment with emodin partially induced apoptosis, reduced cell invasiveness, and sensitized GSCs to ionizing radiation. Intriguingly, emodin induced proteosomal degradation of epidermal growth factor receptor (EGFR)/EGFR variant III (EGFRvIII) by interfering with the association of EGFR/EGFRvIII with heat shock protein 90, resulting in the suppression of stemness pathways. Based on these data, we propose that emodin could be considered as a potent therapeutic adjuvant that targets GSCs.
Introduction
Glioblastoma multiforme (gbm) is a highly aggressive primary brain tumor in adults, and most GBM patients have a median survival of ∼2 years after diagnosis [1]. Despite optimal treatment strategies, including radiotherapy and chemotherapy, the survival rate of patients has not improved. Therefore, more intensive investigation is needed to obtain insight into the mechanism of therapeutic resistance and malignancy of this devastating cancer. One of the main causes of therapeutic failure is the presence of resistant glioma cells that lead to relapse of the cancer after therapy [2,3]. Recently, this phenomenon has been associated with a new concept in cancer biology, the cancer stem cell (CSC) theory. Many studies have shown the existence of CSCs or cancer initiating cells that, like normal stem cells, have a high capacity for multilineage differentiation. Such cells propagate tumor formation as a self-renewing growth and are considered to be responsible for tumorigenesis, therapeutic resistance, metastasis, and recurrence of cancer [4]. The existence of CSCs was first confirmed in leukemia [5] and subsequently demonstrated in many solid tumors, including brain tumors [6–9]. Because glioma stem cells (GSCs) are thought to be the major cause of therapeutic failure, these cells are considered good candidates as potential therapeutic targets.
It has been widely suggested that conventional chemotherapy and radiotherapy would eradicate most of the differentiated cells within glioma, but would fail to remove GSCs because they are relatively quiescent and, therefore, more resistant to these therapies than differentiated cells [3,10,11]. Failure to eliminate these cells is thought to be one of the major causes of tumor relapse. Thus, to overcome the resistance of gliomas to conventional therapy, a combination therapy, including drugs that predominantly target GSCs, is necessary to achieve better clinical outcomes. Some natural dietary compounds have been shown to sensitize CSCs to conventional therapy [12]. Such compounds target the self-renewal pathways of CSCs, induce differentiation of these cells, and make them susceptible to conventional therapies [12]. Therefore, identification of effective natural compounds that preferentially target GSCs with minimal toxicity to normal cells would be a promising approach to improving tumor therapy.
Emodin (1,3,8-trihydroxy-6-methylanthraquinone), an active ingredient isolated from the traditional Chinese herb Rheum palmatum, is a natural anthraquinone derivative with a tyrosine kinase inhibitory activity [13]. This compound has been reported to have antitumor effects in various cancers, including glioma [14–17]. In addition, our group and others reported that emodin exerts antiangiogenic effects by inhibiting vascular endothelial growth factor-A (VEGF-A)-induced activation of VEGF receptor in human umbilical vein endothelial cells and colon cancer cells, respectively [18,19]. Given that numerous natural dietary compounds with antitumorigenic potential also target self-renewal pathways of CSCs [20–25], it seems highly possible that emodin might affect crucial signaling pathways of CSCs. There is only one report addressing the effect of emodin on stem-like cancer cells derived from a side population of gallbladder cancer cells [26]. Therefore, we investigated whether emodin has the potential to target GSCs, and in particular, the specific self-renewal pathways of these cells.
Our data show that emodin significantly inhibits the stemness of GSCs. Emodin effectively blocked the self-renewal activity of GSCs by suppressing crucial stemness signaling pathways involving Notch-1, β-catenin, and STAT3. Importantly, emodin induced proteosomal degradation of epidermal growth factor receptor (EGFR)/EGFR variant III (EGFRvIII) by interfering with its association of heat shock protein 90 (Hsp90), leading to partial induction of apoptosis, inhibition of invasion, and sensitization of GSCs to ionizing radiation (IR). These results strongly suggest the efficacy of this compound in targeting GSCs with EGFR/EGFRvIII and its potential application in adjuvant therapy of malignant glioma.
Materials and Methods
Reagents
Emodin was obtained from Calbiochem. Antibodies against Notch-1, Nanog, and γ-H2AX were obtained from Millipore. Antibodies against Oct4, nestin, c-myc, ubiquitin, and β-actin were purchased from Santa Cruz Biotechnology. Antibodies against phospho-EGFR [p-EGFR (Y 1173)], p-Akt (T308), p-STAT3 (Y705 and S727), p-ERK1/2, p-p38, p-JNK, nonphosphorylated-β-catenin (n-p-β-catenin), snail, slug, and Musashi were obtained from Cell Signaling Biotechnology. Anti-neuron-specific class III beta-tubulin (Tuj1), Sox2, and GFAP were purchased from R&D Systems. Antibodies against EGFR and Hsp90 were obtained from BD Sciences (Becton Dickinson). Epidermal growth factor (EGF), basic fibroblast growth factor (bFGF), and bovine fibronectin (bFN) were obtained from R&D Systems. Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 594 goat anti-mouse immunoglobulin Gs were from Invitrogen.
Cell culture
The human GSC lines X01 and X03, and CSC2 were generously gifted by Dr. Akio Soeda [27] (Gifu University School of Medicin) and raised in the Korea Institute of Radiological and Medical Sciences (KIRAMS), respectively. For sphere culture, cells were cultured in the plates coated with poly(2-hydroxyethyl methacrylate) (10 mg/mL in 95% ethanol) with the serum-free Dulbecco's modified Eagle's medium (DMEM)/F-12 medium (Cellgro) supplemented with 20 ng/mL of EGF, 10 ng/mL of bFGF, B27 (GIBCO), and 1% penicillin/streptomycin. Cells were maintained at 37°C in a 5% CO2 incubator.
Sphere-forming, limiting dilution, and soft agar clonogenic assays
For sphere-forming assay, cells were dissociated with accutase (Millipore) and resuspended in a complete medium. A single-cell suspension (1×103 cells/mL) was cultured in six-well plates with the complete medium with or without indicated concentrations of emodin in the figures. After 10 days of incubation, spheres were attached to the plate by adding fetal bovine serum (FBS, 10%) overnight, stained with 0.05% crystal violet containing 20% methanol, and counted.
For limiting dilution assay, cells were dissociated with accutase, diluted serially, and plated in 96-well plates in 200 μL of culture medium used for sphere-forming assay. After 10 days of incubation, wells with or without spheres were counted and plotted.
For soft agar clonogenic assay, single-cell suspensions (5×103 cells/mL) were obtained in 2× DMEM/F12 with EGF (20 ng/mL) and bFGF (10 ng/mL) without serum, resuspended in a same volume of 0.7% low-melting agar (final 0.35%), and poured onto 24-well plates coated with bottom agar (1:1 mixture of 2× DMEM/F12 and 1% low-melting agar, final 0.5%). After 14 days of incubation, colonies in five random fields per each well were counted under a microscope.
Immunocytochemistry
GSCs were seeded in a bFN (10 μg/mL)-coated chamber slide with the complete medium treated with or without emodin. After 48 h of incubation, cells were fixed with 4% paraformaldehyde for 20 min at 4°C and washed thrice with phosphate-buffered saline (PBS) at room temperature. Cells were then incubated in a blocking solution [5% bovine serum albumin (BSA) and 0.5% Triton X-100 in PBS] for 1 h at room temperature. Cells were stained with primary antibodies in the blocking solution (1:100) for 2 h at 4°C and washed thrice with PBS. Staining was visualized using Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 594 goat anti-mouse (Invitrogen) secondary antibodies (1:1,000) in dark condition for 1 h and washed thrice with PBS. Nuclei were stained using 4′,6-diamidino-2-phenylindole (contained a mounting solution), and stained cells were viewed under a confocal laser scanning microscope.
Flow cytometric analysis
To examine cell cycle distribution, GSCs dissociated into single cells were fixed with 75% ethanol, incubated −20°C for at least 1 h, washed with PBS thrice, resuspended in PBS with 0.1 mg/mL RNase-A, 50 μg/mL propidium iodide, and 0.05% Triton X-100 for 40 min at room temperature, washed with PBS, and analyzed on the FACSCalibur (Becton Dickinson).
Irradiation and drug treatment
For measuring IR resistance, GSCs were seeded in six-well plates (103 cells/mL) and incubated at 37°C under 5% CO2 incubator for 24 h. Cells were pretreated with or without emodin for 6 h and the cells were exposed to γ-rays from a 137Cs γ-ray source (Atomic Energy of Canada, KIRAMS) at the indicated dose rate (Gy/min). After 10 days of incubation, spheres were attached by adding FBS (10%), washed with PBS, and stained with 0.05% crystal violet containing 20% methanol. After washing thrice with distilled water, colonies were counted.
Reverse transcriptase–polymerase chain reaction
Total RNA(1 μg/μL) was isolated from GSCs by using TRIzol (Molecular Research Center) as described in the manufacturer's protocol. Complimentary DNA (cDNA) was synthesized using the cDNA synthesis kit (Bioline). Reverse transcriptase–polymerase chain reaction (RT-PCR) for EGFR and EGFRvIII was performed, as described previously [28]. Oligonucleotide primer sequences used were as follows: EGFR/EGFRvIII forward: 5′ CTTCGGGGAGCAGCGATGCGAC 3′; EGFR/EGFRvIII reverse: 5′ ACCAATACCTATTCCGTTACAC 3′. 18S RNA forward: AAACGGCTACCACATCCAAG; 18S RNA reverse: CGCTCCCAAGATCCAACTAC.
Western blot analysis
GSCs were lysed in 1× RIPA buffer [50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 5 mM ethylenediaminetetraacetic acid (EDTA), 0.5% Nonidet P-40, and protease inhibitor cocktail tablet (Roche)] and the concentration of protein was determined by the Bio-Rad protein assay reagent (Bio-Rad). An aliquot (30 μg protein/lane) of the total protein was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred to nitrocellulose membrane (Millipore) for 2 h at 80 V. The membrane was blocked with 5% BSA in TBST [20 mM Tris-HCl (pH 7.6), 137 mM NaCl, and 0.01% Tween 20] for 1 h at room temperature followed by incubation with a primary antibody for overnight at 4°C. After extensive washing of the membrane with TBST, the membrane was probed with a secondary antibody conjugated with horseradish peroxidase for 1 h at room temperature. After washing thrice with TBST for 15 min, membranes were visualized by enhanced chemiluminescence (Amersham) according to the manufacturer's protocol.
Invasion assays
Invasion assays were performed using transwell chambers with the polycarbonate nucleopore membrane (Costar). Precoated filters [6.5 mm in diameter, 8-μm pore size, Matrigel (100 μg/cm2)] were rehydrated with 100 μL of medium. Next, 1×105 cells in 100 μL of serum-free DMEM/F12 supplemented with 0.1% BSA were seeded into the upper part of each chamber, whereas the lower compartments were filled with 600 μL of the above medium. After incubation for 18 h at 37°C, noninvaded cells on the upper surface of the filter were wiped out with a cotton swab, and the invaded cells on the lower surface of the filter were fixed and stained with the Diff-Quick kit (Sysmex). Invasiveness was determined by counting cells in five random microscopic fields per well, and the extent of invasion was expressed as the average number of cells per microscopic field.
Terminal deoxynucleotidyl transferase dUTP nick-end labeling assays
GSCs were seeded in the bFN (10 μg/mL)-coated chamber slide with the complete medium and treated with or without emodin. After 48 h of incubation, cells were fixed with 4% paraformaldehyde for 10 min at 4°C and washed twice with PBS at room temperature. Cells were then permeabilized with 0.2% Triton X-100 in PBS at room temperature for 5 min and washed twice with PBS. Stained cells were viewed under a confocal laser scanning microscope as described in the manufacturer's protocol (DeadEnd™ Fluorometric TUNEL System; Promega).
Immunoprecipitation
GSCs treated with or without emodin (5 μM) for 24 h were lysed in 1 mL of lysis buffer (20 mM Tris-HCl, pH 8.0, 2 mM EDTA, 137 mM NaCl, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 10% glycerol, and 1% Triton X-100). Lysates were clarified by centrifugation at 15,000 g for 10 min, and the resulting supernatant fractions (500 μg) were immunoprecipitated with 2 μg of anti-EGFR antibody at 4°C for overnight. Protein G agarose beads (20 μL, GE Healthcare) were added to the lysate and incubated with a rotator for 4 h at 4°C. Immunoprecipitates were washed thrice with the lysis buffer, solubilized in the SDS-PAGE sample buffer containing β-mercaptoethanol, and analyzed by western blotting.
Statistical analysis
Statistical analysis was performed using an independent-samples t-test. Differences were considered statistically significant at P<0.05.
Results
Emodin inhibits self-renewal activity of GSCs
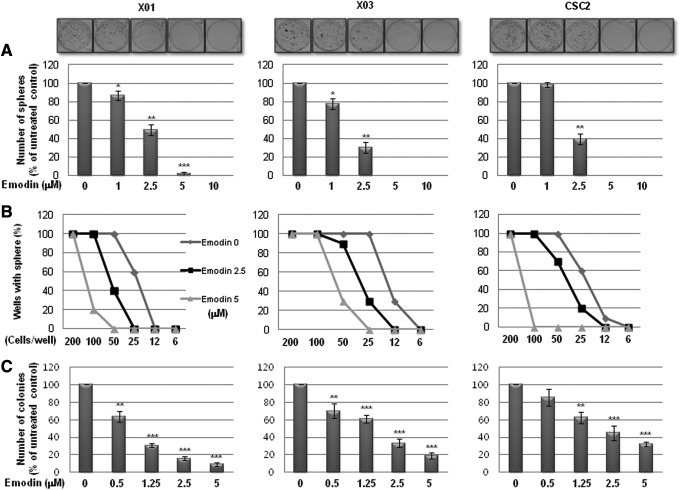
We first examined the effect of emodin on the self-renewal capacity of GSCs. In a sphere-forming assay, a well-known surrogate method for measuring the self-renewal activity of CSCs, emodin significantly decreased the sphere-forming activity of GSCs in a dose-dependent manner (Fig. 1A). Treatment with 5 μM emodin was sufficient to almost completely suppress sphere formation in the three different GSC lines. To further validate the effect of emodin on the self-renewal capacity of GSCs, we performed a limiting dilution assay, a surrogate method to evaluate the self-renewal activity in vitro. As shown in Fig. 1B, emodin significantly increased the number of GSCs required to form sphere(s) in a well. Consistent with this result, a soft agar clonogenic assay also showed that emodin inhibited colony formation of GSCs in a dose-dependent manner (Fig. 1C). These data indicate that the natural compound emodin effectively suppresses the self-renewal capacity of GSCs.
FIG. 1.
Effect of emodin on self-renewal activity of glioma stem cells (GSCs). (A) Sphere-forming assays of GSCs. Cells (1×103 cells/mL) were cultured in serum-free Dulbecco's modified Eagle's medium (DMEM)/F12 medium with growth factors (20 ng/mL of epidermal growth factor and 10 ng/mL of basic fibroblast growth factor) for 10 days and counted the spheres. Representative images of spheres (upper, magnification, ×40) and quantification of the assays (lower). (B) Limiting dilution assays for measuring self-renewal capacity of GSCs. (C) Soft agar colony-forming assays for evaluating tumorigenic capacity of GSCs in vitro. Detailed experimental procedures were described in the section “Materials and Methods.” *P<0.05, **P<0.01, ***P<0.005.
Emodin inhibits stemness signaling of GSCs through downregulation of Notch intracellular domain, n-p-β-catenin, and p-STAT3 (Y705)
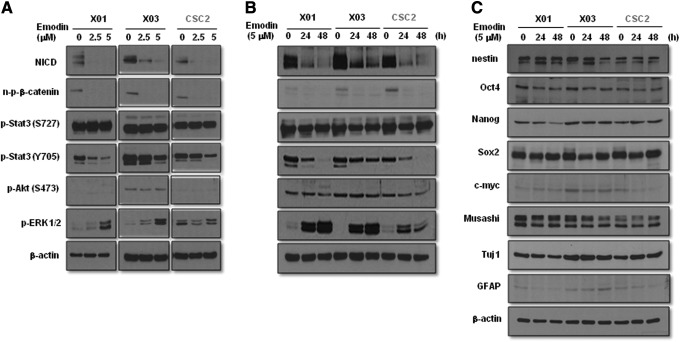
To identify the molecular mechanism underlying the inhibition of self-renewal activity of GSCs by emodin, we investigated the expression pattern of critical signaling molecules involved in the maintenance of stemness in GSCs. Western blot analysis indicated that emodin decreased the expression of stemness related signaling molecules, including notch intracellular domain (NICD), n-p-β-catenin (which indicates nuclear translocation of the molecule), and p-STAT3 (Y705) in a dose- and time-dependent manner in GSCs (Fig. 2A, B). Intriguingly, emodin treatment enhanced the p-ERK1/2 level (Fig. 2A, B), which is the same phenomenon observed when these cells were treated with gefitinib, a powerful EGFR inhibitor (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd). However, emodin had a marginal effect on the expression of stemness factors, including nestin, Oct4, Sox2, Nanog, c-myc, and Musashi, and lineage differentiation factors like Tuj1 and GFAP (Fig. 2C). Immunocytochemical analysis confirmed that emodin downregulated the expression of Notch-1 (Supplementary Fig. S2). These data suggest that emodin is able to suppress the self-renewal capacity of GSCs by blockade of Notch, β-catenin, and STAT3 signaling pathways.
FIG. 2.
Effect of emodin on the self-renewal associated signaling pathways in GSCs. Western blot analysis of GSCs. Cells were treated with or without emodin at the indicated concentrations on the figure for 24 h (A) or 5 μM of emodin at the indicated time points on (B, C). Total cells were lysed in a lysis buffer and subjected to western blot using antibodies described in the figure. β-Actin was used as a loading control.
Emodin induces cell death in GSCs
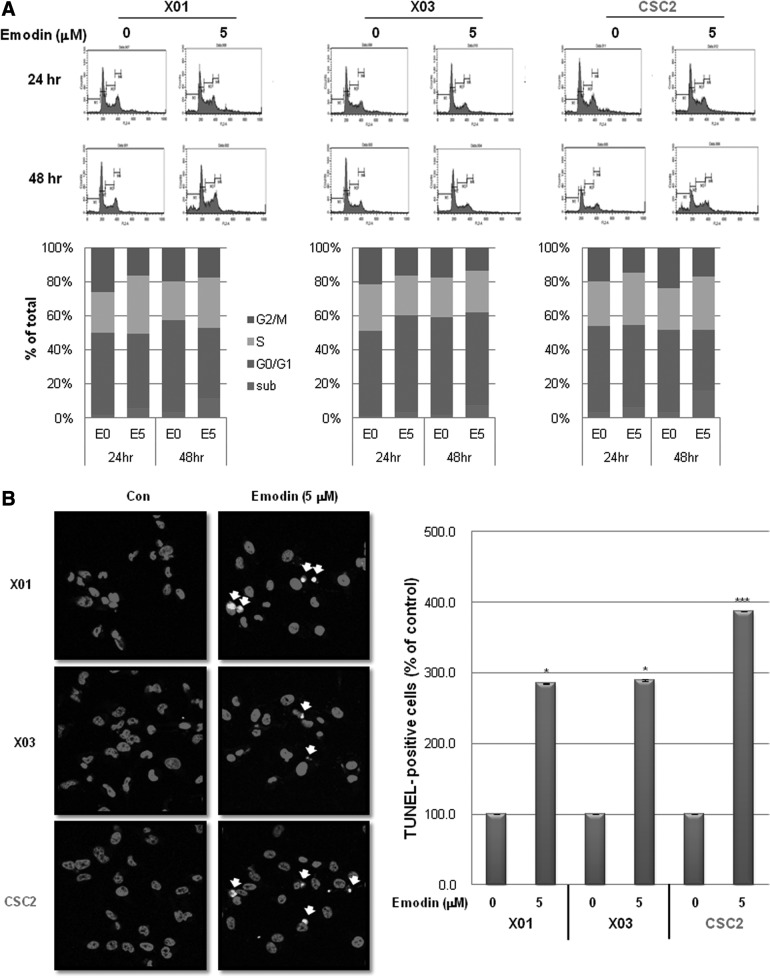
We next examined whether emodin induces cell death in GSCs. As shown in Fig. 3A, cell cycle analysis indicated that treatment with emodin induced S phase arrest and increased the subG1 fraction in GSCs in a time-dependent manner. Immunocytochemical analysis showed that emodin significantly enhanced the number of terminal deoxynucleotidyl transferase dUTP nick-end labeling-positive GSCs (Fig. 3B). These data indicate that emodin induces cell death in GSCs partly at least.
FIG. 3.
Effect of emodin on cell death of GSCs. (A) Cell cycle analysis of GSCs treated with or without emodin (5 μM) for 24 or 48 h. Representative histogram (upper) and quantification of the analysis (lower). (B) Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assays of GSCs. Representative images of immunocytochemistry of GSCs in the assays (left) and quantification of the assays (right). GSCs were pretreated with emodin (5 μM) for 24 h followed by being subjected to the assays. TUNEL positive nuclei were indicated with a white arrow in the figure. Nuclei were visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining (red). Magnification, ×200. E0, emodin 0 μM; E5, emodin 5 μM; Con, untreated control. *P<0.05, ***P<0.005.
Emodin suppresses invasion of GSCs
The hallmark of malignant glioma is infiltration through normal brain parenchyma [29]. We next examined the effect of emodin on the invasion activity of GSCs. As shown in Fig. 4A, emodin strongly suppressed the invasion of GSCs through a Matrigel-coated layer in a transwell assay. Consistent with this result, emodin decreased expression of the transcription factors snail and slug, mesenchymal cell markers that are known to be involved in the invasion of glioma cell lines [30] (Fig. 4B). These data indicate that emodin effectively blocks invasion of GSCs, possibly through downregulation of mesenchymal traits in GSCs.
FIG. 4.
Effect of emodin on the invasiveness of GSCs. (A) Matrigel invasion assays of GSCs. Cells were pretreated with or without emodin (5 μM) for 1 h and performed invasion assays as described in the section “Materials and Methods.” Representative pictures (left, magnification, ×100) and quantification of the assays (right). (B) Western blot analysis of GSCs treated with or without emodin (5 μM) for indicated time points using antibodies indicated in the figure. Con, untreated control. **P<0.01, ***P<0.005.
Emodin sensitizes GSCs to IR
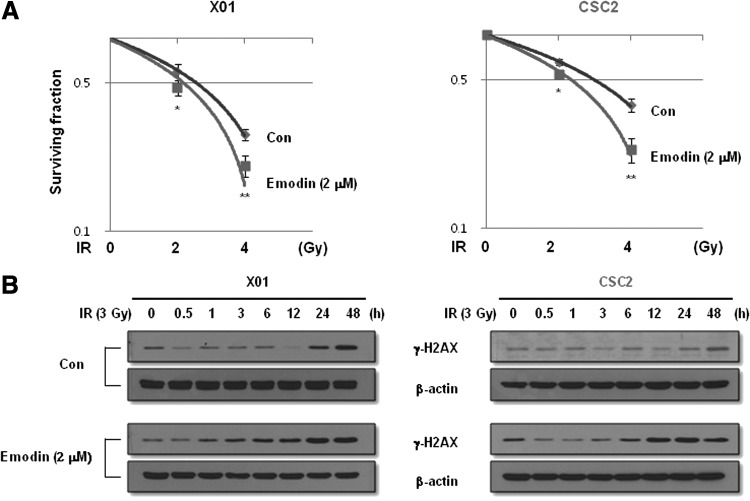
Because GSCs were reported to be resistant to IR and inhibition of the Notch pathway by γ-secretase inhibitors renders GSCs more sensitive to radiation [31], we examined whether emodin treatment sensitizes GSCs to IR. We pretreated GSCs with emodin at the concentration that caused 50% inhibition of sphere formation (2 μM), followed by irradiation with 2 and 4 Gy of IR or no irradiation and incubation in a serum-free complete medium for 10 days. As expected, pretreatment with emodin significantly increased the IR sensitivity of GSCs (Fig. 5A).
FIG. 5.
Effect of emodin on the radiosensitivity of GSCs. (A) Sphere-forming survival assays of GSCs at indicated doses of ionizing radiation (IR) in the figure with or without emodin (2 μM). After 2 weeks of incubation, spheres were attached in the plate by adding 10% of fetal bovine serum, stained with crystal violet, and counted. (B) Western blot analysis of GSCs exposed to IR (3 Gy) at the indicated time points pretreated with or without emodin (2 μM) using antibodies indicated in the figure. Con, untreated control. *P<0.05, **P<0.01.
Although IR exhibits cellular toxicity through the induction of DNA damage, it has been reported that GSCs are resistant to radiotherapy through preferential activation of the DNA damage checkpoint response [3]. We measured γ-H2AX foci formation, a sensitive indicator of DNA double-strand breakage within cells [32]. As shown in Fig. 5B, treatment of GSCs with emodin enhanced the formation of γ-H2AX foci in response to IR. Therefore, emodin enhances DNA damage response to IR, which strongly results in the decreased survival of GSCs after IR.
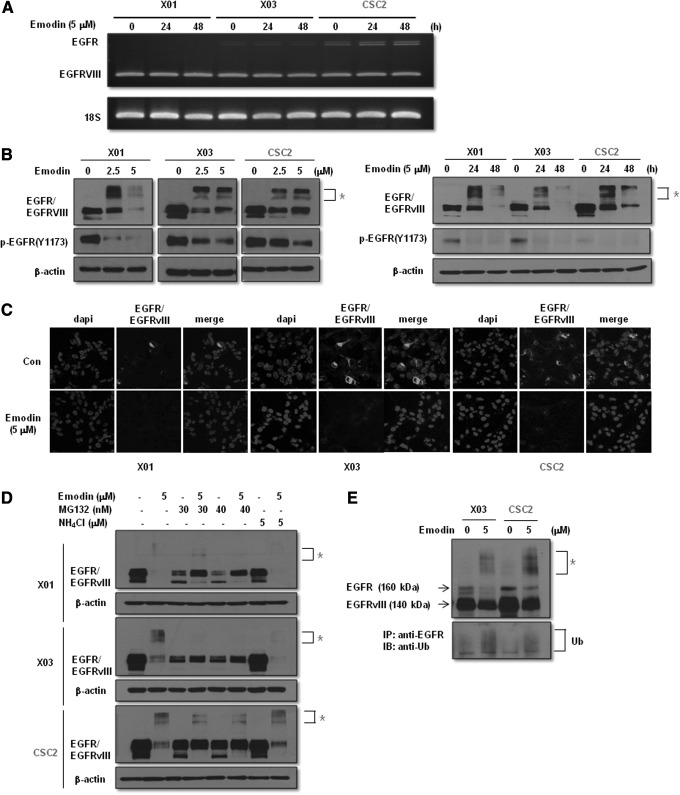
Emodin induces proteosomal degradation of EGFR/EGFRvIII in GSCs
We next examined the possible molecular target of emodin that is responsible for suppressing the self-renewal capacity in GSCs. Because overexpression of EGFR/EGFRvIII is a common feature of GBM and emodin was previously reported to block receptor tyrosine kinases, including VEGFR and ERBB2 [18, 19, and 33], we hypothesized that emodin affects EGFR/EGFRvIII-mediated stemness signaling in GSCs. GSCs naturally express a high level of EGFRvIII (Fig. 6A) that could not be detected in the conventional established cell lines with a low level of wild-type EGFR (Supplementary Fig. S3). As expected, emodin reduced the expression of EGFR/EGFRvIII in a dose- and time-dependent manner (Fig. 6B). Immunocytochemical analysis also showed the suppressive effect of emodin on EGFR/EGFRvIII expression (Fig. 6C). Intriguingly, treatment with emodin generated a slow-migrating EGFR/EGFRvIII band in the western blot analysis that was reminiscent of ubiquitin-mediated degradation (Fig. 6B). Consistent with this finding, emodin had no effect on the transcription level of EGFR/EGFRvIII, as determined by RT-PCR analysis (Fig. 6A). To investigate the mechanism of EGFR/EGFRvIII degradation by emodin, we pretreated GSCs with MG132 and NH4Cl inhibitors of the proteosomal and lysosomal degradation pathways, respectively. As shown in Fig. 6D, pretreatment with MG132, but not NH4Cl, blocked the degradation of EGFR/EGFRvIII by emodin in GSCs. Immunoprecipitation analysis confirmed the effect of emodin on the ubiquitin-mediated degradation of EGFR/EGFRvIII (Fig. 6E). In addition, treatment with gefitinib downregulated the expression level of NICD, n-p-β-catenin, and p-STAT3 (Y705), indicating that these stemness signaling molecules are downstream effectors of EGFR/EGFRvIII signaling (Supplementary Fig. S1). Taken together, these data suggest that emodin induces proteosomal degradation of EGFR/EGFRvIII followed by suppression of the Notch, β-catenin, and STAT3 signaling pathways that are responsible for the self-renewal activity of GSCs.
FIG. 6.
Effect of emodin on the epidermal growth factor receptor (EGFR)/EGFR variant III (EGFRvIII) expression. (A) Semiquantitative reverse transcriptase–polymerase chain reaction analysis of GSCs treated with or without emodin (5 μM) for 24 or 48 h. (B) Western blot analysis of GSCs. Cells were treated with or without emodin at the indicated concentrations in the figure for 24 h (left) or 5 μM of emodin at the indicated time points in the figure (right). Total cells were lysed in a lysis buffer and subjected to western blot using antibodies described in the figure. β-Actin was used as a loading control. (C) Immunocytochemistry of GSCs with the EGFR antibody. GSCs were pretreated with emodin (5 μM) for 24 h followed by being subjected to immunocytochemical analysis. Immunofluorescence staining was visualized by probing with the Alexa Fluor 594 goat anti-mouse secondary antibody (green). Nuclei were visualized by DAPI staining (blue). Magnification, ×200. (D) Western blot analysis of GSCs treated with or without MG132 (30 or 40 nM) or NH4Cl (5 μM) in the presence or absence of emodin (5 μM) using antibodies indicated in the figure. (E) Immunoprecipitation and western blot analysis of GSCs treated with or without emodin (5 μM) using antibodies indicated in the figure. Con, untreated control; Ub, ubiquitin; *slow-migrating EGFR/EGFRvIII bands.
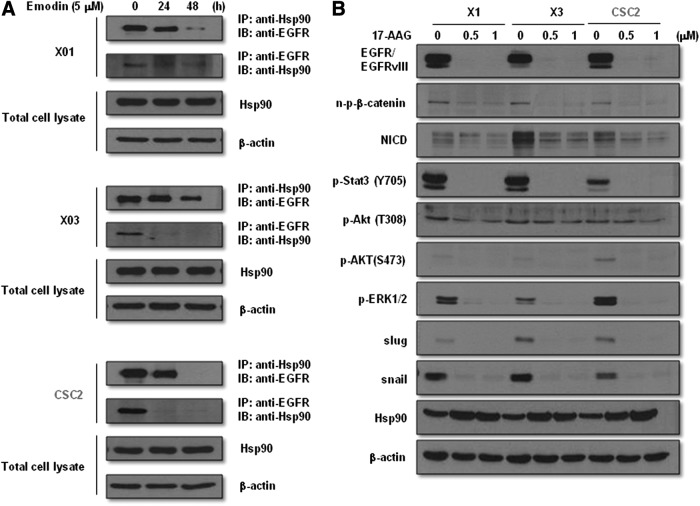
Emodin-induced degradation of EGFR/EGFRvIII is mediated through interference with the interaction between EGFR/EGFRvIII and Hsp90
Association of Hsp90 with ERBB2, wild-type EGFR, and EGFRvIII has been shown to be responsible for the stability of these receptors [34–36]. In addition, an azide methyl anthraquinone derivative of emodin was reported to induce proteosomal degradation of ERBB2 by decreasing its interaction with Hsp90 [33]. We performed immunoprecipitation analysis to examine the role of emodin in the interaction between Hsp90 and EGFR/EGFRvIII and found that emodin effectively disrupted binding of Hsp90 with EGFR/EGFRvIII in GSCs in a time-dependent manner (Fig. 7A). Treatment with 17-AAG, a Hsp90 inhibitor, reduced the expression level of EGFR/EGFRvIII and its downstream stemness signaling molecules NICD, n-p-β-catenin, and p-STAT3 (Y705) (Fig. 7B). These data indicate that emodin-induced degradation of EGFR/EGFRvIII and suppression of downstream stemness signaling molecules is mediated by blocking the interaction between Hsp90 and EGFR/EGFRvIII.
FIG. 7.
Effect of emodin on the interaction of EGFR/EGFRvIII with Hsp90. (A) Immunoprecipitation and western blot analysis of GSCs treated with or without emodin (5 μM) for the indicated time points using antibodies indicated in the figure. (B) Western blot analysis of GSCs treated with or without 17-AAG at the indicated concentrations using antibodies described in the figure.
Overall, our data strongly suggest that emodin effectively suppresses the stemness of GSCs by inducing degradation of EGFR/EGFRvIII and suppression of downstream signaling pathways, and thus sensitizes these cells to IR. Therefore, emodin might be an effective adjuvant agent for combination therapy with IR to target GSCs.
Discussion
The CSC theory postulates that cancers are initiated and maintained by rare stem-like cancer cells with a high tumorigenic potential and resistance to conventional therapy such as chemotherapy and radiotherapy. Therefore, to overcome the therapeutic resistance of CSCs, it is necessary to find compounds that target CSCs for combination therapy with conventional treatments. One promising source of CSC-targeting agents is naturally occurring dietary compounds because several reports have shown that some of these compounds affect the self-renewal capacity of CSCs [20–26]. In the present study, we investigated the effect of emodin, an anthraquinone compound present in rhubarb, on the biology of GSCs. We found that emodin strongly inhibited the self-renewal capacity of GSCs as evidenced by sphere formation, limiting dilution, and soft agar clonogenic assays. Importantly, emodin inhibited the stemness of GSCs by blocking the interaction of EGFR/EGFRvIII with Hsp90, which led to degradation of EGFR/EGFRvIII followed by suppression of stemness signaling pathways involving Notch, β-catenin, and STAT3. In addition, emodin partially induced apoptosis, inhibited invasion, and sensitized GSCs to IR. These results suggest that emodin could be considered as an adjuvant compound for combinational therapy of glioma through targeting the self-renewal activity of GSCs.
Natural dietary compounds have received attention as chemopreventive agents for a long time. These compounds have also been reported to have anticancer effects on various cancer cells. Intriguingly, recent studies have shown that some of these compounds also affect CSCs by inhibiting self-renewal pathways within these cells [20–25]. For example, an Indian dietary polyphenol, curcumin, affects Wnt/β-catenin stem cell signaling by suppressing the β-catenin/TCF transcriptional activity in various cancer cell lines, possibly through downregulation of the Wnt receptor Frizzled-1 [21,22]. Curcumin also inhibits Notch-1 mRNA, which might lead to inactivation of NF-κB DNA binding activity [23]. Furthermore, a recent report demonstrated that curcumin in combination with piperine targeted breast CSCs through potent inhibition of Wnt/β-catenin signaling [24]. Downregulation of Wnt/β-catenin signaling was also observed in breast CSCs treated with sulforaphane, a component of broccoli and broccoli sprouts [25]. Emodin was previously reported to target the stem-like side population of gallbladder carcinoma by suppressing expression of the ATP-binding cassette superfamily G member ABCG2 [26]. Our data showed that emodin effectively suppressed the self-renewal capacity of GSCs and indicated that this occurred through suppression of critical stemness signaling pathways. Emodin potently suppressed the expression of NICD, n-p-β-catenin, and p-STAT3 (T705) in GSCs. The Notch signaling pathway is known to be important for the maintenance of stemness and radioresistance of GSCs [31,37]. In addition, activation of Wnt/β-catenin signaling confers tumorigenesis, invasiveness, and radioresistance on GSCs [38], and STAT3 signaling regulates the Notch pathway and is crucial for the stemness and tumorigenicity of GSCs [39]. Together, our data strongly suggest that emodin acts as a potent suppressor of stemness in GSCs.
Amplification of the EGFR gene is commonly found in cases of GBM and can involve the EGFRvIII deletion mutant, the most common variant form of EGFR [40]. Many studies have shown that EGFRvIII confers chemo- and radioresistance, invasiveness and tumorigenesis, or tumor progression in GBM [40–43]. Moreover, EGFRvIII+ GBM cells are considered putative GSCs [43]. The GSCs used in this study exhibited overexpression of EGFRvIII with marginal expression of EGFR. Treatment of these cells with emodin strongly induced EGFR/EGFRvIII degradation. Blockade of EGFR/EGFRvIII phosphorylation with gefitinib resulted in the suppression of downstream stemness signaling, indicating that the degradation of EGFR/EGFRvIII induced by emodin might similarly lead to inhibition of downstream stemness signaling through Notch, β-catenin, and STAT3 followed by the induction of partial apoptosis, inhibition of invasion, and sensitization to IR.
Hsp90 is a molecular chaperone that is responsible for maintaining the stability of its client proteins [44]. Receptor tyrosine kinases, including EGFR, ErbB2, and PDGFR are known target proteins of Hsp90 [34, 35, and 45]. In addition to wild-type EGFR, mutant type EGFRs such as tyrosine kinase inhibitor resistant EGFR (T790M) and EGFRvIII were also reported to be stabilized by Hsp90 [36 and 46]. Inhibition of Hsp90 with specific inhibitors ablated tumorigenic GSCs through regulation of cell cycle progression and degradation of EGFR and EGFRvIII in these cells [47]. Because the anazide methyl anthraquinone derivative of emodin was reported to induce proteosomal degradation of ErbB2 by blocking its binding to Hsp90 in a breast cancer model [33], we speculate that Hsp90 is involved in emodin-induced proteosomal degradation of EGFR/EGFRvIII. Indeed, immunoprecipitation experiments confirmed that emodin effectively blocked interaction of EGFR/EGFRvIII and Hsp90 and subsequently promoted ubiquitin-mediated proteosomal degradation of EGFR/EGFRvIII. Because emodin has diverse biological activities with no coherent mechanism of action, we could not rule out the possible off-target effects of the compound. Further study will be needed to clarify the specific interaction of emodin on Hsp90.
In conclusion, our data indicate that emodin, a natural anthraquinone, suppresses the self-renewal activity in GSCs through inhibition of stemness signaling, which is augmented by degradation of EGFR/EGFRvIII through blockade of the interaction between EGFR/EGFRvIII and Hsp90.Therefore, we suggest that emodin could be considered as a potent natural agent for targeting stemness signaling in GSCs.
Supplementary Material
Acknowledgments
We thank Dr. Akio Soeda (Gifu University School of Medicine, Gifu, Japan) for providing patient-derived glioma stem cells. This study was supported by a grant from the Nuclear Research and Development Program funded by the Korean Ministry of Science, ICT and Future Planning and Bio-Synergy Research Project (NRF-2014M3A9C4066487) of the Ministry of Science, ICT and Future Planning through the National Research Foundation.
Author Disclosure Statement
The authors declare no conflicts of interest.
References
- 1.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, et al. (2007). Malignant astrocytic glioma: genetics biology, and paths to treatment. Genes Dev 21:2683–2710 [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, et al. (2005). Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996 [DOI] [PubMed] [Google Scholar]
- 3.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD. and Rich JN. (2006). Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444:756–760 [DOI] [PubMed] [Google Scholar]
- 4.Jordan CT, Guzman ML. and Noble M. (2006). Cancer stem cells. N Engl J Med 355:1253–1261 [DOI] [PubMed] [Google Scholar]
- 5.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA. and Dick JE. (1994). A cell initiating human acute myeloid leukemia after transplantation into SCID mice. Nature 367:645–648 [DOI] [PubMed] [Google Scholar]
- 6.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ. and Clarke MF. (2003). Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100:3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD. and Dirks PB. (2004). Identification of human brain tumour initiating cells. Nature 432:396–401 [DOI] [PubMed] [Google Scholar]
- 8.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C. and De Maria R. (2007). Identification and expansion of human colon-cancer-initiating cells. Nature 445:111–115 [DOI] [PubMed] [Google Scholar]
- 9.Visvader JE. and Lindeman GJ. (2008). Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer 8:755–768 [DOI] [PubMed] [Google Scholar]
- 10.Eramo A, Ricci-Vitiani L, Zeuner A, Pallini R, Lotti F, Sette G, Pilozzi E, Larocca LM, Peschle C. and De Maria R. (2006). Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ 13:1238–1241 [DOI] [PubMed] [Google Scholar]
- 11.Rich JN. (2007). Cancer stem cells in brain tumor biology. Cancer Res 67:8980–8984 [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Wicha MS, Schwartz SJ. and Sun D. (2011). Implications of cancer stem cell theory for cancer chemoprevention by natural dietary compounds. J Nutr Biochem 22:799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Q, Lu G, Shen HM, Chung MC. and Ong CN. (2007). Anti-cancer properties of anthraquinones from rhubarb. Med Res Rev 27:609–630 [DOI] [PubMed] [Google Scholar]
- 14.Chen YC, Shen SC, Lee WR, Hsu FL, Lin HY, Ko CH. and Tseng SW. (2002). Emodin induces apoptosis in human promyeloleukemic HL-60 cells accompanied by activation of caspase 3 cascade but independent of reactive oxygen species production. Biochem Pharmacol 64:1713–1724 [DOI] [PubMed] [Google Scholar]
- 15.Acevedo-Duncan M, Russell C, Patel S. and Patel R. (2004). Aloe-emodin modulates PKC isozymes, inhibits proliferation, and induces apoptosis in U-373 MG glioma cells. Int Immunopharmacol 4:1775–1784 [DOI] [PubMed] [Google Scholar]
- 16.Su YT, Chang HL, Shyue SK. and Hsu SL. (2005). Emodin induces apoptosis in human lung adenocarcinoma cells through a reactive oxygen species-dependent mitochondrial signaling pathway. Biochem Pharmacol 70:229–241 [DOI] [PubMed] [Google Scholar]
- 17.Huang Q, Shen HM, Shui G, Wenk MR. and Ong CN. (2006). Emodin inhibits tumor cell adhesion through disruption of the membrane lipid Raft-associated integrin signaling pathway. Cancer Res 66:5807–5815 [DOI] [PubMed] [Google Scholar]
- 18.Kwak HJ, Park MJ, Park CM, Moon SI, Yoo DH, Lee HC, Lee SH, Kim MS, Lee HW, et al. (2006). Emodin inhibits vascular endothelial growth factor-A-induced angiogenesis by blocking receptor-2 (KDR/Flk-1) phosphorylation. Int J Cancer 118:2711–2720 [DOI] [PubMed] [Google Scholar]
- 19.Lu Y, Zhang J. and Qian J. (2008). The effect of emodin on VEGF receptors in human colon cancer cells. Cancer Biother Radiopharm 23:222–228 [DOI] [PubMed] [Google Scholar]
- 20.Kawasaki BT, Hurt EM, Mistree T. and Farrar WL. (2008). Targeting cancer stem cells with phytochemicals. Mol Interv 8:174–184 [DOI] [PubMed] [Google Scholar]
- 21.Park CH, Hahm ER, Park S, Kim HK. and Yang CH. (2005). The inhibitory mechanism of curcumin and its derivative against beta-catenin/Tcf signaling. FEBS Lett 579:2965–2971 [DOI] [PubMed] [Google Scholar]
- 22.Jaiswal AS, Marlow BP, Gupta N. and Narayan S. (2002). Beta-catenin-mediated transactivation and cell-cell adhesion pathways are important in curcumin (diferuylmethane)-induced growth arrest and apoptosis in colon cancer cells. Oncogene 21:8414–8427 [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Zhang Y, Banerjee S, Li Y. and Sarkar FH. (2006). Notch-1 down-regulation by curcumin is associated with the inhibition of cell growth and the induction of apoptosis in pancreatic cancer cells. Cancer 106:2503–2513 [DOI] [PubMed] [Google Scholar]
- 24.Kakarala M, Brenner DE, Korkaya H, Cheng C, Tazi K, Ginestier C, Liu S, Dontu G. and Wicha MS. (2010).Targeting breast stem cells with the cancer preventive compounds curcumin and piperine. Breast Cancer Res Treat 122:777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Zhang T, Korkaya H, Liu S, Lee HF, Newman B, Yu Y, Clouthier SG, Schwartz SJ, Wicha MS. and Sun D. (2010). Sulforaphane, a dietary component of broccoli/broccoli sprouts, inhibits breast cancer stem cells. Clin Cancer Res 16:2580–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li XX, Dong Y, Wang W, Wang HL, Chen YY, Shi GY, Yi J. and Wang J. (2013). Emodin as an effective agent in targeting cancer stem-like side population cells of gallbladder carcinoma. Stem Cells Dev 22:554–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soeda A, Park M, Lee D, Mintz A, Androutsellis-Theotokis A, McKay RD, Engh J, Iwama T, Kunisada T, et al. (2009). Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene 28:3949–3959 [DOI] [PubMed] [Google Scholar]
- 28.Mellinghoff IK, Wang MY, Vivanco I, Kogan Haas-DA, Zhu S, Dia EG, Lu KV, Yoshimoto K, Huang JH, et al. (2006). Molecular determinants of the response of glioblastoma to EGFR kinase inhibitors. N Engl J Med 353:2012–2024 [DOI] [PubMed] [Google Scholar]
- 29.Giese A, Bjerkvig R, Berens ME. and Westphal M. (2003). Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol 21:1624–1636 [DOI] [PubMed] [Google Scholar]
- 30.Yamoutpour F, Bodempudi V, Park SE, Pan W, Mauzy MJ, Kratzke RA, Dudek A, Potter DA, Woo RA, et al. (2008). Gene silencing for epidermal growth factor receptor variant III induces cell-specific cytotoxicity. Mol Cancer Ther 7:3586–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Wakeman TP, Lathia JD, Hjelmeland AB, Wang XF, White RR, Rich JN. and Sullenger BA. (2010). Notch promotes radioresistance of glioma stem cells. Stem Cells 28:17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dickey JS, Redon CE, Nakamura AJ, Baird BJ, Sedelinikova OA. and Bonner WM. (2009). H2AX: functional roles and potential applications. Chromosoma 118:683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan YY, Zheng LS, Zhang X, Chen LK, Singh S, Wang F, Zhang JY, Liang YJ, Dai CL, et al. (2011). Blockade of Her2/neu binding to Hsp90 by emodin azide methyl anthraquinone derivative induces proteasomal degradation of Her2/neu. Mol Pharm 8:1687–1697 [DOI] [PubMed] [Google Scholar]
- 34.Xu W, Mimnaugh E, Rosser MF, Nicchitta C, Marcu M, Yarden Y. and Neckers L. (2001). Sensitivity of mature Erbb2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. J Biol Chem 276:3702–3708 [DOI] [PubMed] [Google Scholar]
- 35.Ahsan A, Ramanand SG, Whitehead C, Hiniker SM, Rehemtulla A, Pratt WB, Jolly S, Gouveia C, Truong K, et al. (2012). Wild-type EGFR is stabilized by direct interaction with HSP90 in cancer cells and tumors. Neoplasia 14:670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavictoire SJ, Parolin DA, Klimowicz AC, Kelly JF. and Lorimer IA. (2003). Interaction of Hsp90 with the nascent form of the mutant epidermal growth factor receptor EGFRvIII. J Biol Chem 278:5292–5299 [DOI] [PubMed] [Google Scholar]
- 37.Yoon CH, Kim MJ, Kim RK, Lim EJ, Choi KS, An S, Hwang SG, Kang SG, Suh Y, Park MJ. and Lee SJ. (2012). c-Jun N-terminal kinase has a pivotal role in the maintenance of self-renewal and tumorigenicity in glioma stem-like cells. Oncogene 31:4655–4666 [DOI] [PubMed] [Google Scholar]
- 38.Kim Y, Kim KH, Lee J, Lee YA, Kim M, Lee SJ, Park K, Yang H, Jin J, et al. (2012). Wnt activation is implicated in glioblastoma radioresistance. Lab Invest 92:466–473 [DOI] [PubMed] [Google Scholar]
- 39.Garner JM, Fan M, Yang CH, Du Z, Sims M, Davidoff AM. and Pfeffer LM. (2013). Constitutive activation of signal transducer and activator of transcription 3 (STAT3) and nuclear factor κB signaling in glioblastoma cancer stem cells regulates the Notch pathway. J Biol Chem 288:26167–26176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hatanpaa KJ, Burma S, Zhao D. and Habib AA. (2010). Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia 12:675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukherjee B, McEllin B, Camacho CV, Tomimatsu N, Sirasanagandala S, Nannepaga S, Hatanpaa KJ, Mickey B, Madden C, et al. (2009). EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res 69:4252–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB. and White FM. (2007). Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci U S A 104:12867–12872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Vecchio CA, Giacomini CP, Vogel H, Jensen KC, Florio T, Merlo A, Pollack JR. and Wong AJ. (2013). EGFRvIII gene rearrangement is an early event in glioblastoma tumorigenesis and expression defines a hierarchy modulated by epigenetic mechanisms. Oncogene 32:2670–2681 [DOI] [PubMed] [Google Scholar]
- 44.Trepel J, Mollapour M, Giaccone G. and Neckers L. (2010). Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer 10:537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matei D, Satpathy M, Cao L, Lai YC, Nakshatri H. and Nonner DB. (2007). The platelet-derived growth factor receptor alpha is destabilized by geldanamycins in cancer cells. J Biol Chem 282:445–453 [DOI] [PubMed] [Google Scholar]
- 46.Shimamura T, Li D, Ji H, Haringsma HJ, Liniker E, Borgman CL, Lowell AM, Minami Y, McNamara K, et al. (2008). Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer Res 68:5827–5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sauvageot CM, Weatherbee JL, Kesari S, Winters SE, Barnes J, Dellagatta J, Ramakrishna NR, Stiles CD, Kung AL, et al. (2009). Efficacy of the HSP90 inhibitor 17-AAG in human glioma cell lines and tumorigenic glioma stem cells. Neuro Oncol 11:109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.