Abstract
Repeat proteins have considerable potential for use as modular binding reagents or biomaterials in biomedical and nanotechnology applications. Here we describe a general computational method for building idealized repeats that integrates available family sequences and structural information with Rosetta de novo protein design calculations. Idealized designs from six different repeat families were generated and experimentally characterized; 80% of the proteins were expressed and soluble and more than 40% were folded and monomeric with high thermal stability. Crystal structures determined for members of three families are within 1 Å root-mean-square deviation to the design models. The method provides a general approach for fast and reliable generation of stable modular repeat protein scaffolds.
Keywords: repeat proteins, computational design, thermodynamic stability, idealized proteins, de novo design
Introduction
Repeat proteins play key roles in biological processes ranging from adhesion to signaling to defense mechanisms [1]. These proteins consist of adjacent series of usually non-identical repeated amino acid sequences; in most cases, these repeated units fold cooperatively into either a solenoid-shaped or a toroid-shaped structure [2–4]. Although extremely diverse in structure and sequence, repeat proteins are characterized by short-ranged intra-repeat and inter-repeat interactions between residues proximal in sequence [2]. The intrinsic modularity of repeat proteins allows combination of functionalities in a single domain (e.g., recognition motifs for nucleic acids [5] and peptides [6]) and can be used to generate biomaterials with tunable mechanical properties [7]. However, interactions between neighboring repeats are not always conserved; hence arbitrary extension by repeat insertion is not usually possible.
To allow modular extension, we have generated designed repeat proteins with self-compatible repeating elements using consensus-based approaches [8–18]. Consensus sequences are defined by the most common residue at each position in a multiple sequence alignment (MSA) of the proteins or repeats in a family. This approach is conceptually simple and powerful but does have non-optimal features. First, the consensus sequence can vary depending on the collection size and the selection method for the sequences used in the alignment. Second, residue–residue packing, particularly critical in the formation of a uniquely defined hydrophobic core, is not considered, and hence in some cases, the consensus may have sub-optimal residue–residue interactions. Incorporating amino acid covariation information derived from statistical analysis of naturally occurring sequences can capture some of these residue–residue coupling effects [19–21], but reliable estimates of covariance require large numbers of sequences that are not available for all protein families.
Here we describe a general computational approach for repeat protein design that integrates Rosetta de novo structure generation and design methodology with protein family-based sequence and structural information. By automatically generating very low energy design models compatible with the available sequence and structure information, the method provides increased versatility compared to standard sequence consensus-based approaches and reduces the manual intervention required to achieve stable designs..5
Results and Discussion
We developed a computational approach that integrates sequence and structural information with Rosetta [22] de novo folding and design calculations for the generation of idealized repeat proteins (Fig. 1). Families with α helical, β and mixed α/β secondary structure were chosen for redesign to illustrate the generality of the method. Sets of sequences were designed for six protein families: ankyrin, armadillo, tetratrico peptide repeat (TPR), HEAT, leucine-rich repeats (LRR) and WD40.
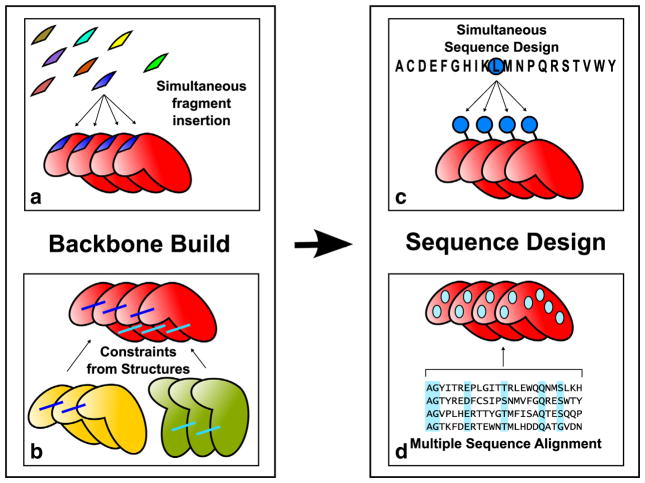
Fig. 1.
Overview of repeat protein design protocol. The protein backbone is built by simultaneous fragment insertion (a) guided by constraints derived from existing structures (b). Rosetta sequence design calculation is carried out enforcing sequence identity between repeats (c) and amino acids at specific positions are favored according to their frequency in the MSA of the target family (d).
Overview of computational method
For a repeat protein family of interest, protein structure and sequence information are extracted from publicly available databases and implemented as constraints in the Rosetta modeling suite [22]. Residue–residue distance constraints are used to guide Rosetta structure generation calculations. Sequence constraints derived from MSAs are used to bias Rosetta design calculations (see Materials and Methods).
Protein backbones are generated de novo [23–25] as poly-valine to avoid bias toward a particular template structure. Structure-based and sequence-based constraints guide the sampling and limit the search space to an area defined by the protein family. We exclude from the sequence and structural alignments sequences that were previously designed with consensus-based approaches. Proteins are represented as a single chain formed by a series of identical repeats. The number of repeats and the secondary structure can be arbitrarily chosen or derived from existing proteins.
Backbone conformations are generated by Monte Carlo fragment insertion using RosettaRemodel [25], with insertions in a single repeat replicated in all other repeats (Fig. 1a). The trajectories optimize a function consisting of the Rosetta energy supplemented with family-specific structural constraints (Fig. 1b). Rosetta sequence design calculations are then carried out on the low-energy backbones that satisfy the constraints. The sequence is designed simultaneously in all the repeats (Fig. 1c), using the Rosetta energy function with a sequence profile term favoring residues observed in the family MSA (Fig. 1d).
Most native repeat proteins form a solenoid-like structure with specialized terminal repeats to avoid aggregation and low solubility. Exposed hydrophobic residues in the terminal repeats were substituted with polar amino acids using Rosetta design calculations (Materials and Methods).
Sequence logos [26,27] of the designed proteins for each family are compared to their native counterparts in Fig. 2. The most conserved positions were recapitulated with a few exceptions in the HEAT family (see below in the family description). Even in the absence of MSA constraints, structural constraints alone allowed recovery of the key residues for each fold (Supplementary Figs. S1 and S2 and Table S1). For experimental evaluation, the full protocol with both sequence and structural constraints was used to generate the final sequences.
Fig. 2.
Comparison of computationally designed and existing sequences. The sequence logo of naturally occurring sequences (top logo for each family) is compared to the sequence logo obtained from the pool of computationally designed sequences with (middle) or without (bottom) family-specific MSA. Blank entries in the logo of naturally occurring sequences are positions not covered by the MSA available in the database. WD40 designs included in both cases an additional sequence bias at positions 14, 24 and 25 (see the family description in the results section), which accounted for the high recovery in the profiles. As noted in the text, the WD40 sequences were rearranged to match the structural unit in the protein.
Synthetic genes were constructed for several low-energy designs from each protein family (Table 1), and the designed proteins were expressed in Escherichia coli. Proteins were purified from the soluble fraction of cell lysate and their oligomeric state was characterized by analytical gel filtration (AGF) with multi-angle light scattering (MALS). Secondary structure and cooperative unfolding during thermal denaturation were measured by circular dichroism (CD) (Table 2 and Fig. 3). We solved crystal structures of several of the designs and their agreement with the models is shown in Fig. 4.
Table 1.
Designed repeat protein families.
| Family | Repeat length | No. of repa | Capsb | Architecturec | No. of resd |
|---|---|---|---|---|---|
| ank | 33 | 3 | des | α sol | 168 |
| TPR | 34 | 4 | no | α sol | 151 |
| arm | 42 | 4 | des | α sol | 252 |
| HEAT | 42 | 4 | des | α sol | 260 |
| LRR | 24 | 7 | adapt | α/β sol | 265 |
| WD_6r | 40 | 6 | no | β tor | 259 |
| WD_7r | 40 | 7 | no | β tor | 301 |
| WD_8r | 40 | 8 | no | β tor | 346 |
Number of internal repeats, excluding capping repeats.
Capping repeats, designed (des), adapted from native (adapt) or not present (no).
Solenoid (sol) or toroid (tor).
Number of residues.
Table 2.
Expression and characterization of repeat proteins.
| Family | Designs tested | Expressed | Soluble | Monomera | Aggregatesb | Foldedc | Monomer Tmd (°C) | Yielde (mg/l) | Crystal structures |
|---|---|---|---|---|---|---|---|---|---|
| Ankyrin | 6 | 6 | 6 | 6 | — | 6 | >95 | 80 | 4 |
| TPR | 6 | 6 | 6 | 4 | — | 5f | 57, > 95 | 60 | — |
| Armadillo | 8 | 8 | 7 | 7 | — | 3 | >95 | 30 | 1 |
| HEAT | 8 | 8 | 5 | 1 | — | 3 | >95 | 30 | — |
| LRR | 2 | 2 | 2 | 2 | 2 | 2 | 78 | 30 | 2 |
| WD40_6rep | 3 | 2 | 2 | 2 | — | — | — | — | — |
| WD40_7rep | 3 | 3 | 3 | — | — | — | — | — | — |
| WD40_8rep | 7 | 4 | 4 | 3 | 3 | 1f | — | 10 | — |
Determined by AGF-MALS.
Soluble aggregates/oligomers were also observed in addition to the monomers.
Proteins were considered folded if they displayed a cooperative transition (sigmoidal shape) or no transition (signal loss of <20% at 95 °C) and if the CD spectrum at 25 °C corresponded to the expected secondary structure.
Midpoint of transition in thermal denaturation for the folded monomeric proteins within the family.
Milligrams of protein per liter of culture, as determined after ion metal affinity chromatography purification and dialysis. No optimization for protein expression was attempted.
Folded protein forms a dimer. Only one TPR design is dimeric.
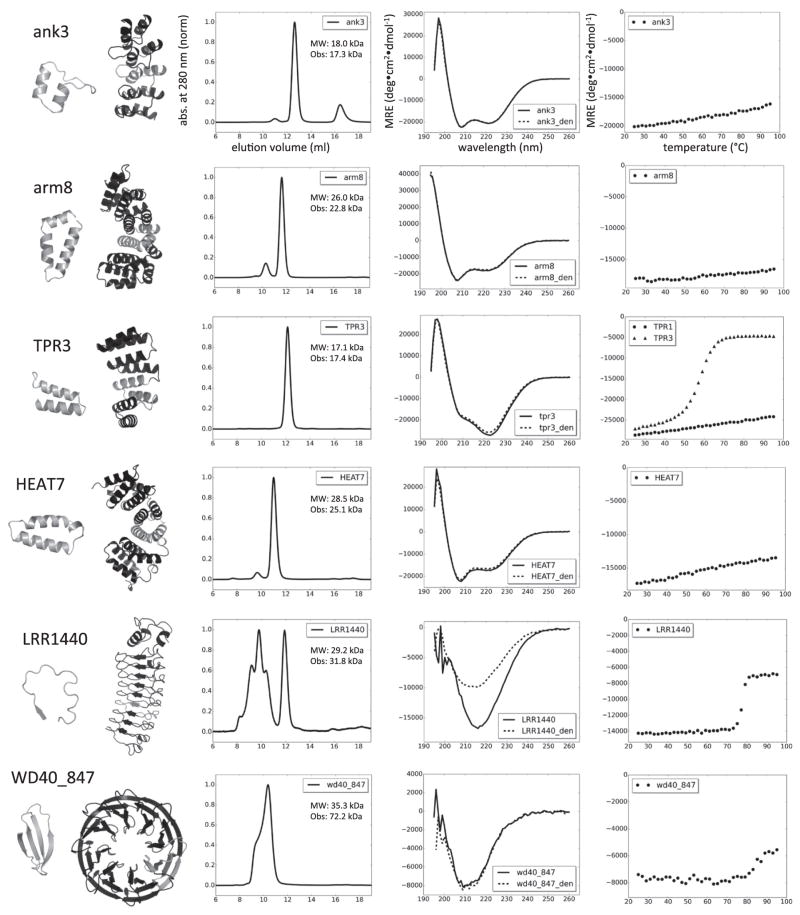
Fig. 3.
Characterization of designed repeat proteins. From left to right, repeating unit and protein model, AGF, CD spectrum and thermal denaturation profile. The repeating unit is highlighted in gray in the models. Axis labels in the first row apply also to the other plots. Abs indicates normalized absorbance measured at 280 nm; MRE is mean residue ellipticity. AGF was performed on a Superdex 75 column (void volume at 8 ml); MW is the expected molecular weight and Obs indicates the observed mass in MALS for the main non-aggregating peak. CD spectra were recorded at 25 °C before and after denaturation (_den). All proteins were able to refold upon thermal denaturation with the exception of LRRs. Thermal denaturation was followed at 220 nm for ank, arm, TPR and HEAT; thermal denaturation was followed at 212 nm for LRR and at 215 nm for WD40. Data shown are from one representative monomeric protein for each family. An additional temperature denaturation curve is displayed for TPR, showing the different behavior observed among monomeric designs within the family.
Fig. 4.
Superposition of models and crystal structures for ank3 (a) (RMSD of 0.9 Å), arm8 (b) (RMSD of 0.9 Å) and LRR_1440 (c) (RMSD of 1.1 Å). Models are in green and crystal structures are in blue. In most cases, the core residues assume the conformation predicted in the models, as shown in (a), (b) and (c) insets for some of the side chains. Parts of the structures have been removed to display the core residues. RMSD was calculated using backbone heavy atoms. For LRR, the N-terminal capping repeat was not included in the RMSD calculation, when considered that the RMSD increases to 1.6 Å. Pictures were realized with PyMOL (Schroedinger).
In the following sections, we describe the computational and experimental results for each family.
Ankyrin (ank)
Ankyrin repeat proteins are characterized by a short β hairpin and two antiparallel α helices (Fig. 3). The design calculations converged on set of sequences with about 50% of the residues conserved (see sequence logo for designed ankyrin; Fig. 2). All six ankyrins selected for experimental characterization were expressed solubly with high yield (>50 mg/l of culture), monomeric, α helical by CD and stable up to 95 °C (Fig. 3). The same properties were observed in previously designed ankyrin repeats (DARPins) [8]. The accuracy of the designs was confirmed by X-ray crystallography. The computational models were within 1 Å root-mean-square deviation (RMSD) from their corresponding crystal structures (Fig. 4a).
Armadillo repeat proteins (arm)
The basic repeat unit of this family is formed by three helices located roughly in the same plane. The designs recapitulated all the sequence features of the family and converged to specific amino acids in several positions (Fig. 2). Surface residues were optimized to reduce the potential electrostatic repulsion from repeated charged amino acids, without affecting significantly the overall energy. Seven out of eight designs tested were expressed and soluble and three were found to be monomeric and folded (Table 2 and Fig. 3). The crystal structure of design arm8 was determined and proved to be very close to the design model (1 Å RMSD; Fig. 4b).
Tetratrico peptide repeats (TPR)
The TPR repeat unit is formed by two α helices connected by a short loop. The design calculations did not converge into a narrow sequence space as in the ankyrin case but did capture the key features of the family (Fig. 2). Capping repeats were not modified, as there were no large exposed hydrophobic residues in the designs. Six 4-repeat designs were selected for experimental testing. All the proteins were expressed at a yield above 30 mg per liter of culture. One displayed molten globule-like properties and five were folded. Four of these were monomeric, while the fifth formed a dimer in solution. TPR3 displayed a cooperative and reversible thermal unfolding, while the other four folded proteins did not denature at 95 °C (Fig. 3).
HEAT repeat proteins (HEAT)
HEAT proteins form a large family of α solenoids characterized by two main helices connected by a loop. The remainder of the repeat can assume multiple conformations, from straight helices to kinked helices to long loops [28]. This variability represents a challenge for consensus-based methods and only one successful design from a small and very conserved subgroup has been reported so far [12]. The designed sequences recapitulate the conserved sequence features with the exception of Pro9 and Arg23; Asp17 was only partially recovered (Fig. 2). The structural features associated with these residues are poorly sampled and energetically disfavored during de novo backbone generation. Instead, the design protocol explored alternative solutions to lower the total energy and improved packing by substituting Pro9 within the first α helix with residues with higher helix propensity and by replacing a buried salt bridge formed by Asp17 and Arg23 with alternative packing solutions. Eight designs were experimentally tested. Three designs were soluble and had the expected CD spectrum, but only HEAT7 was expressed at high yield and monomeric (Table 2 and Fig. 3).
Leucine-rich repeat proteins (LRR)
LRRs are characterized by repeats containing a β strand packed on an α helix (or 3–10 helix or loop) that form a horseshoe-shaped αβ solenoid. The length of the repeat can vary, with 24 residues the most common size. Two short β strands extracted from variable lymphocyte receptor (VLR) structure 2O6S (residues 103–105 and 127–128, chain A) were used to ensure the correct pairing during backbone design. Sequence refinement led to the sequence family depicted in Fig. 2, with all the conserved residues recovered. The two lowest-energy models were similar to natural and designed VLRs [13]. Surface residues were optimized without MSA bias to reduce the electrostatic repulsion from repeated charged amino acids. We used the N-terminal capping repeat of internalin B, successfully employed in VLRs [13], while the C-terminal cap was generated from the internal repeat unit. The two experimentally characterized LRR designs were stable and folded, although a large fraction formed soluble aggregates in addition to monomers (Fig. 3). In contrast to other designs, they aggregated irreversibly at high temperature. We solved crystal structures of two of the designs. These structures obtained are very similar to the design models (~1.1 Å RMSD across backbone heavy atoms in the internal repeats) (Fig. 4c). Although the designs share the same N-terminal cap and are more than 80% identical in the internal repeats, the C-terminal capping repeat is visible only in one structure (4PSJ). The terminal repeat assumes an alternative conformation, forming an α helix that packs against the exposed core of the last internal repeat.
WD40
WD40 proteins form a toroid β propeller, where each “blade” is a four-stranded β sheet that packs against the two neighboring sheets. The fold is characterized by buried polar interactions that were disfavored in the initial design calculations. The definition of the WD40 repeat in the SMART and Pfam databases does not correspond to the structural unit but instead comprises three strands and the fourth strand of the previous blade [29,30]. However, a repeat definition matching the structural unit combined with explicit buried polar residues (Trp24, Asp25 and Ser/Thr14) led to a pool of sequences resembling closely the native proteins (Fig. 2). Separate design calculations for 6-bladed, 7-bladed and 8-bladed propellers were carried out. Thirteen designs were experimentally tested and nine were found to be expressed and soluble; however, only one with eight repeats, WD40_847 appeared folded (Table 2 and Fig. 3). WD40_847 was purified as large soluble aggregates, but upon heating for 10 min at 80 °C, it became mainly dimeric. The CD spectrum was closely resembled by the spectra observed for existing β propellers [31]. WD40_847 was expressed also as a half-propeller, with only four repeats. AGF-MALS analysis identified the species in solution as dimer, suggesting that it might indeed form an 8-repeat β propeller, but we were unable to confirm this with an X-ray crystal structure.
Contribution of protein family information to designed sequences
To investigate the contributions of the different information sources to the design models, we carried out calculations in which either the structural constraints or the sequence constraints were eliminated. At the backbone building stage, structural constraints significantly increase the fraction of models with structural similarity to the naturally occurring family members (Materials and Methods and Supplementary Fig. S1). For each repeat protein family, a subset of the models satisfies the topology requirements, even in absence of structural constraints. The gain is family dependent, with only marginal improvements for simple two-helix topologies such as TPR to more than 90% for a more complex three-helix topology such as armadillo.
The effect of the MSA-derived sequence constraints on the designs generated by our protocol was evaluated by comparing the sequence profiles of models obtained with or without sequence constraints (Materials and Methods). Profile–profile comparison to Pfam families using HHsuite [32] shows that the constraints increase the similarity of the profile to the original family from 35% to 80%, on average (Supplementary Fig. S2). With the exception of the HEAT family, where alternative combinations of core residues were explored, even in the absence of sequence information, the closest match to the designed protein sequence profile in the Pfam database is the naturally occurring corresponding repeat protein family (Supplementary Table S1).
Conclusions
The approach presented here generalizes the current MSA-based methods for repeat protein design with an automated pipeline that integrates sequence, structure and energetic information. Designing backbones de novo avoids potential bias due to the use of a single or few template structures.
Forty percent of the proteins designed with our method were folded and had a melting temperature (Tm) of 57 °C or greater (Table 2). The crystal structures we were able to solve had an RMSD of about 1 Å to the design models. Rosetta calculations recapitulate the majority of the sequence features of all the six families and generate models with excellent core packing and backbone geometry. Family-specific structures, such as proline kinks and conserved buried hydrogen bond networks or charge interactions, can be enforced using specific local conformation of the backbone or a set of residue identities.
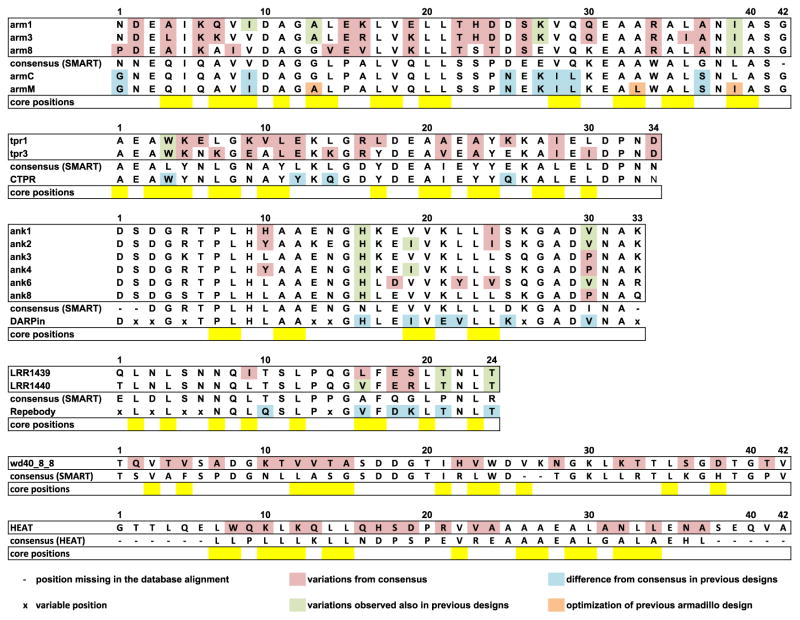
The use of general sequence and structural constraints allows greater exploration of the sequence space available for repeat protein families than strict consensus-based approaches. As shown in Fig. 5, the method generates low-energy sequences that differ from the consensus and were not explored in previous successful designs, in particular, for TPR and armadillo families [10,11], where variations also occur in several hydrophobic core residues. Although the selected amino acid at each position is usually among the three most frequent, the discrimination of viable sequences among the vast number of potential combinations represents a challenge for simple consensus-based approaches. In the armadillo case, the first consensus design (armC in Fig. 6) had molten globule-like characteristics and stabilization required a second round of refinement (armM in Fig. 6) [10]. In contrast, three of our designed sequences (arm1, arm3 and arm8 in Fig. 6) with only ~60% sequence identity to the consensus are stable up to 95 °C, compared to a Tm of 70 °C observed for armM. Overall, our protocol expands the traditional consensus-based approaches, reproducing the finding for highly conserved families, and offers a general solution to the design of idealized repeat proteins, producing a broader range of sequences than what would be available through consensus design.
Fig. 5.
Comparison of designed repeat protein sequences to strict consensus sequences and previous consensus-based designs. Armadillo, TPR, ankyrin and LRR designed sequences are compared to consensus sequences from SMART database [29,33] and to previous successful designs armC and armM [10], CTPR [11], DARPin [8] and Repebody [13]. HEAT and WD40 designed sequences are compared to the consensus from Pfam and SMART, respectively. Positions missing in the SMART and Pfam alignments are represented as dashes and were not considered. Positions identified as variable and included in library designs are labeled with x and were also not considered. Pink: positions that vary between the designs presented here and the SMART consensus; green: positions that differ from the consensus but present in previous designs; blue: differences between the previous design and the consensus; orange: stabilizing mutations introduced in the armadillo consensus design [10]; yellow: hydrophobic core positions.
The extension of Rosetta de novo design methods [23,24] to repeat protein architectures allows the computational design of extended modular non-globular structures for a wide range of applications. In the limit of no available data for a particular topology, it should be possible to use this approach to design completely new types of repeat protein structures and sequences.
Materials and Methods
Generation of sequence and structural constraints
The repeat consensus sequences were obtained from family alignments in the SMART database [29,33]. For the HEAT family, not present in SMART, the Pfam [30] seed alignment was used. A double-repeat sequence was generated by duplication of the consensus. When the consensus sequence did not cover the whole repeat, connecting fragments were added using alanine residues as placeholders. The length of this linking sequence was based on the shortest connection observed, with at least a 10% frequency, in repeats from crystal structures of family members. Using this sequence, we obtained an improved double-repeat consensus and a sequence profile after five rounds of PSI-BLAST [34,35]. Sequences previously designed were excluded from the database. The PSI-BLAST profile was implemented in Rosetta as Sequence-Profile constraints. According to Eq. (1) below, at each position, the amino acid frequencies (f, ranging from 0 to 100) were converted to arbitrary Rosetta energy units (REU) using pseudocounts to allow cases when particular amino acids were not observed.
| (1) |
The values are positive, representing an energy penalty, with higher values for less frequent amino acids, with a maximum value of 2.08 REU.
We selected 100 template structures from the PDB repository‡ (from Refs. [36] and [37]) using HHsearch [38]. Previously designed structures were excluded from the template list. The repeat consensus was threaded on the templates and the resulting models were clustered with 2 Å RMSD cluster radius using Rosetta [22]. For each model within the largest cluster, distances of carbon α to carbon α (Cα–Cα) between residues were measured. Contacts were defined as distances shorter or equal to 10 Å (distance cutoff) between amino acids at least 4 residues apart (sequence separation). Cα–Cα contacts present in all models were considered as conserved and the average distance was calculated. The conserved intra-repeat and inter-repeat contacts were implemented in Rosetta as AtomPair constraints, described by harmonic functions centered at the average Cα–Cα distances, with a spring constant of 10. For each double-repeat model, about 190 AtomPair constraints were used on average, including intra-repeat and inter-repeat constraints.
Secondary structures were assigned as helix, strand or loop, using dssp [39], to the templates belonging to the selected cluster employed for constrained generation. The most common secondary structure at each position was chosen and the information was implemented as Rosetta blueprint file.
Backbone design and refinement
Protein backbones were generated by insertion of fragments of 3 and 9 residues using RosettaRemodel [25]. A definition of the repeat secondary structure derived from the existing protein was used as starting input. The chain was represented as poly-valine during this initial stage, followed then by sequence design. Generation of repeat proteins required the implementation of simultaneous insertion of fragments in each repeat, as well as coupling of dihedral angles and side chains identities between repeats. The structure-based and sequence-based restraints were used to guide the search space toward the desired fold. About 5000 backbone models per family were generated and clustered by RMSD using the cluster application in Rosetta [22] with a cutoff of 3 Å. The quality of the models was evaluated in comparison to the average conserved interatomic distances within proteins in the family, expressed as AtomPair constraints as described above, using a harmonic function with flat bottom between average ± standard deviation and spring constant of 10. Violations of these constraints were calculated as energetic penalties. Structures with violations up to 5 REU per repeat, corresponding to less than 10% of the total energy of a correct full atom model or crystal structure, were accepted and their frequency is depicted in Supplementary Fig. S1.
Protocols described as Rosetta_scripts [40] were used for refining the sequences. Three cycles of sequence design and backbone minimization were performed while optimizing the packing interactions and the total energy. The available amino acids for each position were restricted based on the secondary structure and the solvent accessibility [24,25]. Cysteines were excluded to prevent formation of oligomers upon oxidation. The lowest-energy models from the 30 most populated clusters were selected for refinement, with 500 trajectories each.
Designs were filtered according to total energy (designs within 15% of lowest-energy model), and values of chi2 dihedral angles of aromatic residues (70° < chi2_dh < 110°) [24]. Final designs with lowest energy and RosettaHoles score [41] of <0 were selected. RosettaHoles values up to 1 were accepted for WD40 to increase the number of potential candidates. When several designs with similar values were available, sequence composition (e.g., low number of alanines in the core) was used as discrimination factor.
Hidden Markov model profile–profile comparison was carried out using HHsuite [32] with default settings and the Pfam database [30] (Supplementary Table T1). To visualize the influence of sequence constraints on the selected sequence pool, we normalized the score values by the maximum scores obtained by self-alignment of the reference Pfam family, according to Kamisetty et al. [42] (Supplementary Fig. S2).
Design of capping repeats
Solenoid-like repeat proteins are often characterized by N-terminal and C-terminal specialized capping repeats that protect the hydrophobic core from solvent exposure. The low sequence conservation and the variability in conformation of capping repeats within families prevented the use of a reliable sequence profile as a general strategy for design of capping repeats; hence the Rosetta energy function alone was used to guide selection of surface residue identities. Capping repeats were designed from the internal repeat by mutating exposed hydrophobic residues into polar residues, except for leucine-rich repeat (LRR) N-terminal cap where internalin B (residues 25–110) was used as in VLR designs [13]. The first internal repeat was modified to be compatible with the grafted cap. The backbones of the final models within ankyrin and armadillo families were all very similar. The most frequent amino acids replacing the exposed hydrophobic residues in the simulations were used in all models of the family to generate the capping repeats.
As observed in native sequences and in previous designs, the N-terminal ankyrin capping repeat contains 3 helical N-terminal residues [8,43], and the N-terminal armadillo repeat contains only two helices [10]; therefore, these capping repeats were modifying accordingly. TPR repeats did not possess large exposed hydrophobic residue; therefore, no specialized capping repeats were introduced.
Surface refinement and manual intervention
LRR and armadillo final sequences were characterized by a few charged residues in close proximity. A final design round of the surface residues was performed without sequence constraints, but the positions were selected following structure examination, instead of automatically. For LRRs, positions 1, 3 and 19 were redesigned, while one or two selected positions per repeat (26, 30 or 34) were changed in armadillo sequences from glutamate, lysine or arginine to glutamine in 6 out of 8 models.
Molecular biology and biochemistry
Gene synthesis and cloning
Genes were synthesized and cloned in vector pET21_NESG by GenScript (Piscataway, NJ). gBlocks for TPR genes and oligonucleotides were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA) and cloned into pET21_NESG. WD40 genes were synthesized by gen9 (Cambridge, MA) and cloned into pet15_NESG vector via Gibson cloning [44]. Cloning strains used were XL1-blue and XL10-gold (Agilent Technologies).
Protein expression and purification
Proteins were expressed in BL21(DE3) E. coli cells (Life Technologies) at 37 °C and induced with 250 μM isopropyl-β,D-thiogalactopyranoside (IPTG), either overnight at 22 °C or for 4 h at 37 °C, without significant difference in yield. For cases with low growth or lack of expression, BL21(DE3) pLysS (Life Technologies) was used, without any significant improvement. Cells were lysed by sonication and the clarified lysate was loaded on a Ni-NTA superflow column (Qiagen). Lysis and washing buffer was 50 mM Tris (pH 8), 500 mM NaCl, 30 mM imidazole and 5% (v/v) glycerol. Lysozyme (2 mg/ml), DNase I (0.2 mg/ml) and protease inhibitor cocktail (Roche) were added to the lysis buffer before sonication. Proteins were eluted in 50 mM Tris (pH 8), 500 mM NaCl, 250 mM imidazole and 5% (v/v) glycerol and were dialyzed overnight in 20 mM Tris (pH 8), 50 mM NaCl or PBS (12 mM phosphate, 137 mM NaCl, 2.7 mM KCl, pH 7.4). Protein concentrations were determined using a NanoDrop spectrophotometer (Thermo Scientific). Except indicated above, enzymes and chemical were purchased from Sigma-Aldrich.
Biophysical characterization
Secondary structure content and thermal stability were monitored by CD using an AVIV 62S DA spectrometer (Aviv Biomedical, Lakewood, NJ). Thermal denaturation was followed at 220 nm for structures containing α helices, at 212 nm for LRR and at 215 nm for WD40. Oligomeric state was assessed by AGF coupled to MALS. A Superdex 75 10/300 GL column (GE Healthcare) equilibrated in PBS was used on a HPLC LC 1200 Series (Agilent Technologies) connected to a miniDAWN TREOS (Wyatt Technologies). The chromatograms shown in Fig. 3 were recorded for 100 μl samples at 1–4 mg/ml concentration, with a flow rate of 0.5 ml/min. Protein molecular weights were confirmed by mass spectrometry on a LCQ Fleet Ion Trap Mass Spectrometer (Thermo Scientific).
Preparation of protein samples for crystallography
Crystallization was attempted for all the folded designed repeat proteins. The plasmids were transformed into E. coli BL21(DE3) pMgK-competent cells. All proteins were expressed and purified based on the standard procedures of Northeast Structural Genomics to produce selenomethionine-labeled samples for X-ray crystallography [45]. The selenomethionine-labeled proteins were grown at 37 °C in MJ9 minimal media [46]. When the OD600 reached 0.6, selenomethionine, lysine, phenylalanine, threonine, isoleucine, leucine and valine were added 10 min before induction with 1.0 mM IPTG [47]. Protein expression was carried out at 17 °C. Following overnight incubation, we harvested the cells by centrifugation and stored at −80 °C.
Cells were resuspended in 30 ml of lysis buffer [50 mM Tris (pH 7.5), 500 mM NaCl, 40 mM imidazole, 1 mM TCEP and 0.02% (w/v) NaN3]. Following lysis by sonication, we loaded the supernatant onto an ÄKTAxpress system (GE Health-care) using a two-step protocol consisting of ion metal affinity chromatography (HisTrap HP, 5 ml) chromatography followed by gel-filtration (HiLoad 26/60 Superdex 75) chromatography. Protein-containing fractions were pooled and concentrated to a range of 7.8–12.35 mg/ml. Protein purity and molecular mass were evaluated using SDS-PAGE and matrix-assisted laser desorption/ionization/time of flight mass spectrometry. These pET expression vectors (OR264-21.1, OR265-21.1, OR266-21.1, OR267-21.1, OR329-21.1, OR464-15.1 and OR465-15.1) have been deposited in the PSI Materials Repository§.
Samples for crystallization were assessed by AGF with static light-scattering detection (AGF-MALS). Protein samples at 20 mM Tris–HCl (pH 7.0), 100 mM NaCl, 5 mM DTT and 0.02% NaN3 were injected onto an analytical gel-filtration column (Shodex KW-802.5; Shodex, Japan) at room temperature. The HPLC was run on an Agilent series 1200 system at a flow rate of 0.5 ml/min. Data were then collected on a miniDAWN (TREOS) light-scattering instrument (Wyatt Technology) and refractive index (Optilab rEX). The data were analyzed with ASTRA software (Wyatt Technology, version 6.1.1.17).
Structure solution and refinement
Crystallization screening was performed using a microbatch-under-oil crystallization method at 4 °C (OR265, OR267 and OR329) or 18 °C (OR264, OR266, OR464 and OR465) [48]. After optimization, protein crystals useful for structure determination were grown in drops composed of 1.0 μl of protein and 1.0 μl of precipitant solution [40% polyethylene glycol (PEG) 1000, 0.1 M lithium chloride and 0.1 M Taps (3-{[2-hydroxy-1,1-bis(hydroxy-methyl)ethyl]amino}-1-propanesulfonic acid), pH 9.0 (OR264); 40% PEG 400, 0.1 M lithium sulfate and 0.1 M Hepes, pH 7.5 (OR265); 40% PEG 1000, 0.1 M potassium nitrate and 0.1 M Hepes, pH 7.5 (OR266); 20% PEG 4000, 0.1 M magnesium sulfate and 0.1 M Tris–HCl, pH 8.0 (OR267); 20% PEG 8000, 0.1 M magnesium nitrate and 0.1 M sodium citrate, pH 4.2 (OR329); 25% PEG 3350 and 0.1 M Bistris (2-[bis(2-hydroxyethyl)amino]-2-(hydroxy-methyl)propane-1,3-diol), pH 5.5 (OR464); 40% PEG 4000, 0.1 M potassium phosphate monobasic and 0.1 M Mes (4-morpholineethanesulfonic acid), pH 6.0 (OR465)] under paraffin oil (Hampton Research). Data sets were collected at beamline X4A (OR265, OR266, OR329 and OR464) or X4C (OR264, OR267 and OR465) at the National Synchrotron Light Source at 100 K. The diffraction data from single crystals were processed with the HKL2000 package [49]. The structures were solved by molecular replacement using program MolRep [50] and models 2XEE (OR266) and 4GPM (OR267) or using BALBES [51] and models 2XEE (OR264), 4HB5 (OR265), 4DB8 (OR329) and 3RFJ (OR464 and OR465). The models were completed using iterative cycles of manual rebuilding in Coot [52] and were refined with the program PHENIX [53]. The quality of the model was inspected by the program PROCHECK [54]. The data processing and refinement statistics are provided as Supplementary Materials.
Supplementary Material
Acknowledgments
We thank the members of the protein production facility at the Institute for Protein Design, Seattle, WA, and James Thompson and Justin Ashworth for SequenceProfile implementation in Rosetta. This work was facilitated though the use of advanced computational, storage and networking infrastructure provided by the Hyak supercomputer system at the University of Washington. For technical assistance and coordination of efforts at Northeast Structural Genomics, we thank John Everett, Gregory Kornhaber, Melissa Maglaqui and Dan Lee. This work was supported in part by Defense Threat Reduction Agency (HDTRA1-11-1-0041) and by a grant from the Protein Structure Initiative of the National Institutes of Health (U54-GM094597). F.P. was the recipient of a Swiss National Science Postdoc Fellowship and a Human Frontier Science Program Long-Term Fellowship.
Abbreviations used
- MSA
multiple sequence alignment
- AGF
analytical gel filtration
- MALS
multi-angle light scattering
- VLR
variable lymphocyte receptor
- PEG
polyethylene glycol
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2014.11.005.
Footnotes
Accession numbers
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession codes 4GPM (OR264 ank1), 4HQD (OR265 ank2), 4GMR (OR266 ank3), 4HB5 (OR267 ank4), 4HXT (OR329 arm8), 4PSJ (OR464 LRR1439) and 4PQ8 (OR465 LRR1440).
References
- 1.Jorda J, Baudrand T, Kajava AV. PRDB: Protein Repeat DataBase. Proteomics. 2012;12:1333–6. doi: 10.1002/pmic.201100534. http://dx.doi.org/10.1002/pmic.201100534. [DOI] [PubMed] [Google Scholar]
- 2.Kajava AV. Tandem repeats in proteins: from sequence to structure. J Struct Biol. 2012;179:279–88. doi: 10.1016/j.jsb.2011.08.009. http://dx.doi.org/10.1016/j.jsb.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Kobe B, Kajava AV. When protein folding is simplified to protein coiling: the continuum of solenoid protein structures. Trends Biochem Sci. 2000;25:509–15. doi: 10.1016/s0968-0004(00)01667-4. http://dx.doi.org/10.1016/S0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- 4.Kajava AV. What curves α-solenoids? Evidence for an α-helical toroid structure of Rpn1 and Rpn2 proteins of the 26S proteasome. J Biol Chem. 2002;277:49791–8. doi: 10.1074/jbc.M204982200. http://dx.doi.org/10.1074/jbc.M204982200. [DOI] [PubMed] [Google Scholar]
- 5.Filipovska A, Rackham O. Modular recognition of nucleic acids by PUF, TALE and PPR proteins. Mol Biosyst. 2012;8:699. doi: 10.1039/c2mb05392f. http://dx.doi.org/10.1039/c2mb05392f. [DOI] [PubMed] [Google Scholar]
- 6.Reichen C, Hansen S, Plückthun A. Modular peptide binding: from a comparison of natural binders to designed armadillo repeat proteins. J Struct Biol. n.d doi: 10.1016/j.jsb.2013.07.012. http://dx.doi.org/10.1016/j.jsb.2013.07.012. [DOI] [PubMed]
- 7.Grove TZ, Forster J, Pimienta G, Dufresne E, Regan L. A modular approach to the design of protein-based smart gels. Biopolymers. 2012;97:508–17. doi: 10.1002/bip.22033. http://dx.doi.org/10.1002/bip.22033. [DOI] [PubMed] [Google Scholar]
- 8.Binz HK, Stumpp MT, Forrer P, Amstutz P, Plückthun A. Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J Mol Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. http://dx.doi.org/10.1016/S0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 9.Mosavi LK, Minor DL, Peng Z. Consensus-derived structural determinants of the ankyrin repeat motif. Proc Natl Acad Sci. 2002;99:16029–34. doi: 10.1073/pnas.252537899. http://dx.doi.org/10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parmeggiani F, Pellarin R, Larsen AP, Varadamsetty G, Stumpp MT, Zerbe O, et al. Designed armadillo repeat proteins as general peptide-binding scaffolds: consensus design and computational optimization of the hydrophobic core. J Mol Biol. 2008;376:1282–304. doi: 10.1016/j.jmb.2007.12.014. http://dx.doi.org/10.1016/j.jmb.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 11.Main ERG, Xiong Y, Cocco MJ, D’Andrea L, Regan L. Design of stable α-helical arrays from an idealized TPR motif. Structure. 2003;11:497–508. doi: 10.1016/s0969-2126(03)00076-5. http://dx.doi.org/10.1016/S0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- 12.Urvoas A, Guellouz A, Valerio-Lepiniec M, Graille M, Durand D, Desravines DC, et al. Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins (αRep) based on thermostable HEAT-like repeats. J Mol Biol. 2010;404:307–27. doi: 10.1016/j.jmb.2010.09.048. http://dx.doi.org/10.1016/j.jmb.2010.09.048. [DOI] [PubMed] [Google Scholar]
- 13.Lee S-C, Park K, Han J, Lee J, Kim HJ, Hong S, et al. Design of a binding scaffold based on variable lymphocyte receptors of jawless vertebrates by module engineering. Proc Natl Acad Sci. 2012;109:3299–304. doi: 10.1073/pnas.1113193109. http://dx.doi.org/10.1073/pnas.1113193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aksel T, Majumdar A, Barrick D. The contribution of entropy, enthalpy, and hydrophobic desolvation to cooperativity in repeat-protein folding. Structure. 2011;19:349–60. doi: 10.1016/j.str.2010.12.018. http://dx.doi.org/10.1016/j.str.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker R, Mercedes-Camacho A, Grove TZ. Consensus design of a NOD receptor leucine rich repeat domain with binding affinity for a muramyl dipeptide, a bacterial cell wall fragment. Protein Sci. 2014;23:790–800. doi: 10.1002/pro.2461. http://dx.doi.org/10.1002/pro.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stumpp MT, Forrer P, Binz HK, Plückthun A. Designing repeat proteins: modular leucine-rich repeat protein libraries based on the mammalian ribonuclease inhibitor family. J Mol Biol. 2003;332:471–87. doi: 10.1016/s0022-2836(03)00897-0. http://dx.doi.org/10.1016/S0022-2836(03)00897-0. [DOI] [PubMed] [Google Scholar]
- 17.Nikkhah M, Jawad-Alami Z, Demydchuk M, Ribbons D, Paoli M. Engineering of beta-propeller protein scaffolds by multiple gene duplication and fusion of an idealized WD repeat. Biomol Eng. 2006;23:185–94. doi: 10.1016/j.bioeng.2006.02.002. http://dx.doi.org/10.1016/j.bioeng.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Baabur-Cohen H, Dayalan S, Shumacher I, Cohen-Luria R, Ashkenasy G. Artificial leucine rich repeats as new scaffolds for protein design. Bioorg Med Chem Lett. 2011;21:2372–5. doi: 10.1016/j.bmcl.2011.02.093. http://dx.doi.org/10.1016/j.bmcl.2011.02.093. [DOI] [PubMed] [Google Scholar]
- 19.Magliery TJ, Regan L. Beyond consensus: statistical free energies reveal hidden interactions in the design of a TPR motif. J Mol Biol. 2004;343:731–45. doi: 10.1016/j.jmb.2004.08.026. http://dx.doi.org/10.1016/j.jmb.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan BJ, Nguyen T, Durani V, Mathur D, Rojas S, Thomas M, et al. Stabilizing proteins from sequence statistics: the interplay of conservation and correlation in triosephosphate isomerase stability. J Mol Biol. 2012;420:384–99. doi: 10.1016/j.jmb.2012.04.025. http://dx.doi.org/10.1016/j.jmb.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. Evolutionary information for specifying a protein fold. Nature. 2005;437:512–8. doi: 10.1038/nature03991. http://dx.doi.org/10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 22.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–74. doi: 10.1016/B978-0-12-381270-4.00019-6. http://dx.doi.org/10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–8. doi: 10.1126/science.1089427. http://dx.doi.org/10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 24.Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, et al. Principles for designing ideal protein structures. Nature. 2012;491:222–7. doi: 10.1038/nature11600. http://dx.doi.org/10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang P-S, Ban Y-EA, Richter F, Andre I, Vernon R, Schief WR, et al. RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS ONE. 2011;6:e24109. doi: 10.1371/journal.pone.0024109. http://dx.doi.org/10.1371/journal.pone.0024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. http://dx.doi.org/10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrade MA, Petosa C, O’Donoghue SI, Müller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. http://dx.doi.org/10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 29.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–5. doi: 10.1093/nar/gkr931. http://dx.doi.org/10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. http://dx.doi.org/10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X-H, Chen R-C, Gao Y, Wu Y-D. The effect of Asp-His-Ser/Thr-Trp tetrad on the thermostability of WD40-repeat proteins. Biochemistry (Mosc) 2010;49:10237–45. doi: 10.1021/bi101321y. http://dx.doi.org/10.1021/bi101321y. [DOI] [PubMed] [Google Scholar]
- 32.Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2012;9:173–5. doi: 10.1038/nmeth.1818. http://dx.doi.org/10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 33.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci. 1998;95:5857–64. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. http://dx.doi.org/10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. http://dx.doi.org/10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Mol Biol. 2003;10:980–980. doi: 10.1038/nsb1203-980. http://dx.doi.org/10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 37.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. http://dx.doi.org/10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Söding J. Protein homology detection by HMM–HMM comparison. Bioinformatics. 2005;21:951–60. doi: 10.1093/bioinformatics/bti125. http://dx.doi.org/10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–637. doi: 10.1002/bip.360221211. http://dx.doi.org/10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 40.Fleishman SJ, Leaver-Fay A, Corn JE, Strauch E-M, Khare SD, Koga N, et al. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS ONE. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. http://dx.doi.org/10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheffler W, Baker D. RosettaHoles: rapid assessment of protein core packing for structure prediction, refinement, design, and validation. Protein Sci. 2009;18:229–39. doi: 10.1002/pro.8. http://dx.doi.org/10.1002/pro.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue–residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci. 2013;110:15674–9. doi: 10.1073/pnas.1314045110. http://dx.doi.org/10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Batchelor AH, Piper DE, de la Brousse FC, McKnight SL, Wolberger C. The structure of GABPα/β: an ETS domain—ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–41. doi: 10.1126/science.279.5353.1037. http://dx.doi.org/10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 44.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. http://dx.doi.org/10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 45.Xiao R, Anderson S, Aramini J, Belote R, Buchwald WA, Ciccosanti C, et al. The high-throughput protein sample production platform of the Northeast Structural Genomics Consortium. J Struct Biol. 2010;172:21–33. doi: 10.1016/j.jsb.2010.07.011. http://dx.doi.org/10.1016/j.jsb.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jansson M, Li YC, Jendeberg L, Anderson S, Montelione GT, Nilsson B. High-level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. J Biomol NMR. 1996;7:131–41. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- 47.Doublié S, Kapp U, Åberg A, Brown K, Strub K, Cusack S. Crystallization and preliminary X-ray analysis of the 9 kDa protein of the mouse signal recognition particle and the selenomethionyl-SRP9. FEBS Lett. 1996;384:219–21. doi: 10.1016/0014-5793(96)00316-x. http://dx.doi.org/10.1016/0014-5793(96)00316-X. [DOI] [PubMed] [Google Scholar]
- 48.Chayen NE, Shaw Stewart PD, Maeder DL, Blow DM. An automated system for micro-batch protein crystallization and screening. J Appl Crystallogr. 1990;23:297–302. http://dx.doi.org/10.1107/S0021889890003260. [Google Scholar]
- 49.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. In: Charles W, Carter J, editors. Methods Enzymol. Vol. 276. Academic Press; 1997. pp. 307–26. [DOI] [PubMed] [Google Scholar]
- 50.Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J Appl Crystallogr. 1997;30:1022–5. http://dx.doi.org/10.1107/S0021889897006766. [Google Scholar]
- 51.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr Sect D Biol Crystallogr. 2008;64:125–32. doi: 10.1107/S0907444907050172. http://dx.doi.org/10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. http://dx.doi.org/10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr Sect D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. http://dx.doi.org/10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 54.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–91. http://dx.doi.org/10.1107/S0021889892009944. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.