Abstract
Purpose.
Intraocular pressure (IOP) is an important clinical parameter in the evaluation of ocular health. Elevated IOP is a major risk factor for primary open-angle glaucoma (POAG). The goal of this study was to identify rare and less common variants that influence IOP.
Methods.
We performed an exome array analysis in a subset of 1660 individuals from a population-based cohort, the Beaver Dam Eye Study. Associations with IOP were tested on 45,849 single nucleotide variants and 12,390 autosomal genes across the genome.
Results.
Intraocular pressure was suggestively associated with novel variants located in FAR2 at 12p11.22 (rs4931170, P = 1.2 × 10−5), in GGA3 at 17q25.1 (rs52809447, P = 6.7 × 10−5), and in PKDREJ at 22q13.31 (rs7291444, P = 7.4 × 10−5). Gene-based analysis found suggestive associations between IOP and the genes HAP1, MTBP, FREM3, and PHF12. We successfully replicated the associations with GAS7 (P = 7.4 × 10−3) for IOP, and also identified a previously reported POAG locus in the CAV1/CAV2 region to be associated with IOP (P = 3.3 × 10−3). This association was confirmed in a meta-analysis with three published genome-wide association studies (Pcombined = 4.0 × 10−11).
Conclusions.
Our results suggest that novel genetic variants and genes with multiple, less common variants may play a role in the control of IOP. The implication of the caveolin genes, CAV1/CAV2, as a common genetic factor influencing both IOP variations and POAG may provide new insights of the underlying mechanism leading to glaucoma and glaucomatous visual field loss.
Keywords: exome, SNP, intraocular pressure
Using an exome array, we evaluated rare and low-frequency variants for association with intraocular pressure. We identified an association with the CAV1/CAV2 susceptibility locus, a region known to be associated with primary open angle glaucoma.
Introduction
Intraocular pressure (IOP) is an important physiologic characteristic in maintaining the structure and function of the eye. Normal IOP is regulated by the balance between the production and the drainage of aqueous humor through the trabecular meshwork in the anterior chamber angle of human eyes. Elevated IOP contributes to the accelerated death of retinal ganglion cells and the damage of the optic nerve, and ultimately results in progressive visual field loss and irreversible blindness in glaucoma patients.1 It is a major risk factor of glaucomatous optic neuropathy, and the only target for current glaucoma therapy. Clinical trials have shown that IOP-lowering treatments in eyes with ocular hypertension are effective both in preventing glaucoma onset and delaying its progression.2,3
Intraocular pressure is a heritable polygenic trait with environmental influences.4 The heritability for IOP is estimated to range from 0.30 to 0.42 in populations of European ancestry,4–7 suggesting genetics may play an important role in IOP. Genome-wide studies have located regions on chromosome 10q22 and chromosome 19p with significant genetic linkage to IOP in family-based studies.8,9 In addition, genome-wide association studies (GWAS) have identified multiple common single nucleotide polymorphisms (SNPs) associated with IOP,10–13 some of which are in the overlapping GWAS regions for primary open-angle glaucoma (POAG).14,15 The shared association variants for IOP and glaucoma suggest a possible common pathway for this endophenotype and disease.
The development of exome arrays that target protein-coding variation offers new opportunities to assess the role of rare and low-frequency coding variants in human complex traits. The primary goal of this study was to identify rare (minor allele frequency (MAF) < 1.0%) and less common (1.0% ≤ MAF ≤ 5%)16 variants associated with IOP and to replicate previously reported GWAS loci for IOP in a population-based cohort of European ancestry from the Beaver Dam Eye Study (BDES).
Methods
Study Population
The BDES is a population-based cohort study established in Beaver Dam, Wisconsin, in 1987.17 From 1987 to 1988, the BDES completed a census of the city and township of Beaver Dam and identified 3715 households with at least one occupant aged from 43 to 84 years. Of the 5925 individuals eligible to enroll, 4926 (83.14%) individuals underwent an ocular examination and a personal history questionnaire during 1988–1990.18 Pedigrees have been constructed for 2783 participants with known familial relationships in the catchment area of the study.4 To improve the sampling efficiency while maintaining the full spectrum of the phenotypes, we selected individuals from the full BDES cohort and included those with extreme baseline IOP or refractive error measurements. This sampling resulted in a random sample with the full spectrum of IOP phenotype represented, since refractive error extremes were also included (Supplementary Fig. S1). The study followed the recommendations of the Declaration of Helsinki. Informed consent was obtained from all study participants and the institutional review board at the University of Wisconsin approved all protocols.
Clinical Evaluation
Intraocular pressure was measured with a Goldmann applanation tonometer.18 Unreliable measurements, as assessed by the trained observers, were excluded from the analysis. Blood pressure was recorded as the mean of the second and third measurements according to the Hypertension Detection and Follow-up Program protocol.19 A detailed medical history, including information about hypertension, diabetes, and other medical conditions, and history of medication use were obtained from each participant.
Phenotype
The higher IOP measurement of the two eyes at the baseline visit was used in these analyses. The distribution of intraocular pressure was approximately normal but slightly right skewed (Supplementary Fig. S1). To reduce the impact of extreme outliers, we winsorized the data.20 All individuals with the trait more extreme than 5 standard deviations from the mean were assigned the values at 5 standard deviations from the mean (Supplementary Fig. S1). Before the association analysis, the winsorized trait was first adjusted in a multiple linear regression model for potential confounding variables including age, sex, systolic blood pressure, and IOP treatment. The residual IOP values were used for the association analysis. In this study, we present the adjusted IOP values (sum of the residuals and the intercept from the regression model) with mean centered age and systolic blood pressure.
Genotyping and Quality Controls
A total of 1908 BDES participants underwent genotyping with the Illumina HumanExome BeadChip (San Diego, CA, USA). Genotyping was performed at the Genetic Resources Core Facility at Johns Hopkins Institute of Genetic Medicine. Genotyping calling was done with Illumina's GenTrain clustering algorithm in GenomeStudio.
Standard quality control measures were used. Samples with sex inconsistencies that could not be easily resolved (n = 15) were excluded. All samples had a call rate > 98%. We assessed the cryptic relatedness on the basis of pairwise identity by descent (IBD) sharing and removed one individual from each pair of samples that represented unexpected duplicates or first- and second-degree relatives (IBD sharing > 20%, n = 126). Samples with unavailable IOP measurement of either eye or missing values of any covariates were also excluded (n = 107). Of the 242,901 genotyped variants, we excluded nonautosomal variants (n = 5465), variants with genotyping call rate < 98% (n = 6038), as well as monomorphic variants (n = 130,783). The concordance rate with 44 HapMap controls was 99.82% and the genotype concordance among 21 masked duplicate sample pairs was 99.99%. Since departures from Hardy-Weinberg equilibrium (HWE) can be a result of evolutionary forces, genotyping errors, population admixture, and marker-trait associations,21 we evaluated HWE on the final set of variants, and reported the P value estimated from the HWE exact test for significant associations. Our final data set includes 1660 individuals and 100,615 polymorphic variants.
The BDES population self-reported as primarily European-American. To evaluate the genetic ancestry we performed principal component analysis (PCA) by using SMARTPCA in EIGENSTRAT22 with six HapMap phase III populations (Supplementary Fig. S2). There was no increase in genomic inflation (λ = 1.007) (Supplementary Fig. S3).
Single-Variant Analysis
We tested single variants for association with IOP under an additive linear regression model in PLINK.23 We considered rare (0.3% ≤ MAF < 1%, n = 10,744) as well as low-frequency and common variants (MAF ≥ 1%, n = 35,093) in the association analysis. However, given the sample size of this study, variants with a MAF < 1% may be prone to bias. Variants with a MAF ≥ 1% correspond to at least 33 copies of the minor allele in the analyzed cohort, which should allow stable estimates of sampling errors and provides adequate statistical power to detect low-frequency variants with effect size (β) greater than 3.5 at α = 0.0001 (estimated in QUANTO24).
Linkage disequilibrium (LD)–based pruning identified 33,056 variants with pairwise correlation < 0.20. We set the significance threshold across the exome to P < 1.5 × 10−6, corresponding to a Bonferroni correction for these 33,056 independent tests.
Replication of Published GWAS Loci
Genome-wide association studies have identified seven loci associated with IOP and/or POAG. For IOP these include TMCO1 (1q22-q25),10,11,14 GLCCI1/ICA1 (7p21),12 MVB12B (9q33.3),13 and GAS7 (17p13.1).10,11 For POAG these include CAV1/CAV2 (7q31),15,25,26 CDKN2B-AS1 (9p21.3),14,27–29 and SIX1/SIX6 (14q23).27,29 For replication, we considered (1) the exact variant, and (2) variants in strong LD (r2 > 0.70) with the reported variant in the same GWAS loci. For variants in previously reported IOP loci, associations were considered as replicated if the P value was <0.01. For variants in previously reported POAG loci, we evaluated their associations with IOP in our study and conducted a meta-analysis on those with P < 0.01, combining our results with published studies for IOP in a fixed-effect model using an inverse sample-size weighted approach implemented in METAL.30
Gene-Based Analysis
We performed gene-based analysis by using SKAT-O implemented in R.31–33 The residual IOP values were regressed on rare and less common variants (MAF ≤ 5%) in a gene region, allowing them to have different directions and magnitude of effects. Gene regions with at least two variants in desired MAF range were included. We defined the cumulative minor allele frequency (CMAF) as the sum of the MAF of all variants in a gene region. To ensure accurate statistical inference for this sample size, we focused this analysis on gene regions with CMAF ≥ 1.0%, although we present associations for all CMAF. A total of 12,390 genes (75,936 variants) across the autosomal chromosomes were tested in gene-based analysis. To optimize the statistical power, we applied the default weight
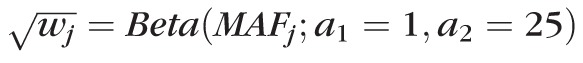 in SKAT-O, which up-weights the rare variants. The significance threshold across all identified gene regions was set to a Bonferroni corrected P < 4.0 × 10−6 (n = 12,390 tests).
in SKAT-O, which up-weights the rare variants. The significance threshold across all identified gene regions was set to a Bonferroni corrected P < 4.0 × 10−6 (n = 12,390 tests).
Annotations
Annotation was performed by using SeattleSeq Annotation Server 138 version 9.01 under GRCh37/hg19 (http://snp.gs.washington.edu/SeattleSeqAnnotation138/; provided in the public domain by the University of Washington, Seattle, WA, USA). Except for 57 insertions and deletions, all variants that passed quality control filters were annotated.
Results
Study Participants
The characteristics of the BDES participants for the analyzed cohort and for the entire population are summarized in Table 1. Females constituted 57.3% of the analyzed cohort and the mean age was 60 years for both men and women. The distribution of IOP in our cohort had a mean 15.6 mm Hg with standard deviation 4.1 mm Hg, which is comparable to other large population-based cohorts of European descent and to the full BDES study. The mean IOP was 15.9 mm Hg in the full BDES study (Table 1); it was 16 and 15.4 mm Hg in the Blue Mountains Eye Study34 and the NEIGHBOR consortium,35 respectively. Intraocular pressure was normally distributed with a similar mean and median measurement (15.6 mm Hg versus 15.0 mm Hg, respectively). After winsorization and covariate adjustment, the distribution of adjusted IOP remained approximately normal (Supplementary Fig. S1). In all, 8.3% of the individuals had elevated IOP, defined as >21 mm Hg, and 2.5% had reported IOP medical treatment. There were 45 individuals with self-reported glaucoma diagnosis at either eye.
Table 1.
The Characteristics of the Beaver Dam Eye Study Participants
|
Characteristic |
BDES Exome |
BDES Full* |
PValue |
| No. of individuals | 1660 | 4580 | -- |
| Age, mean ± SD (range), y | 60.1 ± 10.8 (43–86) | 61.7 ± 11.0 (43–86) | <0.001 |
| Female sex, % (N) | 57.3 (951) | 56.1 (2572) | 0.40 |
| IOP†, mean ± SD (range), mm Hg | 15.6 ± 4.1 (6–63) | 15.9 ± 3.5 (4–63) | 0.003 |
| IOP > 21 mm Hg†, % (N) | 8.3 (138) | 5.7 (261) | <0.001 |
| Participants with IOP treatment, % (N) | 2.5 (42) | 2.6 (120) | 0.92 |
| Participants with glaucoma‡, % (N) | 2.7 (45) | 2.7 (124) | 1.00 |
| Systolic BP, mean ± SD (range), mm Hg | 130.1 ± 20.6 (89–248) | 131.8 ± 20.3 (71–248) | 0.004 |
| Diastolic BP, mean ± SD (range), mm Hg | 77.4 ± 10.8 (44–123) | 77.4 ± 10.9 (42–126) | 0.86 |
BP, blood pressure.
All BDES individuals with reliable IOP measurements of both eyes at baseline visit.
Higher baseline IOP measurements of the two eyes.
Self-reported glaucoma diagnosis at either eye.
Single-Variant Associations
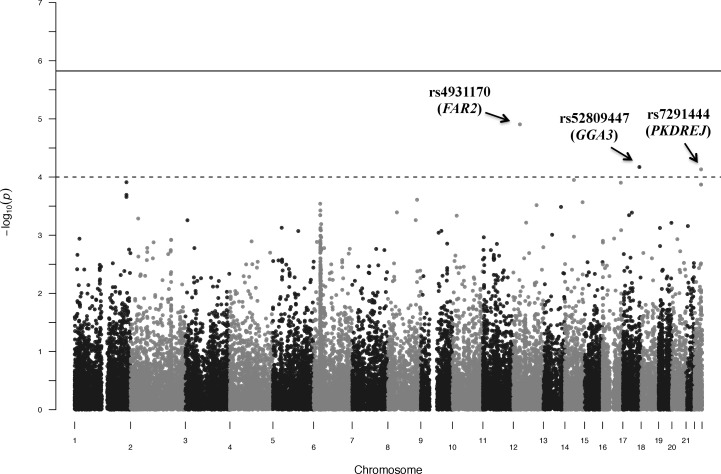
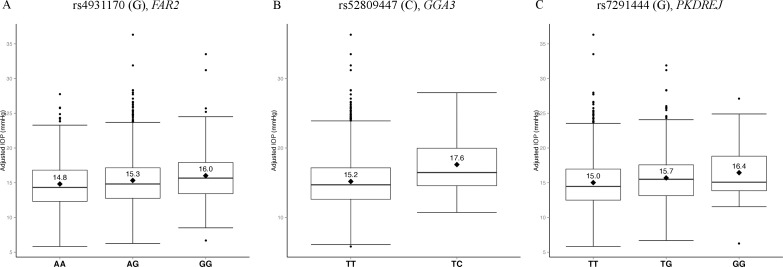
We tested 35,093 low-frequency and common variants (MAF ≥ 1%) individually for association with IOP (Fig. 1) and no variants met the threshold for significance (P < 1.5 × 10−6) (Table 2). However, we did identify suggestive associations. A common variant (rs4931170, MAF = 38.3%) on chromosome 12p11.22 in the intronic region of the fatty acyl-CoA reductase 2 gene (FAR2) was associated with elevated IOP. Each copy of the minor allele G of rs4931170 led to 0.58–mm Hg (95% confidence interval [CI] = 0.33–0.83, P = 1.2 × 10−5) increase in IOP (Fig. 2A).
Figure 1.
Manhattan plot of the single-variant associations for IOP (MAF ≥ 1.0%). A total of 35,093 variants with MAF ≥ 1.0% were tested for single-variant associations. The significance threshold across the exome was set to P < 1.5 × 10−6 (solid line). The associations of three variants reached the suggestive threshold of P < 10−4 (dash line): rs4931170 (P = 1.2 × 10−5), rs52809447 (P = 6.7 × 10−5), and rs7291444 (P = 7.4 × 10−5).
Table 2.
Top Single-Variant Association Results (P < 10−4)
|
Gene |
Chr:Position* |
dbSNP ID |
Amino Acid Alternation† |
MAF |
β (SE)‡ |
PValue |
HWEP |
| MAF ≥ 1.0% | |||||||
| FAR2 | 12:29388772 | rs4931170 | Intron | 38.3% [G] | 0.58 (0.13) | 1.2 × 10−5 | 0.1907 |
| GGA3 | 17:73238508 | rs52809447 | p.Glu97Gly | 1.08% [C] | 2.43 (0.61) | 6.7 × 10−5 | 1.0000 |
| PKDREJ | 22:46656246 | rs7291444 | p.Thr992Pro | 15.8% [G] | 0.69 (0.17) | 7.4 × 10−5 | 0.7195 |
| MAF 0.3%–1.0% | |||||||
| FAM83H | 8:144811340 | rs189033490 | p.His201Tyr | 0.30% [A] | 5.25 (1.15) | 4.9 × 10−6 | 1.0000 |
| MTBP | 8:121530092 | rs61753798 | p.Leu750Phe | 0.63% [T] | 3.28 (0.76) | 1.6 × 10−5 | 0.0617 |
| FREM3 | 4:144532644 | rs72938299 | p.Phe1939Ser | 0.48% [G] | 3.89 (0.91) | 1.9 × 10−5 | 1.0000 |
| FREM3 | 4:144542213 | rs17017968 | p.Ile1919Thr | 0.48% [G] | 3.89 (0.91) | 1.9 × 10−5 | 1.0000 |
| FREM3 | 4:144546704 | rs7679078 | Intron | 0.45% [G] | 4.01 (0.94) | 2.0 × 10−5 | 1.0000 |
| WNK2 | 9:96070669 | rs61753907 | p.Val2144Ile | 0.30% [A] | 4.65 (1.15) | 5.2 × 10−5 | 1.0000 |
| RNASET2 | 6:167343204 | rs41269593 | p.Glu215Lys | 0.48% [T] | 3.63 (0.91) | 6.7 × 10−5 | 1.0000 |
| RBFOX1 | 16:5781710 | rs8060283 | Intron | 0.48% [G] | 3.59 (0.91) | 8.2 × 10−5 | 1.0000 |
dbSNP, Single Nucleotide Polymorphism Database; Chr, chromosome; SE, standard error.
Chr:Position reported in GRCh37.
Amino acid alteration reported in dbSNP.
Intraocular pressure measurements were winsorized and adjusted for age, sex, systolic blood pressure, and IOP treatment.
Figure 2.
Box plots of IOP by genotype at three associated loci. Distributions of adjusted IOP values were plotted for each observed genotype at three suggestive loci: (A) rs4931170 of FAR2, (B) rs52809447 of GGA3, and (C) rs7291444 of PKDREJ. Genotypes are shown on x-axis. The mean adjusted IOP value for each genotype is marked by a diamond with the value listed above. The three horizontal bars from the bottom to the top indicate the 25th, 50th, and 75th percentile of the adjusted IOP values. The adjusted IOP values beyond the 25th and 75th percentile range are represented by black dots. For all three loci, each copy of its minor allele is associated with increased mean IOP.
Additionally, a low-frequency coding variant (rs52809447, MAF = 1.08%) that results in the substitution of glutamic acid with glycine (p.Glu97Gly) on chromosome 17q25.1 in the exonic region of Golgi-associated, gamma adaptin ear containing, ARF-binding protein 3 gene (GGA3) was associated with IOP. Individuals with one copy of the minor allele C have on average an IOP 2.43 mm Hg (95% CI = 1.23–3.63, P = 6.7 × 10−5) higher than noncarriers (Fig. 2B). For this low-frequency variant there were no individuals with two copies of the C allele in this study.
The third suggestive locus for IOP was a common variant on chromosome 22q13.31 (rs7291444, MAF = 15.8%), substituting a threonine with a proline (p.Thr992Pro) in the polycystin (PKD) family receptor for egg jelly gene (PKDREJ). Each copy of the minor allele G of rs7291444 was associated with 0.69–mm Hg (95% CI = 0.36–1.02, P = 7.4 × 10−5) increase in IOP (Fig. 2C).
Analysis of rare variants (0.3% ≤ MAF < 1.0%) identified eight variants in six genes that had large effects on IOP (β > 3.0) with suggestive significance (Table 2). Variants in three of these gene regions, MTBP (rs61753798), FREM3 (rs72938299, rs17017968, and rs7679078), and RNASET2 (rs41269593), were also identified in gene-based analyses.
Replication of Published GWAS Loci for IOP
Except for the MVB12B and GLCCI1/ICA1 region, our exome array contained either the exact variant or at least one variant in strong LD for five published GWAS loci: TMCO1 and GAS7 for IOP, and CAV1/CAV2, CDKN2B-AS1, and SIX1/SIX6 for POAG (Table 3).
Table 3.
Association Results for Genetic Variants at Published IOP/POAG GWAS Loci
|
Chr* |
GWAS Loci |
Reported Variant† |
Correlated Variant (r2)‡ |
MAF |
β (SE) |
PValue |
| IOP | ||||||
| 1q22-q25 | TMCO1 | rs4656461 | rs4656461 (1.00) | 12.9% [G] | 0.35 (0.19) | 6.8 × 10−2 |
| 7p21 | GLCCI1/ICA1 | rs59072263 | -- | -- | -- | -- |
| 9q33.3 | MVB12B | rs2286885 | -- | -- | -- | -- |
| 17p13.1 | GAS7 | rs12150284 | rs9897130 (0.70) | 48.4% [T] | −0.33 (0.12) | 7.4 × 10−3 |
| POAG | ||||||
| 7q31 | CAV1/CAV2 | rs4236601 | rs4236601 (1.00) | 25.7% [A] | 0.42 (0.14) | 3.3 × 10−3 |
| 9p21.3 | CDKN2B-AS1 | rs4977756 | rs2157719 (0.75) | 45.2% [C] | 0.02 (0.13) | 8.9 × 10−1 |
| 14q23 | SIX1/SIX6 | rs10483727 | rs10483727 (1.00) | 39.1% [T] | 0.10 (0.13) | 4.3 × 10−1 |
Gene locations reported in GRCh37.
The top variant reported in published GWAS.
Exact variant or top variant in strong LD with the reported GWAS variant (r2 > 0.70 in 1000 Genome Pilot 1 population of European ancestry).
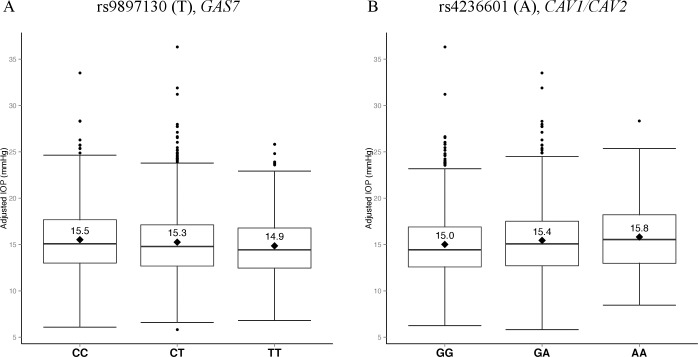
We successfully replicated the previously reported association in growth arrest–specific 7 gene (GAS7) region for IOP. GAS7 on chromosome 17p13.1 was first identified as a susceptibility locus for IOP in a meta-analysis of a large discovery cohort from the Netherlands.10 The minor allele A of the lead variant rs11656696 in GAS7 region was associated with lower IOP (MAF = 43%, β = −0.26, P = 9.8 × 10−9).10 An independent meta-analysis later identified an association in the GAS7 region on a different variant rs12150284 (β = −0.49, P = 2.4 × 10−6), a locus 2.6-kb upstream from rs11656696 (r2 = 0.62).11 Neither of these variants was genotyped in our data set. In our study, rs9897130 was the top GAS7-associated SNP (Fig. 3A), a locus that is highly correlated with rs12150284 (r2 = 0.70) and is also associated with lower IOP (MAF = 48.4%, β = −0.33, P = 7.4 × 10−3). In these independent studies, all three variants in the GAS7 region showed associations with lower IOP and similar magnitudes of effects.
Figure 3.
Box plots of IOP for genotypes at rs9897130 (GAS7) and rs4236601 (CAV1/CAV2). Distributions of the adjusted IOP values were plotted for each observed genotypes at rs9897130 of GAS7 and rs4236601 of CAV1/CAV2. After adjusting for age, sex, systolic blood pressure, and treatment for IOP, (A) the minor allele T of rs9897130 is associated with a 0.33–mm Hg decrease in mean IOP (P = 7.4 × 10−3), and (B) the minor allele A of rs4236601 is associated with a 0.42–mm Hg increase in mean IOP (P = 3.3 × 10−3).
Meta-Analysis of Published GWAS Loci for POAG
Among the variants in published GWAS loci for POAG, rs4236601 on chromosome 7q31, which contains the caveolin genes CAV1 and CAV2, showed a significant association with IOP (Table 3). This locus was first discovered with POAG in an Icelandic population (odds ratio [OR] = 1.36, 95% CI = 1.23–1.50, P = 5.0 × 10−10)15 and was later replicated in two independent studies of POAG in populations of European ancestry.25,26 In these POAG studies, cases consist of both normal-tension and high-tension POAG patients, but Wiggs et al.25 have shown no association among high-tension POAG individuals for rs4236601. In our study the minor allele A of variant rs4236601 was associated with elevated IOP (Fig. 3B; β = 0.42, P = 3.3 × 10−3) and is consistent in direction with these other studies.
Our finding is in agreement with three GWAS of IOP for rs4236601 in European ancestry populations (van Koolwijk et al.10: MAF = 29%, β = 0.19, P = 1.1 × 10−4; Ozel et al.11: β = 0.38, P = 2.8 × 10−4; Strange et al.12: P = 1.5 × 10−3). We performed a meta-analysis combining our results with these three studies and identified an overall genome-wide significant association for IOP at rs4236601 (Table 4, Pcombined = 4.0 × 10−11) in the CAV1/CAV2 gene region (Supplementary Fig. S4).
Table 4.
Meta-Analysis for SNP rs4236601 at the CAV1/CAV2 Locus
|
dbSNP ID |
Chr:Position* |
Strange et al.12† |
van Koolwijk et al.10† |
Ozel et al.11 |
BDES Exome |
Pcombined |
||||
|
N |
PValue |
N |
PValue |
N |
PValue |
N |
PValue |
|||
| rs4236601 | 7:116162729 | 2,765 | 1.5 × 10−3 | 11,972 | 1.1 × 10−4 | 6,236 | 2.8 × 10−4 | 1,660 | 3.3 × 10−3 | 4.0 × 10−11 |
Chr:Position reported in GRCh37.
Association reported in the discovery cohort.
Gene-Based Associations
Gene-based tests offer an alternative to single-variant analysis, which is often underpowered to detect associations with rare variants. Among the 6900 genes with a CMAF ≥ 1%, no gene regions were significant (P < 4.0 ×10−6). Suggestive associations between IOP and four gene regions were identified (Table 5): the huntingtin-associated protein 1 gene (HAP1) on chromosome 17q21.2-q21.3 (CMAF = 2.11%, P = 9.3 × 10−6), the MDM2-binding protein gene (MTBP) on chromosome 8q24.12 (CMAF = 1.26%, P = 2.6 × 10−5), the FRAS1-related extracellular matrix 3 gene (FREM3) on chromosome 4q31.21 (CMAF = 4.49%, P = 9.4 × 10−5), and the PHD finger protein 12 gene (PHF12) on chromosome 17q11.2 (CMAF = 1.14%, P = 9.6 × 10−5).
Table 5.
Top Gene-Based Association Results (P < 10−4)
|
Gene |
Location* |
PValue |
CMAF, %† |
Amino Acid Alteration (N)‡ |
| CMAF ≥ 1.0% | ||||
| HAP1 | 17q21.2-q21.3 | 9.3 × 10−6 | 2.11 | p.Ala480Val (38), p.Arg116Trp (20), p.Pro187His (9), p.Ala22Ser (2), p.Pro504Leu (1) |
| MTBP | 8q24.12 | 2.6 × 10−5 | 1.26 | p.Leu750Phe (21), p.Leu478Val (7), p.Tyr144Cys (6), p.Arg736Trp (3), p.Ser711Leu (2), p.Arg411Lys (1), p.Asn431Ser (1), p.Lys179Glu (1) |
| FREM3 | 4q31.21 | 9.4 × 10−5 | 4.49 | p.Leu82Phe (47), p.Thr746Ile (18), p.Phe1939Ser (16), p.Ile1919Thr (16), Intron (15), p.Tyr1478Asn (9), p.Gln480Lys (9), p.Ile2127Val (7), p.Val546Phe (7), p.Ile731Thr (2), p.Phe1073Leu (1), p.Val2077Met (1), p.Arg1945Cys (1) |
| PHF12 | 17q11.2 | 9.6 × 10−5 | 1.14 | p.Asn737Ser (36), p.Leu979Pro (2) |
| CMAF < 1.0%§ | ||||
| MFAP2 | 1p36.1-p35 | 6.2 × 10−8 | 0.06 | p.Gln33Pro (1), p.Val130Ile (1) |
| SLC30A2 | 1p35.3 | 1.6 × 10−5 | 0.09 | p.Arg291His (2), p.Arg340His (1) |
| STXBP4 | 17q22 | 2.1 × 10−5 | 0.09 | p.Ile146Val (2), p.Pro143Ser (1) |
| TFEB | 6p21 | 4.4 × 10−5 | 0.12 | p.Glu240Lys (2), p.Ser415Asn (1), p.Phe439Leu (1) |
| ZFP36L2 | 2p22.3-p21 | 4.9 × 10−5 | 0.06 | p.Pro247 (1), p.His62Gln (1) |
| LDB1 | 10q24-q25 | 5.0 × 10−5 | 0.06 | p.Asn87Ser (1), p.Ala386Thr (1) |
| MNT | 17p13.3 | 5.1 × 10−5 | 0.06 | p.Thr319Met (1), p.Lys365Thr (1) |
| DPEP1 | 16q24.3 | 6.8 × 10−5 | 0.27 | p.Ala402Thr (7), p.Arg378Ser (1), p.Gly4Arg (1) |
| RNASET2 | 6q27 | 9.6 × 10−5 | 0.60 | p.Glu177Lys (16), p.Pro178 (4) |
Gene locations reported in GRCh37.
Minor allele frequency sum over all variants in the gene region.
Amino acid alteration reported in dbSNP. N = No. of minor allele copies in the analyzed cohort.
Gene regions contain at least two variants with MAF ≤ 5% (CMAF > 0.05%).
For the 5490 gene regions with a CMAF < 1.0% (Table 5), the microfibrillar-associated protein 2 gene (MFAP2) on chromosome 1p36.1–p35 reached significance (CMAF = 0.6%, P = 6.2 × 10−8). Two singleton variants were identified in this gene: one missense mutation (p.Val130Ile) in an individual with an IOP of 36 mm Hg and the other missense mutation (p.Gln33Pro) in an individual with an IOP of 10 mm Hg. As with MFAP2, other reported gene regions with a CMAF < 1.0% contained mostly singleton and doubleton variants that were seen in only a few individuals in our study (Table 5).
Discussion
In this study, we performed an exome array analysis on 1660 BDES participants to identify rare and less common variants influencing intraocular pressure. We successfully replicated the associations of GAS7 in our analysis. Our meta-analysis identified a novel significant association of IOP with CAV1/CAV2, a region previously identified to be a POAG locus. We also reported suggestive associations for three novel variants in FAR2, GGA3, and PKDREJ and four gene regions (HAP1, MTBP, FREM3, and PHF12) with rare and low-frequency variants.
Our results lend support to a role for CAV1/CAV2 in the pathogenesis of elevated IOP. Proteins encoded by caveolin-1 (CAV1) and caveolin-2 (CAV2) genes are major components of the caveolae plasma membranes. Caveolin-1 is expressed in several retinal cell types, including photoreceptor, retinal vascular endothelia cells, Müller glia, and retinal pigment epithelium cells, and has been linked to ocular pathologic processes including autoimmune uveitis, diabetic retinopathy, and POAG.36 The role of caveolin-1 in the retina is largely unknown. Genetic ablation of caveolin-1 might cause retinal functional deficits due to disruptions in microenvironmental homeostasis36 and blood–retina barrier breakdown.37 A cytotoxic agent that increases IOP and aqueous outflow resistance in mice can also increase the expression of caveolin-1 in the treated human trabecular meshwork cells.38 In a study by Elliot and colleagues (Elliott MH, et al. IOVS 2014;55:ARVO E-Abstract 2888), Cav-1 knockout mice experience prolonged elevation of IOP and significant reduction in pressure-dependent outflow, accompanied with morphologic change of caveolae in endothelial cells, suggesting CAV-1 may have a role in IOP regulation. Further investigations are necessary to elucidate the potential mechanisms of caveolin-1 and caveolin-2 in controlling IOP and its relationship with POAG. But this finding of a gene significantly associated with both IOP, an endophenotype, and POAG may yield new understandings of the underlying mechanisms leading to glaucoma and glaucomatous visual field loss.
Our suggestive SNP-associated genes may also have biologic relevance. FAR2 belongs to the short-chain dehydrogenase/reductase superfamily whose protein products convert fatty acyl-CoA into fatty alcohols in wax biosynthesis, and the expression of FAR2 was found to be highest in the eyelid.39 The products of the GGA family, including GGA3, regulate the protein trafficking between the trans-Golgi network and the lysosome, and have a significant role in Alzheimer disease pathogenesis, contributing to the increased accumulation of the amyloid-β protein (Aβ).40
The gene-based tests are helpful in evaluating a gene whose causal variants may have different directionality and frequency. Interestingly, another gene identified in our gene-based analysis, HAP1, which encodes a Huntington's disease–associated protein, may also play a role in the regulation of Aβ protein levels in neurons by controlling intracellular trafficking.41 FREM3 is a member of an extracellular matrix protein family and has a strong implication in Fraser syndrome. Its analogous protein in mice presents high expression levels in a variety of retinal cells during eye development,42 lending biologic plausibility to the associations in our study. The MFAP2 gene association is most relevant. As a component of elastic microfibrils, MFAP2 is involved in the pathogenesis of exfoliation syndrome and exfoliative glaucoma.43 However, the cumulative minor allele frequency for MFAP2 is low (<1%) and we are cautious in interpreting the significance of these associations in the BDES cohort. These gene-based associations reflect extremely rare variants in this population (singletons and doubletons), and require much larger sample sizes, as well as exome or whole-genome sequencing, to fully capture the dispersion of variants.
The lack of significant associations in our study may be due to insufficient power. At a significance level of 0.0001, the single-variant analysis had >80% power to detect large effects (β > 3.5) for variants with 1% MAF, but was underpowered for low-frequency variants with moderate or small effect sizes. The design of the exome array, which is constrained to protein-altering variants, has resulted in a limited number of polymorphic variants when the sample size is moderate. In our analyzed cohort of 1660 individuals, almost 70% of the variants in the genotyping assay were monomorphic or singletons. The remaining variants across the whole genome led to a mean coverage of six variants in each gene region. Although statistical approaches such as SKAT and SKAT-O are designed to detect the effects of low-frequency and rare variants,32 the power of these approaches are diminished when the small number of variants in the sample population cannot provide adequate gene coverage.
Additional studies and data across populations are warranted and encouraged for a comprehensive assessment of the rare and low-frequency variants associated with IOP. These studies, coupled with existing genome-wide association studies of common variants, may start to elucidate the causal alleles and mechanisms for IOP and its association with POAG.
Acknowledgments
We thank the participants of the Beaver Dam Eye Study.
Supported by the National Eye Institute of the National Institutes of Health under Grants R01EY021531 and U10006594.
Disclosure: F. Chen, None; A.P. Klein, None; B.E.K. Klein, None; K.E. Lee, None; B. Truitt, None; R. Klein, None; S.K. Iyengar, None; P. Duggal, None
References
- 1. Garcia-Valenzuela E, Shareef S, Walsh J, Sharma SC. Programmed cell death of retinal ganglion cells during experimental glaucoma. Exp Eye Res. 1995; 61: 33–44. [DOI] [PubMed] [Google Scholar]
- 2. Leske MC, Heijl A, Hussein M, et al. Factors for glaucoma progression and the effect of treatment: the early manifest glaucoma trial. Arch Ophthalmol. 2003; 121: 48–56. [DOI] [PubMed] [Google Scholar]
- 3. The Advanced Glaucoma Intervention Study (AGIS): 4—comparison of treatment outcomes within race: seven-year results. Ophthalmology. 1998; 105: 1146–1164. [DOI] [PubMed] [Google Scholar]
- 4. Duggal P, Klein AP, Lee KE, et al. A genetic contribution to intraocular pressure: the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 2005; 46: 555–560. [DOI] [PubMed] [Google Scholar]
- 5. van Koolwijk LM, Despriet DD, Van duijn CM, et al. Genetic contributions to glaucoma: heritability of intraocular pressure, retinal nerve fiber layer thickness, and optic disc morphology. Invest Ophthalmol Vis Sci. 2007; 48: 3669–3676. [DOI] [PubMed] [Google Scholar]
- 6. Klein BE, Klein R, Lee KE. Heritability of risk factors for primary open-angle glaucoma: the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 2004; 45: 59–62. [DOI] [PubMed] [Google Scholar]
- 7. Charlesworth J, Kramer PL, Dyer T, et al. The path to open-angle glaucoma gene discovery: endophenotypic status of intraocular pressure, cup-to-disc ratio, and central corneal thickness. Invest Ophthalmol Vis Sci. 2010; 51: 3509–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Charlesworth JC, Dyer TD, Stankovich JM, et al. Linkage to 10q22 for maximum intraocular pressure and 1p32 for maximum cup-to-disc ratio in an extended primary open-angle glaucoma pedigree. Invest Ophthalmol Vis Sci. 2005; 46: 3723–3729. [DOI] [PubMed] [Google Scholar]
- 9. Duggal P, Klein AP, Lee KE, Klein R, Klein BE, Bailey-Wilson JE. Identification of novel genetic loci for intraocular pressure: a genomewide scan of the Beaver Dam Eye Study. Arch Ophthalmol. 2007; 125: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Koolwijk LM, Ramdas WD, Ikram MK, et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012; 8: e1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ozel AB, Moroi SE, Reed DM, et al. Genome-wide association study and meta-analysis of intraocular pressure. Hum Genet. 2014; 133: 41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Strange A, Bellenguez C, Sim X, et al. Genome-wide association study of intraocular pressure identifies the GLCCI1/ICA1 region as a glaucoma susceptibility locus. Hum Mol Genet. 2013; 22: 4653–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nag A, Venturini C, Small KS, et al. A genome-wide association study of intra-ocular pressure suggests a novel association in the gene FAM125B in the TwinsUK cohort. Hum Mol Genet. 2014; 23: 3343–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burdon KP, Macgregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011; 43: 574–578. [DOI] [PubMed] [Google Scholar]
- 15. Thorleifsson G, Walters GB, Hewitt AW, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010; 42: 906–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010; 11: 415–425. [DOI] [PubMed] [Google Scholar]
- 17. Linton KL, Klein BE, Klein R. The validity of self-reported and surrogate-reported cataract and age-related macular degeneration in the Beaver Dam Eye Study. Am J Epidemiol. 1991; 134: 1438–1446. [DOI] [PubMed] [Google Scholar]
- 18. Klein BE, Klein R, Linton KL. Intraocular pressure in an American community: The Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 1992; 33: 2224–2228. [PubMed] [Google Scholar]
- 19. The Hypertension Detection and Follow-up Program: Hypertension Detection and Follow-up Program Cooperative Group. Prev Med. 1976; 5: 207–215. [DOI] [PubMed] [Google Scholar]
- 20. Fernández JR, Etzel C, Beasley TM, Shete S, Amos CI, Allison DB. Improving the power of sib pair quantitative trait loci detection by phenotype winsorization. Hum Hered. 2002; 53: 59–67. [DOI] [PubMed] [Google Scholar]
- 21. Wang J, Shete S. Testing departure from Hardy-Weinberg proportions. Methods Mol Biol. 2012; 850: 77–102. [DOI] [PubMed] [Google Scholar]
- 22. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006; 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 23. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gauderman WJ, Morrison JM. QUANTO. 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies. 2006. [Google Scholar]
- 25. Wiggs JL, Kang JH, Yaspan BL, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet. 2011; 20: 4707–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loomis SJ, Kang JH, Weinreb RN, et al. Association of CAV1/CAV2 genomic variants with primary open-angle glaucoma overall and by gender and pattern of visual field loss. Ophthalmology. 2014; 121: 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Osman W, Low SK, Takahashi A, Kubo M, Nakamura Y. A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum Mol Genet. 2012; 21: 2836–2842. [DOI] [PubMed] [Google Scholar]
- 28. Nakano M, Ikeda Y, Tokuda Y, et al. Common variants in CDKN2B-AS1 associated with optic-nerve vulnerability of glaucoma identified by genome-wide association studies in Japanese. PLoS One. 2012; 7: e33389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wiggs JL, Yaspan BL, Hauser MA, et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012; 8: e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010; 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seunggeun L, Miropolsky L, Wu M. SKAT: SNP-set (Sequence) Kernel Association Test: R package version 0.95. 2014. [Google Scholar]
- 32. R Development Core Team. R Foundation for Statistical Computing. R: a language and environment for statistical computing. 2008. [Google Scholar]
- 33. Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012; 13: 762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rochtchina E, Mitchell P, Wang JJ. Relationship between age and intraocular pressure: the Blue Mountains Eye Study. Clin Experiment Ophthalmol. 2002; 30: 173–175. [DOI] [PubMed] [Google Scholar]
- 35. Wiggs JL, Hauser MA, Abdrabou W, et al. The NEIGHBOR consortium primary open-angle glaucoma genome-wide association study: rationale, study design, and clinical variables. J Glaucoma. 2013; 22: 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li X, Mcclellan ME, Tanito M, et al. Loss of caveolin-1 impairs retinal function due to disturbance of subretinal microenvironment. J Biol Chem. 2012; 287: 16424–16434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gu X, Fliesler SJ, Zhao YY, Stallcup WB, Cohen AW, Elliott MH. Loss of caveolin-1 causes blood-retinal barrier breakdown, venous enlargement, and mural cell alteration. Am J Pathol. 2014; 184: 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Giovingo M, Nolan M, Mccarty R, et al. sCD44 overexpression increases intraocular pressure and aqueous outflow resistance. Mol Vis. 2013; 19: 2151–2164. [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng JB, Russell DW. Mammalian wax biosynthesis, I: identification of two fatty acyl-Coenzyme A reductases with different substrate specificities and tissue distributions. J Biol Chem. 2004; 279: 37789–37797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tesco G, Koh YH, Kang EL, et al. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007; 54: 721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang GZ, Yang M, Lim Y, et al. Huntingtin associated protein 1 regulates trafficking of the amyloid precursor protein and modulates amyloid beta levels in neurons. J Neurochem. 2012; 122: 1010–1022. [DOI] [PubMed] [Google Scholar]
- 42. Kiyozumi D, Sugimoto N, Nakano I, Sekiguchi K. Frem3, a member of the 12 CSPG repeats-containing extracellular matrix protein family, is a basement membrane protein with tissue distribution patterns distinct from those of Fras1, Frem2, and QBRICK/Frem1. Matrix Biol. 2007; 26: 456–462. [DOI] [PubMed] [Google Scholar]
- 43. Ritch R. The management of exfoliative glaucoma. Prog Brain Res. 2008; 173: 211–224. [DOI] [PubMed] [Google Scholar]