Abstract
The endomembrane system in mammalian cells has evolved over the past two billion years from a simple endocytic pathway in a single-celled primordial ancestor to complex networks supporting multicellular structures that form metazoan tissue and organ systems. The increased organellar complexity of metazoan cells requires additional trafficking machinery absent in yeast or other unicellular organisms to maintain organ homoeostasis and to process the signals that control proliferation, differentiation or the execution of cell death programmes. The PACS (phosphofurin acidic cluster sorting) proteins are one such family of multifunctional membrane traffic regulators that mediate organ homoeostasis and have important roles in diverse pathologies and disease states. This review summarizes our current knowledge of the PACS proteins, including their structure and regulation in cargo binding, their genetics, their roles in secretory and endocytic pathway traffic, interorganellar communication and how cell-death signals reprogramme the PACS proteins to regulate apoptosis. We also summarize our current understanding of how PACS genes are dysregulated in cancer and how viral pathogens ranging from HIV-1 to herpesviruses have evolved to usurp the PACS sorting machinery to promote virus assembly, viral spread and immunoevasion.
Keywords: Akt, cancer, furin, herpesvirus, HIV-1, Nef, phosphofurin acidic cluster sorting (PACS)-1/2, polycystin-2, secretory pathway, tumour-necrosis-factor-related apoptosis-inducing ligand (TRAIL), 14-3-3 protein
INTRODUCTION
Metazoan cells represent dynamic and adaptable information processing centres. They receive and integrate signals from neighbouring or distant cells to affect responses ranging from maintaining organ homoeostasis to the control of cellular proliferation and differentiation or the execution of cell death. These varied responses require the membrane trafficking machinery to sort and localize a diverse set of proteins, lipids and various small molecules.
The organelles of the mammalian secretory and endocytic pathways are central to the machinery underlying information processing and communicate largely by vesicular or tubulovesicular carriers [1,2]. The mammalian secretory pathway includes the ER (endoplasmic reticulum), the Golgi cisternae and the TGN (trans-Golgi network) [3]. The ER houses the chaperone-assisted folding of newly synthesized soluble and membrane proteins, which are either localized to the ER or exported to the Golgi and distal compartments [4]. The TGN is the principal organelle responsible for sorting secretory pathway proteins to their final destinations, including the sorting of cargo to lysosomes, secretory granules and the surfaces of polarized cells [5]. The endocytic pathway, which also includes the TGN, is composed of a collection of membranous organelles, including sorting and recycling early endosomes, late endosomes and lysosomes [6]. These organelles collaborate to mediate the uptake of proteins, lipids and nutrients from the extracellular environment and sort internalized cargo to lysosomes, the TGN or back to the cell surface [7,8].
Metazoan cells must co-ordinate the complex endomembrane trafficking pathways to control the many cellular activities that regulate cell and organ homoeostasis. To accommodate these unique demands, metazoan cells have evolved protein machinery beyond the core machines present in unicellular organisms [9,10]. In this review we summarize studies on one such family of trafficking molecules, the PACS (phosphofurin acidic cluster sorting) proteins. We describe our current knowledge about their structure and function in endomembrane traffic, interorganellar communication and apoptosis, as well as their regulation by phosphorylation and how their function is hijacked by pathogens to cause disease.
EVOLUTION OF THE PACS FAMILY
Evolution of the endomembrane system
The appearance of the PACS proteins coincided with the evolution of complex endomembrane systems and multicellularity. Three milestones mark this evolution, which began over two billion years ago. The first milestone was the autogenous generation of the endomembrane system that enabled the primordial phagocyte to pursue and engulf prey, and target biomaterial to lysosomes for degradation or metabolism [10]. Next, acquisition of mitochondria, possibly by engulfment of an α-proteobacterium, gave the phagocyte a metabolic advantage within the developing oxygenated atmosphere. These endomembrane systems enabled the primitive cells to increase their volume and to spatially and temporally control their membrane surface composition and complexity [9]. The subsequent coalescence of phagocytes into multicellular structures permitted formation of organ systems containing polarized epithelium and underlying mesenchyme [11].
The formation of organ systems posed new challenges as cells became integrated into multicellular structures. Membrane trafficking in yeast largely emphasizes biosynthetic transport [12], but metazoans have evolved a complex set of recycling compartments to accommodate the flux of ligand- and signal-based communication between cells. In addition, metazoans required sophisticated programmes to regulate cell death. Whereas death in single-celled organisms occurs by a necrosis-like process, metazoan cells have evolved apoptotic, or programmed-cell-death mechanisms, permitting the resorptive elimination of unwanted cells while preventing the tissue damage caused by necrosis.
Phylogenetic studies suggest that the majority of membrane trafficking machinery arose autogenously [11]. Common to all eukaryotic lineages are clathrin, COPI (coatamer protein I), COPII, heterotetrameric adaptors, a subset of the Rab and Arf families of GTPases, SNAREs (soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptors) and ESCRT (endosomal sorting complex required for transport) proteins [9]. In fungi and metazoans, which evolved along a common pathway [9], the endomembrane system increased in complexity due to gene duplications and paralogous expansion leading to additional members of each family [9]. In addition, fungi and metazoans further share additional trafficking proteins, including vps27/hrs (vacuolar protein sorting 27) and the GGA [Golgi-associated γ-adaptin ear homology domain ARF (ADP-ribosylation factor)-interacting protein] monomeric adaptors. The increased number of membrane cargo that must be trafficked in metazoans requires yet additional trafficking machinery absent in yeast. The PACS proteins, which bind cargo containing acidic cluster sorting motifs, are representative of this class of protein.
Sorting motifs in membrane cargo proteins
Eukaryotic sorting adaptors evolved to recognize diverse sorting motifs on the cytosolic domains of membrane cargo. The canonical tyrosine- (Tyr-X-X-Φ) and leucine-based ([DE]XXX[LI]) sorting signals direct clathrin-mediated trafficking in the late secretory pathway by linking cargo to heterotetrameric adaptors, whereas -KKXX and FF-motifs direct sorting in the early secretory pathway by linking cargo to COPI and COPII respectively [2]. Numerous membrane cargo in metazoan cells contain additional sorting signals, including a complex signal composed of clusters of acidic residues that often contain serine or threonine residues, which are phosphorylated by protein kinase CK2 (casein kinase 2) and dephosphorylated by isoforms of PP2A (protein phosphatase 2A) (for reviews see [5,13]). Importantly, the phosphorylation state of these acidic cluster sites controls the subcellular distribution of their membrane cargoes. For example, CK2 phosphorylation of a pair of serine residues within the acidic cluster of the furin cytosolic domain (EECPpS773DpSEED) localizes the endoprotease to the TGN [14], mediates retrieval of furin from immature secretory granules in neuroendocrine cells [15,16], and mediates the recycling of endocytosed furin molecules from early endosomes to the cell surface [17]. Studies on furin led to the identification of acidic cluster motifs that mediate numerous endomembrane trafficking events, including those used by other proprotein convertases, receptors, ion channels, SNARE proteins and proteins targeted to the primary cilium. In addition, viral pathogens have rapidly evolved to include key proteins containing acidic cluster motifs that usurp the PACS proteins to mediate processes ranging from virus envelopment to immunoevasion.
BIOCHEMISTRY OF THE PACS PROTEINS
PACS genes and proteins
The identification of the PACS family of sorting proteins was grounded by investigation of the mechanism controlling the subcellular localization of the proprotein convertase furin, which is regulated by phosphorylation of the acidic cluster sorting motif in its cytosolic domain ([13,14,18–20] and described below). The human PACS-1 gene is located on chromosome 11q13.1 and encodes a 963 residue cytosolic protein, PACS-1a [15,19].
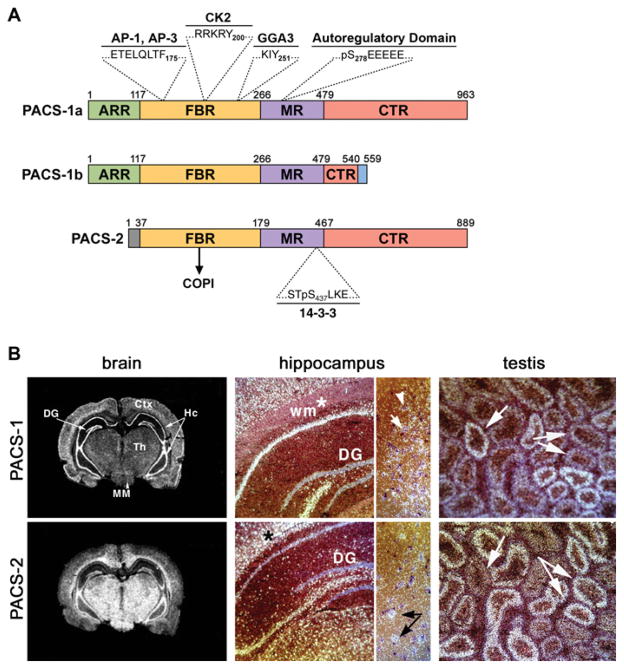
The human PACS-1 gene exhibits complex architecture, spanning nearly 175 000 bp and containing 25 exons. In addition to the canonical transcript, there is evidence for at least seven additional transcripts generated by alternative splicing events [21]. The mouse PACS-1 gene shows similar complexity, containing 25 exons that are spliced to produce six putative transcripts [21]. In rodents, one transcript produces a smaller PACS-1 isoform, PACS-1b, which encodes a 559 residue protein, is expressed in tissues enriched in epithelial cells, such as small intestine and kidney [22]. PACS-1b is identical with PACS-1a through the first 540 amino acids, but contains a unique 19 amino acid C-terminus, which is encoded by a read-through into intron 12. Based upon the overall sequence similarity between PACS-1a and PACS-1b, four regions were identified (Figure 1A). An N-terminal region enriched in proline/glutamine and serine/alanine stretches that share low sequence identity with a region of the atrophin-1 transcriptional repressor called the ARR (atrophin-1-related region) but the function of this region is not yet known. The ARR is followed by a 140 residue FBR [furin (cargo)-binding region] identified in the original yeast two-hybrid screen [15,19], a MR (middle region) containing an autoregulatory domain ([23] and see below), and a CTR (C-terminal region), which is absent in PACS-1b. Like the ARR, the function of the CTR is not yet known. A database search for PACS-1 homologues identified the human PACS-2 gene, which is located near the telomere on chromosome 14 at 14q32.33 and the canonical protein encodes an 889 amino acid protein that shares 54 % overall identity with PACS-1. PACS-2 lacks an ARR but the PACS-2 FBR shares 75 % identity within the PACS-1 FBR [24]. The human PACS-2 gene locus, similar to PACS-1, is highly complex, spanning 83 000 bp and 25 exons. Alternative splicing events are predicted to give rise to ten additional putative transcripts of various sizes [21]. The mouse PACS-2 gene is predicted to transcribe 12 putative transcripts. Several predicted isoforms in human and mouse have alternative or deleted N-terminal regions.
Figure 1. Domain organization of the PACS proteins.
(A) Schematic of PACS-1 and PACS-2 illustrating the proposed domains and residues important for partner protein binding. (B) In situ hybridization of PACS-1 and PACS-2. Left-hand panels, coronal sections of rat brain stained with PACS-1 or PACS-2 cRNA probes. DG, dentate gyrus; Hc, hippocampus; Ctx, cortex; Th, thalamus; MM, medial mammillary nucleus. Middle panels, darkfield staining of hippocampus showing neuronal and glial labelling. (*), alignment marker; white arrows, neurons; black arrows, glia. Right-hand panels, PACS-1 and PACS-2 in consecutive serial sections of the testis. Arrows mark seminiferous tubules with inverse staining of PACS-1 and PACS-2. Sense probes showed no staining.
The PACS-1 and PACS-2 genes are broadly expressed in all tissues examined, including heart, brain, kidney, liver and pancreas, but PACS-1 is selectively enriched in peripheral blood lymphocytes, whereas PACS-2 is selectively enriched in skeletal muscle [22]. Moreover, underlying tissues of organs with high levels of PACS-1 and PACS-2 have differential expression patterns, as revealed by in situ hybridization studies (Figure 1B). In brain tissue, PACS-1 is most highly expressed in neuronal centres, whereas PACS-2 is most highly expressed in white matter enriched in glial cells. In agreement with these findings, gene array analysis revealed PACS-1 is broadly expressed in the brain and that PACS-2 expression is increased approx. 30-fold following oligodendrocyte differentiation [25]. In testes tissue, the patterns of PACS-1 and PACS-2 expression in serial sections of seminiferous tubules of young rats are inverse and largely non-overlapping, which correlates with the waves of differentiation and apoptosis that occur during spermatogenesis [26].
PACS-1 possesses approx. 40 % well-defined secondary structure as determined by CD spectroscopy (R. T. Youker, U. Shinde and G. Thomas, unpublished work). Consistent with this finding, protein disorder algorithms suggest that several regions in the PACS proteins lack defined secondary structure [27,28], characteristic of IUPs (intrinsically unfolded proteins). The IUPs contain regions lacking a definitive protein fold but can attain tertiary structure upon binding to a client protein or ligand [29,30]. Examples include protein kinases, transcriptional regulators and protein translation initiation factors [30]. Consistent with this possibility, ab initio modelling predicts that the PACS-1 FBR is a loosely packed structure composed of long flexible loops interspersed with several defined α-helical and β-sheet elements (see Figure 4 and discussion below).
Figure 4. Ab initio model of PACS-1 FBR.
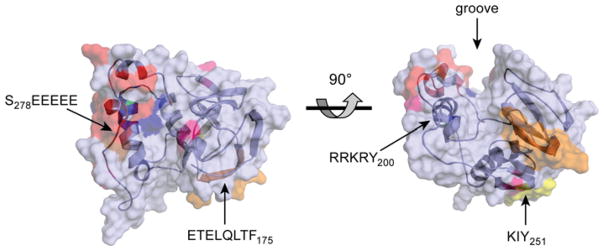
Ab initio modelling of the PACS-1 FBR (residues 117–300) was performed using Rosetta++ [177] and the predicted structure was energy minimized using Insight II (Accelrys Insight II modelling software). The tertiary structure is represented as a ribbon diagram surrounded by a semi-transparent surface projection. Acidic residues are coloured red, basic residues are coloured blue, residues required for adaptor binding are coloured orange, residues required for GGA binding are coloured yellow and residues positively selected through evolution are coloured pink. Left-hand panel, top view looking down on the major groove. Right-hand panel, the image is rotated 90° towards the viewer. Images were created using PyMOL (DeLano Scientific; http://pymol.sourceforge.net/). An interactive three-dimensional version of this Figure is available at http://www.BiochemJ.org/bj/421/0001/bj4210001add.htm.
Characteristic of an IUP, the PACS-1 and PACS-2 FBRs bind cargo proteins, adaptors and signalling molecules with low affinity but with high selectivity. For example, binding of the PACS-1 and PACS-2 FBRs to the acidic cluster sites in furin (EECPpS773DpSEEDE), in the CI-MPR (cation-independent mannose-6-phosphate receptor; HDDpS2492DEDLLHI) or polycystin-2 (also called TRPP2, DDpS812EEDDDEDS) is strongly dependent on the phosphorylation of Ser773, Ser2492 or Ser812 respectively [31]. Despite the high selectivity in cargo recognition, PACS-1 and PACS-2 FBRs bind acidic clusters relatively weakly. For example, the phosphorylated polycystin-2 acidic cluster bound the PACS-1 and PACS-2 FBRs with micromolar affinity (PACS-1, Kd = 14.8 μM; PACS-2, Kd = 15.4 μM, see [24]).
Not all residues in acidic cluster sites are necessarily required for binding PACS proteins. For example, binding to the furin acidic cluster requires only pSer773 and two adjacent residues ([19] and L. Wan and G. Thomas, unpublished work). Other segments of the furin acidic cluster have different roles in sorting. For example, the EEDE779 segment is required for basolateral sorting of furin in polarized cells [32]. The lack of binding of PACS proteins to EEDE779 may explain why others failed to observe a role for PACS proteins in the trafficking of furin reporter proteins mutated in the EEDE779 site in an attempt to specifically block PACS binding [33]. Similarly, the CI-MPR acidic cluster binds PACS proteins and GGAs [34,35], and the regulated interaction between PACS-1 and GGA3 contributes to the localization of the CI-MPR to the TGN (see below). Importantly, some acidic clusters must be de-phosphorylated to bind PACS proteins, including the acidic clusters in the ER chaperone calnexin (KS554DAEE) as well as in the PACS-1 autoregulatory domain (S278EEEEE) [23,36], as described below.
The PACS FBRs also bind components of vesicular coats. PACS-1 interacts in vivo with AP-1 (adaptor protein complex 1) and AP-3, but not AP-2, and this interaction requires the FBR [15]. The PACS-1 FBR binds purified AP-1 and can link the adaptor to cargo proteins to form a ternary complex in vitro [15]. An eight-amino-acid segment of the PACS-1 FBR, E168TELQLTF, was identified to be important for AP-1 binding and mutation of this sequence generated a PACS-1 interfering mutant, PACS-1Admut, which binds cargo but not AP-1 or AP-3 [15]. The PACS-2 FBR selectively interacts with COPI in vivo and, similar to PACS-1, can link purified cargo to COPI to form a ternary complex in vitro [24]. Mutation of the PACS-2 FBR at residues corresponding to those in PACS-1 required for adaptor binding, generated a PACS-2 interfering mutant which binds cargo but not COPI [24]. Precisely how PACS-2 binds COPI and where on the heteromeric adaptors and COPI the FBRs bind is not yet known.
Despite the ability to form ternary complexes with cargo and coat proteins in vitro, PACS proteins have so far not been detected in coated vesicles (L. Wan and G. Thomas, unpublished work). These findings suggest that PACS proteins may mediate the recruitment and organization of trafficking machinery to regulate trafficking steps in the endomembrane system. Such a possibility would suggest that acidic clusters form part of a sensing mechanism that regulates recruitment of adaptors and vesicular coat proteins to canonical tyrosine- or di-leucine-adaptor-binding sites to organize specific sorting steps. This possibility is consistent with the determination that the VAMP-4 (vesicle-associated membrane protein 4) acidic cluster, which binds PACS-1, and its proximal LL motif, which binds AP-1, are both required for the PACS-1- and AP-1-dependent trafficking of this SNARE protein [37].
Regulation of cargo binding
Similar to furin, PACS-1 itself contains an acidic cluster, S278EEEEE, which is located in the MR and is phosphorylated at Ser278 by CK2 and dephosphorylated by PP2A [17,23]. This PACS-1 acidic cluster is part of an autoregulatory domain that controls cargo binding to the FBR as dictated by the phosphorylation state of Ser278 [23]. However, opposite of the binding of furin to the PACS-1 FBR, non-phosphorylated Ser278 binds the FBR, thereby inhibiting cargo binding, whereas pSer278 relaxes binding to the FBR, thereby promoting cargo binding [23]. Consistent with this model, a PACS-1 S278A mutant blocks trafficking of PACS-1 cargo, whereas a PACS-1 S278D phosphomimic mutant is permissive to PACS-1 sorting [23,34].
PACS-1 recruits CK2 to phosphorylate Ser278, thereby regulating cargo binding [34]. This mechanism is manifested by the binding of the CK2β (CK2 regulatory β-subunit) to a polybasic sequence in the PACS-1 FBR, R196RKRY, which stimulates the bound CK2 activity [34]. The ability of the PACS-1 polybasic site to bind and stimulate CK2 is similar to the ability of polyamines to stimulate CK2 activity by binding an acidic groove on CK2β [38–42]. The stimulated CK2 bound to the FBR then phosphorylates Ser278 to promote cargo binding [34]. Mutation of the PACS-1 polybasic site to alanine residues blocks binding to CK2β, which prevents Ser278 phosphorylation and cargo binding [34]. The mechanism regulating cargo binding to PACS-2 is currently under investigation.
Evolutionary and ab initio modelling of PACS proteins
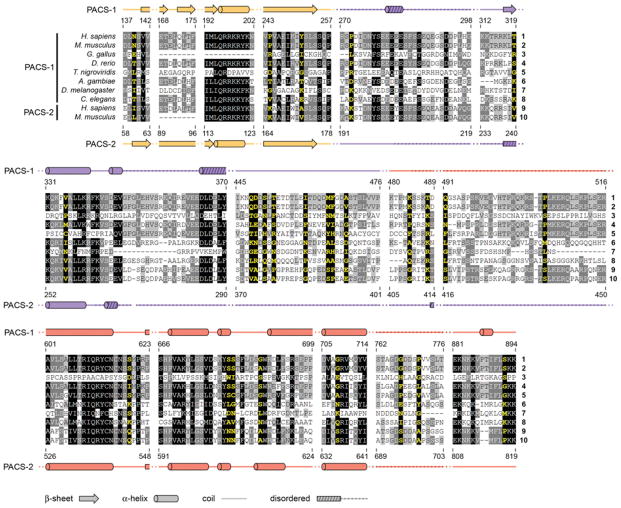
A phylogenetic dendrogram of the human PACS-1 gene suggests that the PACS family emerged late in evolution, arising in metazoans by gene duplication from a common ancestor protein that evolved along a separate path to also form the AP-3 adaptor (Figure 2). Lower metazoans, including Caenorhabditis elegans and Drosophila melanogaster express a single PACS gene. The PACS gene in lower metazoans probably underwent duplication, generating the PACS-1 and PACS-2 genes in higher metazoans. The appearance of two PACS genes in metazoans correlates with the increased complexity of membrane traffic in mammalian cells compared with lower eukaryotes [9]. This would suggest that the PACS proteins are evolutionally adaptive proteins, undergoing positive selection of amino acids in positions that assume new functions. A phylogeny-based statistical analysis on the PACS family shows that 2.7 % of the residues in PACS-1 are under strong positive selection, supporting this model and suggesting a significant adaptive evolution. Hence, although PACS family members share several well-conserved domains, a significant adaptive evolution followed the duplication of the PACS gene.
Figure 2. Phylogenetic analysis of PACS genes.
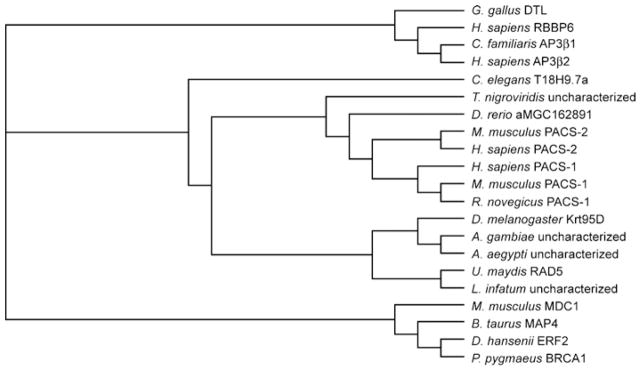
Non-redundant protein sequences of the PACS family members were obtained from the SwissProt and NCBI databases (Accession numbers used: A0NG63, Q5ZJW8, Q95QA3, Q7YRF1, P36225, Q8MRS5, Q4PGG5, Q13367, Q6VY07, Q86VP3, Q7Z6E9, A4HVI7, Q17EX1, Q5PSV9, Q8K212, Q3V3Q7, Q6J6J0, Q4SQP4, Q9PVV4, Q9HGI6, A5PKS9). The protein sequences were aligned using ClustalW and were manually examined/modified for their accuracy within the non-conserved domains that flank conserved domains. The protein alignments were used to obtain nucleotide alignments, using a PERL script, as described previously [172]. The tree topology was estimated from the DNA alignment using the DNAML module from the PHYLIP package and our data set was analysed in PAML using appropriate models (M8, M8a) [172]. A LRT (likelihood ratio test) was used to search for positive selection. Positive selection (Ω > 1) was tested by applying an empirical Bayesian approach which allows for all types of selection (purifying, neutral and positive), including nesting of models which do not allow positive selection [173–175]. The statistical significance of our tree was estimated through a LRT test on a null tree (M8a), which does not allow for positive selection [176]. Consistent with studies on other protein families, trees generated using different programs (DNAPARS and DNADIST) do not affect the analysis. A. aegypti, Aedes aegypti; A. gambiae, Anopheles gambiae; AP3β1, AP-3 complex subunit β-1; AP3β2, AP-3 complex subunit β-2; BRCA1, breast cancer type 1 susceptibility protein homologue; B. taurus, Bos taurus; C. familiaris, Canis familiaris; D. hansenii, Debaryomyces hansenii; D. rerio, Danio rerio; DTL, denticleless protein homologue; ERF2, eukaryotic peptide chain release factor GTP-binding subunit; G. gallus, Gallus gallus; L. infantum, Leishmania infantum; H. sapiens, Homo sapiens; MAP4, microtubule-associated protein 4; MDC1, mediator of DNA damage checkpoint protein 1; M. musculus, Mus musculus; P. pygmaeus, Pipistrellus pygmaeus; RBBP6, retinoblastoma-binding protein 6R. norvegicus, Rattus norvegicus; T. nigroviridis, Tetraodon nigroviridis; U. maydis, Ustilago maydis.
Residues in PACS-1 and PACS-2 required for binding partner proteins can be used to test the hypothesis that the PACS proteins have undergone adaptive evolution. For example, PACS-1, but not PACS-2, binds to GGA3 and this interaction requires PACS-1 residues KIY251 ([34] and discussed below). Consistent with the selective binding of GGA3 to PACS-1 is the positive selection of Tyr251 in PACS-1 orthologues by non-synonymous substitutions (Figure 3). Conversely, PACS-2 Ser437, which is phosphorylated in vivo by the survival kinase Akt to regulate PACS-2 apoptotic activity ([43] and discussed below), also arose by positive selection and is absent in PACS-1 orthologues. Several additional regions in PACS-1 and PACS-2 arose by positive selection. For example, a 44-residue intrinsically unstructured region (PACS-1 residues 448–491) contains 11 positively selected residues and has probably evolved to discriminate between interacting partners. Interestingly, several positions throughout PACS-1 appear to be positively selected to either incorporate or replace serine residues (Figures 3 and 4). Phosphorylation plays a vital role in PACS protein functions [23,34,43], and these positions may represent putative phosphorylation sites that modulate PACS–cargo interactions.
Figure 3. Positively selected residues of PACS-1 and PACS-2.
Multiple-sequence alignment of conserved regions in PACS-1 and PACS-2 based on the phylogenetic analysis of the PACS proteins (Figure 2). For simplicity, eight PACS-1 and two PACS-2 sequences were used to generate the alignment. Residues shaded in black are > 80 % conserved. Residues shaded dark grey ~ 60–65 % conserved and residues shaded in light grey are ~ 50 % conserved. Residues highlighted in yellow are positively selected, which was determined by measuring the rate of non-synonymous to synonymous substitutions (Ω), which is an unequivocal indicator of evolution [172]. Residues with a value Ω = 1 are under neutral selection whereas residues with Ω values < 1 or > 1 suggest that the residues are under a purifying or positive selection respectively [174]. Of the residues, 2.7 % in PACS-1 exhibit strong positive selection (Ω > 1). Residues were shaded using genedoc. Secondary structure prediction (PsiPredict) for PACS-1 (above alignment) and PACS-2 (below alignment) was colour-coded the same as the constructs in Figure 1(A). Predicted regions of disorder (PONDR) for PACS-1 and PACS-2 are stippled on the schematic of the secondary structure. A. gambiae, Anopheles gambiae; D. rerio, Danio rerio; G. gallus; Gallus gallus; H. sapiens, Homo sapiens; M. musculus, Mus musculus; T. nigroviridis, Tetraodon nigroviridis.
Interestingly, the amino acids in PACS-1 important for binding to AP-1 or AP-3, GGA3 adaptor or to CK2, each reside in ordered regions located on different surfaces of the predicted structure (Figure 4). This is in agreement with in vitro experiments that demonstrated that the PACS-1 FBR forms a ternary complex with adaptor and cargo proteins [15], suggesting the existence of distinct binding surfaces. The glutamate residues in the acidic cluster S278EEEEE are predicted to reside in an α-helix on a ridge parallel to a prominent groove spanning the middle of the FBR. The acidic cluster in its dephosphorylated state inhibits cargo binding to the FBR. The ab initio model suggests that the PACS-1 acidic cluster may fold to interact directly with a potential surface for binding acidic clusters in cargo proteins. The basic amino acid cluster R196RKRY required for CK2 binding is predicted to reside in an α-helix, but at the base of the groove in close proximity to the acidic cluster and positioning the kinase adjacent to Ser278, whose phosphorylation is required for cargo binding. This ab initio model of the PACS-1 FBR provides new insight into the possible mechanism of cargo binding and a platform for future genetic and biochemical experiments to further dissect the structural basis for cargo binding and selectivity.
PACS PROTEINS AS MEMBRANE TRAFFIC REGULATORS
Genetic analyses of PACS sorting function
Genetic disruption of PACS expression in diverse metazoans demonstrates the importance of the PACS proteins in secretory pathway sorting, interorganellar communication and apoptosis. RNAi (RNA interference) knockdown of C. elegans PACS, which is expressed from a single gene and localizes to presynaptic endosomes, blocks synaptic transmission leading to paralysis [44], demonstrating genetically an essential role for this trafficking protein. Studies in human cells using antisense RNA or siRNA (small interfering RNA) knockdown reveal that PACS-1 is required for the TGN localization of furin and a subset of other cellular proteins that contain acidic cluster sites, as well as envelope glycoproteins from a number of pathogenic viruses [15,19,45,46]. PACS-1 has additional roles in late secretory trafficking steps, among them the recycling of internalized cargo from early endosomes to the cell surface [17], and the trafficking of acidic-cluster-containing cargo to the primary cilium in epithelial cells [47]. Similarly, siRNA studies or analysis of cells from cells from gene trap mice mice reveal PACS-2 is required for localizing membrane-cargo-containing acidic cluster sites to the ER [24,48,49], and combines with PACS-1 to mediate the step-wise movement of some cargo from the ER to the TGN and to the cell surface in model systems ranging from Xenopus oocytes to zebrafish [24,50]. PACS-1 and PACS-2 also combine to mediate the sorting of internalized cargo from early endosomes [49]. Studies using siRNA knockdown and cells from gene trap mice reveal that the secretory pathway sorting activities of PACS-1 and PACS-2 are pirated by pathogenic viruses, including HIV-1 and herpesviruses, to mediate steps ranging from immunoevasion to virus assembly [17,46,49,51]. In addition, PACS-2 mediates the interaction between ER and mitochondria, which promotes communication between these organelles and, in response to death cues, becomes pro-apoptotic, promoting the translocation of apoptotic activators to mitochondria [22]. Indeed, studies with gene trap mice show that PACS-2 is a homoeostatic regulator that integrates membrane traffic with TRAIL [TNF (tumour necrosis factor)-related apoptosis-inducing ligand]-induced apoptosis [43,49]. These multifunctional roles of the PACS proteins are summarized below. The sections are organized to reflect the flow of membrane traffic from the ER, which is mediated by PACS-2, to the TGN/endosomal system, which is mediated by PACS-1 and PACS-2.
PACS-2 mediates the ER localization of polycystin-2 and a mobile pool of calnexin
Membrane proteins synthesized in the ER possess trafficking motifs that regulate their localization to the ER and their trafficking to late secretory pathway compartments where they assume a highly compartmentalized distribution [52]. These cytosolic trafficking motifs range from simple -RXR- motifs that regulate the ER localization and assembly of the KATP channel [53,54], to more complex motifs including CK2-phosphorylatable acidic clusters such as DDpS812EEDDDEDS, which regulates the ER localization and subcellular distribution of the calcium-selective transient receptor potential channel polycystin-2 [31,50,55–57].
The function of polycystin-2 is dependent on its subcellular localization, which is regulated by the phosphorylation state of its acidic cluster. At the ER, polycystin-2 collaborates with IP3 (inositol 1,4,5-trisphosphate) or ryanodine receptors to regulate intracellular calcium concentrations, which appear to mediate L/R-axis symmetry and limit the amount of ER calcium available to promote apoptosis ([31,50,55,56,58] and see below). When localized to the primary cilium, polycystin-2 combines with polycystin-1 to form a mechanosensor that detects fluid flow in tubules and modulates multiple signalling pathways to arrest cell proliferation and promote tubulogenesis [59–63]. Mutations or deletion of the polycystin-2 acidic cluster cause ADPKD (autosomal dominant polycystic kidney disease), which is characterized by the development of fluid-filled cysts in the kidney epithelium that lead to end-stage renal failure [31,64,65].
The PACS proteins bind the CK2-phosphorylated acidic cluster in polycystin-2 (see above and [24]) and combine to mediate ER localization and the stepwise trafficking of the ion channel between the ER and the cell surface [24]. PACS-2 is required for localizing polycystin-2 to the ER. Expression of PACS-2Admut, which binds cargo but not COPI, caused a polycystin-2 reporter to redistribute from the ER to the Golgi/TGN. PACS-1 localized the polycystin-2 reporter to the TGN since combined expression of PACS-2Admut with PACS-1Admut, which binds cargo but not AP-1, caused the polycystin-2 reporter to redistribute to the cell surface [24]. The combined roles of PACS-2 and PACS-1 in mediating the stepwise trafficking of the active polycystin-2 channel to the cell surface could be recapitulated by siRNA knockdown of the PACS proteins in Xenopus oocytes, demonstrating the efficacy of the PACS-interfering mutants to dissect the complex trafficking itinerary of the ion channel [24]. Studies in zebrafish confirm and extend these findings [50]. Overexpression of PACS-2 or the phosphomimic polycystin-2-S812D in zebrafish, both of which restrict polycystin-2 to the ER and prevent it from reaching the cell surface, increases pronephric cysts as well as other defects, including hydrocephalous, increased body axis curvature and pericardial oedema. Pronephric cysts also resulted from morpholino-mediated knockdown of PACS-1, which similarly causes an imbalance of PACS protein expression resulting in increased levels of PACS-2.
The role of PACS proteins in the kidney epithelium is not restricted to polycystin-2. PACS-1 targets nephrocystin to the base of the primary cilium where it triggers ERK (extracellular-signal-regulated kinase) 1 and ERK2 signalling pathways [47,66,67]. Localization of this protein to the primary cilium is lost in patients suffering from nephronophthisis [68]. Additional ion channels, including TRPV4 (vanilloid transient receptor potential 4) and CLC7, and the G-protein-coupled receptor mGLUR5 (metabotropic glutamate subtype 5 receptor) interact with the PACS proteins, suggesting that the two-step model described for polycystin-2 trafficking may be broadly applicable [24,69]. PACS-2/COPI are also required for the ER localization of other cellular proteins, including a mobile pool of the ER chaperone calnexin, which in the absence of PACS-2 is redistributed to the cell surface, as well as an ER-restricted profurin mutant [36,48].
PACS-2 mediates ER–mitochondria communication
High-resolution three-dimensional electron tomography reveals that the expansive, reticulated ER forms close contacts with mitochondria [70]; as much as 20 % of the mitochondrial surface is in direct contact with the ER, underscoring the dynamic and highly regulated communication between these organelles [71–74]. The ER forms close contacts with mitochondria, called MAMs (mitochondria-associated membranes), that mediate lipid transfer and calcium exchange between these organelles and, in response to death inducers, co-ordinates steps in the apoptotic elimination of cells [73,75–79]. Several molecules partition to the MAM, including a subpopulation of the ER pool of calnexin [36], the IP3 receptor and SERCA2a (sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 2a), which mediate calcium exchange, and FACL4 (fatty acyl-CoA ligase-4), which mediates formation of lipid esters required for β-oxidation in mitochondria, and PSS-1 (phosphatidylserine synthase-1) which promotes the partitioning of phosphatidylserine between the ER and mitochondria [80]. PACS-2 partitions between the ER, mitochondria and MAMs, suggesting that this sorting protein may mediate MAM integrity [22,36]. In support of this possibility, siRNA knockdown of PACS-2 or expression of PACS-2 mutants uncouple mitochondria from the ER, mislocalize FACL4 from MAMs and disrupt histamine-inducible calcium release from the ER [22]. Furthermore, loss of PACS-2 by siRNA knockdown causes mitochondrial uncoupling from the ER due to cleavage of the ER stress sensor BAP31 (B-cell receptor-associated protein 31) which in turn stimulates Drp1 (dynamin-related protein-1) to fissure mitochondria [22]. Expression of a cleavage-resistant BAP31 represses mitochondria uncoupling [22], suggesting PACS-2 and BAP31 co-operate to maintain MAM integrity.
PACS-1 regulates the TGN localization of furin and other membrane proteins
The proprotein convertase furin localizes to the TGN and follows a highly regulated trafficking itinerary through multiple processing compartments in the TGN/endosomal system, including early endosomes and the cell surface [13,17,20]. This complex itinerary, which is controlled in large part by the phosphorylation state of the furin acidic cluster [14,81], enables furin to proteolytically activate diverse proprotein substrates. In the TGN/biosynthetic pathway, furin activates bioactive proteins ranging from neurotrophins and TGF-β (transforming growth factor β) family members to viral envelope glycoproteins, including HIV-1 gp160 and HCMV (human cytomegalovirus) gB (glycoprotein B) and H5N1 HA [20]. At the cell surface, furin activates several bacterial toxins, including anthrax protective antigen, and in the mildly acidic early endosomes furin activates other bacterial toxins, including pseudomonas exotoxin A [20].
Similar to polycystin-2, localization of furin to the TGN requires a CK2 phosphorylatable acidic cluster (EECPpS773-DpSEEDE) that must be phosphorylated to act as a TGN-localization signal [14,18,19,81–83]. Antisense knockdown of PACS-1 or expression of PACS-1 interfering mutants cause furin to mislocalize from the TGN in vivo and block the endosome-to-TGN trafficking of PACS-1 cargo in vitro [19]. PACS-1 is not exclusively dedicated to furin; it mediates the TGN localization of a number of membrane proteins that contain acidic cluster motifs. These include cellular proteins, such as the CI-MPR, the furin homologue PC6B, CPD (carboxypeptidase D) and SorLA/LR11 (sorting protein-related receptor), which mediates the trafficking and proteolysis of the β-amyloid precursor protein ([15,84,85] and L. Thomas and G. Thomas, unpublished work). PACS cargo also includes many viral envelope glycoproteins as discussed below.
In neuroendocrine cells, furin buds from the TGN into nascent ISGs (immature secretory granules) together with hormones and other molecules destined for dense core MSGs (mature secretory granules) [16,38]. ISGs are short-lived AP-1/clathrin-coated compartments that undergo homotypic fusions and extensive membrane remodelling during microtubule-based transport to the cell periphery [86,87]. At the periphery, furin is removed from the ISG, apparently during the BFA (brefeldin A)-sensitive, ARF1-dependent remodelling of the ISG membrane [88], and is returned to the TGN [16,87]. BFA blockage of ISG remodelling is due to inhibition of the ARF1-mediated recruitment of AP-1/clathrin. Removal of furin from ISGs requires its phosphorylated acidic cluster and is blocked by PACS-1 mutants [15,16]. Similar results have been reported for the retrieval of VAMP-4, CPD and VMAT2 (vesicular monoamine transporter-2), suggesting roles for CK2 and PACS-1 in MSG maturation [89–91].
Distinct roles for PACS-1 and PACS-2 in sorting cargo from early endosomes
After endocytosis of furin from the cell surface to early endosomes, the endoprotease is either recycled to the surface or trafficked to the TGN [6,13,17]. Recycling of furin to the cell surface requires PACS-1 binding to the phosphorylated acidic cluster of furin [19], whereas transport from early endosomes to the TGN requires dephosphorylation of the furin acidic cluster by PP2A isoforms [17], and transit through a late endosome intermediate [92]. Maintenance of furin at the TGN then requires PACS-1 binding to the phosphorylated acidic cluster [19]. Thus PACS-1 and CK2 appear to localize furin in one of two local cycling loops, one between the TGN and an associated late endosomal compartment [17], and one between the plasma membrane and early endosomes. Sorting between the two loops requires dephosphorylation of furin by PP2A [17]. The PACS-1-dependent sorting of CPD and HCMV gB between the TGN and cell surface are similarly controlled by the phosphorylation state of their acidic clusters [46,90].
Whereas endocytosed furin traffics from early endosomes to the TGN via a late endosomal intermediate [92], endocytosed CI-MPR, which binds both PACS-1 and PACS-2, follows a different itinerary, sorting from early endosomes to the TGN [49]. Knockdown of PACS-2, but not PACS-1, inhibits the early endosome-to-TGN sorting of an endocytosed CI-MPR reporter [49]. These results suggest that the PACS proteins mediate distinct trafficking steps of endocytosed cargo from the early endosome; PACS-1 directs retrieval of cargo to the cell surface [17], whereas PACS-2 directs sorting deeper into the endocytic pathway [49]. These opposing roles of early endosomal PACS-1 (recycling) and PACS-2 (TGN sorting) suggest why specific PACS-1 mutants block the sorting of endocytosed CI-MPR to the TGN [34]. They also suggest why PACS-1 siRNA fails to disrupt the endocytic itinerary of internalized SorLa [93]. Whether PACS-1 and PACS-2 act independently or sequentially as part of a stepwise sensing process to regulate sorting of cargo from early endosomes to either the cell surface or deeper into endocytic compartments and whether PACS-2 acts in the same pathway as retromer, which is an evolutionally conserved sorting complex that directs early endosome-to-TGN trafficking of several cargo, including CI-MPR warrants further investigation [94].
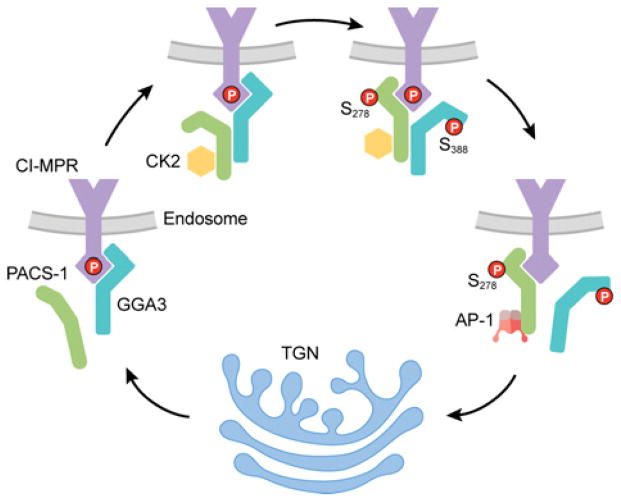
PACS-1 and CK2 regulate GGA3 binding to cargo
The mammalian GGAs are monomeric clathrin adaptors that direct anterograde trafficking of CI-MPR from the TGN to the endosomes [95,96], whereas PACS-1 mediates the local retrieval of CI-MPR to the TGN [15,34]. The CI-MPR cytosolic domain contains overlapping binding sites for GGA3 and PACS-1 (..H2489DDpSDEDLLHI, residues required for maximal binding to PACS-1 are in bold and residues required for binding GGA3 are underlined) and their binding to CI-MPR are mutually exclusive, suggesting separable roles for GGA3 and PACS-1 in mediating CI-MPR traffic [34,97]. Despite the roles of GGA3 and PACS-1 in mediating opposing legs of a CI-MPR local trafficking loop between the TGN and endosomes, GGA3 binds PACS-1 at K249IY and this site is required for localization of CI-MPR to the TGN [34]. Recent studies suggest a model whereby following delivery of CI-MPR to endosomes, retrieval to the TGN requires that PACS-1 bind and displace the bound GGA3. This sorting protein ‘switch’ appears to be controlled by the autoregulatory domains in GGA3 and PACS-1 [34]. Similar to PACS-1, mammalian GGAs contain a CK2-phosphorylatable autoregulatory domain (Ser388 in GGA3) that controls cargo binding [98]. However, whereas phosphorylation of the PACS-1 autoregulatory site at Ser278 promotes binding to CI-MPR, phosphorylation of Ser388 in GGA3 inhibits its binding to CI-MPR [34,98]. In addition, the CK2 bound to the PACS-1 polybasic site not only stimulates Ser278 phosphorylation but also stimulates GGA3 phosphorylation at Ser388, suggesting the presence of a CK2–PACS-1–GGA3 phosphorylation relay that regulates the local cycling of CI-MPR at the TGN (Figure 5). In this model, GGA3 mediates the anterograde traffic of CI-MPR to endosomes, and then PACS-1 is recruited to the endosome where its K249IY motif binds GGA3. CK2 bound to the PACS-1 RRKRY200 motif then phosphorylates GGA3 Ser388 and PACS-1 Ser278 to close and open respectively, their autoregulatory domains, allowing PACS-1 to displace GGA3 and mediate the endosome-to-TGN retrieval of CI-MPR (Figure 5).
Figure 5. Working model of PACS-1/GGA3-regulated trafficking of CI-MPR.
GGA3 transports phosphorylated CI-MPR from the TGN to endosomes [180]. At the endosome, PACS-1 binds to the VHS domain of GGA3 through its FBR and recruits CK2 to phosphorylate Ser388 on GGA3 and phosphorylate Ser278 on PACS-1, thus releasing the autoregulatory domain and activating PACS-1 for cargo binding. CK2 may also phosphorylate additional sites on GGA3 [181] to promote release from endosomal membranes. Phosphorylated GGA3 dissociates, and activated PACS-1 binds to CI-MPR. Bound PACS-1 then recruits AP-1 to transport CI-MPR back to the TGN. It is not known whether CK2 bound to PACS-1 can phosphorylate the CI-MPR tail or other cargo molecules. An animated version of this Figure is available at http://www.BiochemJ.org/bj/421/0001/bj4210001add.htm.
PATHOGENIC VIRUSES TARGET THE PACS PROTEINS
Pathogenic viruses have evolved to express genes that exploit components of the host endomembrane system to replicate, increase tropism and to counterattack the antiviral response. Summarized here are examples of how pathogenic viruses, ranging from HIV-1 to HCMV and KSHV (Kaposi’s sarcoma-associated herpesvirus), exploit the PACS proteins to regulate processes ranging from immunoevasion to virus assembly.
HIV-1 immunoevasion requires the PACS-2 endocytic sorting pathway
The early HIV-1 gene product Nef is required for the efficient onset of AIDS following HIV-1-infection [99–102]. Nef affects cells in many ways, including alteration of T-cell activation and maturation [103–106], subversion of the apoptotic machinery, and the down-regulation of cell-surface molecules, notably CD4 and MHC-I proteins [107–109]. Down-regulation of CD4 to lysosomes through the clathrin/AP-2 pathway eliminates interference of the viral receptor with HIV-1 envelopment or release, whereas down-regulation of cell-surface MHC-I molecules (HLA-A and -B) to paranuclear compartments allows HIV-1 to evade the CD8+ immune surveillance system [110–112].
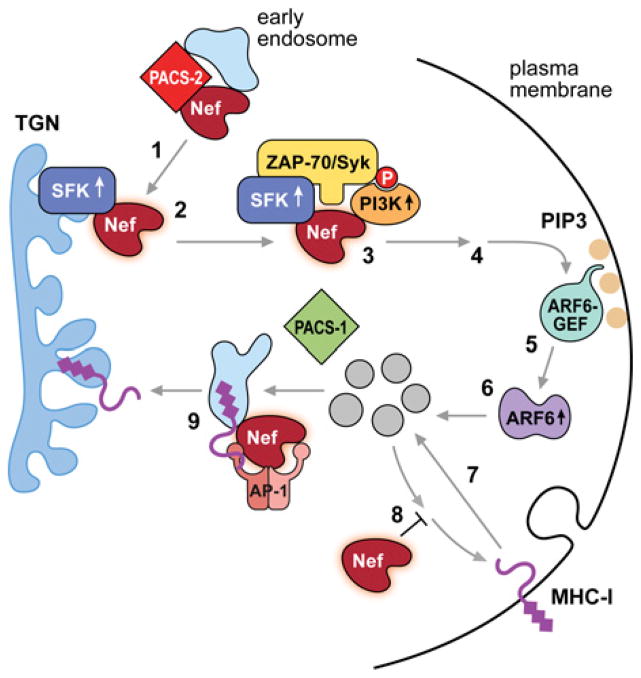
Nef uses separate mechanisms to down-regulate cell-surface CD4 and MHC-I [108,113]. In the case of CD4, Nef acts as a ‘connector’, binding cell-surface CD4 and linking it to the clathrin adaptor AP-2 for delivery to lysosomes for degradation [112,114–116]. In contrast, Nef does not bind cell-surface MHC-I, but rather directs internalization of HLA-A and -B molecules through a clathrin/AP-2/dynamin-independent, ARF6-controlled endocytic pathway [110,117]. This MHC-I down-regulation pathway requires three sites in Nef distinct from those that down-regulate CD4: an N-proximal α-helical region (residues 7–26, see [118]) containing a critical methionine residue (Met20) [119], which mediates interaction with the AP-1 adaptor [120]; an acidic cluster (EEEE65; see [112]), required for maximal binding to PACS-1 and PACS-2 [49,51]; and an SH3-binding domain formed by a polyproline helix (PXXP75, [112,118]) that promotes association of Nef with several kinases, including SFKs (Src family tyrosine kinases) [121–123]. These three sites are conserved in the pandemic M group HIV-1, that account for over 90 % of AIDS cases worldwide, suggesting they control an essential pathway for HIV-1 pathogenesis [124,125].
The EEEE65 and PXXP75 sites act sequentially to assemble a multi-kinase cascade to trigger the MHC-I down-regulation pathway [51]. This signalling cascade begins with the Nef EEEE65-dependent binding to PACS-2 for trafficking Nef to the TGN region, apparently by usurping the PACS-2-dependent early endosome-to-TGN trafficking pathway ([49]; and see Figure 6). This enables Nef PXXP75 to bind and activate a TGN-localized SFK. The activated SFK then recruits and phosphorylates the tyrosine kinase ZAP-70 [ζ-chain (T-cell receptor)-associated protein kinase of 70 kDa] in CD4+ T-cells, or the ZAP-70 homologue Syk in monocytic cells [49,51]. Tyrosine phosphorylated ZAP-70/Syk then ligates the second SH2 domain in the class I PI3K (phosphoinositide 3-kinase) p85α regulatory subunit, thereby recruiting and stimulating the bound class I PI3K p110 catalytic subunit. This pSyk–p85 ligation is not unique to HIV-1 as it is used by phagocytic cells to trigger ARF6-mediated Fc receptor internalization, suggesting that Nef subverts a common cellular pathway triggering ARF6 action [126]. The recruited class I PI3K triggers the ARF6-mediated endocytosis of cell-surface MHC-I. Delivery of internalized MHC-I to the paranuclear region then requires the Nef Met20 site as well as AP-1 [117,120]. Expression of a Nef M20A mutant, which is blocked in AP-1 binding [118], promotes efficient MHC-I endocytosis, but fails to sequester the internalized molecules, which are rapidly recycled to the cell surface leaving the steady-state level of cell-surface MHC-I unaltered [117]. In the absence of PACS-2, either by siRNA knockdown in primary CD4+ T-cells or splenocytes from PACS-2−/− MEFs, Nef is unable to assemble the multi-kinase cascade, thereby repressing MHC-I down-regulation [49]. The required role of PACS-2 for Nef-mediated MHC-I down-regulation is phenocopied in cells from PACS-2−/− mice, confirming genetically an essential role for PACS-2 in this pathway [49]. The ability of isoform-selective class I PI3K inhibitors or small molecules that disrupt the interaction of Nef with SFKs to repress MHC-I down-regulation in primary CD4+ T-cells suggests that Nef may be a candidate target for the development of HIV-1 therapeutics [49,51,127].
Figure 6. ‘Signalling’ model of Nef-mediated down-regulation of cell-surface MHC-I.
Step 1: Nef binds PACS-2 through its acidic cluster (EEEE65) and is targeted to the late Golgi/TGN. Step 2: at the TGN, Nef PXXP75 binds and activates a TGN-localized SFK. Step 3: the activated Nef–SFK complex recruits and activates ZAP-70/Syk. Tyrosine-phosphorylated ZAP-70/Syk then binds a class I PI3K. Step 4: Nef-stimulated class I PI3K generates PIP3 on the inner leaflet of the plasma membrane. Step 5: an ARF6 guanine-nucleotide-exchange factor (ARF6-GEF) is recruited to PIP3 at the plasma membrane. Step 6: the recruited ARF6-GEF in turn activates ARF6. Step 7: MHC-I is rapidly endocytosed from the plasma membrane to internal endosomal compartments. Steps 8 and 9: sequestration of newly internalized MHC-I molecules to a paranuclear region requires the Nef Met20 and its interaction with AP-1. The precise step of Nef-mediated MHC-I down-regulation requiring PACS-1 is unclear but the ability of PACS-1 to bind Nef and AP-1 raises the possibility that PACS-1 may contribute to the Met20-dependent internalization of MHC-I molecules.
An alternative model suggests that Nef acts as a stoichiometric inhibitor to divert newly synthesized MHC-I molecules directly to degradative compartments by a PI3K-independent pathway [113,128,129]. Although these studies explain the importance of Nef Met20 in mediating MHC-I down-regulation, a finding consistent with both models, they do not address the importance of the EEEE65 and PXXP75 sites. The discrepancy between the models was suggested to result from differences in the efficiency by which Nef can impede ER-to-cell-surface transport of overexpressed MHC-I in T-cells compared with HeLa cells [129]. Nef, however, negligibly affects the rate of cell-surface delivery of endogenous MHC-I molecules in CD4+ T-cells under conditions that induce MHC-I down-regulation, suggesting that other factors contribute to the differing results [51]. Possibilities include the use of PTEN (phosphatase and tensin homologue deleted on chromosome 10)-deficient cell lines, which possess inordinate levels of PIP3 (phosphatidylinositol 1,4,5-trisphosphate) and are thus poorly responsive to PI3K inhibitors [51,130,131], and the use of overexpressed MHC-I molecules in studies that described the stoichiometric model of Nef action. Consistent with this possibility, overexpression of MHC-I is coupled with alterations in the Nef sites and partner proteins seen to be required for down-regulation of endogenous MHC-I [33,132]. Thus, despite the assertion that the models are mutually exclusive and controversial [133–135], we suggest that when analysing endogenous MHC-I in normal CD4+ T-cells with a replete PI3K signalling pathway, the two models simply represent the priming (PACS-2-dependent SFK–ZAP-70/Syk–PI3K cascade) and execution (AP-1 interaction) phases of a single pathway Nef uses to enable HIV-1 to escape immune surveillance.
The Nef-mediated, PACS-2-dependent assembly of the SFK–ZAP-70/Syk–PI3K cascade may extend beyond the down-regulation of cell-surface MHC-I. For example, binding of the SFK Hck to Nef at the Golgi region of macrophages is required for down-regulation of the M-CSF (macrophage colony stimulating factor) receptor [136]. In addition, Nef interaction with SFKs and PI3K induces superoxide release from macrophages to enhance HIV-1 pathogenesis [137], and Nef stimulates class I PI3K to drive the TLR4 (Toll-like receptor 4)-mediated suppression of the innate immune system, possibly contributing to secondary mycobacterial infections suffered by AIDS victims [138]. In addition, Nef-mediated ZAP-70 activation is required for PAK2 (p21-activated kinase 2) activation, Fas-ligand up-regulation and formation of virological synapses [139–142]. It will be important to determine whether PACS-2 contributes to these various Nef-controlled pathways.
KSHV pirates the PACS-2 ER trafficking machinery
KSHV is the main cause of Kaposi’s sarcoma, which is the most common AIDS-associated malignancy and is characterized by lesions that contain networks of endothelial-derived spindle-shaped cells [143,144]. KSHV encodes two ubiquitin ligases, K3 and K5, which target cell-surface MHC-I molecules for lysosomal degradation, thereby impairing antigen presentation to CD8+ T-cells [145,146]. Whereas K3 appears to selectively target MHC-I [147], K5 targets additional molecules for destruction, including PECAM-1 (platelet-endothelial cell adhesion molecule-1; also called CD31), which disrupts endothelial cell contacts thereby increasing virus spread [148,149]. The K5-mediated down-regulation of CD31 requires a pair of acidic clusters in its cytosolic domain, EDNIE182 and DEEPTD199, which interact with PACS-2. Dominant-negative PACS-2Admut, but not PACS-1Admut, prevents K5-mediated down-regulation of CD31, suggesting that PACS-2 enables K5 to ubiquitinylate newly synthesized CD31 molecules at the ER, tagging them for proteosomal degradation [149].
The PACS-1 TGN sorting pathway is usurped to regulate virus assembly
HCMV, the prototypical β-herpesvirus, is a major cause of morbidity and mortality in organ transplant recipients and patients with AIDS [150,151], and has evolved to usurp the PACS-1-mediated TGN localization pathway. Like other herpesviruses, HCMV DNA replication, encapsidation and capsid assembly occur in the nucleus [152]. The nucleocapsid traverses the double-membrane of the nuclear envelope to release the core capsid into the cytosol. The cytosolic nucleocapsid obtains the tegument and viral envelope by budding into an assembly compartment formed from TGN and associated endosomal membranes that contain tegument proteins and envelope glycoproteins, including gB [152,153]. Like other herpesvirus envelope glycoproteins, HCMV gB contains a CK2-phosphorylatable acidic cluster in its cytosolic domain, and phosphorylation of this gB acidic cluster at Ser900 directs binding to PACS-1 [46,154]. Disruption of PACS-1 by siRNA knockdown or expression of PACS-1Admut mislocalizes gB from the TGN to endosomal compartments and reduces the titre of infectious progeny [46]. In contrast, overexpression of PACS-1 increases the titre of progeny virus [46]. Consistent with these findings, an HCMV mutant containing a S900A substitution, reduced binding to PACS-1 [46] and, correspondingly, reduced virus titre [154]. These results suggest that PACS-1 mediates localization of gB to TGN-associated compartments to promote virus envelopment.
Whereas herpesvirus assembles by budding into TGN-derived compartments, retroviruses undergo envelopment by budding into MVBs (multivesicular bodies) by an ESCRT/Tg101-dependent process [155]. The interaction of the retroviral capsid with the MVB-localized envelope glycoprotein is regulated by the virus-specific interaction of the envelope protein with different cytosolic sorting proteins [156]. For example, TIP47 (47 kDa tail-interacting protein) binds to an YW motif in the cytosolic domain of the HIV-1 envelope glycoprotein, gp120/41, and links the glycoprotein to Gag in a ternary complex [157,158]. In contrast, PACS-1 regulates envelopment of the feline oncovirus RD114 by its envelope glycoprotein, which possesses a cytosolic acidic cluster [45]. Knockdown of PACS-1 by shRNA (small hairpin RNA) redistributed RD114 from the TGN to late endosomes and increased virus envelopment [45]. By contrast, PACS-1 overexpression concentrated RD114 in the TGN and inhibited virus envelopment [45]. Thus the endosome-to-TGN sorting activity of PACS-1 is used by HCMV to promote virus assembly in TGN-derived compartments, but is used by RD114 to regulate the efficiency of retrovirus assembly in endosomes.
PACS PROTEINS IN CANCER AND APOPTOSIS
The PACS-2 gene on chromosome 14q32.33 is frequently deleted in sporadic cases of colorectal cancer and immunohistochemical studies of PACS-2 protein expression support these findings, suggesting a link between the loss of PACS-2 and the development of colorectal cancer [43,159]. In agreement with this observation, transformed cells depleted of PACS-2 are resistant to apoptosis induced by treatment with staurosporine or by treatment of cells with death ligands such as TNFα or TRAIL [22,43]. Genetic studies show that PACS-2 is a TRAIL effector, required for killing virally infected hepatocytes in vivo in a mouse model of viral hepatitis and colorectal cancer cells in vitro [43] Together with the determination that PACS-2 expression is frequently lost in colorectal cancer [43,159], these findings raise the possibility that loss of PACS-2 expression may contribute to the colorectal cancer sequence and may serve as a biomarker for TRAIL-resistant cancer.
Biochemical studies show that PACS-2 is required by TRAIL for the release of cytochrome c from mitochondria, which allows for the activation of executioner caspases, including caspase 3 [22,43]. In response to apoptotic stimuli, PACS-2 binds the pro-apoptotic Bcl-2 family protein Bid, and mediates its translocation to mitochondria [22]. Like many PACS cargo proteins, Bid contains an acidic cluster and is phosphorylated by CK1 and CK2 in healthy cells and becomes dephosphorylated in response to apoptotic stimuli [160,161]. Similar to the binding of PACS-2 to calnexin [36], PACS-2 binds preferentially to non-phosphorylated Bid and it is this form of Bid that appears to translocate to mitochondria [22,162].
Key to understanding the roles of PACS-2 in integrating membrane traffic and interorganellar communication with apoptosis is a determination of the factors that regulate these varied roles. Recent studies show that PACS-2 is bound to 14-3-3 proteins, which are known to sequester pro-apoptotic molecules [163,164]. 14-3-3 binding to PACS-2 is regulated by the Akt/PKB (protein kinase B) phosphorylation of PACS-2 at Ser437 [43]. In healthy cells, pSer437-PACS-2 is bound to 14-3-3. Upon apoptotic induction by TRAIL or TNFα or other death ligands, pSer437 becomes dephosphorylated, releasing PACS-2 from 14-3-3 and allowing PACS-2 to execute apoptotic programmes [43]. The binding of 14-3-3 to pSer437 may not simply repress PACS-2 activity. Rather, in healthy cells it appears to be required for the ability of PACS-2 to mediate the ER localization of polycystin-2 [43]. Consistent with this finding, TRAIL disrupts the PACS-2-dependent localization of polycystin-2 to the ER, which may augment apoptosis by repressing the anti-apoptotic properties of the ion channel [43,165]. The suppression of PACS-2 apoptotic activity by Akt/14-3-3 binding parallels the regulation of the pro-apoptotic protein Bad ([166]; see Figure 7). Like PACS-2, previous studies suggest that Bad is multifunctional, integrating cellular metabolism in healthy cells with the execution of death programmes in response to apoptotic inducers [167,168].
Figure 7. Working model of Akt/14-3-3/PP2A regulation of PACS-2 apoptotic activity.
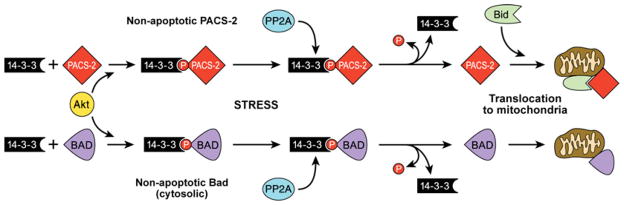
Under non-stressed conditions, apoptotic PACS-2 is phosphorylated on Ser437 and held inactive in the cytosol through binding to 14-3-3 isoforms. Loss of Akt signalling or apoptotic activation of PP2A leads to dephosphorylation of PACS-2 and release from 14-3-3. ‘Activated’ PACS-2 binds and facilitates translocation of full-length Bid to mitochondria, inducing cytochrome c release. This pathway parallels the previously described 14-3-3-regulated translocation of the pro-apoptotic protein Bad [182,183].
Much less is known about the role of PACS-1 in apoptosis and cancer. Interestingly, however, mutation of chromosome 11q13.1, where the PACS-1 gene is located, is associated with the development of several cancers, including cervical carcinoma [164,169,170]. In addition, cervical cancers express elevated levels of an altered PACS-1 transcript, suggesting that dysregulation of PACS-1 might be coupled with the development of cervical cancer [171].
SUMMARY
The initial identification of a role for PACS-1 in mediating the TGN localization of furin suggested the PACS family of membrane traffic regulators would fit neatly into a category of secretory pathway trafficking proteins. However, over the past 10 years, the PACS proteins have become recognized for their roles in integrating membrane traffic with interorganellar communication and, in response to death signals, regulation of apoptotic programmes. These integrative roles of the PACS proteins suggest why they evolved late in evolution, with the formation of metazoan organisms that rely on complex membrane trafficking events to regulate processes ranging from organ homoeostasis to the control of cell death programmes. Their role in key membrane trafficking steps also suggests why pathogenic viruses ranging from herpesviruses to HIV-1 have evolved membrane proteins that usurp PACS functions in the ER, the TGN and in endosomal compartments. Future studies will require a precise determination of the molecular basis of cargo and coat selection and in vivo models to delineate the physiological roles of the PACS proteins.
Acknowledgments
We are greatly indebted to Lori Vaskalis for the illustrations. We thank Joseph Aslan, Matthew Brush and Caroline Enns for helpful discussions and critical reading of the manuscript. We also thank Franco Pissani and Danielle Williamson for assistance with the phylogenetic analysis.
FUNDING
This work was supported by the National Institutes of Health [grant numbers DK37274, AI49793, P50CA97186]; OCTRI (Oregon Clinical and Translational Research Institute) [grant number RR0241 (to G. T.)]; the NRSA (National Research Service Award) fellowship [grant number DK076343 (to R. T. Y.)].
Abbreviations used
- AP-1 etc
adaptor protein complex 1 etc
- ARF
ADP-ribosylation factor
- ARR
atrophin-1-related region
- BAP31
B-cell receptor-associated protein 31
- BFA
brefeldin A
- CI-MPR
cation-independent mannose-6-phosphate receptor
- CK2
casein kinase 2
- CK2β
CK2 regulatory β-subunit
- COPI etc
coatamer protein I etc
- CPD
carboxypeptidase D
- CTR
C-terminal region
- ER
endoplasmic reticulum
- ESCRT
endosomal sorting complex required for transport
- FACL4
fatty acyl-CoA ligase-4
- FBR
furin (cargo)-binding region
- gB
glycoprotein B
- GGA
Golgi-associated γ-adaptin ear homology domain ARF-interacting protein
- HCMV
human cytomegalovirus
- IP3
inositol 1,4,5-trisphosphate
- ISG
immature secretory granule
- IUP
intrinsically unfolded protein
- KSHV
Kaposi’s sarcoma-associated herpesvirus
- MAM
mitochondria-associated membrane
- MR
middle region
- MSG
mature secretory granule
- MVB
multivesicular body
- PACS
phosphofurin acidic cluster sorting protein
- PI3K
phosphoinositide 3-kinase
- PIP3
phosphatidylinositol 1,4,5-trisphosphate
- PP2A
protein phosphatase 2A
- SFK
Src family tyrosine kinase
- siRNA
small interfering RNA
- SNARE
soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptor
- TGN
trans-Golgi network
- TNF
tumour necrosis factor
- TRAIL
TNF-related apoptosis-inducing ligand
- VAMP-4
vesicle-associated membrane protein 4
- ZAP-70
ζ-chain (T-cell receptor)-associated protein kinase of 70 kDa
References
- 1.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 2.McMahon HT, Mills IG. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr Opin Cell Biol. 2004;16:379–391. doi: 10.1016/j.ceb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 4.Fewell SW, Travers KJ, Weissman JS, Brodsky JL. The action of molecular chaperones in the early secretory pathway. Annu Rev Genet. 2001;35:149–191. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- 5.Gu F, Crump CM, Thomas G. Trans-Golgi network sorting. Cell Mol Life Sci. 2001;58:1067–1084. doi: 10.1007/PL00000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 7.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 8.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 9.Dacks JB, Field MC. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J Cell Sci. 2007;120:2977–2985. doi: 10.1242/jcs.013250. [DOI] [PubMed] [Google Scholar]
- 10.de Duve C. The origin of eukaryotes: a reappraisal. Nat Rev Genet. 2007;8:395–403. doi: 10.1038/nrg2071. [DOI] [PubMed] [Google Scholar]
- 11.Dacks JB, Field MC. Eukaryotic cell evolution from a genomic perspective: the endomembrane system. In: Hirt R, Homer D, editors. Organelles, Genomes and Eukaryote Phylogenetics: an Evolutionary Synthesis in the Age of Genomics. CRC; London: 2004. pp. 309–334. [Google Scholar]
- 12.Saraste J, Goud B. Functional symmetry of endomembranes. Mol Biol Cell. 2007;18:1430–1436. doi: 10.1091/mbc.E06-10-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molloy SS, Anderson ED, Jean F, Thomas G. Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 1999;9:28–35. doi: 10.1016/s0962-8924(98)01382-8. [DOI] [PubMed] [Google Scholar]
- 14.Jones BG, Thomas L, Molloy SS, Thulin CD, Fry MD, Walsh KA, Thomas G. Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO J. 1995;14:5869–5883. doi: 10.1002/j.1460-2075.1995.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crump CM, Xiang Y, Thomas L, Gu F, Austin C, Tooze SA, Thomas G. PACS-1 binding to adaptors is required for acidic cluster motif-mediated protein traffic. EMBO J. 2001;20:2191–2201. doi: 10.1093/emboj/20.9.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dittie AS, Thomas L, Thomas G, Tooze SA. Interaction of furin in immature secretory granules from neuroendocrine cells with the AP-1 adaptor complex is modulated by casein kinase II phosphorylation. EMBO J. 1997;16:4859–4870. doi: 10.1093/emboj/16.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molloy SS, Thomas L, Kamibayashi C, Mumby MC, Thomas G. Regulation of endosome sorting by a specific PP2A isoform. J Cell Biol. 1998;142:1399–1411. doi: 10.1083/jcb.142.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molloy SS, Thomas L, VanSlyke JK, Stenberg PE, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL, Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- 20.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thanaraj TA, Stamm S, Clark F, Riethoven JJ, Le Texier V, Muilu J. ASD: the Alternative Splicing Database. Nucleic Acids Res. 2004;32:D64–D69. doi: 10.1093/nar/gkh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simmen T, Aslan JE, Blagoveshchenskaya AD, Thomas L, Wan L, Xiang Y, Feliciangeli SF, Hung CH, Crump CM, Thomas G. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 2005;24:717–729. doi: 10.1038/sj.emboj.7600559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott GK, Gu F, Crump CM, Thomas L, Wan L, Xiang Y, Thomas G. The phosphorylation state of an autoregulatory domain controls PACS-1-directed protein traffic. EMBO J. 2003;22:6234–6244. doi: 10.1093/emboj/cdg596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kottgen M, Benzing T, Simmen T, Tauber R, Buchholz B, Feliciangeli S, Huber TB, Schermer B, Kramer-Zucker A, Hopker K, et al. Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. EMBO J. 2005;24:705–716. doi: 10.1038/sj.emboj.7600566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jahnukainen K, Chrysis D, Hou M, Parvinen M, Eksborg S, Soder O. Increased apoptosis occurring during the first wave of spermatogenesis is stage-specific and primarily affects midpachytene spermatocytes in the rat testis. Biol Reprod. 2004;70:290–296. doi: 10.1095/biolreprod.103.018390. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 28.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 29.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 30.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 31.Cai Y, Maeda Y, Cedzich A, Torres VE, Wu G, Hayashi T, Mochizuki T, Park JH, Witzgall R, Somlo S. Identification and characterization of polycystin-2, the PKD2 gene product. J Biol Chem. 1999;274:28557–28565. doi: 10.1074/jbc.274.40.28557. [DOI] [PubMed] [Google Scholar]
- 32.Simmen T, Nobile M, Bonifacino JS, Hunziker W. Basolateral sorting of furin in MDCK cells requires a phenylalanine-isoleucine motif together with an acidic amino acid cluster. Mol Cell Biol. 1999;19:3136–3144. doi: 10.1128/mcb.19.4.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lubben NB, Sahlender DA, Motley AM, Lehner PJ, Benaroch P, Robinson MS. HIV-1 Nef-induced down-regulation of MHC class I requires AP-1 and clathrin but not PACS-1 and is impeded by AP-2. Mol Biol Cell. 2007;18:3351–3365. doi: 10.1091/mbc.E07-03-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott GK, Fei H, Thomas L, Medigeshi GR, Thomas G. A PACS-1, GGA3 and CK2 complex regulates CI-MPR trafficking. EMBO J. 2006;25:4423–4435. doi: 10.1038/sj.emboj.7601336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y, Doray B, Poussu A, Lehto VP, Kornfeld S. Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science. 2001;292:1716–1718. doi: 10.1126/science.1060896. [DOI] [PubMed] [Google Scholar]
- 36.Myhill N, Lynes EM, Nanji JA, Blagoveshchenskaya AD, Fei H, Simmen KC, Cooper TJ, Thomas G, Simmen T. The Subcellular distribution of calnexin is mediated by PACS-2. Mol Biol Cell. 2008;19:2777–2788. doi: 10.1091/mbc.E07-10-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinners I, Wendler F, Fei H, Thomas L, Thomas G, Tooze SA. AP-1 recruitment to VAMP4 is modulated by phosphorylation-dependent binding of PACS-1. EMBO Rep. 2003;4:1182–1189. doi: 10.1038/sj.embor.7400018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Litchfield DW. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J. 2003;369:1–15. doi: 10.1042/BJ20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonnet H, Filhol O, Truchet I, Brethenou P, Cochet C, Amalric F, Bouche G. Fibroblast growth factor-2 binds to the regulatory beta subunit of CK2 and directly stimulates CK2 activity toward nucleolin. J Biol Chem. 1996;271:24781–24787. doi: 10.1074/jbc.271.40.24781. [DOI] [PubMed] [Google Scholar]
- 40.Leroy D, Heriche JK, Filhol O, Chambaz EM, Cochet C. Binding of polyamines to an autonomous domain of the regulatory subunit of protein kinase CK2 induces a conformational change in the holoenzyme. A proposed role for the kinase stimulation. J Biol Chem. 1997;272:20820–20827. doi: 10.1074/jbc.272.33.20820. [DOI] [PubMed] [Google Scholar]
- 41.Sayed M, Kim SO, Salh BS, Issinger OG, Pelech SL. Stress-induced activation of protein kinase CK2 by direct interaction with p38 mitogen-activated protein kinase. J Biol Chem. 2000;275:16569–16573. doi: 10.1074/jbc.M000312200. [DOI] [PubMed] [Google Scholar]
- 42.Koffa MD, Kean J, Zachos G, Rice SA, Clements JB. CK2 protein kinase is stimulated and redistributed by functional herpes simplex virus ICP27 protein. J Virol. 2003;77:4315–4325. doi: 10.1128/JVI.77.7.4315-4325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aslan JE, You H, Williamson DM, Endig J, Youker RT, Thomas L, Shu H, Du Y, Milewski RL, Brush MH, et al. Akt and 14-3-3 control a PACS-2 homeostatic switch that integrates membrane traffic with TRAIL-induced apoptosis. Mol Cell. 2009 doi: 10.1016/j.molcel.2009.04.011. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sieburth D, Ch’ng Q, Dybbs M, Tavazoie M, Kennedy S, Wang D, Dupuy D, Rual JF, Hill DE, Vidal M, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 45.Bouard D, Sandrin V, Boson B, Negre D, Thomas G, Granier C, Cosset FL. An acidic cluster of the cytoplasmic tail of the RD114 virus glycoprotein controls assembly of retroviral envelopes. Traffic. 2007;8:835–847. doi: 10.1111/j.1600-0854.2007.00581.x. [DOI] [PubMed] [Google Scholar]
- 46.Crump CM, Hung CH, Thomas L, Wan L, Thomas G. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J Virol. 2003;77:11105–11113. doi: 10.1128/JVI.77.20.11105-11113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schermer B, Hopker K, Omran H, Ghenoiu C, Fliegauf M, Fekete A, Horvath J, Kottgen M, Hackl M, Zschiedrich S, et al. Phosphorylation by casein kinase 2 induces PACS-1 binding of nephrocystin and targeting to cilia. EMBO J. 2005;24:4415–4424. doi: 10.1038/sj.emboj.7600885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feliciangeli SF, Thomas L, Scott GK, Subbian E, Hung CH, Molloy SS, Jean F, Shinde U, Thomas G. Identification of a pH sensor in the furin propeptide that regulates enzyme activation. J Biol Chem. 2006;281:16108–16116. doi: 10.1074/jbc.M600760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atkins KM, Thomas L, Youker RT, Harriff MJ, Pissani F, You H, Thomas G. HIV-1 Nef binds PACS-2 to assemble a multikinase cascade that triggers major histocompatibility complex class I (MHC-I) down-regulation: analysis using short interfering RNA and knock-out mice. J Biol Chem. 2008;283:11772–11784. doi: 10.1074/jbc.M707572200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu X, Wang Y, Schetle N, Gao H, Putz M, von Gersdorff G, Walz G, Kramer-Zucker AG. The subcellular localization of TRPP2 modulates its function. J Am Soc Nephrol. 2008;19:1342–1351. doi: 10.1681/ASN.2007070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hung CH, Thomas L, Ruby CE, Atkins KM, Morris NP, Knight ZA, Scholz I, Barklis E, Weinberg AD, Shokat KM, Thomas G. HIV-1 Nef assembles a Src family kinase-ZAP-70/Syk-PI3K cascade to downregulate cell-surface MHC-I. Cell Host Microbe. 2007;1:121–133. doi: 10.1016/j.chom.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Deutsch C. The birth of a channel. Neuron. 2003;40:265–276. doi: 10.1016/s0896-6273(03)00506-3. [DOI] [PubMed] [Google Scholar]
- 53.Zerangue N, Schwappach B, Jan YN, Jan LY. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 54.Michelsen K, Yuan H, Schwappach B. Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 2005;6:717–722. doi: 10.1038/sj.embor.7400480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koulen P, Cai Y, Geng L, Maeda Y, Nishimura S, Witzgall R, Ehrlich BE, Somlo S. Polycystin-2 is an intracellular calcium release channel. Nat Cell Biol. 2002;4:191–197. doi: 10.1038/ncb754. [DOI] [PubMed] [Google Scholar]
- 56.Li Y, Wright JM, Qian F, Germino GG, Guggino WB. Polycystin 2 interacts with type I inositol 1,4,5-trisphosphate receptor to modulate intracellular Ca2+ signaling. J Biol Chem. 2005;280:41298–41306. doi: 10.1074/jbc.M510082200. [DOI] [PubMed] [Google Scholar]
- 57.Giamarchi A, Padilla F, Coste B, Raoux M, Crest M, Honore E, Delmas P. The versatile nature of the calcium-permeable cation channel TRPP2. EMBO Rep. 2006;7:787–793. doi: 10.1038/sj.embor.7400745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anyatonwu GI, Estrada M, Tian X, Somlo S, Ehrlich BE. Regulation of ryanodine receptor-dependent calcium signaling by polycystin-2. Proc Natl Acad Sci USA. 2007;104:6454–6459. doi: 10.1073/pnas.0610324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhunia AK, Piontek K, Boletta A, Liu L, Qian F, Xu PN, Germino FJ, Germino GG. PKD1 induces p21(waf1) and regulation of the cell cycle via direct activation of the JAK-STAT signaling pathway in a process requiring PKD2. Cell. 2002;109:157–168. doi: 10.1016/s0092-8674(02)00716-x. [DOI] [PubMed] [Google Scholar]
- 60.Li X, Luo Y, Starremans PG, McNamara CA, Pei Y, Zhou J. Polycystin-1 and polycystin-2 regulate the cell cycle through the helix-loop-helix inhibitor Id2. Nat Cell Biol. 2005;7:1202–1212. doi: 10.1038/ncb1326. [DOI] [PubMed] [Google Scholar]
- 61.Boletta A, Qian F, Onuchic LF, Bhunia AK, Phakdeekitcharoen B, Hanaoka K, Guggino W, Monaco L, Germino GG. Polycystin-1, the gene product of PKD1, induces resistance to apoptosis and spontaneous tubulogenesis in MDCK cells. Mol Cell. 2000;6:1267–1273. doi: 10.1016/s1097-2765(00)00123-4. [DOI] [PubMed] [Google Scholar]
- 62.Low SH, Vasanth S, Larson CH, Mukherjee S, Sharma N, Kinter MT, Kane ME, Obara T, Weimbs T. Polycystin-1, STAT6, and P100 function in a pathway that transduces ciliary mechanosensation and is activated in polycystic kidney disease. Dev Cell. 2006;10:57–69. doi: 10.1016/j.devcel.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 63.Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AE, Lu W, Brown EM, Quinn SJ, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 64.Sutters M, Germino GG. Autosomal dominant polycystic kidney disease: molecular genetics and pathophysiology. J Lab Clin Med. 2003;141:91–101. doi: 10.1067/mlc.2003.13. [DOI] [PubMed] [Google Scholar]
- 65.Delmas P, Padilla F, Osorio N, Coste B, Raoux M, Crest M. Polycystins, calcium signaling, and human diseases. Biochem Biophys Res Commun. 2004;322:1374–1383. doi: 10.1016/j.bbrc.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 66.Benzing T, Gerke P, Hopker K, Hildebrandt F, Kim E, Walz G. Nephrocystin interacts with Pyk2, p130(Cas), and tensin and triggers phosphorylation of Pyk2. Proc Natl Acad Sci USA. 2001;98:9784–9789. doi: 10.1073/pnas.171269898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fliegauf M, Horvath J, von Schnakenburg C, Olbrich H, Muller D, Thumfart J, Schermer B, Pazour GJ, Neumann HP, Zentgraf H, et al. Nephrocystin specifically localizes to the transition zone of renal and respiratory cilia and photoreceptor connecting cilia. J Am Soc Nephrol. 2006;17:2424–2433. doi: 10.1681/ASN.2005121351. [DOI] [PubMed] [Google Scholar]
- 68.Hildebrandt F, Otto E. Molecular genetics of nephronophthisis and medullary cystic kidney disease. J Am Soc Nephrol. 2000;11:1753–1761. doi: 10.1681/ASN.V1191753. [DOI] [PubMed] [Google Scholar]
- 69.Farr CD, Gafken PR, Norbeck AD, Doneanu CE, Stapels MD, Barofsky DF, Minami M, Saugstad JA. Proteomic analysis of native metabotropic glutamate receptor 5 protein complexes reveals novel molecular constituents. J Neurochem. 2004;91:438–450. doi: 10.1111/j.1471-4159.2004.02735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marsh BJ, Mastronarde DN, Buttle KF, Howell KE, McIntosh JR. Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc Natl Acad Sci USA. 2001;98:2399–2406. doi: 10.1073/pnas.051631998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 72.Mannella CA, Buttle K, Rath BK, Marko M. Electron microscopic tomography of rat-liver mitochondria and their interaction with the endoplasmic reticulum. Biofactors. 1998;8:225–228. doi: 10.1002/biof.5520080309. [DOI] [PubMed] [Google Scholar]
- 73.Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol. 2009;19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berridge MJ. The endoplasmic reticulum: a multifunctional signaling organelle. Cell Calcium. 2002;32:235–249. doi: 10.1016/s0143416002001823. [DOI] [PubMed] [Google Scholar]
- 76.Franke WW, Kartenbeck J. Outer mitochondrial membrane continuous with endoplasmic reticulum. Protoplasma. 1971;73:35–41. doi: 10.1007/BF01286409. [DOI] [PubMed] [Google Scholar]
- 77.Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- 78.Achleitner G, Gaigg B, Krasser A, Kainersdorfer E, Kohlwein SD, Perktold A, Zellnig G, Daum G. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem. 1999;264:545–553. doi: 10.1046/j.1432-1327.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 79.Gaigg B, Simbeni R, Hrastnik C, Paltauf F, Daum G. Characterization of a microsomal subfraction associated with mitochondria of the yeast, Saccharomyces cerevisiae Involvement in synthesis and import of phospholipids into mitochondria. Biochim Biophys Acta. 1995;1234:214–220. doi: 10.1016/0005-2736(94)00287-y. [DOI] [PubMed] [Google Scholar]
- 80.Vance JE. Molecular and cell biology of phosphatidylserine and phosphatidylethanolamine metabolism. Prog Nucleic Acid Res Mol Biol. 2003;75:69–111. doi: 10.1016/s0079-6603(03)75003-x. [DOI] [PubMed] [Google Scholar]
- 81.Takahashi S, Nakagawa T, Banno T, Watanabe T, Murakami K, Nakayama K. Localization of furin to the trans-Golgi network and recycling from the cell surface involves Ser and Tyr residues within the cytoplasmic domain. J Biol Chem. 1995;270:28397–28401. doi: 10.1074/jbc.270.47.28397. [DOI] [PubMed] [Google Scholar]
- 82.Bosshart H, Humphrey J, Deignan E, Davidson J, Drazba J, Yuan L, Oorschot V, Peters PJ, Bonifacino JS. The cytoplasmic domain mediates localization of furin to the trans-Golgi network en route to the endosomal/lysosomal system. J Cell Biol. 1994;126:1157–1172. doi: 10.1083/jcb.126.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schafer W, Stroh A, Berghofer S, Seiler J, Vey M, Kruse ML, Kern HF, Klenk HD, Garten W. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO J. 1995;14:2424–2435. doi: 10.1002/j.1460-2075.1995.tb07240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, Willnow TE. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem. 2007;282:32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- 85.Xiang Y, Molloy SS, Thomas L, Thomas G. The PC6B cytoplasmic domain contains two acidic clusters that direct sorting to distinct trans-Golgi network/endosomal compartments. Mol Biol Cell. 2000;11:1257–1273. doi: 10.1091/mbc.11.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tooze SA. Biogenesis of secretory granules in the trans-Golgi network of neuroendocrine and endocrine cells. Biochim Biophys Acta. 1998;1404:231–244. doi: 10.1016/S0167-4889(98)00059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rudolf R, Salm T, Rustom A, Gerdes HH. Dynamics of immature secretory granules: role of cytoskeletal elements during transport, cortical restriction, and F-actin-dependent tethering. Mol Biol Cell. 2001;12:1353–1365. doi: 10.1091/mbc.12.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Littleton JT, Pallanck L, Ganetzky B. Mechanisms of neurotransmitter release. Int Rev Neurobiol. 1999;43:139–161. doi: 10.1016/s0074-7742(08)60544-9. [DOI] [PubMed] [Google Scholar]
- 89.Varlamov O, Fricker LD. Intracellular trafficking of metallocarboxypeptidase D in AtT-20 cells: localization to the trans-Golgi network and recycling from the cell surface. J Cell Sci. 1998;111:877–885. doi: 10.1242/jcs.111.7.877. [DOI] [PubMed] [Google Scholar]
- 90.Varlamov O, Eng FJ, Novikova EG, Fricker LD. Localization of metallocarboxypeptidase D in AtT-20 cells. Potential role in prohormone processing. J Biol Chem. 1999;274:14759–14767. doi: 10.1074/jbc.274.21.14759. [DOI] [PubMed] [Google Scholar]
- 91.Waites CL, Mehta A, Tan PK, Thomas G, Edwards RH, Krantz DE. An acidic motif retains vesicular monoamine transporter 2 on large dense core vesicles. J Cell Biol. 2001;152:1159–1168. doi: 10.1083/jcb.152.6.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mallet WG, Maxfield FR. Chimeric forms of furin and TGN38 are transported with the plasma membrane in the trans-Golgi network via distinct endosomal pathways. J Cell Biol. 1999;146:345–359. doi: 10.1083/jcb.146.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nielsen MS, Gustafsen C, Madsen P, Nyengaard JR, Hermey G, Bakke O, Mari M, Schu P, Pohlmann R, Dennes A, Petersen CM. Sorting by the cytoplasmic domain of the amyloid precursor protein binding receptor SorLA. Mol Cell Biol. 2007;27:6842–6851. doi: 10.1128/MCB.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Doray B, Ghosh P, Griffith J, Geuze HJ, Kornfeld S. Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science. 2002;297:1700–1703. doi: 10.1126/science.1075327. [DOI] [PubMed] [Google Scholar]
- 96.Klumperman J, Hille A, Veenendaal T, Oorschot V, Stoorvogel W, von Figura K, Geuze HJ. Differences in the endosomal distributions of the two mannose 6-phosphate receptors. J Cell Biol. 1993;121:997–1010. doi: 10.1083/jcb.121.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Puertollano R, Aguilar RC, Gorshkova I, Crouch RJ, Bonifacino JS. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science. 2001;292:1712–1716. doi: 10.1126/science.1060750. [DOI] [PubMed] [Google Scholar]
- 98.Ghosh P, Kornfeld S. Phosphorylation-induced conformational changes regulate GGAs 1 and 3 function at the trans-Golgi network. J Biol Chem. 2003;278:14543–14549. doi: 10.1074/jbc.M212543200. [DOI] [PubMed] [Google Scholar]
- 99.Kestler HW, 3rd, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 100.Sinclair E, Barbosa P, Feinberg MB. The nef gene products of both simian and human immunodeficiency viruses enhance virus infectivity and are functionally interchangeable. J Virol. 1997;71:3641–3651. doi: 10.1128/jvi.71.5.3641-3651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hanna Z, Kay DG, Rebai N, Guimond A, Jothy S, Jolicoeur P. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell. 1998;95:163–175. doi: 10.1016/s0092-8674(00)81748-1. [DOI] [PubMed] [Google Scholar]
- 102.Quaranta MG, Mattioli B, Giordani L, Viora M. The immunoregulatory effects of HIV-1 Nef on dendritic cells and the pathogenesis of AIDS. FASEB J. 2006;20:2198–2208. doi: 10.1096/fj.06-6260rev. [DOI] [PubMed] [Google Scholar]
- 103.Rhee SS, Marsh JW. HIV-1 Nef activity in murine T cells. CD4 modulation and positive enhancement. J Immunol. 1994;152:5128–5134. [PubMed] [Google Scholar]
- 104.Schrager JA, Marsh JW. HIV-1 Nef increases T cell activation in a stimulus-dependent manner. Proc Natl Acad Sci USA. 1999;96:8167–8172. doi: 10.1073/pnas.96.14.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Swingler S, Brichacek B, Jacque JM, Ulich C, Zhou J, Stevenson M. HIV-1 Nef intersects the macrophage CD40L signalling pathway to promote resting-cell infection. Nature. 2003;424:213–219. doi: 10.1038/nature01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stove V, Naessens E, Stove C, Swigut T, Plum J, Verhasselt B. Signaling but not trafficking function of HIV-1 protein Nef is essential for Nef-induced defects in human intrathymic T-cell development. Blood. 2003;102:2925–2932. doi: 10.1182/blood-2003-03-0833. [DOI] [PubMed] [Google Scholar]
- 107.Piguet V, Gu F, Foti M, Demaurex N, Gruenberg J, Carpentier JL, Trono D. Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of β-COP in endosomes. Cell. 1999;97:63–73. doi: 10.1016/s0092-8674(00)80715-1. [DOI] [PubMed] [Google Scholar]
- 108.Lindwasser OW, Chaudhuri R, Bonifacino JS. Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Curr Mol Med. 2007;7:171–184. doi: 10.2174/156652407780059177. [DOI] [PubMed] [Google Scholar]
- 109.Foster JL, Garcia JV. Role of Nef in HIV-1 replication and pathogenesis. Adv Pharmacol. 2007;55:389–409. doi: 10.1016/S1054-3589(07)55011-8. [DOI] [PubMed] [Google Scholar]
- 110.Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol. 2007;81:3877–3890. doi: 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 112.Greenberg M, DeTulleo L, Rapoport I, Skowronski J, Kirchhausen T. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr Biol. 1998;8:1239–1242. doi: 10.1016/s0960-9822(07)00518-0. [DOI] [PubMed] [Google Scholar]
- 113.Roeth JF, Collins KL. Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways. Microbiol Mol Biol Rev. 2006;70:548–563. doi: 10.1128/MMBR.00042-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 115.Jin YJ, Cai CY, Zhang X, Zhang HT, Hirst JA, Burakoff SJ. HIV Nef-mediated CD4 down-regulation is adaptor protein complex 2 dependent. J Immunol. 2005;175:3157–3164. doi: 10.4049/jimmunol.175.5.3157. [DOI] [PubMed] [Google Scholar]
- 116.Lindwasser OW, Smith WJ, Chaudhuri R, Yang P, Hurley JH, Bonifacino JS. A diacidic motif in human immunodeficiency virus type 1 Nef is a novel determinant of binding to AP-2. J Virol. 2008;82:1166–1174. doi: 10.1128/JVI.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Blagoveshchenskaya AD, Thomas L, Feliciangeli SF, Hung CH, Thomas G. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell. 2002;111:853–866. doi: 10.1016/s0092-8674(02)01162-5. [DOI] [PubMed] [Google Scholar]
- 118.Mangasarian A, Piguet V, Wang JK, Chen YL, Trono D. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal α helix and proline repeat of Nef selectively regulate MHC-I trafficking. J Virol. 1999;73:1964–1973. doi: 10.1128/jvi.73.3.1964-1973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Akari H, Arold S, Fukumori T, Okazaki T, Strebel K, Adachi A. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J Virol. 2000;74:2907–2912. doi: 10.1128/jvi.74.6.2907-2912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roeth JF, Williams M, Kasper MR, Filzen TM, Collins KL. HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J Cell Biol. 2004;167:903–913. doi: 10.1083/jcb.200407031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Trible RP, Emert-Sedlak L, Smithgall TE. HIV-1 Nef selectively activates Src family kinases Hck, Lyn, and c-Src through direct SH3 domain interaction. J Biol Chem. 2006;281:27029–27038. doi: 10.1074/jbc.M601128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee CH, Leung B, Lemmon MA, Zheng J, Cowburn D, Kuriyan J, Saksela K. A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J. 1995;14:5006–5015. doi: 10.1002/j.1460-2075.1995.tb00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Renkema GH, Saksela K. Interactions of HIV-1 NEF with cellular signal transducing proteins. Front Biosci. 2000;5:D268–D283. doi: 10.2741/renkema. [DOI] [PubMed] [Google Scholar]
- 124.Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Agopian K, Wei BL, Garcia JV, Gabuzda D. CD4 and MHC-I downregulation are conserved in primary HIV-1 Nef alleles from brain and lymphoid tissues, but Pak2 activation is highly variable. Virology. 2007;358:119–135. doi: 10.1016/j.virol.2006.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moon KD, Post CB, Durden DL, Zhou Q, De P, Harrison ML, Geahlen RL. Molecular basis for a direct interaction between the Syk protein-tyrosine kinase and phosphoinositide 3-kinase. J Biol Chem. 2005;280:1543–1551. doi: 10.1074/jbc.M407805200. [DOI] [PubMed] [Google Scholar]
- 127.Betzi S, Restouin A, Opi S, Arold ST, Parrot I, Guerlesquin F, Morelli X, Collette Y. Protein protein interaction inhibition (2P2I) combining high throughput and virtual screening: application to the HIV-1 Nef protein. Proc Natl Acad Sci USA. 2007;104:19256–19261. doi: 10.1073/pnas.0707130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kasper MR, Collins KL. Nef-mediated disruption of HLA-A2 transport to the cell surface in T cells. J Virol. 2003;77:3041–3049. doi: 10.1128/JVI.77.5.3041-3049.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kasper MR, Roeth JF, Williams M, Filzen TM, Fleis RI, Collins KL. HIV-1 Nef disrupts antigen presentation early in the secretory pathway. J Biol Chem. 2005;280:12840–12848. doi: 10.1074/jbc.M413538200. [DOI] [PubMed] [Google Scholar]
- 130.Astoul E, Edmunds C, Cantrell DA, Ward SG. PI 3-K and T-cell activation: limitations of T-leukemic cell lines as signaling models. Trends Immunol. 2001;22:490–496. doi: 10.1016/s1471-4906(01)01973-1. [DOI] [PubMed] [Google Scholar]
- 131.Deane JA, Fruman DA. Phosphoinositide 3-kinase: diverse roles in immune cell activation. Annu Rev Immunol. 2004;22:563–598. doi: 10.1146/annurev.immunol.22.012703.104721. [DOI] [PubMed] [Google Scholar]
- 132.Fackler OT, Moris A, Tibroni N, Giese SI, Glass B, Schwartz O, Krausslich HG. Functional characterization of HIV-1 Nef mutants in the context of viral infection. Virology. 2006;351:322–339. doi: 10.1016/j.virol.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 133.Baugh LL, Garcia JV, Foster JL. Functional characterization of the human immunodeficiency virus type 1 Nef acidic domain. J Virol. 2008;82:9657–9667. doi: 10.1128/JVI.00107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wonderlich ER, Williams M, Collins KL. The tyrosine binding pocket in the adaptor protein 1 (AP-1) mu1 subunit is necessary for Nef to recruit AP-1 to the major histocompatibility complex class I cytoplasmic tail. J Biol Chem. 2008;283:3011–3022. doi: 10.1074/jbc.M707760200. [DOI] [PubMed] [Google Scholar]
- 135.Noviello CM, Benichou S, Guatelli JC. Cooperative binding of the class I major histocompatibility complex cytoplasmic domain and human immunodeficiency virus type 1 Nef to the endosomal AP-1 complex via its mu subunit. J Virol. 2008;82:1249–1258. doi: 10.1128/JVI.00660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hiyoshi M, Suzu S, Yoshidomi Y, Hassan R, Harada H, Sakashita N, Akari H, Motoyoshi K, Okada S. Interaction between Hck and HIV-1 Nef negatively regulates cell surface expression of M-CSF receptor. Blood. 2008;111:243–250. doi: 10.1182/blood-2007-04-086017. [DOI] [PubMed] [Google Scholar]
- 137.Olivetta E, Mallozzi C, Ruggieri V, Pietraforte D, Federico M, Sanchez M. HIV-1 Nef induces p47(phox) phosphorylation leading to a rapid superoxide anion release from the U937 human monoblastic cell line. J Cell Biochem. 2009;106:812–822. doi: 10.1002/jcb.22041. [DOI] [PubMed] [Google Scholar]
- 138.Tachado SD, Li X, Swan K, Patel N, Koziel H. Constitutive activation of phosphatidylinositol 3-kinase signaling pathway down-regulates TLR4-mediated tumor necrosis factor-α release in alveolar macrophages from asymptomatic HIV-positive persons in vitro. J Biol Chem. 2008;283:33191–33198. doi: 10.1074/jbc.M805067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sol-Foulon N, Sourisseau M, Porrot F, Thoulouze MI, Trouillet C, Nobile C, Blanchet F, di Bartolo V, Noraz N, Taylor N, et al. ZAP-70 kinase regulates HIV cell-to-cell spread and virological synapse formation. EMBO J. 2007;26:516–526. doi: 10.1038/sj.emboj.7601509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zauli G, Gibellini D, Secchiero P, Dutartre H, Olive D, Capitani S, Collette Y. Human immunodeficiency virus type 1 Nef protein sensitizes CD4(+) T lymphoid cells to apoptosis via functional upregulation of the CD95/CD95 ligand pathway. Blood. 1999;93:1000–1010. [PubMed] [Google Scholar]
- 141.Das SR, Jameel S. Biology of the HIV Nef protein. Indian J Med Res. 2005;121:315–332. [PubMed] [Google Scholar]
- 142.Peterlin BM, Trono D. Hide, shield and strike back: how HIV-infected cells avoid immune eradication. Nat Rev Immunol. 2003;3:97–107. doi: 10.1038/nri998. [DOI] [PubMed] [Google Scholar]
- 143.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 144.Ganem D. KSHV infection and the pathogenesis of Kaposi’s sarcoma. Annu Rev Pathol. 2006;1:273–296. doi: 10.1146/annurev.pathol.1.110304.100133. [DOI] [PubMed] [Google Scholar]
- 145.Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo HG, Reitz MS, Hayward GS. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Moore PS, Chang Y. Molecular virology of Kaposi’s sarcoma-associated herpesvirus. Philos Trans R Soc London Ser B. 2001;356:499–516. doi: 10.1098/rstb.2000.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ishido S, Wang C, Lee BS, Cohen GB, Jung JU. Downregulation of major histocompatibility complex class I molecules by Kaposi’s sarcoma-associated herpesvirus K3 and K5 proteins. J Virol. 2000;74:5300–5309. doi: 10.1128/jvi.74.11.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Means RE, Lang SM, Jung JU. The Kaposi’s sarcoma-associated herpesvirus K5 E3 ubiquitin ligase modulates targets by multiple molecular mechanisms. J Virol. 2007;81:6573–6583. doi: 10.1128/JVI.02751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mansouri M, Douglas J, Rose PP, Gouveia K, Thomas G, Means RE, Moses AV, Fruh K. Kaposi sarcoma herpesvirus K5 removes CD31/PECAM from endothelial cells. Blood. 2006;108:1932–1940. doi: 10.1182/blood-2005-11-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Britt WJ, Alford CA. Cytomegalovirus. In: Fieldsm BN, Knipe DM, Howley PM, editors. Fields Virology. Lippincott-Raven Publishers; Philadelphia, PA: 1996. pp. 2493–2523. [Google Scholar]
- 151.Vancikova Z, Dvorak P. Cytomegalovirus infection in immunocompetent and immunocompromised individuals: a review. Curr Drug Targets Immune Endocr Metabol Disord. 2001;1:179–187. [PubMed] [Google Scholar]
- 152.Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: a tale of two membranes. Curr Opin Microbiol. 2006;9:423–429. doi: 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 153.Sanchez V, Greis KD, Sztul E, Britt WJ. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol. 2000;74:975–986. doi: 10.1128/jvi.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jarvis MA, Jones TR, Drummond DD, Smith PP, Britt WJ, Nelson JA, Baldick CJ. Phosphorylation of human cytomegalovirus glycoprotein B (gB) at the acidic cluster casein kinase 2 site (Ser900) is required for localization of gB to the trans-Golgi network and efficient virus replication. J Virol. 2004;78:285–293. doi: 10.1128/JVI.78.1.285-293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol. 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- 156.Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 157.Blot G, Janvier K, Le Panse S, Benarous R, Berlioz-Torrent C. Targeting of the human immunodeficiency virus type 1 envelope to the trans-Golgi network through binding to TIP47 is required for env incorporation into virions and infectivity. J Virol. 2003;77:6931–6945. doi: 10.1128/JVI.77.12.6931-6945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lopez-Verges S, Camus G, Blot G, Beauvoir R, Benarous R, Berlioz-Torrent C. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc Natl Acad Sci USA. 2006;103:14947–14952. doi: 10.1073/pnas.0602941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Anderson GR, Brenner BM, Swede H, Chen N, Henry WM, Conroy JM, Karpenko MJ, Issa JP, Bartos JD, Brunelle JK, et al. Intrachromosomal genomic instability in human sporadic colorectal cancer measured by genome-wide allelotyping and inter-(simple sequence repeat) PCR. Cancer Res. 2001;61:8274–8283. [PubMed] [Google Scholar]
- 160.Desagher S, Osen-Sand A, Montessuit S, Magnenat E, Vilbois F, Hochmann A, Journot L, Antonsson B, Martinou JC. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol Cell. 2001;8:601–611. doi: 10.1016/s1097-2765(01)00335-5. [DOI] [PubMed] [Google Scholar]
- 161.Vogel A, Aslan JE, Willenbring H, Klein C, Finegold M, Mount H, Thomas G, Grompe M. Sustained phosphorylation of Bid is a marker for resistance to Fas-induced apoptosis during chronic liver diseases. Gastroenterology. 2006;130:104–119. doi: 10.1053/j.gastro.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tafani M, Karpinich NO, Hurster KA, Pastorino JG, Schneider T, Russo MA, Farber JL. Cytochrome c release upon Fas receptor activation depends on translocation of full-length bid and the induction of the mitochondrial permeability transition. J Biol Chem. 2002;277:10073–10082. doi: 10.1074/jbc.M111350200. [DOI] [PubMed] [Google Scholar]
- 163.Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Aslan JE, Thomas G. Death by committee: organellar trafficking and communication in apoptosis. Traffic. 2009 doi: 10.1111/j.1600-0854.2009.00951.x. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wegierski T, Steffl D, Kopp C, Tauber R, Buchholz B, Nitschke R, Kuehn EW, Walz G, Kottgen M. TRPP2 channels regulate apoptosis through the Ca2+ concentration in the endoplasmic reticulum. EMBO J. 2009;28:490–499. doi: 10.1038/emboj.2008.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 167.Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- 168.Danial NN, Walensky LD, Zhang CY, Choi CS, Fisher JK, Molina AJ, Datta SR, Pitter KL, Bird GH, Wikstrom JD, et al. Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat Med. 2008;14:144–153. doi: 10.1038/nm1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Srivatsan ES, Chakrabarti R, Zainabadi K, Pack SD, Benyamini P, Mendonca MS, Yang PK, Kang K, Motamedi D, Sawicki MP, et al. Localization of deletion to a 300 Kb interval of chromosome 11q13 in cervical cancer. Oncogene. 2002;21:5631–5642. doi: 10.1038/sj.onc.1205698. [DOI] [PubMed] [Google Scholar]
- 170.Cheng Y, Chakrabarti R, Garcia-Barcelo M, Ha TJ, Srivatsan ES, Stanbridge EJ, Lung ML. Mapping of nasopharyngeal carcinoma tumor-suppressive activity to a 1.8-megabase region of chromosome band 11q13. Genes Chromosomes Cancer. 2002;34:97–103. doi: 10.1002/gcc.10048. [DOI] [PubMed] [Google Scholar]
- 171.Zainabadi K, Benyamini P, Chakrabarti R, Veena MS, Chandrasekharappa SC, Gatti RA, Srivatsan ES. A 700-kb physical and transcription map of the cervical cancer tumor suppressor gene locus on chromosome 11q13. Genomics. 2005;85:704–714. doi: 10.1016/j.ygeno.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 172.Subbian E, Yabuta Y, Shinde U. Positive selection dictates the choice between kinetic and thermodynamic protein folding and stability in subtilases. Biochemistry. 2004;43:14348–14360. doi: 10.1021/bi048397x. [DOI] [PubMed] [Google Scholar]
- 173.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 174.Yang Z. Inference of selection from multiple species alignments. Curr Opin Genet Dev. 2002;12:688–694. doi: 10.1016/s0959-437x(02)00348-9. [DOI] [PubMed] [Google Scholar]
- 175.Bielawski JP, Yang Z. Maximum likelihood methods for detecting adaptive evolution after gene duplication. J Struct Funct Genomics. 2003;3:201–212. [PubMed] [Google Scholar]
- 176.Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18:1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
- 177.Das R, Baker D. Macromolecular Modeling with Rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 178.Reference deleted
- 179.Reference deleted
- 180.Meresse S, Hoflack B. Phosphorylation of the cation-independent mannose 6-phosphate receptor is closely associated with its exit from the trans-Golgi network. J Cell Biol. 1993;120:67–75. doi: 10.1083/jcb.120.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kametaka S, Mattera R, Bonifacino JS. Epidermal growth factor-dependent phosphorylation of the GGA3 adaptor protein regulates its recruitment to membranes. Mol Cell Biol. 2005;25:7988–8000. doi: 10.1128/MCB.25.18.7988-8000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, Greenberg ME. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 183.Masters SC, Yang H, Datta SR, Greenberg ME, Fu H. 14-3-3 inhibits Bad-induced cell death through interaction with serine-136. Mol Pharmacol. 2001;60:1325–1331. doi: 10.1124/mol.60.6.1325. [DOI] [PubMed] [Google Scholar]