Abstract
The purpose of this review is to explore recombination strategies in DNA viruses. Homologous recombination is a universal genetic process that plays multiple roles in the biology of all organisms, including viruses. Recombination and DNA replication are interconnected, with recombination being essential for repairing DNA damage and supporting replication of the viral genome. Recombination also creates genetic diversity, and viral recombination mechanisms have important implications for understanding viral origins as well as the dynamic nature of viral-host interactions. Both bacteriophage λ and herpes simplex virus (HSV) display high rates of recombination, both utilizing their own proteins and commandeering cellular proteins to promote recombination reactions. We focus primarily on λ and HSV, as they have proven amenable to both genetic and biochemical analysis and have recently been shown to exhibit some surprising similarities that will guide future studies.
Keywords: DNA replication, concatemer formation, single-strand annealing, exonucleases, single-strand annealing proteins, recombineering
INTRODUCTION
Homologous recombination plays many important roles in the biology of all living organisms including DNA replication and repair of DNA damage. Recombination is essential for genetic diversification required to enable organisms to adapt and evolve (65, 149). Given the central role of recombination in all living organisms, it is not surprising that viruses have also evolved to rely heavily on recombination for DNA replication and repair and also to promote viral diversity. Both viruses and their hosts are under strong evolutionary pressure, and over time, host cells have evolved sophisticated antiviral strategies that are countered by the ability of viruses to evade or disarm these cellular defenses. Moreover, understanding the origins of viral diversity affects human health, owing to the emergence of new viral species and the ability of viruses to evade vaccine and other antiviral chemotherapeutic strategies. Viruses, from seemingly simple bacteriophage to complex eukaryotic viruses, have adopted sophisticated mechanisms that not only promote genetic diversity but also promote viral replication.
OVERVIEW OF SIMILARITIES BETWEEN λ AND HSV
Since its discovery in 1950 by Esther Lederberg (68), phage λ and its relatives have been intensely studied, providing paradigms for gene regulation, replication, and recombination. Much of the foundation of modern molecular biology stems from research on λ. The Herpesviridae are a large family of eukaryotic DNA viruses responsible for lifelong debilitating infections including some cancers. Three subfamilies of herpesviruses have been described (α, β, and γ) that exhibit considerable diversity in cell and tissue tropism, length of productive cycles, and other properties related to pathogenesis. Herpes simplex viruses (HSV-1 and -2) are α herpesviruses that are associated with cold sores, genital lesions, kerititis, corneal blindness, and encephalitis. HSV affects between 60% and 95% of the world’s population (14). In this review we compare and contrast recombination mechanisms in λ and HSV and explore how each virus utilizes recombination during its life cycle, especially with respect to viral DNA replication.
Recombination between lambdoid phage occurs frequently in nature and leads to diversity in phage populations (reviewed in 13). A successful life cycle for phage λ requires the generation of multiple-length DNA molecules (concatemers), either by rolling circle replication or by recombination. λ recombination was shown to be active even in hosts deficient in recA (the main Escherichia coli recombination function), leading to the identification of a phage-encoded homologous recombination system, Red. Two λ genes were identified as responsible for recombination in recA mutant cells, exo and bet, encoding Exo and β (28, 36, 123). Later, a third gene, gam, which encodes the Gam protein, was found to be necessary to promote maximal recombination levels (31, 162). λ Exo is a member of a family of 5′-to-3′ exonucleases that are encoded by many, if not all, linear double-stranded viruses that replicate by forming concatemers and are found in plants, insects, bacteria, and mammals (105, 148). Another member of the λ Exo family is the RecE protein, encoded by a cryptic defective lambdoid prophage, rac, present in many E. coli strains. Nucleases λ Exo and RecE associate with single-strand annealing proteins (SSAP) β and RecT, respectively (42, 43, 102, 103, 123). RedExo/β and RecE/T are the founding members of a large family of two-component recombinases (49); other members of this family include Chu/35 in phage SPP1, AN/LEF-3 in the insect virus Autographa californica multinucleocapsid nucleopolyhedrovirus, and as we describe below, UL12/ICP8 from HSV (105, 148). The λ RedExo/β and the RecE/T recombinases are of considerable interest for their ability to promote in vivo recombination-mediated genetic engineering using short homologies—recombineering in bacteria (18, 30, 88, 91, 161).
Recombination is also important for HSV; rates of recombination are high between coinfecting HSVs in cultured cells, in animal infection models, and in human populations (11, 12, 44, 59, 74, 114, 120). HSV encodes a two-component recombinase consisting of a 5′-to-3′ exonuclease (UL12) and a SSAP (ICP8) that is reminiscent of λ Exo/β. UL12 and ICP8 can also work together to mediate robust strand-exchange activity in vitro (104, 106). In addition, ICP8 and UL12 interact with components of the host repair/recombination machinery (1, 135). Thus by analogy with λ Exo/β, UL12/ICP8 may function alone or in conjunction with host cell recombination machinery to promote recombination in infected cells. Understanding recombination mechanisms in HSV and λ will shed light on how these viruses replicate their genomes and generate genetic diversity and may lead to new tools for establishing recombineering and gene therapy in mammalian cells.
Another important similarity between λ and HSV is the choice they must make between two different lifestyles. λ is a temperate phage; upon infection the λ chromosome can integrate into the host chromosome via site-specific recombination to form a lysogen. In the lysogenic form, the integrated viral chromosome expresses the CI repressor protein that in turn prevents expression of most of the other λ genes; however, the repressed state can be reversed under DNA-damaging conditions that induce lytic growth. During lytic infection, the λ chromosome replicates, viral genes are expressed, and infectious progeny are produced. The choice between entering into the lytic or lysogenic life cycle is determined by many factors, including multiplicity of infection (62), temperature (39), and physiology of E. coli (131). In a similar fashion, herpesviruses have the ability to establish lifelong latent infections. During latent infection, the HSV genome is found in a circular episomal state characterized by heterochromatin and reduced viral gene expression, analogous to lysogenic λ. One of the key players in the decision to establish a lytic or latent infection cycle is immediate early protein ICP0, an E3 ubiquitin ligase that can induce the reactivation of HSV from a latent or quiescent infection and can also influence the decision to establish a lytic or latent infection (10, 33, 71).
GENERAL RECOMBINATION PATHWAYS
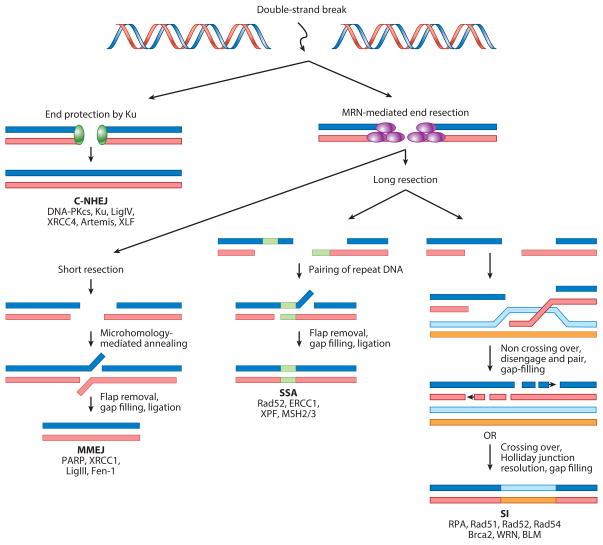
Several recombination pathways for the repair of broken DNA have been described, starting with homologous recombination in E. coli in the mid-1940s (69). Recombination is known to initiate at double-strand breaks (DSBs) and at single-stranded-DNA (ssDNA) gaps. Several events can lead to the formation of double-stranded-DNA (dsDNA) ends, including damage by ionizing radiation, completion of the replication of a linear genome, nuclease cleavage (e.g., restriction enzymes or the viral-encoded packaging enzyme, terminase), or replication through a nick or ssDNA gap. ssDNA gaps can be generated during replication at a blocked replication fork, by an inefficient primase, or during repair of DNA damage. If not repaired correctly, DSBs can cause deletions, translocations, and other deleterious genome rearrangements. Four recombination models that initiate at DSBs have been described; these function in varying degrees for both prokaryotes and eukaryotes (56, 156). Three require some degree of homology and one does not. Although the term homologous recombination (HR) is often used to refer to events mediated by strand invasion (SI), in this review we refer to these HR events as SI to distinguish them from microhomology-mediated end-joining (MMEJ) and single-strand annealing (SSA) that also use homology to promote recombination (Figure 1). The fourth pathway, classical nonhomologous end-joining (C-NHEJ), does not require homology. In eukaryotes, pathway choice following a DSB is tightly regulated and is controlled in part by the availability of a homologous sequence to repair the break. Pathway choice is also controlled by proteins recruited to the DSB: Nucleases that carry out end resection such as the damage-sensing MRN complex promote homologous recombination, whereas proteins that prevent DNA end resection such as Ku and 53BP1 appear to favor C-NHEJ (16). The recombinases λ RedExo/β and HSVUL12/ICP8 both utilize SSA (116, 129); however, λ RedExo/β can also utilize SI in the presence of RecA (129). Although it is not known whether HSV utilizes SI, knock-down of RecA-like Rad51, necessary for SI, has little effect on HSV growth (89).
Figure 1.
Major pathways for double-strand break (DSB) repair: classical nonhomologous end-joining (C-NHEJ), microhomology-mediated end-joining (MMEJ), single-strand annealing (SSA), and strand invasion (SI). This diagram depicts the four major DSB repair pathways and lists some key components for each pathway. MMEJ, SSA, and SI all require some degree of DNA homology and require end resection stimulated by the MRN complex (Mre11, Rad50, and Nbs1). MRN in conjunction with other cellular exonucleases such as Exo1 resect DNA at a DSB in the 5′ to 3′ direction, leaving a free 3′ overhang to participate in various types of homology-driven events. The requirements for SI-mediated recombination include a specialized ATP-dependent recombinase that can perform SI (Rad51 in eukaryotes, RecA in bacteria, and UvsX in T4). Although SI reactions are generally referred to in the literature as HR, in this review we refer to them as SI to distinguish them from MMEJ and SSA, which also utilize homology. SSA reactions do not require ATP and are mediated by a single-strand annealing protein (SSAP) (β in λ, ICP8 in HSV, and Rad52 in eukaryotic cells).
TWO-COMPONENT RECOMBINASES
The Exonucleases
Both Exo (24 kDa) and RecE (140 kDa) are dsDNA-dependent 5′-to-3′ exonucleases that bind tightly to dsDNA ends and degrade one strand (53, 75). They degrade ssDNA inefficiently. λ Exo requires Mg2+ but not ATP and can be highly processive, degrading an average of 18 kb per event (145). The high degree of processivity observed in λ Exo (63) and RecE (159) may be explained by their structure. They both form multisubunit toroidal proteins that contain a central funnel-shaped channel; one side is large enough to allow dsDNA to enter, and the other side is only able to accommodate the exiting ssDNA. Exo interacts with β (103), and RecE interacts with RecT (91); interestingly, however, these protein pairs cannot be mixed to make a functional complex (91).
The HSV-1 alkaline nuclease, encoded by the UL12 gene, was first reported in the early 1960s as a DNase induced in HSV-infected cells (57). It has a high pH optimum and a requirement for a divalent metal cation but not ATP (46, 57, 133). UL12 exhibits both endonuclease and exonuclease activities (45, 46, 61); however, the endonuclease activity is approximately tenfold less active than the exonuclease (133). UL12 is a relatively abundant 85-kDa phosphoprotein expressed very early in infected cells (2, 3, 19). UL12 interacts with ICP8 (104, 138, 147) and has also been shown to interact with MRN, the primary sensor of DSBs and MSH3 (1, 84).
The Single-Strand Annealing Proteins
One common feature of SSAPs is their ability to bind to ssDNA and anneal complementary DNA strands (60, 87). Although β (29 kDa) and RecT (33 kDa) share little overall sequence identity, they contain conserved residues that are also present in Rad52, a SSAP that functions in homologous recombination in eukaryotes (32, 83). Electron microscopy (EM) and atomic force microscopy (AFM) studies have suggested that β forms rings of ~12 subunits in the absence of DNA (96). In the presence of ssDNA, larger rings of ~15–18 subunits are observed, and left-handed helices are seen after complementary ssDNA are annealed. RecT can also form higher-order structures such as tetramers (42), rings (141), and filaments. In the presence of circular ssDNA, helical rod-shaped nucleoprotein structures can be observed. In general, β and RecT proteins bind ssDNA weakly, anneal complementary DNA molecules, and remain more tightly bound to the resulting dsDNA, protecting it from nuclease digestion (43, 55, 92). Unlike RecA (64), β and RecT do not require ATP for SSA.
The HSV SSAP, ICP8, is multifunctional, playing roles in DNA replication, recombination, and gene expression. It was one of the first viral proteins shown to be absolutely essential for viral DNA synthesis (17, 153). It has helix-destabilizing activity, consistent with a role in unwinding duplex DNA during DNA synthesis (24), and interacts with several viral proteins (reviewed in 150, 152). ICP8 is a 130-kDa zinc metalloprotein that binds nonspecifically to ssDNA (41) and exhibits potent annealing activity of homologous ssDNAs (9, 27). In the absence of DNA, ICP8 forms double-helical protein filaments (79); however, in the presence of ssDNA, ICP8 forms thin helical filaments and oligomeric rings, similar to those observed with RecT, β, and Rad52 (77, 78, 96, 121, 141). Structures believed to represent intermediates of the annealing reaction have been observed by EM and 3-D reconstruction and consist of two nonameric rings, one on top of the other (142). It appears that several SSAPs share the ability to form rings and filaments, and further analysis of these quaternary structures is expected to shed light on the mechanism of DNA strand annealing.
LINKAGES BETWEEN DNA REPLICATION AND RECOMBINATION
Phage λ
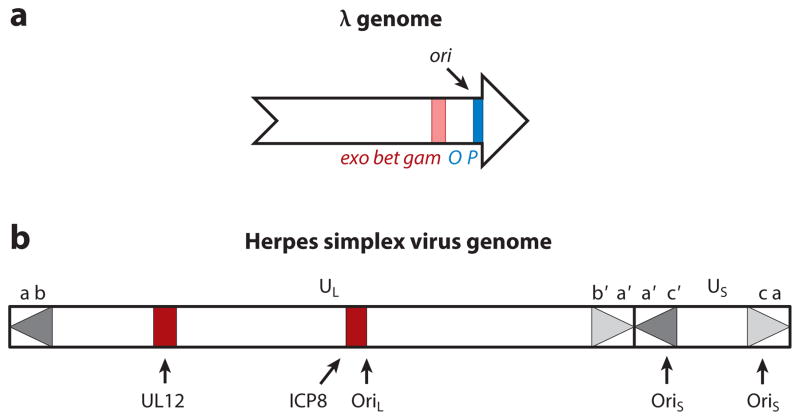
The λ chromosome contains unique 12-base ssDNA ends that are complementary to each other (Figure 2a). After infection, these ends, the cohesive end site (cos), anneal and ligate, and early λ DNA replication initiates with a few rounds of circle-to-circle DNA replication, termed θ replication. In this mode of replication, closed circular chromosomes generate duplicate circular molecules by bidirectional replication from a unique origin (115). λ only encodes two known replication proteins, O and P, and the remaining replication functions are performed by host (E. coli) proteins (93). The O protein binds to ori, which maps within the O gene (143), and the P protein interacts with O and facilitates initiation by recruiting the E. coli DnaB helicase (154). Fifteen minutes after infection (or induction), θ replication ceases (54).
Figure 2.
Phage λ and herpes simplex virus (HSV) genomes. (a) The 48.5-kb phage λ chromosome as it exists in the phage head, before it circularizes after infection. The recombination genes, Red exo, bet, and gam (located in the red region), and replication genes (located in the blue region), O and P, are indicated, as is the origin of replication, ori. The arrow’s head and tail signify cos, the 12-base-pair self-complementary ends that anneal after infection. (b) Herpes simplex virus-1 (HSV-1) has a 152-kb linear genome consisting of two unique sequences, UL and US, flanked by inverted sequences ab-b′a′ and a′c′-ca, respectively. The “a” region contains the packaging sequences and is repeated directly at genome termini and in an inverted orientation at the UL-US junction. The position of the UL12 and ICP8 genes (shown in red) as well as OriL and both copies of OriS are shown.
Late replication, also known as rolling circle or σ replication, starts about 15 minutes after infection. An alternate view is that σ replication starts when θ replication does but continues after θ replication stops (7). σ replication is characterized by the formation of concatemers that are two to eight times the length of a linear λ monomer (31, 124). Replication of these molecules is still bidirectional, with some circles replicating clockwise and others replicating counterclockwise. Concatemer formation is necessary to produce the substrate required for λ DNA packaging, two tandem cos sites (31, 140). How the switch is made between θ and σ replication is still not understood, but it could result from breakage of one replication fork. The Gam protein is important to allow σ replication (31), as it binds to RecB and protects tails of rolling circles from being degraded by the RecBCD exonuclease (81). By the end of the lytic cycle, there are enough λ genomes to fill ~100 phage heads.
λ Red-mediated recombination is associated with DNA replication. In a red mutant phage infection, production of phage DNA is slow and only about 50% of the wild-type level is made, indicating that Red recombination influences DNA replication (31). How it does so is yet to be fully understood. Although infection of red mutant phage produces less intracellular phage DNA, the DNA appears normal in structure. During DNA replication, recombination is stimulated (128) and is believed to be initiated by the tips of tails of rolling circle replication intermediates (127, 137). Recombination events occur throughout the phage chromosome. Interestingly, the λ P protein was shown to interact with Redβ by two-hybrid analysis, suggesting an additional link between DNA replication and recombination (103). In the absence of DNA replication, Red-mediated recombination occurs near the end of the λ chromosome, cos, or near any introduced DSB (127, 136).
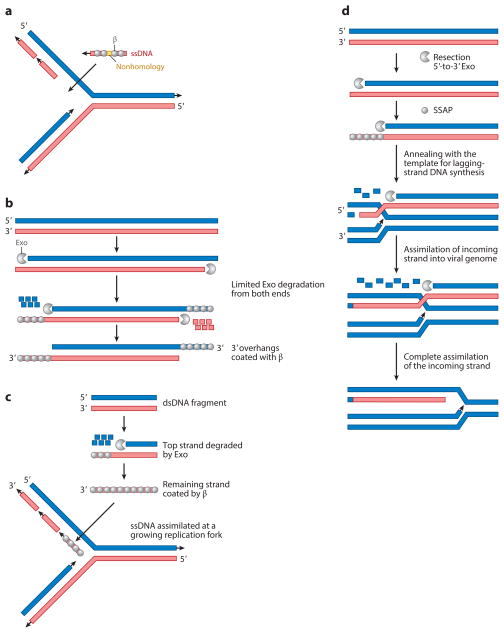
Investigation into the mechanism(s) of Red-mediated recombination via recombineering (see sidebar, Recombineering) has provided additional insight into the interconnection between replication and recombination in λ. For instance, during ssDNA recombination, mutations in DNA replication proteins Pol I, Pol III, and DNA primase alter the frequency and/or final product of recombination (67, 72, 101). Current models for ssDNA recombination suggest that a β-coated ss-oligo is annealed at the DNA replication fork (Figure 3a) and that this occurs most readily when the oligo can anneal on the lagging-strand template (20, 30, 47).
Figure 3.
Red-mediated single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) recombination. (a) Current model for ssDNA recombination by recombineering. An oligo is coated by β and inserted at the DNA replication fork. Although recombination on the lagging strand is depicted, it also occurs at a reduced frequency on the leading strand. The gold patch on the ssDNA represents a region of nonhomology such as a mutation that is being introduced by recombineering. Arrowheads represent the 3′ end of a DNA molecule. (b) Limited resection model. Exo performs limited degradation to expose 3′ overhangs, which are subsequently coated with β, thereby promoting annealing at regions of homology. (c) Alternative model for dsDNA recombination. Exo binds one end of a dsDNA fragment and degrades the entire strand. The remaining strand is coated with β and incorporated into a replication fork on the lagging-strand template. (d) Simultaneous resection and annealing model. Resection from a DSB by an exonuclease occurs simultaneously with annealing of the other strand at the DNA replication fork. In this model the Exo and single-strand annealing protein (SSAP) (λExo/β or UL12/ICP8) likely work in conjunction to regulate each other’s activities. In fact, RedExo/β have already been shown to modulate each other’s activities. This model was originally proposed by Kuzminov (66).
An initial model for dsDNA recombination suggested that a dsDNA fragment could undergo limited degradation by Exo at each end, resulting in a double-stranded region flanked by two 3′ overhangs that could be coated with β and participate in SSA (21, 75) (Figure 3b). Two observations, however, have cast doubt on this mechanism. First, when a DNA substrate with 3′ overhangs was tested directly for recombination, very low efficiencies were observed, suggesting that limited resection from each end may not produce a preferred substrate for dsDNA recombination (158). On the other hand, a substrate with 5′ ssDNA overhangs was shown to recombine at frequencies nearly as high as a ssDNA. Second, it has now been shown that dsDNA recombination requires replication of the target molecule (80, 100). An emerging model posits that dsDNA recombination occurs by a mechanism similar to that for ssDNA recombination (Figure 3a). Instead of limited degradation at a dsDNA end, it is possible that Exo degrades one entire strand of the dsDNA substrate and that the remaining strand is coated with β and incorporated into a replication fork (158) (Figure 3c). According to this scenario, the high efficiency of the 5′ overhang substrate is due to the fact that recessed 3′ ends can be extended by DNA polymerase to generate blunt dsDNA ends, which are excellent substrates for Exo. Support for this model was provided by experiments that examined recombination of a dsDNA containing a drug-resistance cassette (80, 85) and additional genetic markers. It was shown that after recombination of a dsDNA, most of the markers in a recombinant came from one strand, consistent with the assimilation of an entire strand at a replication fork. Additional experimentation will be required before this model can be adopted with confidence, however.
RECOMBINEERING.
In the last approximately 15 years, Red (91, 157), RecET (91, 161), and a growing list of other bacteriophage-encoded recombination systems (22, 134, 144, 146) have been used in bacteria for in vivo, recombination-mediated genetic engineering, also known as recombineering. These systems are independent of host recombination proteins and require only short homologies, ~50 bp, which can be included in a synthesized oligonucleotide. Recombineering can be performed efficiently with either ssDNA or dsDNA when introducing only a few base changes. Under optimized conditions, no selection is required, and up to 75% of colonies contain a recombinant (111, 112). Recombineering with ssDNA to remove a large DNA segment or with dsDNA to insert a large DNA segment (e.g., drug cassette) is up to 1000-fold less efficient and requires selection for recovery of desired recombinant. Recombineering has been used to introduce base changes, deletions (1 bp to >50 kb), insertions (1 bp to ~4 kb), duplications, inversions, and gene fusions into bacterial genomes (~5 Mb). Recently, a similar system has been developed in the eukaryote Saccharomyces cerevisiae (23). We are intrigued by the possibility that UL12/ICP8 might promote similar recombination-dependent genetic engineering in human cells. For reviews on recombineering see 88, 113, 139, and references within.
Herpes Simplex Virus
At the outset of infection, enveloped virions fuse with cellular membranes and viral capsids translocate along microtubules (125) to nuclear pores, where they dock and eject their 152-kb linear genomes into the nucleus (94). The HSV-1 genome consists of unique components (UL and US) flanked by inverted repeat sequences, and the UL and US regions invert relative to one another during replication (44, 120) (Figure 2b). The HSV genome contains three origins of replication, two copies of OriS and one of OriL (reviewed in 150, 152). Deletions of one or two of these origins are tolerated as long as one remains intact (48, 98). Another feature of the HSV genome likely relevant for its mode of replication is that both the replicating DNA and encapsi-dated viral genomes contain nicks and gaps that are randomly located and present on both strands (58, 155).
HSV encodes seven essential replication proteins: an origin-binding protein (UL9) plus six core or replication-fork proteins. The core complex consists of ICP8, a two-subunit DNA polymerase (UL30 and its accessory subunit, UL42), and a three-subunit helicase/primase complex (UL5, UL8, and UL52) (reviewed in 90, 150, 152). Despite identification of these cis- and transacting factors essential for DNA replication, the overall mechanism remains poorly understood. Considerable controversy exists surrounding the fate of the incoming HSV genome at the outset of infection. According to one model, the viral genome circularizes, leading to a θ replication mode followed by σ replication (108). This model is supported by the disappearance of terminal fragments and the appearance of joint fragments that could be a result of ligation of the ends of the viral genome (29, 37, 97, 132). There is no direct proof, however, for either circularization or θ replication in infected cells. Gardella gel electrophoresis, reported to distinguish circular from linear forms of large viral genomes (38), has been used to determine whether the viral genome circularizes in infected cells. Interestingly, Jackson & Deluca (50) reported that circular genomes could not be detected during lytic infection with wild-type virus; however, circles could be seen in cells infected with a mutant virus lacking the immediate early protein, ICP0. Also, in cells infected with a virus that is deleted for all immediate early genes and thus fails to induce any viral gene expression (109), only circular genomes are detected (50). Infection with this mutant is thought to mimic latent infection, and thus it is possible that circularization is associated with the establishment of a quiescent state or latency.
Current models for initiation of HSV DNA replication posit that UL9, in conjunction with ICP8, distorts or destabilizes one of the viral origins of replication (reviewed in 90, 150, 152). The subsequent recruitment of the helicase/primase complex and the two-subunit polymerase is expected to unwind duplex DNA, synthesize short RNA primers, and catalyze leading- and lagging-strand DNA synthesis. On the one hand, if the viral genome circularizes, bidirectional replication would be expected to lead to θ replication; on the other hand, if initiation occurs on a linear molecule, bidirectional replication would proceed to the end of the molecule. By analogy to the bacteriophage T4, the ends of the daughter molecules could participate in recombination (76, 86). The role of the HSVUL12/ICP8 recombinase in DNA replication is not clear. Because ICP8 is essential for viral DNA replication, ICP8 mutants are not viable and do not synthesize viral DNA (17, 153). The phenotype of a UL12-null virus is more complex. Although viral DNA synthesis occurs at wild-type levels, UL12 is essential for the production of infectious progeny (119). Although some encapsidation can occur, DNA packaged in cells infected with UL12 mutant viruses is not infectious and appears to be structurally aberrant (40, 99). Although we initially proposed that UL12 was needed to process viral genomes prior to packaging (40), we now favor a role in directing the replication machinery toward a pathway that can generate concatemers that can be packaged into infectious virions.
Several lines of evidence support the notion that DNA replication and recombination in HSV are linked (25, 26, 151). Genomic inversions are observed as soon as newly replicated DNA can be detected (4, 5, 117, 118, 160) and are mediated by ICP8 and the other HSV replication proteins (110, 151). EM analysis suggests that viral DNA replication results in the accumulation of head-to-tail concatemers that are highly branched, with X- and Y-junctions (51, 52, 118, 122). In addition, pulsed-field gel electrophoresis suggests that replicating HSV DNA adopts a complex, branched structure that cannot enter the gel (remains in the well) even after DNA restriction digestion with a single cutting enzyme (82, 117, 160). Replicating DNA appears to be held together by branches, generating a network of DNA reminiscent of the replication/recombination intermediates of bacteriophage T4. These structures are not consistent with a simple rolling circle mechanism of replication.
Additional evidence suggesting that HSV has evolved to utilize recombination-dependent replication was provided by the analysis of replication intermediates generated by another DNA virus, SV40. In SV40-infected cells, the small circular viral genome is replicated by the origin-binding protein T-antigen and cellular replication proteins to generate a θ structure and produce two circular daughter molecules, much like early λ replication. However, if SV40 DNA is introduced in HSV-infected cells, the SV40 genome is replicated by SV40 large T-antigen and the six core HSV-encoded replication factors (8). In these cells, the SV40 genome adopts complex structures reminiscent of HSV replication intermediates. Furthermore, a high frequency of homologous recombination between SV40 genomes was observed when the SV40 genomes were replicated using the HSV replication machinery (8). Thus, there appears to be an intrinsic herpesvirus-specific replication mode that generates complex branched structures, and this mode of replication correlates with increased recombination frequencies.
MECHANISMS OF RECOMBINATION
Phage λ
λ recombination can occur in the absence or presence of DNA replication. For λ recombination studied in the absence of DNA replication, both SI and SSA have been demonstrated. When RecA protein is absent, recombination proceeds by SSA (129) and recombinants that are found contain a long stretch of heteroduplex DNA, as expected. When RecA protein is present in these replication-blocked crosses, SI is the most prevalent recombination pathway.
When the DNA can replicate, replication stimulates recombination levels and creates DNA substrates that are favorable for SSA, such as the tips of rolling-circle tails, DSBs that arise during packaging, and lagging-strand gaps such as those at the replication fork (126). Red-mediated recombination can proceed by two different pathways, SI or SSA (129) (Figure 1). With both pathways, Exo processes the DSB, creating a 3′ single-strand overhang. For SI, this ssDNA overhang then synapses with and invades an intact homologous chromosome in a reaction that requires a recombinase with synaptase activity, in this case, a β/RecA complex (64, 107). In the absence of RecA, β promotes SSA. For example, the tips of rolling-circle tails can be processed by Exo, and β can anneal the end to complementary single-strand regions such as lagging-strand gaps at a replication fork (60, 87). During DNA replication, Red recombination becomes less RecA dependent.
Herpes Simplex Virus
Recombination is known to initiate at DSBs and at ssDNA gaps. DSBs are likely to occur during HSV DNA replication if DNA replication occurs through a nick or gap or if HSV replication occurs on a linear rather than circular molecule. Furthermore, during the encapsidation phase it is expected that terminase will cleave concatemeric DNA, packaging a viral genome on one side of the packaging sequence and leaving a DSB on the other. HSV also encodes a viral primase that at low concentrations is inefficient (15, 130), providing a possible mechanism for the formation of nicks and gaps. Thus, HSV DNA replication is likely to produce DSBs and DNA molecules with gaps, conditions that would be expected to stimulate recombination events. As depicted in Figure 1, several recombination mechanisms are available to HSV to repair DSBs. To determine which pathways were active in cells infected with HSV, chromosomally integrated reporter assays have been used to measure activation of C-NHEJ, MMEJ, SI, and SSA (6). Interestingly, SSA was increased in HSV-infected cells, whereas SI, MMEJ, and NHEJ were inhibited (116) (see sidebar, Why Has HSV Evolved to Inactivate NHEJ and SI?). This increase in SSA was abolished when cells were infected with a viral mutant lacking UL12, and expression of UL12 alone caused an increase in SSA (116). UL12 and its partner ICP8 may be responsible for the stimulation of SSA in HSV-infected cells; however, we have not ruled out the contribution of cellular proteins such as Rad52 and other interaction partners of UL12 and ICP8 (1, 135). We are intrigued by the observation that UL12 interacts directly with MSH3, one of the players in cellular SSA reactions (84). The activation of SSA and apparent inhibition of NHEJ and SI in HSV-infected cells have important implications for the DNA replication mechanism of this virus. HSV may have evolved to utilize the two-component recombinase UL12/ICP8 to promote recombination-dependent replication by SSA because this pathway is conducive to the production of concatemeric DNA, which can be packaged into an infectious virus. Although DNA synthesis occurs in the absence of UL12, the production of aberrant genomes suggests that alternate pathways may be deleterious for the production of an infectious virus.
WHY HAS HERPES SIMPLEX VIRUS EVOLVED TO INACTIVATE NONHOMOLOGOUS END-JOINING AND STRAND INVASION?
When a virus infects a cell, it encounters a hostile environment, as cells have evolved sophisticated intrinsic mechanisms to counter viral infections. Viruses, in turn, have evolved to evade or disarm these cellular defenses; in particular, the immediate early protein ICP0 has been shown to degrade several cellular proteins involved in antiviral responses (10, 34, 35). Interestingly, HSV replication is more efficient in cells lacking the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) (95); and in Ku-deficient murine embryonic fibroblasts, viral yields are increased by almost 50-fold (135). Given that DNA-PKcs and Ku are known components of the NHEJ pathway, these results suggest that this pathway is antiviral. ICP0 is known to induce the degradation of DNA-PKcs (70, 95), consistent with the observation that NHEJ is decreased in HSV-infected cells (116). By inhibiting NHEJ, ICP0 may prevent circularization and promote lytic infection. ICP0 has also been shown to degrade two cellular ubiquitin ligases involved in repair by SI, RNF8 and RNF168 (73). It is possible that HSV has evolved to control the pathway by which DSBs are repaired because NHEJ and SI result in negative outcomes such as genome silencing. Repair by SSA may be conducive to the production of concatemeric DNA that can be packaged into an infectious virus during lytic infection. Additional experimentation will be required to test these predictions.
IMPLICATIONS OF SSA FOR λ AND HERPES SIMPLEX VIRUS
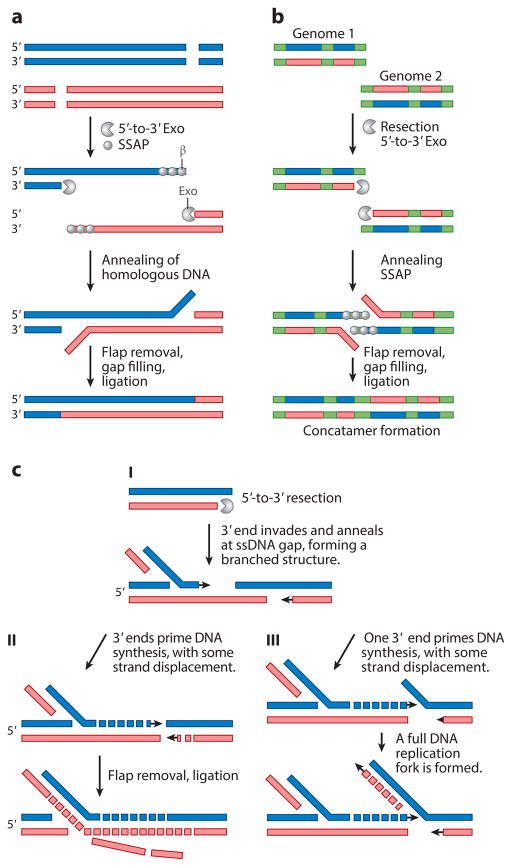
SSA could theoretically function in λ, HSV, or both to repair DSBs, create concatemers, prime DNA synthesis, and generate branched replication intermediates. DSBs that arise following replication through a nick or a gap, as a result of terminase activity, or as a result of bidirectional replication of a linear genome could be repaired by SSA, as outlined in Figure 4a. Under this scenario, DSBs at different locations would be resected by the exonuclease activity of Exo or UL12, and homologous regions would be annealed by β or ICP8. This mechanism could be used to produce concatemers from DNA fragments generated by DSBs; however, it may also generate deletions, especially in a genome with repeated elements, such as HSV. Figure 4b shows a similar mechanism that might also result in the formation of concatemers from two viral genomes by resection and annealing of the directly repeated “a” sequence at HSV termini (Figure 2b).
Figure 4.
Implications for single-strand annealing (SSA) during λ and herpes simplex virus (HSV) infection. These models depict potential roles for SSA during λ and HSV infection. (a) SSA at double-strand breaks (DSBs). SSA can be used when replication through a nick or gap leads to a DSB, or when a DSB occurs by another mechanism (see text for examples). After limited resection by Exo or UL12, a SSA protein (SSAP) such as β, ICP8, or Rad52 binds to single-stranded DNA (ssDNA) regions and induces annealing of complementary ssDNA. Unpaired ssDNA overhangs after annealing are removed by a structure-specific endonuclease such as ERCC1/XPF; remaining gaps are filled and ligated. In the case of λ, Exo is known to degrade until the entire strand is assimilated, and then ligation occurs; thus, unpaired ssDNA overhangs (flaps) would not be expected. This reaction is potentially mutagenic, as it can lead to deletions. (b) SSA at viral termini. Resection and annealing of the directly repeated “a” sequences found at HSV termini (shown in green) could lead to concatemer formation. (c) Formation of branched replication intermediates. A DSB is processed by a 5′-to-3′ Exo, and the 3′ overhang can anneal at a single-strand gap on another genome. In scenario I, a branched structure is generated. In scenario II, 3′ ends denoted in black can be extended by DNA polymerase, with accompanying strand displacement to complete a recombinant. In scenario III, a full replication fork is established. This can replicate out to the end, resulting in a linear chromosome fragment that could either invade another genome or participate in reactions shown in panel a or b, possibly leading to concatemer formation. We have depicted a simple case, but additional annealing events could occur at gaps along each strand of the HSV genome, such as the one depicted in scenario III.
If SSA pathways are active during viral DNA replication, as we have argued, additional scenarios can be envisioned. As discussed above, dsDNA can be degraded by Exo or UL12, leaving a ssDNA molecule that can be coated by β or ICP8, and assimilated at a DNA replication fork, predominantly on the lagging-strand template (Figure 3c). A modification of this scenario (Figure 3d) was originally proposed by Kuzminov (66). According to this model, degradation occurs simultaneously with assimilation of the other strand at the DNA replication fork by the SSAP. In this model the Exo and SSAP work in conjunction to regulate each other’s activities, as is known to occur with RedExo/β. A final scenario suggests a mechanism by which branched replication/recombination intermediates could form (Figure 4c). According to this scenario, a DSB could be resected by Exo and the exposed 3′ overhangs coated with SSAP. The SSAP-coated overhang could anneal at a ssDNA gap and initiate formation of a DNA replication fork (Figure 4c, scenario III). Two different outcomes of this type of reaction are described in the figure legend (scenarios II and III), and either could generate the complex branched molecules seen among the replication intermediates during HSV infection (51, 52, 118). These reactions coupled with reactions depicted in Figure 4b could create branched, longer-than-unit-length concatemeric molecules that could be resolved by cellular Holliday junction resolvases or perhaps during encapsidation. Thus, it is possible that SSA results in the generation of the longer-than-unit-length molecules that are required to produce the substrate for encapsidation.
The extent to which the various models for SSA are used by either λ or HSV during DNA replication to generate concatemeric DNA is unclear. Genetic and biochemical experiments have confirmed that the RedExo/β and RecE/T systems are capable of many of the reactions shown in Figures 3 and 4. Models shown in panels 3a, 3b, 3c, and 4a have experimental support in the λ system. λ does not contain the repeat sequences required for the reactions in 4b, and it is therefore unlikely that end resection leads to concatemer formation in cells infected with λ. The model outlined in Figure 4c is consistent with the known biochemical properties of the RedExo/β and UL12/ICP8 systems, and it will be of interest to test whether this mechanism contributes to concatemer formation in λ- or HSV-infected cells. Comparison of the viral recombination systems encoded by λ and HSV demonstrates striking similarities and fascinating differences. The models shown in Figures 3 and 4 suggest testable hypotheses for future experimentation that will lead to a better understanding of whether λ and HSV utilize θ and σ replication mechanisms, recombination-dependent replication, or a combination of both.
SUMMARY POINTS AND FUTURE ISSUES.
Both λ and HSV encode a two-component recombinase capable of stimulating SSA, and both generate concatemers during DNA replication that are packaged into infectious progeny.
Both λ and HSV genomes have been reported to circularize and replicate through θ and σ replication; however, in the case of HSV, concatemer formation may also involve recombination-dependent replication. The observation that recombineering requires DNA replication raises the interesting possibility that Red recombination also functions at the replication fork during the λ life cycle. It will be of considerable interest to determine the extent to which the various models for SSA are used by either λ or HSV during DNA replication to generate concatemeric DNA.
λ RedExo/β can function on its own to promote SSA or in conjunction with RecA to promote SI. It is unclear whether the HSVUL12/ICP8 system functions on its own or in conjunction with cellular repair/recombination proteins. Although UL12 interacts with MRN and both UL12 and ICP8 interact with host repair/recombination proteins, further experimentation will be required to determine the biological significance of these interactions.
It is clear that λ Red and RecE/T are very powerful tools for genetic engineering in bacteria. It will be very interesting to determine whether UL12 and ICP8 can be utilized to perform recombineering in eukaryotic cells.
Acknowledgments
We thank Rik Myers for originally suggesting that UL12/ICP8 is part of the superfamily of two-component recombinases defined by λ Exo/β. We thank Don Court, Lynn Thomason, Jack Griffith, and Rob Kuchta, for many discussions and careful reading of the manuscript. We also thank Adam Parks, Nina Costantino, Brandon Albright, Samantha Smith, Anthar Darwish, and Lorry Grady for suggestions on the manuscript and April Schumacher for the suggestions on the construction of Figure 1. This project was supported by the following National Institutes of Health grants awarded to SKW: AI069136 and AI021747 and to Don Court: from the National Cancer Institute, National Institutes of Health (NIH), under contract HHSN261200800001E. This research was also supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Glossary
- SSAP
single-strand annealing proteins make up a family of proteins that are able to anneal complementary strands; their quaternary structures, rings, and filaments are believed to be important for their annealing activities
- DSB
a double-strand break in DNA can form via ionizing radiation, replication of a linear genome, nuclease cleavage, or replication through a nick or ssDNA gap
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- SSA
single-strand annealing is DSB repair whereby ends are processed by a 5′-to-3′ exonuclease and complementary single-strand regions are annealed by a SSAP
- MRN
a protein complex consisting of Mre11, Rad50, and Nbs1 in eukaryotes binds to DSBs and is believed to initiate resection prior to repair by homologous recombination
- EM
electron microscopy
- θ
a mode of bidirectional DNA replication whereby the molecules look like a θ by EM
- σ
a mode of DNA replication that generates linear multimer concatemers; the molecules look like a σ by EM
Footnotes
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
RELATED RESOURCES
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Sandra K. Weller, Email: weller@uchc.edu.
James A. Sawitzke, Email: sawitzkj@mail.nih.gov.
LITERATURE CITED
- 1.Balasubramanian N, Bai P, Buchek G, Korza G, Weller SK. Physical interaction between the herpes simplex virus type 1 exonuclease, UL12, and the DNA double-strand break-sensing MRN complex. J Virol. 2010;84:12504–14. doi: 10.1128/JVI.01506-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banks L, Purifoy DJ, Hurst PF, Killington RA, Powell KL. Herpes simplex virus non-structural proteins. IV. Purification of the virus-induced deoxyribonuclease and characterization of the enzyme using monoclonal antibodies. J Gen Virol. 1983;64:2249–60. doi: 10.1099/0022-1317-64-10-2249. [DOI] [PubMed] [Google Scholar]
- 3.Banks LM, Halliburton IW, Purifoy DJM, Killington RA, Powell KL. Studies on the herpes simplex virus alkaline nuclease: detection of type-common and type-specific epitopes on the enzyme. J Gen Virol. 1985;66:1–14. doi: 10.1099/0022-1317-66-1-1. [DOI] [PubMed] [Google Scholar]
- 4.Bataille D, Epstein A. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology. 1994;203:384–88. doi: 10.1006/viro.1994.1498. [DOI] [PubMed] [Google Scholar]
- 5.Bataille D, Epstein AL. Equimolar generation of the four possible arrangements of adjacent L components in herpes simplex virus type 1 replicative intermediates. J Virol. 1997;71:7736–43. doi: 10.1128/jvi.71.10.7736-7743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennardo N, Stark JM. ATM limits incorrect end utilization during non-homologous end joining of multiple chromosome breaks. PLoS Genet. 2010;6:e1001194. doi: 10.1371/journal.pgen.1001194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Better M, Freifelder D. Studies on the replication of Escherichia coli phage λ DNA. I. The kinetics of DNA replication and requirements for the generation of rolling circles. Virology. 1983;126:168–82. doi: 10.1016/0042-6822(83)90469-5. [DOI] [PubMed] [Google Scholar]
- 8.Blumel J, Graper S, Matz B. Structure of simian virus 40 DNA replicated by herpes simplex virus type 1. Virology. 2000;276:445–54. doi: 10.1006/viro.2000.0574. [DOI] [PubMed] [Google Scholar]
- 9.Bortner C, Hernandez TR, Lehman IR, Griffith J. Herpes simplex virus 1 single-strand DNA-binding protein (ICP8) will promote homologous pairing and strand transfer. J Mol Biol. 1993;231:241–50. doi: 10.1006/jmbi.1993.1279. [DOI] [PubMed] [Google Scholar]
- 10.Boutell C, Everett RD. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol. 2013;94:465–81. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- 11.Bowden R, Sakaoka H, Donnelly P, Ward R. High recombination rate in herpes simplex virus type 1 natural populations suggests significant co-infection. Infect Genet Evol. 2004;4:115–23. doi: 10.1016/j.meegid.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Brown SM, Ritchie DA, Subak SJH. Genetic interactions between temperature-sensitive mutants of types 1 and 2 herpes simplex viruses. J Gen Virol. 1973;18:347–57. doi: 10.1099/0022-1317-18-3-347. [DOI] [PubMed] [Google Scholar]
- 13.Campbell A. Comparative molecular biology of lambdoid phages. Annu Rev Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- 14.Chayavichitsilp P, Buckwalter JV, Krakowski AC, Friedlander SF. Herpes simplex. Pediatr Rev. 2009;30:119–29. doi: 10.1542/pir.30-4-119. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Bai P, Mackay S, Korza G, Carson JH, et al. Herpes simplex virus type 1 helicase-primase: DNA binding and consequent protein oligomerization and primase activation. J Virol. 2011;85:968–78. doi: 10.1128/JVI.01690-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conley AJ, Knipe DM, Jones PC, Roizman B. Molecular genetics of herpes simplex virus. VII. Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of γ polypeptides. J Virol. 1981;37:191–206. doi: 10.1128/jvi.37.1.191-206.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–79. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 19.Costa RH, Draper KG, Banks L, Powell KL, Cohen G, et al. High-resolution characterization of herpes simplex virus type 1 transcripts encoding alkaline exonuclease and a 50,000-dalton protein tentatively identified as a capsid protein. J Virol. 1983;48:591–603. doi: 10.1128/jvi.48.3.591-603.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costantino N, Court DL. Enhanced levels of λ Red-mediated recombinants in mismatch repair mutants. Proc Natl Acad Sci USA. 2003;100:15748–53. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–88. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 22.Datta S, Costantino N, Zhou X, Court DL. Identification and analysis of recombineering functions from gram-negative and gram-positive bacteria and their phages. Proc Natl Acad Sci USA. 2008;105:1626–31. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiCarlo JE, Conley AJ, Penttilä M, Jäntti J, Wang HH, Church GM. Yeast oligo-mediated genome engineering (YOGE) ACS Synth Biol. 2013;2:741–49. doi: 10.1021/sb400117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudas KC, Ruyechan WT. Identification of a region of the herpes simplex virus single-stranded DNA-binding protein involved in cooperative binding. J Virol. 1998;72:257–65. doi: 10.1128/jvi.72.1.257-265.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutch RE, Bianchi V, Lehman IR. Herpes simplex virus type 1 DNA replication is specifically required for high-frequency homologous recombination between repeated sequences. J Virol. 1995;69:3084–89. doi: 10.1128/jvi.69.5.3084-3089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dutch RE, Bruckner RC, Mocarski ES, Lehman IR. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J Virol. 1992;66:277–85. doi: 10.1128/jvi.66.1.277-285.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutch RE, Lehman IR. Renaturation of complementary DNA strands by herpes simplex virus type 1 ICP8. J Virol. 1993;67:6945–49. doi: 10.1128/jvi.67.12.6945-6949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Echols H, Gingery R. Mutants of bacteriophage λ defective in vegetative genetic recombination. J Mol Biol. 1968;34:239–49. doi: 10.1016/0022-2836(68)90249-0. [DOI] [PubMed] [Google Scholar]
- 29.Efstathiou S, Minson AC, Field HJ, Anderson JR, Wildy P. Detection of herpes simplex virus-specific DNA sequences in latently infected mice and in humans. J Virol. 1986;57:446–55. doi: 10.1128/jvi.57.2.446-455.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci USA. 2001;98:6742–46. doi: 10.1073/pnas.121164898. First demonstration of recombination via ssDNA in E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enquist LW, Skalka A. Replication of bacteriophage λ DNA dependent on the function of host and viral genes. I. Interaction of red, gam, and rec. J Mol Biol. 1973;75:185–212. doi: 10.1016/0022-2836(73)90016-8. Suggests λ replication and recombination are linked. [DOI] [PubMed] [Google Scholar]
- 32.Erler A, Wegmann S, Elie-Caille C, Bradshaw CR, Maresca M, et al. Conformational adaptability of Redβ during DNA annealing and implications for its structural relationship with Rad52. J Mol Biol. 2009;391:586–98. doi: 10.1016/j.jmb.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 33.Everett RD. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays. 2000;22:761–70. doi: 10.1002/1521-1878(200008)22:8<761::AID-BIES10>3.0.CO;2-A. Review shows that ICP0 is important for regulating lytic and latent infection. [DOI] [PubMed] [Google Scholar]
- 34.Everett RD, Freemont P, Saitoh H, Dasso M, Orr A, et al. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–91. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Everett RD, Parada C, Gripon P, Sirma H, Orr A. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol. 2008;82:2661–72. doi: 10.1128/JVI.02308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franklin NC. Deletions and functions of the center of the Φ80-λ phage genome. Evidence for a phage function promoting genetic recombination. Genetics. 1967;57:301–18. doi: 10.1093/genetics/57.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garber DA, Beverley SM, Coen DM. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology. 1993;197:459–62. doi: 10.1006/viro.1993.1612. [DOI] [PubMed] [Google Scholar]
- 38.Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J Virol. 1984;50:248–54. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giladi H, Goldenberg D, Koby S, Oppenheim AB. Enhanced activity of the bacteriophage λ PL promoter at low temperature. Proc Natl Acad Sci USA. 1995;92:2184–88. doi: 10.1073/pnas.92.6.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein JN, Weller SK. In vitro processing of HSV-1 DNA replication intermediates by the viral alkaline nuclease, UL12. J Virol. 1998;72:8772–81. doi: 10.1128/jvi.72.11.8772-8781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupte SS, Olson JW, Ruyechan WT. The major herpes simplex virus type-1 DNA-binding protein is a zinc metalloprotein. J Biol Chem. 1991;266:11413–16. [PubMed] [Google Scholar]
- 42.Hall SD, Kane MF, Kolodner RD. Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. J Bacteriol. 1993;175:277–87. doi: 10.1128/jb.175.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall SD, Kolodner RD. Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc Natl Acad Sci USA. 1994;91:3205–9. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayward GS, Frenkel N, Roizman B. Anatomy of herpes simplex virus DNA: strain differences and heterogeneity in the locations of restriction endonuclease cleavage sites. Proc Natl Acad Sci USA. 1975;72:1768–72. doi: 10.1073/pnas.72.5.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffmann PJ. Mechanism of degradation of duplex DNA by the DNase induced by herpes simplex virus. J Virol. 1981;38:1005–14. doi: 10.1128/jvi.38.3.1005-1014.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann PJ, Cheng Y-C. The deoxyribonuclease induced after infection of KB cells by herpes simplex virus type 1 or type 2. I. Purification and characterization of the enzyme. J Biol Chem. 1978;253:3557–62. [PubMed] [Google Scholar]
- 47.Huen MS, Li XT, Lu LY, Watt RM, Liu DP, Huang JD. The involvement of replication in single stranded oligonucleotide-mediated gene repair. Nucleic Acids Res. 2006;34:6183–94. doi: 10.1093/nar/gkl852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Igarashi K, Fawl R, Roller RJ, Roizman B. Construction and properties of a recombinant herpes simplex virus 1 lacking both S-component origins of DNA synthesis. J Virol. 1993;67:2123–32. doi: 10.1128/jvi.67.4.2123-2132.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iyer LM, Koonin EV, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redβ, ERF and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackson SA, DeLuca NA. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc Natl Acad Sci USA. 2003;100:7871–76. doi: 10.1073/pnas.1230643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacob RJ, Roizman B. Anatomy of herpes simplex virus DNA. VIII. Properties of the replicating DNA. J Virol. 1977;23:394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jean JH, Blankenship ML, Ben-Porat T. Replication of herpesvirus DNA. I. Electron microscopic analysis of replicative structures. Virology. 1977;79:281–91. doi: 10.1016/0042-6822(77)90355-5. [DOI] [PubMed] [Google Scholar]
- 53.Joseph JW, Kolodner R. Exonuclease VIII of Escherichia coli. II. Mechanism of action. J Biol Chem. 1983;258:10418–24. [PubMed] [Google Scholar]
- 54.Joyner A, Isaacs LN, Echols H, Sly WS. DNA replication and messenger RNA production after induction of wild-type λ bacteriophage and λ mutants. J Mol Biol. 1966;19:174–86. doi: 10.1016/s0022-2836(66)80059-1. [DOI] [PubMed] [Google Scholar]
- 55.Karakousis G, Ye N, Li Z, Chiu SK, Reddy G, Radding CM. The β protein of phage λ binds preferentially to an intermediate in DNA renaturation. J Mol Biol. 1998;276:721–31. doi: 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- 56.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–8. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keir HM, Gold E. Deoxyribonucleic acid nucleotidyltransferase and deoxyribonuclease from cultured cells infected with herpes simplex virus. Biochim Biophys Acta. 1963;72:263–76. [Google Scholar]
- 58.Kieff ED, Bachenheimer SL, Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971;8:125–32. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kintner RL, Allan RW, Brandt CR. Recombinants are isolated at high frequency following in vivo mixed ocular infection with two avirulent herpes simplex virus type 1 strains. Arch Virol. 1995;140:231–44. doi: 10.1007/BF01309859. [DOI] [PubMed] [Google Scholar]
- 60.Kmiec E, Holloman WK. β protein of bacteriophage λ promotes renaturation of DNA. J Biol Chem. 1981;256:12636–39. [PubMed] [Google Scholar]
- 61.Knopf CW, Weisshart K. Comparison of exonucleolytic activities of herpes simplex virus type-1 DNA polymerase and DNase. Eur J Biochem. 1990;191:263–73. doi: 10.1111/j.1432-1033.1990.tb19119.x. [DOI] [PubMed] [Google Scholar]
- 62.Kourilsky P. Lysogenization by bacteriophage lambda. II. Identification of genes involved in the multiplicity dependent processes. Biochimie. 1974;56:1511–16. [PubMed] [Google Scholar]
- 63.Kovall R, Matthews BW. Toroidal structure of λ-exonuclease. Science. 1997;277:1824–27. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- 64.Kowalczykowski SC, Eggleston AK. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 65.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage. λ. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. Excellent review with a comprehensive set of models for recombinational repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lajoie MJ, Gregg CJ, Mosberg JA, Washington GC, Church GM. Manipulating replisome dynamics to enhance lambda Red-mediated multiplex genome engineering. Nucleic Acids Res. 2012;40:e170. doi: 10.1093/nar/gks751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lederberg EM, Lederberg J. Genetic studies of lysogenicity in Escherichia coli. Genetics. 1953;38:51–64. doi: 10.1093/genetics/38.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lederberg J. Gene recombination and linked segregations in Escherichia coli. Genetics. 1947;32:505–25. doi: 10.1093/genetics/32.5.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lees-Miller SP, Long MC, Kilvert MA, Lam V, Rice SA, Spencer CA. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–77. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leib DA, Coen DM, Bogard CL, Hicks KA, Yager DR, et al. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–68. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li XT, Thomason LC, Sawitzke JA, Costantino N, Court DL. Bacterial DNA polymerases participate in oligonucleotide recombination. Mol Microbiol. 2013;88:906–20. doi: 10.1111/mmi.12231. Shows direct involvement of DNA polymerases with ssDNA recombination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lilley CE, Chaurushiya MS, Boutell C, Landry S, Suh J, et al. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 2010;29:943–55. doi: 10.1038/emboj.2009.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lingen M, Hengerer F, Falke D. Mixed vaginal infections of Balb/c mice with low virulent herpes simplex type 1 strains result in restoration of virulence properties: vaginitis/vulvitis and neuroinvasiveness. Med Microbiol Immunol. 1997;185:217–22. doi: 10.1007/s004300050033. [DOI] [PubMed] [Google Scholar]
- 75.Little JW. An exonuclease induced by bacteriophage λ. II. Nature of the enzymatic reaction. J Biol Chem. 1967;242:679–86. [PubMed] [Google Scholar]
- 76.Luder A, Mosig G. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: priming by RNA polymerase and by recombination. Proc Natl Acad Sci USA. 1982;79:1101–5. doi: 10.1073/pnas.79.4.1101. Demonstrates how recombination can initiate a DNA replication fork. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Makhov AM, Boehmer PE, Lehman IR, Griffith JD. Visualization of the unwinding of long DNA chains by the herpes simplex virus type 1 UL9 protein and ICP8. J Mol Biol. 1996;258:789–99. doi: 10.1006/jmbi.1996.0287. [DOI] [PubMed] [Google Scholar]
- 78.Makhov AM, Griffith JD. Visualization of the annealing of complementary single-stranded DNA catalyzed by the herpes simplex virus type 1 ICP8 SSB/recombinase. J Mol Biol. 2006;355:911–22. doi: 10.1016/j.jmb.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 79.Makhov AM, Sen A, Yu X, Simon MN, Griffith JD, Egelman EH. The bipolar filaments formed by herpes simplex virus type 1 SSB/recombination protein (ICP8) suggest a mechanism for DNA annealing. J Mol Biol. 2009;386:273–79. doi: 10.1016/j.jmb.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maresca M, Erler A, Fu J, Friedrich A, Zhang Y, Stewart AF. Single-stranded heteroduplex intermediates in λ Red homologous recombination. BMC Mol Biol. 2010;11:54. doi: 10.1186/1471-2199-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marsić N, Roje S, Stojiljković I, Salaj-Smic E, Trgovcević Z. In vivo studies on the interaction of RecBCD enzyme and λ Gam protein. J Bacteriol. 1993;175:4738–43. doi: 10.1128/jb.175.15.4738-4743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martinez R, Sarisky RT, Weber PC, Weller SK. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–85. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsubara K, Malay AD, Curtis FA, Sharples GJ, Heddle JG. Structural and functional characterization of the Redβ recombinase from bacteriophageλ. PLoS ONE. 2013;8:e78869. doi: 10.1371/journal.pone.0078869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mohni KN, Mastrocola AS, Bai P, Weller SK, Heinen CD. DNA mismatch repair proteins are required for efficient herpes simplex virus type I replication. J Virol. 2011;85:12241–53. doi: 10.1128/JVI.05487-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mosberg JA, Lajoie MJ, Church GM. Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics. 2010;186:791–99. doi: 10.1534/genetics.110.120782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mosig G. Homologous recombination. In: Karam JD, editor. Bacteriophage T4. Washington, DC: ASM; 1994. pp. 54–82. [Google Scholar]
- 87.Muniyappa K, Radding CM. The homologous recombination system of phage λ: pairing activities of β protein. J Biol Chem. 1986;261:7472–78. [PubMed] [Google Scholar]
- 88.Murphy KC. Phage recombinases and their applications. Adv Virus Res. 2012;83:367–414. doi: 10.1016/B978-0-12-394438-2.00008-6. [DOI] [PubMed] [Google Scholar]
- 89.Muylaert I, Elias P. Contributions of nucleotide excision repair, DNA polymerase η, and homologous recombination to replication of UV-irradiated herpes simplex virus type 1. J Biol Chem. 2010;285:13761–68. doi: 10.1074/jbc.M110.107920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Muylaert I, Tang KW, Elias P. Replication and recombination of herpes simplex virus DNA. J Biol Chem. 2011;286:15619–24. doi: 10.1074/jbc.R111.233981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Muyrers JP, Zhang Y, Buchholz F, Stewart AF. RecE/RecT and Redα/Redβ initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 2000;14:1971–82. [PMC free article] [PubMed] [Google Scholar]
- 92.Mythili E, Kumar KA, Muniyappa K. Characterization of the DNA-binding domain of β protein, a component of phage λ Red-pathway, by UV catalyzed cross-linking. Gene. 1996;182:81–87. doi: 10.1016/s0378-1119(96)00518-5. [DOI] [PubMed] [Google Scholar]
- 93.Ogawa T, Tomizawa J. Replication of bacteriophage DNA. I. Replication of DNA of lambda phage defective in early functions. J Mol Biol. 1968;38:217–25. doi: 10.1016/0022-2836(68)90407-5. [DOI] [PubMed] [Google Scholar]
- 94.Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol Cell Biol. 2000;20:4922–31. doi: 10.1128/mcb.20.13.4922-4931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parkinson J, Lees-Miller SP, Everett RD. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73:650–57. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Passy SI, Yu X, Li Z, Radding CM, Egelman EH. Rings and filaments of β protein from bacteriophage λ suggest a superfamily of recombination proteins. Proc Natl Acad Sci USA. 1999;96:4279–84. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Poffenberger KL, Roizman B. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985;53:587–95. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Polvino-Bodnar M, Orberg PK, Schaffer PA. Herpes simplex virus type 1 oriL is not required for virus replication or for the establishment and reactivation of latent infection in mice. J Virol. 1987;61:3528–35. doi: 10.1128/jvi.61.11.3528-3535.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Porter IM, Stow ND. Virus particles produced by the herpes simplex virus type 1 alkaline nuclease null mutant ambUL12 contain abnormal genomes. J Gen Virol. 2004;85:583–91. doi: 10.1099/vir.0.19657-0. [DOI] [PubMed] [Google Scholar]
- 100.Poteete AR. Involvement of DNA replication in phage lambda Red-mediated homologous recombination. Mol Microbiol. 2008;68:66–74. doi: 10.1111/j.1365-2958.2008.06133.x. [DOI] [PubMed] [Google Scholar]
- 101.Poteete AR. Involvement of Escherichia coli DNA replication proteins in phage lambda Red-mediated homologous recombination. PLoS ONE. 2013;8:e67440. doi: 10.1371/journal.pone.0067440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Radding CM, Rosenzweig J, Richards F, Cassuto E. Separation and characterization of exonuclease, β protein, and a complex of both. J Biol Chem. 1971;246:2510–12. [Google Scholar]
- 103.Rajagopala SV, Casjens S, Uetz P. The protein interaction map of bacteriophage lambda. BMC Microbiol. 2011;11:213. doi: 10.1186/1471-2180-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reuven NB, Antoku S, Weller SK. The UL12.5 gene product of herpes simplex virus type 1 exhibits nuclease and strand exchange activities but does not localize to the nucleus. J Virol. 2004;78:4599–608. doi: 10.1128/JVI.78.9.4599-4608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reuven NB, Staire AE, Myers RS, Weller SK. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J Virol. 2003;77:7425–33. doi: 10.1128/JVI.77.13.7425-7433.2003. First report that UL12/ICP8 could act as a recombinase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reuven NB, Willcox S, Griffith JD, Weller SK. Catalysis of strand exchange by the HSV-1 UL12 and ICP8 proteins: Potent ICP8 recombinase activity is revealed upon resection of dsDNA substrate by nuclease. J Mol Biol. 2004;342:57–71. doi: 10.1016/j.jmb.2004.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roca AI, Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 108.Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979;16:481–94. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- 109.Samaniego LA, Neiderhiser L, DeLuca NA. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–20. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sarisky RT, Weber PC. Requirement for double-strand breaks but not for specific DNA sequences in herpes simplex virus type 1 genome isomerization events. J Virol. 1994;68:34–47. doi: 10.1128/jvi.68.1.34-47.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sawitzke JA, Costantino N, Li XT, Thomason LC, Bubunenko M, et al. Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J Mol Biol. 2011;407:45–59. doi: 10.1016/j.jmb.2011.01.030. Shows optimization. Defines conditions where recombineering can be done without selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sawitzke JA, Thomason LC, Bubunenko M, Li X, Costantino N, Court DL. Recombineering: highly efficient in vivo genetic engineering using single-strand oligos. Methods Enzymol. 2013;533:157–77. doi: 10.1016/B978-0-12-420067-8.00010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, Court DL. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 2007;421:171–99. doi: 10.1016/S0076-6879(06)21015-2. [DOI] [PubMed] [Google Scholar]
- 114.Schaffer PA, Tevethia MJ, Benyesh MM. Recombination between temperature-sensitive mutants of herpes simplex virus type 1. Virology. 1974;58:219–28. doi: 10.1016/0042-6822(74)90156-1. [DOI] [PubMed] [Google Scholar]
- 115.Scherer G. Nucleotide sequence of the O gene and of the origin of replication in bacteriophage lambda DNA. Nucleic Acids Res. 1978;5:3141–56. doi: 10.1093/nar/5.9.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schumacher AJ, Mohni KN, Kan Y, Hendrickson EA, Stark JM, Weller SK. The HSV-1 exonuclease, UL12, stimulates recombination by a single strand annealing mechanism. PLoS Pathog. 2012;8:e1002862. doi: 10.1371/journal.ppat.1002862. First report that HSV can stimulate SSA in a UL12 dependent fashion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Severini A, Morgan AR, Tovell DR, Tyrrell DL. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200:428–35. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 118.Severini A, Scraba DG, Tyrrell DL. Branched structures in the intracellular DNA of herpes simplex virus type 1. J Virol. 1996;70:3169–75. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shao L, Rapp LM, Weller SK. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology. 1993;196:146–62. doi: 10.1006/viro.1993.1463. [DOI] [PubMed] [Google Scholar]
- 120.Sheldrick P, Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Part 2):667–78. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- 121.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–56. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 122.Shlomai J, Friedmann A, Becker Y. Replication intermediates of herpes simplex virus DNA. Virology. 1976;69:647–59. doi: 10.1016/0042-6822(76)90493-1. [DOI] [PubMed] [Google Scholar]
- 123.Shulman MJ, Hallick LM, Echols H, Signer ER. Properties of recombination-deficient mutants of bacteriophage λ. J Mol Biol. 1970;52:501–20. doi: 10.1016/0022-2836(70)90416-x. [DOI] [PubMed] [Google Scholar]
- 124.Smith MG, Skalka A. Some properties of DNA from phage-infected bacteria. J Gen Physiol. 1966;49:127–42. doi: 10.1085/jgp.49.6.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sodeik B, Ebersold MW, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–21. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stahl FW, Fox MS, Faulds D, Stahl MM. Break-join recombination in phage λ. Genetics. 1990;125:463–74. doi: 10.1093/genetics/125.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stahl FW, Kobayashi I, Stahl MM. In phage λ, cos is a recombinator in the Red pathway. J Mol Biol. 1985;181:199–209. doi: 10.1016/0022-2836(85)90085-3. [DOI] [PubMed] [Google Scholar]
- 128.Stahl FW, McMilin KD, Stahl MM, Nozu Y. An enhancing role for DNA synthesis in formation of bacteriophage λ recombinants. Proc Natl Acad Sci USA. 1972;69:3598–601. doi: 10.1073/pnas.69.12.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stahl MM, Thomason L, Poteete AR, Tarkowski T, Kuzminov A, Stahl FW. Annealing vs. invasion in phage λ recombination. Genetics. 1997;147:961–77. doi: 10.1093/genetics/147.3.961. Key paper defining what types of recombination λ Red can do. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stengel G, Kuchta RD. Coordinated leading and lagging strand DNA synthesis by using the herpes simplex virus 1 replication complex and minicircle DNA templates. J Virol. 2011;85:957–67. doi: 10.1128/JVI.01688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.St-Pierre F, Endy D. Determination of cell fate selection during phage lambda infection. Proc Natl Acad Sci USA. 2008;105:20705–10. doi: 10.1073/pnas.0808831105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Strang BL, Stow ND. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J Virol. 2005;79:12487–94. doi: 10.1128/JVI.79.19.12487-12494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Strobel-Fidler M, Francke B. Alkaline deoxyribonuclease induced by herpes simplex virus type 1: composition and properties of the purified enzyme. Virology. 1980;103:493–501. doi: 10.1016/0042-6822(80)90206-8. [DOI] [PubMed] [Google Scholar]
- 134.Swingle B, Bao Z, Markel E, Chambers A, Cartinhour S. Recombineering using RecTE from Pseudomonas syringae. Appl Environ Microbiol. 2010;76:4960–68. doi: 10.1128/AEM.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Taylor TJ, Knipe DM. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J Virol. 2004;78:5856–66. doi: 10.1128/JVI.78.11.5856-5866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Thaler DS, Stahl MM, Stahl FW. Double-chain-cut sites are recombination hotspots in the Red pathway of phage λ. J Mol Biol. 1987;195:75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- 137.Thaler DS, Stahl MM, Stahl FW. Evidence that the normal route of replication-allowed Red-mediated recombination involves double-chain ends. EMBO J. 1987;6:3171–76. doi: 10.1002/j.1460-2075.1987.tb02628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Thomas MS, Gao M, Knipe DM, Powell KL. Association between the herpes simplex virus major DNA-binding protein and alkaline nuclease. J Virol. 1992;66:1152–61. doi: 10.1128/jvi.66.2.1152-1161.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Thomason LC, Sawitzke JA, Li X, Costantino N, Court DL. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol. 2014;106:1.16.1–39. doi: 10.1002/0471142727.mb0116s106. [DOI] [PubMed] [Google Scholar]
- 140.Thomason LC, Thaler DS, Stahl MM, Stahl FW. In vivo packaging of bacteriophage λ monomeric chromosomes. J Mol Biol. 1997;267:75–87. doi: 10.1006/jmbi.1996.0870. [DOI] [PubMed] [Google Scholar]
- 141.Thresher RJ, Makhov AM, Hall SD, Kolodner R, Griffith JD. Electron microscopic visualization of RecT protein and its complexes with DNA. J Mol Biol. 1995;254:364–71. doi: 10.1006/jmbi.1995.0623. [DOI] [PubMed] [Google Scholar]
- 142.Tolun G, Makhov AM, Ludtke SJ, Griffith JD. Details of ssDNA annealing revealed by an HSV-1 ICP8-ssDNA binary complex. Nucleic Acids Res. 2013;41:5927–37. doi: 10.1093/nar/gkt266. Recent paper describing ssDNA annealing by ICP8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tsurimoto T, Matsubara K. Purified bacteriophage λ O protein binds to four repeating sequences at the λ replication origin. Nucleic Acids Res. 1981;9:1789–99. doi: 10.1093/nar/9.8.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4:147–52. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 145.van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Single-molecule kinetics of λ exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–38. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 146.van Pijkeren J-P, Britton RA. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 2012;40:e76. doi: 10.1093/nar/gks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vaughan PJ, Banks LM, Purifoy DJ, Powell KL. Interactions between herpes simplex virus DNA-binding proteins. J Gen Virol. 1984;65:2033–41. doi: 10.1099/0022-1317-65-11-2033. [DOI] [PubMed] [Google Scholar]
- 148.Vellani TS, Myers RS. Bacteriophage SPP1 Chu is an alkaline exonuclease in the SynExo family of viral two-component recombinases. J Bacteriol. 2003;185:2465–74. doi: 10.1128/JB.185.8.2465-2474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vos M. Why do bacteria engage in homologous recombination? Trends Microbiol. 2009;17:226–32. doi: 10.1016/j.tim.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 150.Ward S, Weller S. HSV-1 DNA replication. In: Weller SK, editor. Alphaherpesviruses: Molecular Virology. Norfolk, UK: Caister Academic; 2011. pp. 89–112. [Google Scholar]
- 151.Weber PC, Challberg MD, Nelson NJ, Levine M, Glorioso JC. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988;54:369–81. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- 152.Weller SK, Coen DM. Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb Perspect Biol. 2012;4:a013011. doi: 10.1101/cshperspect.a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Weller SK, Lee KJ, Sabourin DJ, Schaffer PA. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J Virol. 1983;45:354–66. doi: 10.1128/jvi.45.1.354-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wickner SH. DNA replication proteins of Escherichia coli and phage λ. Cold Spring Harb Symp Quant Biol. 1979;43(Part 1):303–10. doi: 10.1101/sqb.1979.043.01.037. [DOI] [PubMed] [Google Scholar]
- 155.Wilkie NM. The synthesis and substructure of herpesvirus DNA: the distribution of alkali-labile single strand interruptions in HSV-1 DNA. J Gen Virol. 1973;21:453–67. doi: 10.1099/0022-1317-21-3-453. [DOI] [PubMed] [Google Scholar]
- 156.Wyman C, Kanaar R. DNA double-strand break repair: All’s well that ends well. Annu Rev Genet. 2006;40:363–83. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 157.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–83. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yu D, Sawitzke JA, Ellis H, Court DL. Recombineering with overlapping single-stranded DNA oligonucleotides: testing a recombination intermediate. Proc Natl Acad Sci USA. 2003;100:7207–12. doi: 10.1073/pnas.1232375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang J, Xing X, Herr AB, Bell CE. Crystal structure of E. coli RecE protein reveals a toroidal tetramer for processing double-stranded DNA breaks. Structure. 2009;17:690–702. doi: 10.1016/j.str.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202:530–39. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]
- 161.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–28. doi: 10.1038/2417. First demonstration of RecET-promoted recombineering using short homologies. [DOI] [PubMed] [Google Scholar]
- 162.Zissler J, Signer E, Schaefer F. The role of recombination in growth of bacteriophage λ. I. The gamma gene. In: Hershey AD, editor. The Bacteriophage Lambda. New York: Cold Spring Harb. Lab; 1971. pp. 455–68. [Google Scholar]