Abstract
Mammalian circadian timekeeping arises from a transcription-based feedback loop driven by a set of dedicated clock proteins. At its core, the heterodimeric transcription factor CLOCK:BMAL1 activates expression of Period, Cryptochrome, and Rev-Erb genes, which feed back to repress transcription and create oscillations in gene expression that confer circadian timing cues to cellular processes. The formation of different clock protein complexes throughout this transcriptional cycle helps to establish the intrinsic ∼24 h periodicity of the clock; however, current models of circadian timekeeping lack the explanatory power to fully describe this process. Recent studies confirm the presence of at least three distinct regulatory complexes: a transcriptionally active state comprising the CLOCK:BMAL1 heterodimer with its coactivator CBP/p300, an early repressive state containing PER:CRY complexes, and a late repressive state marked by a poised but inactive, DNA-bound CLOCK:BMAL1:CRY1 complex. In this review, we analyze high-resolution structures of core circadian transcriptional regulators and integrate biochemical data to suggest how remodeling of clock protein complexes may be achieved throughout the 24 h cycle. Defining these detailed mechanisms will provide a foundation for understanding the molecular basis of circadian timing and help to establish new platforms for the discovery of therapeutics to manipulate the clock.
Predictable environmental changes arising from the rotation of Earth about its axis set the fundamental diurnal tempo of our lives. Intrinsic molecular clocks synchronize behavior and physiology into circadian (about a day) rhythms that are coordinated with the solar day to provide stability within an ever-changing environment. While modern society allows us to live outside of these ancient routines through the use of artificial lights, discrepancies between our behavior and intrinsic circadian clock elicit comorbid metabolic syndromes, cardiovascular disease, and/or cancer.1 Understanding how circadian timekeeping occurs at the molecular level could inform new strategies for therapeutic discovery that aim to reinforce circadian systemic synchronization for its numerous health benefits.
While the past 15 years has seen major advances in our understanding of the genetic basis of circadian timing, we have not yet achieved a level of mechanistic insight comparable to those of other important global regulatory processes such as the cell cycle. A recent explosion of high-resolution structures of mammalian clock proteins, cistrome mapping, and biochemical data support some aspects of the canonical model for clock function and challenge others. Our goal in this review is to present an analysis of these data to suggest how remodeling of clock protein complexes throughout the 24 h cycle might regulate circadian timing. We start with an overview of the current model of the transcription/translation feedback mechanism and then discuss this clock mechanism from a biochemical and structural perspective.
Transcription Feedback Loops of the Mammalian Clock
At the heart of the mammalian circadian timekeeping system, a group of dedicated clock proteins work in concert with one another to generate transcription/translation-based feedback loops (TTFL) with ∼24 h periodicity (Figure 1A). The basic helix–loop–helix Per-Arnt-Sim (bHLH-PAS) proteins CLOCK and BMAL1 constitute a heterodimeric transcription factor that is the driving force behind the molecular clock in mammals.2,3 Transcriptional activation by CLOCK:BMAL1 at E/E′ boxes in the promoters of the core clock genes Period, Cryptochrome, and Rev-Erb initiates the beginning of negative feedback within the loop, which is ultimately closed when these factors enter the nucleus and repress transcription.4−7 Subsequent cycles of activation and repression generate circadian oscillations (i.e., one peak per day) in the expression of clock proteins to determine the intrinsic timing of the molecular clock. In addition to the core clock genes, the TTFL regulates the expression of output genes to confer circadian timing to physiological processes in a tissue-specific manner (Figure 1B).8,9 Post-transcriptional regulation also plays an important role through RNA processing and stabilization to establish intrinsic circadian timing and clock-controlled output genes10−13 (see DOI: 10.1021/bi500707c).
Figure 1.
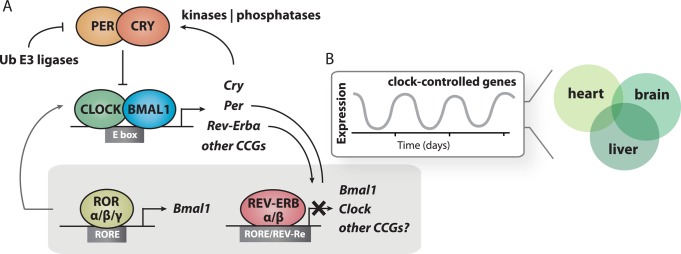
Transcription/translation feedback loops control mammalian circadian timing. (A) Core clock proteins interact with one another to regulate transcription. (B) The circadian feedback loop creates oscillations in the tissue-specific expression of clock-controlled genes with 24 h periodicity.
The current model of the core feedback loop is defined by periodic regulation of CLOCK:BMAL1 activity from repressive complexes containing PER and CRY proteins. The intrinsic ∼24 h periodicity of the clock is regulated in large part by controlling the subcellular localization and stability of PER:CRY complexes through the opposing activity of kinases, phosphatases, and ubiquitin E3 ligases.14−23 Loss of a single phosphoacceptor site within PER or modulation of kinase activity (by mutation or pharmacological inhibition) elicits periods ranging from ∼20 to 44 h,24−27 underscoring an amazing flexibility that is inherent within the architecture of the circadian feedback loop. Therefore, post-translational regulation of PER and CRY helps to establish critical delays in feedback regulation that contribute to the 24 h period of the clock.
A second, interlocked feedback loop involves the nuclear receptors ROR and REV-ERB, which generate oscillations in Bmal1 expression and the cyclic repression of other gene targets.28−30 Phenotypes of mice with deletions of individual genes within this interlocked feedback loop are mild;31 however, deletion of both Rev-Erbα and Rev-Erbβ genes disrupts circadian rhythms, demonstrating that cyclical repression at RORE and REV-Re elements by the REV-ERB proteins represents a critical component of circadian timekeeping.32 In addition to ROR/REV-ERB, many other proteins work in concert with core clock proteins to regulate the epigenome and influence circadian transcriptional activity (MLL1, MLL3, SIRT1, EZH2, etc.).33−37
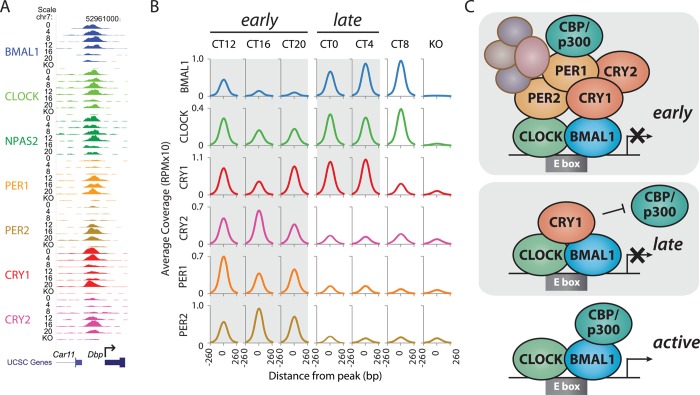
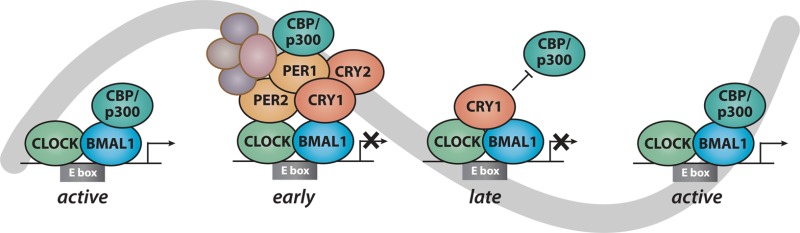
Determining how PER and CRY regulate CLOCK:BMAL1 activity will illuminate regulatory nodes within the feedback loop that can be further targeted for therapeutic intervention. A comprehensive mapping of the circadian cistrome in mouse liver shows that core circadian factors (CRY1/CRY2, PER1/PER2, and CLOCK:BMAL1) are coordinately recruited to nearly 1500 sites in the genome.11,12,38 The temporal pattern of their recruitment to clock-controlled genes such as Dbp (Figure 2A) generally agrees with the canonical TTFL model in which CLOCK:BMAL1 activity is repressed by a PER:CRY complex in the evening from CT12 to CT20. Analysis of native PER:CRY complexes by mass spectrometry and co-IP demonstrates that they contain additional proteins capable of eliciting transcription termination and epigenetic changes that reduce the extent of transcriptional activation (Figure 2C).39,40 PER:CRY complexes are very large, approaching several megadaltons in mass, and recent studies show that constituents of the complex can evolve over time,41,42 expanding the initial, simplified view of a static PER:CRY repressive complex.
Figure 2.
Multiple transcriptional regulatory complexes within the circadian feedback loop. (A) UCSC genome browser view of core clock protein occupancy at a portion of the Dbp locus on chromosome 7 (chr7), representing normalized chromatin immunoprecipitation and massively parallel sequencing read coverage at single time points within the circadian cycle. Six time points are shown every 4 h beginning at circadian time 0 (CT0) and ending at CT20, with knockout (KO) mice as controls. (B) Binding coverage profiles [RPM (reads per million) × 10] from ChIP-Seq data illustrate the temporal basis of clock protein recruitment to their 1444 common sites. Gray boxes behind the data illustrate the composition of the putative clock protein complexes. Parts A and B reproduced from ref (38). Copyright 2012 American Association for the Advancement of Science. (C) Cartoon schematics of distinct clock regulatory complexes based on ChIP-Seq and proteomic data, including the large early repressive PER:CRY complexes, and the late ternary CLOCK:BMAL1:CRY complex.
Perhaps the most striking conclusion to arise from recent studies is the clear evidence of a late repressive complex constituted by DNA-bound CLOCK:BMAL1 with CRY1, independent of PER proteins, that exists from CT0 to CT4 (Figure 2B).38,43,44 The foundation for this emerging model of the clock originates from biochemical studies in the Sancar lab providing the first evidence of a ternary CLOCK:BMAL1:CRY complex in vitro and in vivo that is incompatible with PER binding.44 ChIP-Seq studies in mouse liver indicate that recruitment of the early repressive PER:CRY complex, recruitment of the late repressive CRY1 complex, and recruitment of the transcriptional activators CBP/p300 to CLOCK:BMAL1 are three temporally distinct phases of the molecular cycle. Concomitant analysis of RNA Pol II complexes throughout the circadian cycle indicates that the late, CRY1-containing repressive complex most likely represents a poised but inactive form of CLOCK:BMAL1 (Figure 2C). These studies suggest that binding of CRY1 to CLOCK:BMAL1 may hold off activation by coactivators CBP/p300 and thus transcriptional initiation until CT8.38 This model is consistent with a critical role for the delayed expression of Cry1 and its exclusive ability to support cycling in minimal cellular oscillators.45−47 Accumulating evidence from genetic, chemogenetic, and computational modeling studies further supports the spatiotemporal separation of key PER and CRY functions in the feedback loop.48−50 Recent structural and biochemical data further support this exciting new role for CRY1 and will be discussed below.
Protein Architecture of Core Circadian Transcriptional Regulators
The repertoire of high-resolution structures of mammalian clock proteins has grown dramatically in the past three years to include most of the structured domains of the core positive and negative circadian regulators (Table 1 and Figure 3). These structures provide a foundation for understanding the biochemistry of clock proteins and yield insight into the disruptive power of mutations that arise from forward genetic screens. The conservation of domain architectures between positive and negative regulators suggests that they use common structural motifs to interact with DNA and/or core clock proteins (Figure 3). For example, REV-ERB proteins share the same common nuclear receptor architecture as the RORs, although they bind different ligands within their LBDs and lack a specific helix that is needed to interact with transcriptional coactivators, thus designating them as constitutive repressors.51 By contrast, CLOCK and BMAL1 are defined by their tandem PAS domains, which mediate heterodimerization,3 and disordered C-termini that regulate their activity.33,52−54 PER proteins also have tandem PAS domains that control formation of PER homo- and heterodimers55,56 followed by a long, disordered C-terminus that contains binding sites for kinases and cryptochrome.57,58
Table 1. Mammalian Clock Protein Structures.
| proteina | domain(s) | ligand | method | resolutionb (Å) | PDB entry | yearc |
|---|---|---|---|---|---|---|
| mCLOCK:mBMAL1 | bHLH | DNA | xtal | 2.27 | 4H10 | 2012 |
| mCLOCK:mBMAL1 | bHLH-PAS-AB | xtal | 2.40 | 4F3L | 2012 | |
| mBMAL2 | PAS-B | NMR | 2KDK | 2011 | ||
| mPER1 | PAS-AB | xtal | 2.75 | 4DJ2 | 2012 | |
| mPER2 | PAS-AB | xtal | 2.40 | 3GDI | 2009 | |
| mPER3 | PAS-AB | xtal | 2.50 | 4DJ3 | 2012 | |
| mCRY1 | PHR-CC | xtal | 2.65 | 4K0R | 2013 | |
| mCRY1 | PHR-CC | PER2 CBD | xtal | 2.45 | 4CT0 | 2014 |
| mCRY2 | PHR-CC | xtal | 2.70 | 4I6E | 2013 | |
| mCRY2 | PHR-CC | FAD | xtal | 2.20 | 4I6G | 2013 |
| mCRY2 | PHR-CC | FBXL3-SKP1 | xtal | 2.70 | 4I6J | 2013 |
| mCRY2 | PHR-CC | KL001 | xtal | 1.94 | 4MLP | 2013 |
| hREV-ERBα | LBD | NCoR peptide | xtal | 2.60 | 3N00 | 2010 |
| hREV-ERBα | DBD | DNA | xtal | 2.80 | 1HLZ | 2001 |
| hREV-ERBβ | LBD | xtal | 2.40 | 2V0V | 2007 | |
| hREV-ERBβ | LBD | heme | xtal | 1.90 | 3CQV | 2008 |
| hRORα | LBD | cholesterol | xtal | 1.63 | 1N83 | 2002 |
| rRORβ | LBD | retinoic acid | xtal | 1.50 | 1NQ7 | 2003 |
| hRORγ | LBD | hydroxycholesterol | xtal | 2.35 | 3KYT | 2010 |
Core transcriptional regulators only.
The NMR structure represents 10 lowest-energy structures.
Release date.
Figure 3.
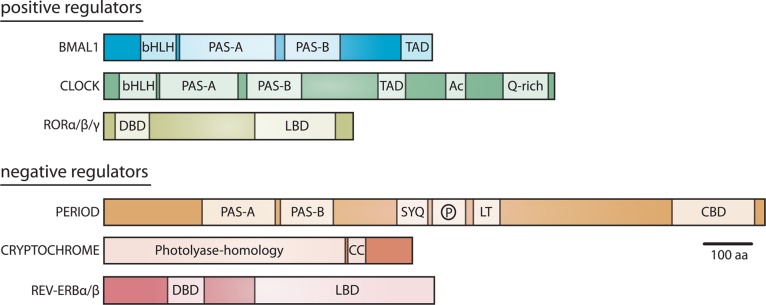
Domain architecture of core circadian transcriptional regulators. The location and size of conserved domains within core clock proteins are indicated according to the scale bar.
Cryptochromes have a protein architecture distinctly different from those of the other core clock proteins as they lack PAS domains. Most of the protein is composed of a photolyase homology region (PHR) that is strongly structurally similar to photolyase.59 In addition to the PHR, cryptochromes possess disordered C-termini that are not present in photolyase ancestors. Also unique to mammalian cryptochromes is a highly conserved helix just after the PHR that has a periodic spacing of nonpolar residues common to coiled coils, thus designated the CC helix.60 The CC helix plays a central role in the biochemistry of the mammalian clock by facilitating competitive interactions between CRY and its E3 ubiquitin ligase FBXL3 and PER61 to regulate its stability and by mediating the ability of CRY to repress the CLOCK:BMAL1 complex.60 The structural basis of competition for the cryptochrome CC helix will be discussed below.
PAS Domains
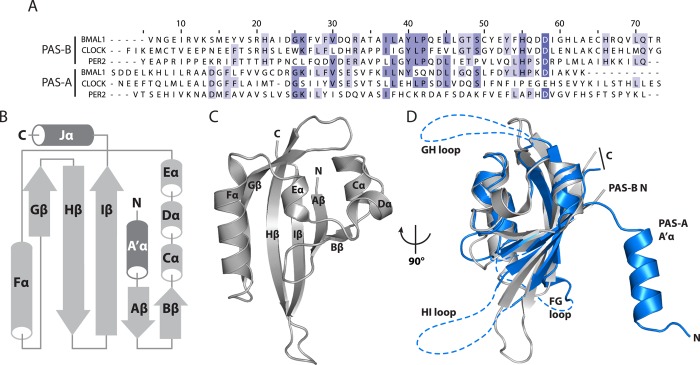
Present in tandem in three of the four core clock proteins, PAS domains have a central role in clock protein function.62 Originally identified through conservation of a minimal sequence motif from Drosophila to mammals (Figure 4A),62 PAS domains function as sensors of chemicals and light and act as oligomerization nodes. Since the original identification of the PAS motif, the definition of a PAS domain has been expanded to include the highly conserved secondary structure topology, consisting of a five-stranded antiparallel β-sheet flanked on one side by a series of α-helices. These strands are named alphabetically from the first (Aβ) to the final β-strand (Iβ) of the domain (Figure 4B).63 The original PAS motif is restricted to the conserved residues that form the short helices that run along the front of the PAS domain, and the accompanying PAS-associated C-terminal (PAC) motif encompasses part of the β-sheet. While acknowledging the historical significance and sequence conservation of PAS motifs, we argue that it is important to define PAS domains as their folded, biologically relevant domains rather than fragmented sequence motifs. A comparison of the boundaries of PAS domains identified in the recent crystal structures of clock proteins to the bioinformatic PAS/PAC predictions is illustrated in Table 2.
Figure 4.
PAS domain. (A) Alignment of UniPROT PAS motifs for mammalian CLOCK, BMAL1, and PER2. (B) Secondary structure topology of PAS domains. The core PAS fold is encoded by the structural elements colored light gray, while the dark gray A′α and Jα helices represent structural elements associated with certain PAS domains. N indicates the N-terminus and C the C-terminus. (C) Tertiary structure of a PAS domain. Secondary structure elements are labeled throughout the mouse BMAL1 PAS-B domain (PDB entry 4F3L). (D) Conservation of the PAS domain core in PAS-A and PAS-B domains despite distinct changes in domain size: gray, BMAL1 PAS-B domain; blue, BMAL1 PAS-A domain (PDB entry 4F3L). Note the lack of structured FG, GH, and HI loops (dashed blue lines) and the presence of an A′α helix for the PAS-A crystal structure.
Table 2. PAS Domain Boundaries in Representative Clock Proteins.
| proteina | domainb | residues | motifc | residues |
|---|---|---|---|---|
| CLOCK | PAS-A | 106–258 | PAS 1 | 107–177 |
| PAS-B | 274–376 | PAS 2 | 262–332 | |
| PAC | 336–379 | |||
| BMAL1 | PAS-A | 142–320 | PAS 1 | 143–215 |
| PAS-B | 353–442 | PAS 2 | 327–397 | |
| PAC | 402–445 | |||
| PER2 | PAS-A | 191–315 | PAS 1 | 179–246 |
| PAS-B | 329–435 | PAS 2 | 319–385 | |
| PAC | 393–436 |
Determined from domain boundaries in crystal structures 4F3L and 3GDI; includes A′α helix (PAS-A) or Jα helix (PAS-B) where appropriate.
From the UniPROT database (http://www.uniprot.org).
Several features distinguish the PAS-A and PAS-B domains of tandem PAS-containing proteins, although they share the same core PAS domain fold (Figure 4D). PAS-A domains frequently possess an N-terminal helix termed A′α that docks onto the β-sheet to mediate PAS–PAS interactions (Figure 4B,D).63 PAS-A domains also tend to have longer flexible loops (∼25–35 residues) between the Gβ–Hβ and Hβ–Iβ strands (known as the GH and HI loops, respectively). The importance of these disordered loops within PAS-A domains is generally not well understood within the bHLH-PAS family. The loops could be regulated by post-translational modification or mediate interactions with transcriptional regulatory proteins, both of which are facilitated by backbone flexibility.64 Along these lines, the BMAL1 PAS-A domain is sumoylated at a conserved lysine in the GH loop to regulate CLOCK:BMAL1 activity through an unknown mechanism.65 By contrast, PAS-B domains are generally much more compact than PAS-A domains with shorter loops (Figure 4D). Some PAS-B domains have a C-terminal helix termed Jα (Figure 4B) that can dock onto either the α-helical or β-sheet face of the PAS-B domain in a reversible manner to regulate PAS domain function.66,67 Therefore, not all PAS domains are alike, although they share a common core fold, suggesting that PAS-A and PAS-B domains could contribute to clock regulation through different mechanisms.
Both the α-helical and β-sheet interfaces of PAS domains can interact with PAS domains and other proteins in a variety of modes to control the architecture of transcriptional regulatory complexes.3,68−70 For example, the ARNT PAS-B domain simultaneously mediates heterodimer formation with the PAS-B domain of its bHLH-PAS partner HIF-2α and recruits coactivators needed to activate gene expression for hypoxia adaptation.71 Notably, the PAS domains of CLOCK, BMAL1, and PER have been implicated in the recruitment of PER to the CLOCK:BMAL1 complex by truncation/co-IP studies from cells,72 but we still lack biochemical confirmation of direct interactions in vitro. Understanding how the PAS domains of mammalian clock proteins assemble transcriptional regulatory complexes will provide important insight into clock function.
The close relationship of mammalian PAS domains to those in plants and bacteria that bind small molecule ligands and possess direct sensory capabilities suggests the intriguing possibility that small molecule metabolites could regulate clock function through the PAS domains.63 Indeed, the PAS domains of PER, CLOCK, and NPAS2 are reported to bind heme in vitro;73−75 however, there is some disagreement over heme binding,76 and the physiological relevance of these interactions has yet to be clearly demonstrated in cell-based assays or in vivo.73−76 The general plasticity of PAS domains that allows this small conserved fold to accommodate chemically diverse ligands has been exploited to find small molecules that bind within the solvated cores of PAS domains in the mammalian hypoxia adaptation response pathway.77 Ligand binding within the PAS-B domains of HIF-2α and ARNT allosterically regulates protein interactions to inhibit transcriptional responses to hypoxia.78−81 While researchers may have yet to identify endogenous ligands for PAS domains in circadian proteins, it may be possible to identify exogenous ligands that target PAS domains of the molecular clock to exploit them for regulation.
Period: A PAS Dimer at the Heart of the Clock
PER proteins have a fundamentally important role within the mammalian circadian clock, as changes in their post-translational modification state throughout the day exert exquisite control over the stability and localization of early PER:CRY repressive complexes.82 Attenuating this process with kinase inhibitors can lengthen the period of the molecular clock,15,27,83,84,26 demonstrating that PER proteins help establish the rather long, circadian period of the feedback loop. Moreover, induction of Per mRNA by external stimuli known as zeitgebers (time givers) controls the phase of the molecular oscillator to synchronize internal clocks with the environment.85 Because PER proteins are stoichiometrically limiting for the assembly of clock protein complexes,86 the introduction of naïve PER protein at different points within the feedback loop appears to advance or delay the molecular oscillator by controlling assembly of the early repressive complex. Therefore, understanding the biochemical basis of PER interactions with other clock proteins and transcriptional regulators will provide key insights into the molecular basis of circadian timing.
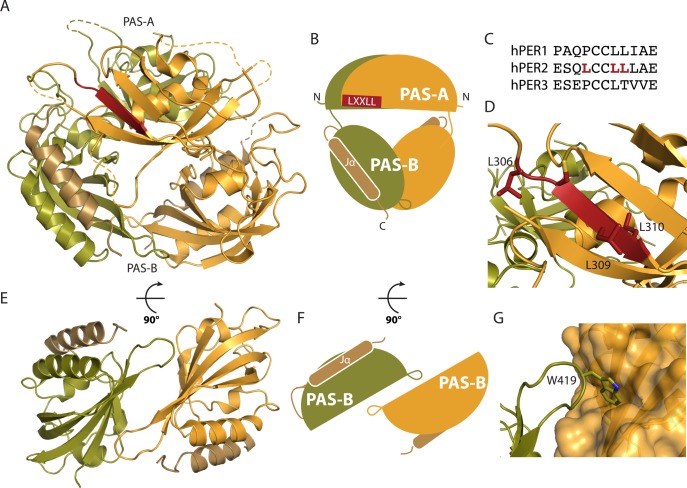
PER proteins exist as homo- and heterodimers in vivo,56 and given the observation of PAS dimers in other systems,62 attention focused on the PAS domains as likely candidates for dimerization domains. Crystal structures have been determined for the tandem PAS domains of all three PER proteins, each displaying the same PAS-B-mediated dimer (Figure 5A,B).67,87 The PAS-B β-sheets interact in an antiparallel orientation (Figure 5E,F) like the HIF-2α:ARNT PAS-B dimer,68 using a highly conserved tryptophan in the HI loop (W419 in PER2) to pack onto a predominantly hydrophobic cleft on the β-sheet of its partner (Figure 5G). This appears to be the predominant dimer interface within full-length proteins, and disruption of these interactions alters the mobility of full-length PER proteins in the cell.87 Moreover, in-frame deletion of the PAS-B domain in the Per2Brdm mutant disrupts clock function,88 but it is unclear whether the phenotype arises from loss of PER dimers or disruption of other functions. One such role could be subcellular localization, as each PAS-B domain has a C-terminal Jα helix that comprises an active nuclear export signal.89 Residues of the export motif also mediate direct interaction with the α-helical face of the PAS-B domain,67,87 suggesting that the structure represents an inhibited NES and that the interaction is regulated and/or reversible in solution. Given that the β-TrCP-dependent PER2 phosphodegron is also located immediately downstream of the Jα helix,21 these structural insights hint that there is still rich biochemistry to explore within the PAS domain structures.
Figure 5.
PER2 PAS-AB dimer structure. (A) Crystal structure of the mouse PER2 tandem PAS-AB domains (PDB entry 3GDI). Monomers are colored orange and olive and the Jα helices tan. The conserved LXXLL motif in PER2 PAS-A is colored red. Dashed lines represent disordered loops absent in the crystal structure. (B) Cartoon of the relative PAS domain orientations in the crystal structure. (C) PER2 has an LXXLL motif not conserved in PER1 or PER3. (D) Close-up of the LXXLL motif in the PER2 PAS-A domain with side chains of the motif leucines labeled. (E) Antiparallel orientation of the PAS-B dimer, shown without PAS-A domains for the sake of clarity. (F) Cartoon of the relative orientation of the PAS-B domains, illustrating contacts between the HI loop of one domain and the β-sheet of the other domain. (G) Close-up of the HI loop with the side chain of residue W419 shown docking into its hydrophobic cleft on the β-sheet of its partner PAS domain.
The PAS-A domains sit above the PAS-B dimer and lack substantial contacts with either the PAS-B domains or each other, suggesting that they have the ability to assume various structures in solution. The PER2 PAS-A domain has an LXXLL motif that is needed to interact with REV-ERBα and other nuclear receptors (Figure 5A–C).90 Canonical LXXLL motifs present the conserved leucine side chains on one face of a helix to interact with a host of transcriptional regulators.91,92 However, the LXXLL motif in PER2 is embedded with the Iβ strand of the PAS-A domain (Figure 5D), suggesting that it may not have a role in direct binding, at least in terms of a canonical LXXLL motif. Nevertheless, mutational analyses of this region show that this PAS-AB interface is important for interaction with nuclear receptors.90 Further study here will help to define how PERs could regulate transcription outside of the core clock loop, consistent with their recruitment to thousands of sites throughout the day outside of CLOCK:BMAL1 control, including nuclear receptor motifs.38
A region comprising the tandem PAS domains of PER has been found to interact with the PAS-containing regions of individual CLOCK and BMAL1 proteins by co-IP53,72 and is generally presumed to represent the basis for recruitment of PER:CRY complexes to CLOCK:BMAL1 early in the repressive phase of the feedback loop. However, full-length PER2 purified from insect or mammalian cells either as a pure protein or in complex with CRY1, respectively, does not interact directly with the CLOCK:BMAL1 complex on DNA in vitro;44,93 either the purified protein(s) lacks essential post-translational modification(s) needed to interact with each other, or other proteins are needed to bridge the interaction. While co-IP assays provide some insight into protein complexes, they lack the ability to elucidate direct protein interactions within complexes or discriminate possible heterogeneity of complexes in solution. Therefore, the question of how early repressive PER:CRY complexes are recruited to DNA-bound CLOCK:BMAL1 remains one of the central unanswered questions in the biochemistry of the mammalian clock.
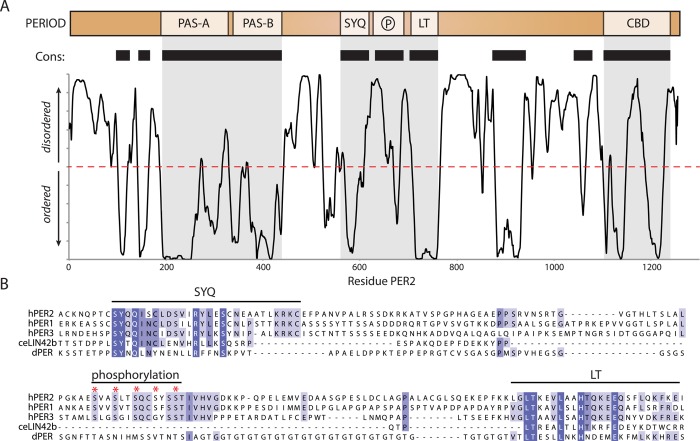
Aside from the tandem PAS domains, PER proteins are largely unstructured, a feature that PER proteins share with their functional analogue FRQ in Neurospora.94 The lack of intrinsic structure in a protein with many interaction partners provides a thermodynamic advantage by allowing specificity and affinity to be tuned for multiple partners.95,96 In addition to binding promiscuity, unstructured regions could allow for variable post-translational modifications and rapid degradation when not in complex with their binding partners. Many examples of how intrinsic disorder modulates protein stability have been noted in other systems,97 including the immediate post-translational degradation of the Neurospora crassa clock protein FRQ when not in complex with its partner, FRH.94 Because the lack of tertiary structure removes purely architectural constraints on conservation, alignment of intrinsically disordered regions tends to highlight functionally important residues. Algorithms such as PONDR use sequence biases to predict disorder,98 but of particular interest are predictions for short regions of order within long stretches of disorder, represented as downward spikes in the PONDR prediction (Figure 6A). These “minima” represent regions with increased propensity to form secondary structure, typically α-helices, which often act to nucleate formation of protein interactions.99 Aside from the ordered PAS domains, many of the minima in PER2 have weak predictions of secondary structure and high levels of sequence conservation, serving as a useful guide for future biochemical studies of PER function.
Figure 6.
Conservation and disorder in PER2. (A) The domain architecture of PER2 is aligned above the PONDR prediction of disorder. Within this plot, curves below the red dashed line are predicted to be ordered while those above the line are flexible; stretches longer than 30 residues are classified as “intrinsic disorder”. Regions exhibiting a high degree of conservation (cons.) between PER1 and PER2 are represented as black bars below the domain schematic. Gray boxes denote known regions of functional importance: PAS dimer, kinase-binding SYQ/LT regions, and the CRY-binding domain (CBD). (B) Alignment of the SYQ-phosphorylation-LT region shows conservation of key sequence motifs among human and Drosophila PER proteins and the C. elegans PER-homologue LIN-42, isoform b.
Binding sites for kinases CKIδ and CKIε represented by the SYQ and LT motifs are minima that are highly conserved in PER across phyla (Figure 6A,B).57 Conservation of these regions with the Caenorhabditis elegans PER homologue LIN-42,100 which controls developmental timing,101,102 and in Drosophila where it impacts PER clock function103 suggests that they represent an ancient site for regulation of PER activity. The multisite phosphorylation cassette that controls subcellular localization of mammalian PERs is located between the SYQ and LT motifs (Figure 6B).24 We speculate that additional conserved minima in PER2 may recruit other proteins to form the native megadalton transcriptional regulatory complexes seen in vivo.39−41 The extreme C-terminus of PER1 and PER2, but not PER3, harbors a conserved CRY-binding domain (CBD) that is essential for clock function.58,72,93 Control of CRY through its interaction with PER at this site represents a critical component of the mammalian feedback loop. Therefore, the absence of this motif in PER3 may explain its nonessential role in the circadian feedback loop,104,105 while still allowing for modulation of circadian timing through heterodimer formation with PER1 or PER2 to titrate CRY recruitment from the early repressive complex.106
CRY1: The Circadian Repressor
Since their discovery 15 years ago, CRY proteins have been acknowledged as a key negative regulator of the core circadian feedback loop. Cryptochromes repress CLOCK:BMAL1 transcriptional activation in reporter assays, and their loss in Cry1–/–Cry2–/– animals leads to an increased level of expression of clock target genes and disruption of the feedback loop, consistent with global derepression of CLOCK:BMAL1.5,6,107,108 However, the mechanism by which cryptochromes repress CLOCK:BMAL1 to “close” the feedback loop on a daily basis is still not known. Early studies of native clock proteins by co-IP demonstrated that PER:CRY complexes interact with CLOCK:BMAL1 in the early phase of repression, helping to establish the canonical model for the feedback loop (Figure 1A).82 Concurrent with these studies, it was observed that CRY can repress CLOCK:BMAL1 after transient transfection in cell-based reporter assays without stoichiometric PER, including in heterologous systems that lack all mammalian PERs.105 While these initial studies provided clear evidence that CRY does not need PER to assert its repressive function on CLOCK:BMAL1, concerns over artifacts from transfection studies proved the need for further studies to resolve the roles of PER and CRY in the clock.
Recent biochemical, structural, and genomic mapping studies highlight several lines of evidence that challenge the canonical feedback loop model. First, CRY interacts with the CLOCK:BMAL1 complex on DNA in vitro and in vivo to form what has been dubbed the “late repressive complex” (Figure 2C).38,43,44 In the mouse liver, CRY is found at the CLOCK:BMAL1 complex on DNA in the early morning without PER; this ternary complex is suggested to represent a poised but repressed state that holds off activation by the histone acetyltransferases p300/CBP until the appropriate time of day.38 Consistent with this, PER proteins are not needed for CRY to repress CLOCK:BMAL1 activation of the Dbp locus in vivo.(44) Second, PER proteins titrate CRY away from ternary CLOCK:BMAL1:CRY complexes in vitro and in vivo.(44,72,93) The C-terminal CRY-binding domain (CBD) of PER1 and PER2 is sufficient for this activity,72,93 and the feedback loop is disrupted when the PER2 CBD is constitutively expressed.72 Thus, one function of early repressive complexes may be to protect and hold CRY stably in reserve, bound to PER, until the initiation of the late repressive phase when CRY interacts directly with CLOCK:BMAL1.38,43 This model may explain the sensitivity of the circadian feedback loop to PER:CRY stoichiometry.86,109,110 Given the body of evidence that points to both independent and combined roles for PER and CRY, it is time to reevaluate the canonical model for the mammalian circadian feedback loop.
Cryptochrome structure is largely based on its homology to photolyase, encompassing an ∼50 kDa photolyase homology region (PHR) with an N-terminal α/β-domain and C-terminal α-helical domain that harbors the canonical flavin-binding site.59 Mammalian CRYs bind FAD weakly because of structural variations that create a flavin-binding site much shallower than that of photolyases.61,111 The predominant structural features that distinguish CRYs from photolyase are their disordered C-terminal tails.112 The CRY C-termini are divergent from one another and dispensable for generating the molecular feedback loop, but their deletion alters the period and amplitude of cycling,47,60 indicating that they can modulate clock protein function. Consistent with this regulatory role, the C-termini possess several phosphorylation sites and a nuclear localization signal.113−115 To date, no CRY structures have included the C-terminal tails, most likely because of their intrinsic flexibility. However, the C-termini interact in trans with the PHR in vitro and are partially protected from proteolysis in the full-length protein, suggesting that interaction with the PHR may impart some order to the tails.112,116 Understanding the cryptic role of CRY C-termini interactions with the PHR and other clock proteins represents a structural biology challenge that may require moving beyond X-ray crystallography.
Two highly conserved motifs on CRY are needed for its function in the clock: a hydrophobic motif adjacent to the flavin-binding site termed the interface loop or C-terminal lid61,117 and the CC helix.60 The PER2 CBD118 and the E3 ubiquitin ligase FBXL361 both engage these motifs, wrapping snugly around the CC helix (Figure 7A). The overlapping nature of binding sites on the CRY CC helix nicely explains the stabilizing effect that PER has on CRY in vivo;82,119 by interacting with the PER2 CBD, CRY is restricted from interacting with its E3 ubiquitin ligase.61 Moreover, these structures demonstrate how the small molecule KL001 stabilizes CRY by competing with the FBXL3 C-terminus for binding at the flavin-binding cleft.48,120 Altogether, an elegant integration of high-resolution structures, biochemistry, and in vivo studies provides a solid model for regulation of CRY stability and localization predicated on antagonistic interactions at overlapping binding sites.
Figure 7.
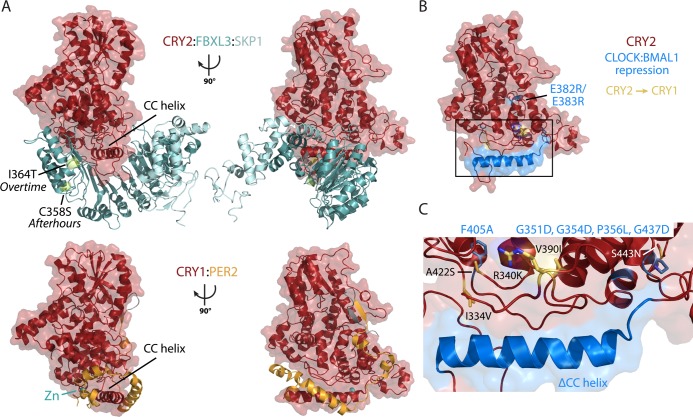
Competition for the CC helix controls CRY function in the feedback loop. (A) Crystal structures of mouse CRY2 in complex with FBXL3:SKP1 (top, PDB entry 4I6J) and mouse CRY1 in complex with PER2 CBD (bottom, PDB entry 4CT0) illustrate their overlapping binding sites. The location of FBXL3 Afterhours and Overtime mutations is depicted with the residues shown as spheres (citron). The location of CRY1:PER2-coordinated zinc is shown as a sphere (teal). (B) Crystal structure of apo mouse CRY2 (PDB entry 4I6E) colored red with residues needed for repression of CLOCK:BMAL1 colored blue. Residues that confer CRY1 activity to CRY2 are colored yellow. (C) Close-up of one face of CRY illustrating residues needed for CLOCK:BMAL1 repression. Side chains for CRY1 residues that confer cycling ability to CRY2 are colored yellow, with black labels; side chains for residues needed to repress CLOCK:BMAL1 are colored blue, with blue labels above the box.
The elusive role of Timeless in the mammalian clock122 may result from its involvement in this competitive, regulatory mechanism through its interactions with the CRY CC helix.123 As a replication fork-associated factor,121 Timeless is involved in DNA damage repair and checkpoint activation, and its expression is strictly controlled by the cell cycle.121,122,124−127 Therefore, its interaction with CRY suggests that it may serve to connect the circadian clock with cellular proliferation.124 Exciting studies also suggest that Timeless could play a role in conveying phase-resetting zeitgebers arising from DNA damage to the core feedback loop.123,128−130 Finally, like PER, cryptochromes also interact with nuclear receptors131 and are recruited to thousands of sites outside of CLOCK:BMAL1 regulation each day to expand the scope of circadian transcriptional regulation.38 Defining the molecular basis of these interactions with CRY will provide further insight into its regulation and activity in the core clock feedback loop and beyond.
Underscoring the importance of competition for CRY regulation, the same interface used as a hot spot to regulate CRY localization and stability is also needed to regulate CLOCK:BMAL1 activity.47,60 The CC helix interacts directly with the transcriptional activation domain (TAD) of BMAL1 in vitro,117 and mutations of the BMAL1 TAD or CRY CC decrease the level of repression by CRY.53,54,132 Additional residues needed to repress CLOCK:BMAL1 activity and drive the feedback loop surround the CC helix (Figure 7B,C). Some of these residues are unique to CRY1 and can confer the ability to drive cycling in minimal cellular oscillators to CRY2, a property it otherwise lacks without compensation from systemic properties.45,47 The presence of distinctive residues within the CRY1 PHR required for repression that do not overlay the PER2 CBD-binding site118 suggests they may be involved in interactions with CLOCK:BMAL1. While the model that holds CRY may function by antagonizing coactivators to hold off transcription has been slowly emerging over the past few years, the mechanism by which cryptochromes interact with and repress CLOCK:BMAL1 has not yet been elucidated.
CLOCK:BMAL1: The Principal Circadian Transcription Factor
Although a detailed model of the core circadian feedback loop is beginning to emerge, many questions remain. Recent structures of the CLOCK:BMAL1 heterodimer provide a solid foundation for these models by elucidating the protein architecture of the primary circadian transcription factor and how it binds DNA. Functional studies augment the crystal structure models and demonstrate that intrinsically disordered regions in both CLOCK and BMAL1 (Figure 8A) play critical roles in regulation of CLOCK:BMAL1 activity in vivo,2,52−54,133,134 as they do within the larger bHLH-PAS family of transcription factors.135
Figure 8.
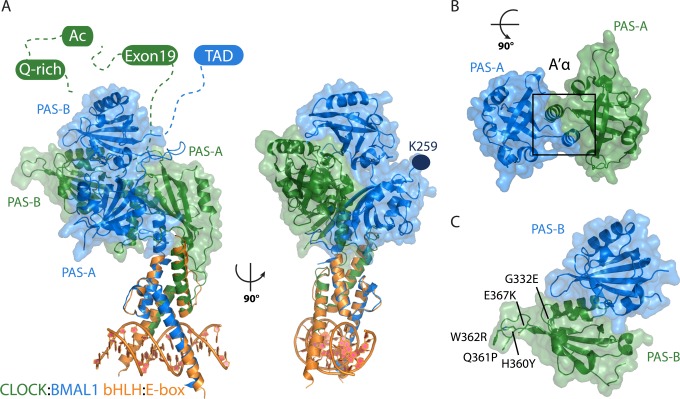
CLOCK:BMAL1 bHLH-PAS dimer structure. (A) Crystal structures of the bHLH dimer:E-box complex (orange, PDB entry 4H10) and the bHLH-PAS-AB dimer (PDB entry 4F3L) overlaid with CLOCK (green) and BMAL1 (blue). Dashed lines indicate intrinsically disordered C-terminal regions containing functionally important regions, including CLOCK exon 19, the acetyl-CoA-binding motif (Ac), the glutamine-rich (Q-rich) region, and the BMAL1 transcriptional activation domain (TAD). K259 (dark blue) is the SUMOylation site on BMAL1 PAS-A. (B) Depiction of interactions between the PAS-A domains, shown in isolation at a 90° rotation relative to the left structure in panel A. (C) Depiction of interactions between the PAS-B domains, shown in isolation, with mutations within the CLOCK PAS-B domain that decrease the level of CRY binding and/or repression of the CLOCK:BMAL1 complex.
During the active phase of the circadian complex, the transcriptional coactivators CBP and p300 interact directly with BMAL1 to initiate transcription through acetylation of histone H3 and recruitment of basal transcriptional machinery at E-box promoter regions.38,136−139 Somewhat perplexingly, CBP and p300 also interact with PER and are found in the early repressive PER:CRY complex, while they are absent from the late repressive complex that contains CRY1.38 CLOCK:BMAL1 also recruits additional coactivators during the active phase of transcription, including MLL and TRAP150.33,34,140 The molecular mechanisms that lead to transcriptional activation at CLOCK:BMAL1 sites, and therefore reinitiation of the feedback loop, are still relatively poorly defined but play an important role in the feedback loop mechanism.
A structure of the bHLH heterodimer in complex with its cognate E-box recognition sequence provides the first detailed view of interactions that determine the specificity of recruitment of CLOCK:BMAL1 to DNA and its regulation by phosphorylation.141,142 In contrast to studies showing a dramatic effect of the redox state of NAD cofactors on DNA binding in vitro,143,144 this new study finds no effect of NAD cofactors on the conformation of the bHLH heterodimers or their affinity for DNA.141 Moreover, the redox ratios of NAD cofactors previously found to influence the CLOCK:BMAL1 complex are not likely to occur in cells,145 indicating that DNA binding is probably not regulated directly by changes in NAD redox status. The structure of the isolated bHLH domains bound to DNA is highly similar to that of bHLH domains within the larger bHLH-PAS-AB heterodimer structure obtained in the absence of DNA (Figure 8A),3 suggesting that large changes in conformation of the bHLH domains upon DNA binding are unlikely.
CLOCK and BMAL1 PAS domains form the major basis for heterodimer recognition and stability of the complex.3 Within the complex, both BMAL1 PAS domains are presented on one face and the CLOCK PAS domains on the other (Figure 8A). The PAS-A domains form the heart of the heterodimer complex by swapping their A′α helices to interact with the β-sheet of the opposing PAS-A domain (Figure 8B), and disruption of this helical embrace significantly disrupts heterodimer stability in cells.3 A mutation in the linker between the CLOCK PAS-A and PAS-B domains (I254N) found in a zebrafish ENU screen causes a short period;146 Ile254 is buried under the CLOCK:BMAL1 PAS-B domains and relatively inaccessible to solvent, suggesting that the mutation may influence the packing or flexibility of the two PAS-B domains in relation to the bHLH-PAS-A core of the complex. The BMAL1 PAS-B domain β-sheet sits atop the α-helical face of the CLOCK PAS-B domain in a parallel orientation (Figure 8C). One consequence of this tandem arrangement is that the β-sheet of CLOCK PAS-B remains largely exposed, in contrast to most other PAS domains studied to date, which seem to have an obligate requirement to engage their β-sheets in protein interactions.63
In addition to mediating heterodimerization of bHLH-PAS proteins, PAS domains can also recruit regulatory proteins to control transcription.71,80,147,148 As noted earlier, it is unclear if PER proteins can interact directly with the CLOCK:BMAL1 complex on DNA.44,72 CRY interacts with the CLOCK PAS-B domain by a yeast two-hybrid form, and mutation of two residues within the HI loop of the PAS-B domain disrupts binding.149 Other mutations within the HI loop or directly adjacent to it on the CLOCK PAS-B domain reduce the level of CRY repression of the CLOCK:BMAL1 complex in vivo (Figure 8C).54,149 Furthermore, mutation of the BMAL1 TAD disrupts interactions with the CRY1 CC helix and synergistically decreases the level of repression by CRY when assayed with CLOCK PAS-B mutants.53,54 These data suggest that CRY interacts with multiple distinct sites on CLOCK:BMAL1, likely a component of its potent repressive activity toward the complex.
Regions downstream of the PAS domains lack ordered structure, and few in vitro biochemical or biophysical studies have been conducted on the isolated CLOCK or BMAL1 C-termini.117 However, we believe that the C-termini represent an exciting frontier that is rich with unexplored biology because they are essential for regulating the activity of CLOCK:BMAL1.2,52−54,133,134,150,151 The activity of the CLOCK:BMAL1 complex is not simply predicated on DNA binding as it bound to E-boxes throughout much of the day (Figure 2B).12,38 Therefore, describing the mechanisms by which positive and negative acting factors are recruited via the C-termini to control the activity of the complex should yield important insight into clock function. As noted earlier, flexibility of the C-termini is likely an important aspect of CLOCK:BMAL1 function, a property that is common to other “malleable machines” involved in transcriptional regulation.95 For example, how the acetyl CoA-binding motif of CLOCK confers acetylation to clock proteins133,134 in the absence of a canonical MYST family HAT domain structure152 remains to be shown. Moreover, short regions of predicted order such as the 51-residue α-helical exon 19 are essential for clock function52 by interacting with positive and negative transcriptional regulators33,149 yet exist within a sea of disordered protein. The intrinsic flexibility of these regions may allow them to interact with a host of binding partners in a temporally dependent manner, allowing intrinsic timekeeping of the circadian clock to be finely tuned by interactions between regulatory proteins, possibly in a tissue-specific manner.
Conclusion
We believe that continuing to integrate in vivo and in vitro studies will provide deep insight into the fundamental, biochemical processes that establish circadian timing in mammals. The recent abundance of high-resolution structures has provided a much-needed visualization of clock proteins and their interactions at the atomic level. However, many important regions of clock proteins are not depicted in these crystal structures because they lack a single, static structure. Protein flexibility is inherent to transcriptional regulators because it dramatically expands their repertoire of protein interactions,95,96 even helping to establish the time scale and/or magnitude of gene activation.153 Therefore, it is important that a wide range of techniques (crystallography, NMR, SAXS, EM, HDX MS, and native MS) be utilized to fully describe protein interactions and their dynamics that give rise to circadian timing.
Looking forward, we believe this is a truly exciting time for the field of circadian biology. We now possess the tools to probe clock protein function from complex animal models down to the resolution of structural details to learn how the clock sets intrinsic timing and interfaces with metabolism, tissue-specific factors, and pathophysiological states. There is no shortage of surprises at the complex and elegant biochemistry of the clock. The recent discovery of ancient, redox-based post-translational circadian oscillators conserved from archaebacteria to humans154,155 (and see review by O’Neill et al. in this issue) highlights that we are still learning ways in which living organisms have evolved to coordinate their activity with the environment.
Acknowledgments
We thank Eva Wolf for sharing coordinates of the mCRY1:PER2 CBD crystal structure (PDB entry 4CT0) prior to its release. We apologize to colleagues whose work could not be cited because of space considerations.
Glossary
Abbreviations
- Ac
acetyl-CoA-binding motif
- ARNT
aryl hydrocarbon nuclear translocator
- ARNTL
ARNT-like 1
- bHLH
basic helix–loop–helix
- BMAL1
brain and muscle ARNT-like 1
- CBD
CRY-binding domain
- CBP
CREB-binding protein
- CC
coiled coil
- CCG
clock-controlled gene
- ChIP-Seq
chromatin immunoprecipitation sequencing
- CKI
casein kinase I
- CLOCK
circadian locomotor output cycles kaput
- co-IP
co-immunoprecipitation
- CREB
cyclic AMP response element-binding protein
- CRY
cryptochrome
- CT
circadian time
- DBD
DNA-binding domain
- DBP
D site-binding protein
- EM
electron microscopy
- EMSA
electrophoretic mobility shift assay
- ENU
N-ethyl-N-nitrosourea
- FRQ
frequency
- GR
glucocorticoid receptor
- HAT
histone acetyltransferase
- HDX MS
hydrogen–deuterium exchange mass spectrometry
- LBD
ligand-binding domain
- NMR
nuclear magnetic resonance
- NR
nuclear receptor
- PAS
PER-ARNT-SIM
- PDB
Protein Data Bank
- PER
period
- REV-ERB
reverse strand of ERB
- REV-Re
REV-ERB response element
- ROR
retinoic acid-related orphan receptor
- RORE
ROR response element
- SAXS
small-angle X-ray scattering
- SIM
single-minded
- TAD
transcriptional activation domain
- TTFL
transcription/translation feedback loop.
This work was supported by a grant from the National Institutes of Health (R01 GM107069) and the University of California Cancer Research Coordinating Committee to C.L.P.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Takahashi J. S.; Hong H.-K.; Ko C. H.; McDearmon E. L. (2008) The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat. Rev. Genet. 9, 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N.; Staknis D.; Nguyen H. B.; Davis F. C.; Wilsbacher L. D.; King D. P.; Takahashi J. S.; Weitz C. J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569. [DOI] [PubMed] [Google Scholar]
- Huang N.; Chelliah Y.; Shan Y.; Taylor C. A.; Yoo S. H.; Partch C.; Green C. B.; Zhang H.; Takahashi J. S. (2012) Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex. Science 337, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B.; Albrecht U.; Kaasik K.; Sage M.; Lu W.; Vaishnav S.; Li Q.; Sun Z. S.; Eichele G.; Bradley A.; Lee C. C. (2001) Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105, 683–694. [DOI] [PubMed] [Google Scholar]
- van der Horst G. T.; Muijtjens M.; Kobayashi K.; Takano R.; Kanno S.; Takao M.; de Wit J.; Verkerk A.; Eker A. P.; van Leenen D.; Buijs R.; Bootsma D.; Hoeijmakers J. H.; Yasui A. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630. [DOI] [PubMed] [Google Scholar]
- Vitaterna M. H.; Selby C. P.; Todo T.; Niwa H.; Thompson C.; Fruechte E. M.; Hitomi K.; Thresher R. J.; Ishikawa T.; Miyazaki J.; Takahashi J. S.; Sancar A. (1999) Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. U.S.A. 96, 12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N.; Damiola F.; Lopez-Molina L.; Zakany J.; Duboule D.; Albrecht U.; Schibler U. (2002) The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260. [DOI] [PubMed] [Google Scholar]
- Storch K.-F.; Lipan O.; Leykin I.; Viswanathan N.; Davis F. C.; Wong W. H.; Weitz C. J. (2002) Extensive and divergent circadian gene expression in liver and heart. Nature 417, 78–83. [DOI] [PubMed] [Google Scholar]
- Panda S.; Hogenesch J. B.; Kay S. A. (2002) Circadian rhythms from flies to human. Nature 417, 329–335. [DOI] [PubMed] [Google Scholar]
- Morf J.; Rey G.; Schneider K.; Stratmann M.; Fujita J.; Naef F.; Schibler U. (2012) Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science 338, 379–383. [DOI] [PubMed] [Google Scholar]
- Le Martelot G.; Canella D.; Symul L.; Migliavacca E.; Gilardi F.; Liechti R.; Martin O.; Harshman K.; Delorenzi M.; Desvergne B.; Herr W.; Deplancke B.; Schibler U.; Rougemont J.; Guex N.; Hernandez N.; Naef F.; (2012) Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 10, e1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet J. S.; Rodriguez J.; Abruzzi K. C.; Rosbash M. (2012) Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife 1, e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S.; Sher-Chen E. L.; Green C. B. (2012) Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 26, 2724–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T.; Lewis W. G.; Liu A. C.; Lee J. W.; Schultz P. G.; Kay S. A. (2008) A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3β. Proc. Natl. Acad. Sci. U.S.A. 105, 20746–20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isojima Y.; Nakajima M.; Ukai H.; Fujishima H.; Yamada R. G.; Masumoto K.-H.; Kiuchi R.; Ishida M.; Ukai-Tadenuma M.; Minami Y.; Kito R.; Nakao K.; Kishimoto W.; Yoo S.-H.; Shimomura K.; Takao T.; Takano A.; Kojima T.; Nagai K.; Sakaki Y.; Takahashi J. S.; Ueda H. R. (2009) CKIε/δ-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 106, 15744–15749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia K. A.; Sachdeva U. M.; DiTacchio L.; Williams E. C.; Alvarez J. G.; Egan D. F.; Vasquez D. S.; Juguilon H.; Panda S.; Shaw R. J.; Thompson C. B.; Evans R. M. (2009) AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-H.; Mohawk J. A.; Siepka S. M.; Shan Y.; Huh S. K.; Hong H.-K.; Kornblum I.; Kumar V.; Koike N.; Xu M.; Nussbaum J.; Liu X.; Chen Z.; Chen Z. J.; Green C. B.; Takahashi J. S. (2013) Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152, 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A.; Yumimoto K.; Tsunematsu R.; Matsumoto M.; Oyama M.; Kozuka-Hata H.; Nakagawa T.; Lanjakornsiripan D.; Nakayama K. I.; Fukada Y. (2013) FBXL21 Regulates Oscillation of the Circadian Clock through Ubiquitination and Stabilization of Cryptochromes. Cell 152, 1106–1118. [DOI] [PubMed] [Google Scholar]
- Siepka S. M.; Yoo S.-H.; Park J.; Song W.; Kumar V.; Hu Y.; Lee C.; Takahashi J. S. (2007) Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L.; Bassermann F.; Maiolica A.; Lee C.; Nolan P. M.; Godinho S. I. H.; Draetta G. F.; Pagano M. (2007) SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316, 900–904. [DOI] [PubMed] [Google Scholar]
- Shirogane T.; Jin J.; Ang X. L.; Harper J. W. (2005) SCFβ-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 280, 26863–26872. [DOI] [PubMed] [Google Scholar]
- Partch C. L.; Shields K. F.; Thompson C. L.; Selby C. P.; Sancar A. (2006) Posttranslational regulation of the mammalian circadian clock by cryptochrome and protein phosphatase 5. Proc. Natl. Acad. Sci. U.S.A. 103, 10467–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.; Chen R.; Lee Y.; Yoo S.; Lee C. (2009) Essential roles of CKIδ and CKIε in the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 106, 21359–21364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh K. L.; Jones C. R.; He Y.; Eide E. J.; Hinz W. A.; Virshup D. M.; Ptácek L. J.; Fu Y. H. (2001) An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291, 1040–1043. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Padiath Q. S.; Shapiro R. E.; Jones C. R.; Wu S. C.; Saigoh N.; Saigoh K.; Ptácek L. J.; Fu Y.-H. (2005) Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature 434, 640–644. [DOI] [PubMed] [Google Scholar]
- Hirota T.; Lee J. W.; Lewis W. G.; Zhang E. E.; Breton G.; Liu X.; Garcia M.; Peters E. C.; Etchegaray J.-P.; Traver D.; Schultz P. G.; Kay S. A. (2010) High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biol. 8, e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Yoo S.-H.; Park Y.-S.; Kim K.-H.; Wei S.; Buhr E.; Ye Z.-Y.; Pan H.-L.; Takahashi J. S. (2012) Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc. Natl. Acad. Sci. U.S.A. 109, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H. R.; Chen W.; Adachi A.; Wakamatsu H.; Hayashi S.; Takasugi T.; Nagano M.; Nakahama K.-I.; Suzuki Y.; Sugano S.; Iino M.; Shigeyoshi Y.; Hashimoto S. (2002) A transcription factor response element for gene expression during circadian night. Nature 418, 534–539. [DOI] [PubMed] [Google Scholar]
- Sato T. K.; Panda S.; Miraglia L. J.; Reyes T. M.; Rudic R. D.; McNamara P.; Naik K. A.; FitzGerald G. A.; Kay S. A.; Hogenesch J. B. (2004) A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43, 527–537. [DOI] [PubMed] [Google Scholar]
- Crumbley C.; Burris T. P. (2011) Direct regulation of CLOCK expression by REV-ERB. PLoS One 6, e17290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A. C.; Tran H. G.; Zhang E. E.; Priest A. A.; Welsh D. K.; Kay S. A. (2008) Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 4, e1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.; Zhao X.; Hatori M.; Yu R. T.; Barish G. D.; Lam M. T.; Chong L.-W.; DiTacchio L.; Atkins A. R.; Glass C. K.; Liddle C.; Auwerx J.; Downes M.; Panda S.; Evans R. M. (2012) Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S.; Sassone-Corsi P. (2010) The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol. 17, 1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valekunja U. K.; Edgar R. S.; Oklejewicz M.; van der Horst G. T. J.; O’Neill J. S.; Tamanini F.; Turner D. J.; Reddy A. B. (2013) Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc. Natl. Acad. Sci. U.S.A. 110, 1554–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y.; Kaluzova M.; Grimaldi B.; Sahar S.; Hirayama J.; Chen D.; Guarente L. P.; Sassone-Corsi P. (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G.; Gatfield D.; Stratmann M.; Reinke H.; Dibner C.; Kreppel F.; Mostoslavsky R.; Alt F. W.; Schibler U. (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328. [DOI] [PubMed] [Google Scholar]
- Etchegaray J.-P.; Yang X.; DeBruyne J. P.; Peters A. H. F. M.; Weaver D. R.; Jenuwein T.; Reppert S. M. (2006) The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem. 281, 21209–21215. [DOI] [PubMed] [Google Scholar]
- Koike N.; Yoo S. H.; Huang H. C.; Kumar V.; Lee C.; Kim T. K.; Takahashi J. S. (2012) Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science 338, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong H. A.; Robles M. S.; Knutti D.; Weitz C. J. (2011) A molecular mechanism for circadian clock negative feedback. Science 332, 1436–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K.; Robles M. S.; Westerling T.; Weitz C. J. (2012) Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 337, 599–602. [DOI] [PubMed] [Google Scholar]
- Duong H. A.; Weitz C. J. (2014) Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat. Struct. Mol. Biol. 21, 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. A.; Ripperger J.; Kadener S.; Fleury-Olela F.; Vilbois F.; Rosbash M.; Schibler U. (2005) PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308, 693–696. [DOI] [PubMed] [Google Scholar]
- Stratmann M.; Stadler F.; Tamanini F.; van der Horst G. T. J.; Ripperger J. A. (2010) Flexible phase adjustment of circadian albumin D site-binding protein (DBP) gene expression by CRYPTOCHROME1. Genes Dev. 24, 1317–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R.; Selby C. P.; Ozturk N.; Annayev Y.; Sancar A. (2011) Biochemical analysis of the canonical model for the mammalian circadian clock. J. Biol. Chem. 286, 25891–25902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A. C.; Welsh D. K.; Ko C. H.; Tran H. G.; Zhang E. E.; Priest A. A.; Buhr E. D.; Singer O.; Meeker K.; Verma I. M.; Doyle F. J.; Takahashi J. S.; Kay S. A. (2007) Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukai-Tadenuma M.; Yamada R. G.; Xu H.; Ripperger J. A.; Liu A. C.; Ueda H. R. (2011) Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144, 268–281. [DOI] [PubMed] [Google Scholar]
- Khan S. K.; Xu H.; Ukai-Tadenuma M.; Burton B.; Wang Y.; Ueda H. R.; Liu A. C. (2012) Identification of a novel cryptochrome differentiating domain required for feedback repression in circadian clock function. J. Biol. Chem. 287, 25917–25926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T.; Lee J. W.; St John P. C.; Sawa M.; Iwaisako K.; Noguchi T.; Pongsawakul P. Y.; Sonntag T.; Welsh D. K.; Brenner D. A.; Doyle F. J.; Schultz P. G.; Kay S. A. (2012) Identification of small molecule activators of cryptochrome. Science 337, 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood E. S.; Chesham J. E.; Meng Q.-J.; Nolan P. M.; Loudon A. S. I.; Hastings M. H. (2011) Tuning the period of the mammalian circadian clock: Additive and independent effects of CK1εTau and Fbxl3Afh mutations on mouse circadian behavior and molecular pacemaking. J. Neurosci. 31, 1539–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John P. C.; Hirota T.; Kay S. A.; Doyle F. J. (2014) Spatiotemporal separation of PER and CRY posttranslational regulation in the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 111, 2040–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojetin D. J.; Burris T. P. (2014) REV-ERB and ROR nuclear receptors as drug targets. Nat. Rev. Drug Discovery 13, 197–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D. P.; Zhao Y.; Sangoram A. M.; Wilsbacher L. D.; Tanaka M.; Antoch M. P.; Steeves T. D.; Vitaterna M. H.; Kornhauser J. M.; Lowrey P. L.; Turek F. W.; Takahashi J. S. (1997) Positional cloning of the mouse circadian clock gene. Cell 89, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara Y. B.; Tagao S.; Tamanini F.; Morita A.; Sugisawa Y.; Yasuda M.; Yamanaka I.; Ueda H. R.; van der Horst G. T. J.; Kondo T.; Yagita K. (2006) The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc. Natl. Acad. Sci. U.S.A. 103, 10074–10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T. K.; Yamada R. G.; Ukai H.; Baggs J. E.; Miraglia L. J.; Kobayashi T. J.; Welsh D. K.; Kay S. A.; Ueda H. R.; Hogenesch J. B. (2006) Feedback repression is required for mammalian circadian clock function. Nat. Genet. 38, 312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei H.; Okamura H.; Shigeyoshi Y.; Fukuhara C.; Ozawa R.; Hirose M.; Sakaki Y. (1997) Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 389, 512–516. [DOI] [PubMed] [Google Scholar]
- Yagita K.; Yamaguchi S.; Tamanini F.; van der Horst G. T.; Hoeijmakers J. H.; Yasui A.; Loros J. J.; Dunlap J. C.; Okamura H. (2000) Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 14, 1353–1363. [PMC free article] [PubMed] [Google Scholar]
- Lee C.; Weaver D. R.; Reppert S. M. (2004) Direct association between mouse PERIOD and CKIε is critical for a functioning circadian clock. Mol. Cell. Biol. 24, 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K.; Mesaki M.; Ishida N. (2001) Nuclear entry mechanism of rat PER2 (rPER2): Role of rPER2 in nuclear localization of CRY protein. Mol. Cell. Biol. 21, 6651–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A. (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103, 2203–2237. [DOI] [PubMed] [Google Scholar]
- Chaves I.; Yagita K.; Barnhoorn S.; Okamura H.; van der Horst G. T. J.; Tamanini F. (2006) Functional evolution of the photolyase/cryptochrome protein family: Importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol. Cell. Biol. 26, 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W.; Busino L.; Hinds T. R.; Marionni S. T.; Saifee N. H.; Bush M. F.; Pagano M.; Zheng N. (2013) SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature 496, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. J.; Edery I.; Rosbash M. (1993) PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature 364, 259–262. [DOI] [PubMed] [Google Scholar]
- Möglich A.; Ayers R. A.; Moffat K. (2009) Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield C. J.; Dunker A. K. (2014) Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 83, 553–584. [DOI] [PubMed] [Google Scholar]
- Cardone L.; Hirayama J.; Giordano F.; Tamaru T.; Palvimo J. J.; Sassone-Corsi P. (2005) Circadian clock control by SUMOylation of BMAL1. Science 309, 1390–1394. [DOI] [PubMed] [Google Scholar]
- Harper S. M.; Neil L. C.; Gardner K. H. (2003) Structural basis of a phototropin light switch. Science 301, 1541–1544. [DOI] [PubMed] [Google Scholar]
- Hennig S.; Strauss H. M.; Vanselow K.; Yildiz O.; Schulze S.; Arens J.; Kramer A.; Wolf E. (2009) Structural and functional analyses of PAS domain interactions of the clock proteins Drosophila PERIOD and mouse PERIOD2. PLoS Biol. 7, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel P. J. A.; Card P. B.; Karakuzu O.; Bruick R. K.; Gardner K. H. (2003) Structural basis for PAS domain heterodimerization in the basic helix–loop–helix-PAS transcription factor hypoxia-inducible factor. Proc. Natl. Acad. Sci. U.S.A. 100, 15504–15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D.; Potluri N.; Kim Y.; Rastinejad F. (2013) Structure and dimerization properties of the aryl hydrocarbon receptor PAS-A domain. Mol. Cell. Biol. 33, 4346–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razeto A.; Ramakrishnan V.; Litterst C. M.; Giller K.; Griesinger C.; Carlomagno T.; Lakomek N.; Heimburg T.; Lodrini M.; Pfitzner E.; Becker S. (2004) Structure of the NCoA-1/SRC-1 PAS-B domain bound to the LXXLL motif of the STAT6 transactivation domain. J. Mol. Biol. 336, 319–329. [DOI] [PubMed] [Google Scholar]
- Partch C. L.; Gardner K. H. (2011) Coactivators necessary for transcriptional output of the hypoxia inducible factor, HIF, are directly recruited by ARNT PAS-B. Proc. Natl. Acad. Sci. U.S.A. 108, 7739–7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.; Schirmer A.; Lee Y.; Lee H.; Kumar V.; Yoo S.-H.; Takahashi J. S.; Lee C. (2009) Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol. Cell 36, 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioum E. M.; Rutter J.; Tuckerman J. R.; Gonzalez G.; Gilles-Gonzalez M.-A.; McKnight S. L. (2002) NPAS2: A gas-responsive transcription factor. Science 298, 2385–2387. [DOI] [PubMed] [Google Scholar]
- Kaasik K.; Lee C. C. (2004) Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430, 467–471. [DOI] [PubMed] [Google Scholar]
- Lukat-Rodgers G. S.; Correia C.; Botuyan M. V.; Mer G.; Rodgers K. R. (2010) Heme-Based Sensing by the Mammalian Circadian Protein CLOCK. Inorg. Chem. 49, 6349–6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airola M. V.; Du J.; Dawson J. H.; Crane B. R. (2010) Heme binding to the mammalian circadian clock protein period 2 is nonspecific. Biochemistry 49, 4327–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amezcua C. A.; Harper S. M.; Rutter J.; Gardner K. H. (2002) Structure and interactions of PAS kinase N-terminal PAS domain: Model for intramolecular kinase regulation. Structure 10, 1349–1361. [DOI] [PubMed] [Google Scholar]
- Scheuermann T. H.; Tomchick D. R.; Machius M.; Guo Y.; Bruick R. K.; Gardner K. H. (2009) Artificial ligand binding within the HIF2α PAS-B domain of the HIF2 transcription factor. Proc. Natl. Acad. Sci. U.S.A. 106, 450–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann T. H.; Li Q.; Ma H.-W.; Key J.; Zhang L.; Chen R.; Garcia J. A.; Naidoo J.; Longgood J.; Frantz D. E.; Tambar U. K.; Gardner K. H.; Bruick R. K. (2013) Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat. Chem. Biol. 9, 271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.; Partch C. L.; Key J.; Card P. B.; Pashkov V.; Patel A.; Bruick R. K.; Wurdak H.; Gardner K. H. (2013) Regulating the ARNT/TACC3 axis: Multiple approaches to manipulating protein/protein interactions with small molecules. ACS Chem. Biol. 8, 626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key J.; Scheuermann T. H.; Anderson P. C.; Daggett V.; Gardner K. H. (2009) Principles of ligand binding within a completely buried cavity in HIF2α PAS-B. J. Am. Chem. Soc. 131, 17647–17654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.; Etchegaray J. P.; Cagampang F. R.; Loudon A. S.; Reppert S. M. (2001) Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867. [DOI] [PubMed] [Google Scholar]
- Eide E. J.; Woolf M. F.; Kang H.; Woolf P.; Hurst W.; Camacho F.; Vielhaber E. L.; Giovanni A.; Virshup D. M. (2005) Control of mammalian circadian rhythm by CKIε-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 25, 2795–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q.-J.; Maywood E. S.; Bechtold D. A.; Lu W.-Q.; Li J.; Gibbs J. E.; Dupré S. M.; Chesham J. E.; Rajamohan F.; Knafels J.; Sneed B.; Zawadzke L. E.; Ohren J. F.; Walton K. M.; Wager T. T.; Hastings M. H.; Loudon A. S. I. (2010) Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc. Natl. Acad. Sci. U.S.A. 107, 15240–15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U.; Ripperger J.; Brown S. A. (2003) Peripheral circadian oscillators in mammals: Time and food. J. Biol. Rhythms 18, 250–260. [DOI] [PubMed] [Google Scholar]
- Lee Y.; Chen R.; Lee H.-M.; Lee C. (2011) Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J. Biol. Chem. 286, 7033–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera N.; Schmalen I.; Hennig S.; Öllinger R.; Strauss H. M.; Grudziecki A.; Wieczorek C.; Kramer A.; Wolf E. (2012) Unwinding the differences of the mammalian PERIOD clock proteins from crystal structure to cellular function. Proc. Natl. Acad. Sci. U.S.A. 109, 3311–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B.; Larkin D. W.; Albrecht U.; Sun Z. S.; Sage M.; Eichele G.; Lee C. C.; Bradley A. (1999) The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400, 169–173. [DOI] [PubMed] [Google Scholar]
- Vielhaber E. L.; Duricka D.; Ullman K. S.; Virshup D. M. (2001) Nuclear export of mammalian PERIOD proteins. J. Biol. Chem. 276, 45921–45927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz I.; Ripperger J. A.; Baeriswyl-Aebischer S.; Albrecht U. (2010) The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 24, 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery D. M.; Kalkhoven E.; Hoare S.; Parker M. G. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387, 733–736. [DOI] [PubMed] [Google Scholar]
- Plevin M. J.; Mills M. M.; Ikura M. (2005) The LxxLL motif: A multifunctional binding sequence in transcriptional regulation. Trends Biochem. Sci. 30, 66–69. [DOI] [PubMed] [Google Scholar]
- Akashi M.; Okamoto A.; Tsuchiya Y.; Todo T.; Nishida E.; Node K. (2014) A Positive Role for PERIOD in Mammalian Circadian Gene Expression. Cell Rep. 7, 1056–1064. [DOI] [PubMed] [Google Scholar]
- Hurley J. M.; Larrondo L. F.; Loros J. J.; Dunlap J. C. (2013) Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered neurospora clock protein FRQ. Mol. Cell 52, 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxreiter M.; Tompa P.; Simon I.; Uversky V. N.; Hansen J. C.; Asturias F. J. (2008) Malleable machines take shape in eukaryotic transcriptional regulation. Nat. Chem. Biol. 4, 728–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M. S.; Uversky V. N.; Dunker A. K. (2008) Intrinsic disorder in scaffold proteins: Getting more from less. Prog. Biophys. Mol. Biol. 98, 85–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H. J.; Komives E. A. (2012) Role of disorder in IκB-NFκB interaction. IUBMB Life 64, 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. P.; Obradovic Z.; Dunker A. K. (2004) Natively Disordered Proteins. Appl. Bioinf. 3, 105–113. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M.; Simon I.; Friedrich P.; Tompa P. (2004) Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 338, 1015–1026. [DOI] [PubMed] [Google Scholar]
- Tennessen J. M.; Gardner H. F.; Volk M. L.; Rougvie A. E. (2006) Novel heterochronic functions of the Caenorhabditis elegans period-related protein LIN-42. Dev. Biol. 289, 30–43. [DOI] [PubMed] [Google Scholar]
- Joen M.; Gardner H. F.; Miller E. A.; Deshler J.; Rougvie A. E. (1999) Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286, 1141–1146. [DOI] [PubMed] [Google Scholar]
- Monsalve G. C.; Van Buskirk C.; Frand A. R. (2011) LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr. Biol. 21, 2033–2045. [DOI] [PubMed] [Google Scholar]
- Nawathean P.; Stoleru D.; Rosbash M. (2007) A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol. Cell. Biol. 27, 5002–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K.; Jin X.; Maywood E. S.; Hastings M. H.; Reppert S. M.; Weaver D. R. (2001) Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30, 525–536. [DOI] [PubMed] [Google Scholar]
- Shearman L. P.; Sriram S.; Weaver D. R.; Maywood E. S.; Chaves I.; Zheng B.; Kume K.; Lee C. C.; van der Horst G. T.; Hastings M. H.; Reppert S. M. (2000) Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019. [DOI] [PubMed] [Google Scholar]
- Pendergast J. S.; Niswender K. D.; Yamazaki S. (2012) Tissue-specific function of Period3 in circadian rhythmicity. PLoS One 7, e30254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E. A.; Staknis D.; Weitz C. J. (1999) Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286, 768–771. [DOI] [PubMed] [Google Scholar]
- Kume K.; Zylka M. J.; Sriram S.; Shearman L. P.; Weaver D. R.; Jin X.; Maywood E. S.; Hastings M. H.; Reppert S. M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205. [DOI] [PubMed] [Google Scholar]
- Oster H.; Yasui A.; van der Horst G. T. J.; Albrecht U. (2002) Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes Dev. 16, 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. K.; Forger D. B. (2012) A mechanism for robust circadian timekeeping via stoichiometric balance. Mol. Syst. Biol. 8, 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S.; Sancar A. (2003) Purification and properties of human blue-light photoreceptor cryptochrome 2. Biochemistry 42, 2926–2932. [DOI] [PubMed] [Google Scholar]
- Partch C. L.; Clarkson M. W.; Ozgur S.; Lee A. L.; Sancar A. (2005) Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry 44, 3795–3805. [DOI] [PubMed] [Google Scholar]
- Kurabayashi N.; Hirota T.; Sakai M.; Sanada K.; Fukada Y. (2010) DYRK1A and glycogen synthase kinase 3β, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol. Cell. Biol. 30, 1757–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P.; Yoo S. H.; Lee K. J.; Rosensweig C.; Takahashi J. S.; Chen B. P.; Green C. B. (2013) Phosphorylation of the Cryptochrome 1 C-terminal Tail Regulates Circadian Period Length. J. Biol. Chem. 288, 35277–35286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.; Conte F.; Green C. B. (2003) Nuclear localization and transcriptional repression are confined to separable domains in the circadian protein CRYPTOCHROME. Curr. Biol. 13, 1653–1658. [DOI] [PubMed] [Google Scholar]
- Czarna A.; Berndt A.; Singh H. R.; Grudziecki A.; Ladurner A. G.; Timinszky G.; Kramer A.; Wolf E. (2013) Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell 153, 1394–1405. [DOI] [PubMed] [Google Scholar]
- Czarna A.; Breitkreuz H.; Mahrenholz C. C.; Arens J.; Strauss H. M.; Wolf E. (2011) Quantitative Analyses of Cryptochrome-mBMAL1 Interactions: Mechanistic Insights into the Transcriptional Regulation of the Mammalian Circadian Clock. J. Biol. Chem. 286, 22414–22425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalen I.; Reischl S.; Wallach T.; Klemz R.; Grudziecki A.; Prabu J. R.; Benda C.; Kramer A.; Wolf E. (2014) Interaction of Circadian Clock Proteins CRY1 and PER2 Is Modulated by Zinc Binding and Disulfide Bond Formation. Cell 157, 1203–1215. [DOI] [PubMed] [Google Scholar]
- Yagita K.; Tamanini F.; Yasuda M.; Hoeijmakers J. H. J.; van der Horst G. T. J.; Okamura H. (2002) Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 21, 1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangle S.; Xing W.; Zheng N. (2013) Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res. 23, 1417–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter A. L.; Suppa C.; Emanuel B. S. (2007) Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J. Mol. Biol. 366, 36–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter A. L. (2006) A Timeless debate: Resolving TIM’s noncircadian roles with possible clock function. NeuroReport 17, 1229–1233. [DOI] [PubMed] [Google Scholar]
- Engelen E.; Janssens R. C.; Yagita K.; Smits V. A. J.; van der Horst G. T. J.; Tamanini F. (2013) Mammalian TIMELESS is involved in period determination and DNA damage-dependent phase advancing of the circadian clock. PLoS One 8, e56623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsal-Kaçmaz K.; Mullen T. E.; Kaufmann W. K.; Sancar A. (2005) Coupling of human circadian and cell cycles by the timeless protein. Mol. Cell. Biol. 25, 3109–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou D. M.; Elledge S. J. (2006) Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc. Natl. Acad. Sci. U.S.A. 103, 18143–18147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman A. R.; Noguchi C.; Lee C. Y.; Noguchi E. (2010) Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J. Cell Sci. 123, 660–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman A. R.; Noguchi E. (2012) Local and global functions of Timeless and Tipin in replication fork protection. Cell Cycle 11, 3945–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oklejewicz M.; Destici E.; Tamanini F.; Hut R. A.; Janssens R.; van der Horst G. T. J. (2008) Phase resetting of the mammalian circadian clock by DNA damage. Curr. Biol. 18, 286–291. [DOI] [PubMed] [Google Scholar]
- Gamsby J. J.; Loros J. J.; Dunlap J. C. (2009) A phylogenetically conserved DNA damage response resets the circadian clock. J. Biol. Rhythms 24, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T.-H.; Leem S.-H. (2014) Modulation of ATR-mediated DNA damage checkpoint response by cryptochrome 1. Nucleic Acids Res. 42, 4427–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia K. A.; Papp S. J.; Yu R. T.; Barish G. D.; Uhlenhaut N. H.; Jonker J. W.; Downes M.; Evans R. M. (2011) Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozber N.; Baris I.; Tatlici G.; Gur I.; Kilinc S.; Unal E. B.; Kavakli I. H. (2010) Identification of two amino acids in the C-terminal domain of mouse CRY2 essential for PER2 interaction. BMC Mol. Biol. 11, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M.; Hirayama J.; Sassone-Corsi P. (2006) Circadian Regulator CLOCK Is a Histone Acetyltransferase. Cell 125, 497–508. [DOI] [PubMed] [Google Scholar]
- Hirayama J.; Sahar S.; Grimaldi B.; Tamaru T. (2007) CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450, 1086–1090. [DOI] [PubMed] [Google Scholar]
- Michael A. K.; Partch C. L. (2014) bHLH-PAS proteins: Functional specification through modular domain architecture. OA Biochemistry 1, 1–6. [Google Scholar]
- Lee Y.; Lee J.; Kwon I.; Nakajima Y.; Ohmiya Y.; Son G. H.; Lee K. H.; Kim K. (2010) Coactivation of the CLOCK-BMAL1 complex by CBP mediates resetting of the circadian clock. J. Cell Sci. 123, 3547–3557. [DOI] [PubMed] [Google Scholar]
- Takahata S.; Ozaki T.; Mimura J.; Kikuchi Y.; Sogawa K.; Fujii Kuriyama Y. (2000) Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3. Genes Cells 5, 739–747. [DOI] [PubMed] [Google Scholar]
- Hosoda H.; Kato K.; Asano H.; Ito M.; Kato H.; Iwamoto T.; Suzuki A.; Masushige S.; Kida S. (2009) CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol. Brain 2, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray J.-P.; Lee C.; Wade P. A.; Reppert S. M. (2003) Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421, 177–182. [DOI] [PubMed] [Google Scholar]
- Lande-Diner L.; Boyault C.; Kim J. Y.; Weitz C. J. (2013) A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc. Natl. Acad. Sci. U.S.A. 110, 16021–16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Wu Y.; Li L.; Su X. D. (2013) Intermolecular recognition revealed by the complex structure of human CLOCK-BMAL1 basic helix-loop-helix domains with E-box DNA. Cell Res. 23, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru T.; Hirayama J.; Isojima Y.; Nagai K. (2009) CK2α phosphorylates BMAL1 to regulate the mammalian clock. Nat. Struct. Mol. Biol. 16, 446–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J.; Reick M.; Wu L. C.; McKnight S. L. (2001) Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293, 510–514. [DOI] [PubMed] [Google Scholar]
- Yoshii K.; Ishijima S.; Sagami I. (2013) Effects of NAD(P)H and its derivatives on the DNA-binding activity of NPAS2, a mammalian circadian transcription factor. Biochem. Biophys. Res. Commun. 437, 386–391. [DOI] [PubMed] [Google Scholar]
- Wise D. D.; Shear J. B. (2004) Circadian tracking of nicotinamide cofactor levels in an immortalized suprachiasmatic nucleus cell line. Neuroscience 128, 263–268. [DOI] [PubMed] [Google Scholar]
- Tan Y.; DeBruyne J.; Cahill G. M.; Wells D. E. (2008) Identification of a mutation in the Clock1 gene affecting zebrafish circadian rhythms. J. Neurogenet. 22, 149–166. [DOI] [PubMed] [Google Scholar]
- Partch C. L.; Card P. B.; Amezcua C. A.; Gardner K. H. (2009) Molecular basis of coiled coil coactivator recruitment by the aryl hydrocarbon receptor nuclear translocator (ARNT). J. Biol. Chem. 284, 15184–15192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrecque M. P.; Prefontaine G. G.; Beischlag T. V. (2013) The aryl hydrocarbon receptor nuclear translocator (ARNT) family of proteins: Transcriptional modifiers with multi-functional protein interfaces. Curr. Mol. Med. 13, 1047–1065. [DOI] [PubMed] [Google Scholar]
- Zhao W.-N.; Malinin N.; Yang F.-C.; Staknis D.; Gekakis N.; Maier B.; Reischl S.; Kramer A.; Weitz C. J. (2007) CIPC is a mammalian circadian clock protein without invertebrate homologues. Nat. Cell Biol. 9, 268–275. [DOI] [PubMed] [Google Scholar]
- Annayev Y.; Adar S.; Chiou Y. Y.; Lieb J. D.; Sancar A.; Ye R. (2014) Gene Model 129 (Gm129) Encodes a Novel Transcriptional Repressor That Modulates Circadian Gene Expression. J. Biol. Chem. 289, 5013–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anafi R. C.; Lee Y.; Sato T. K.; Venkataraman A.; Ramanathan C.; Kavakli I. H.; Hughes M. E.; Baggs J. E.; Growe J.; Liu A. C.; Kim J.; Hogenesch J. B. (2014) Machine learning helps identify CHRONO as a circadian clock component. PLoS Biol. 12, e1001840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H.; Rossetto D.; Mellert H.; Dang W.; Srinivasan M.; Johnson J.; Hodawadekar S.; Ding E. C.; Speicher K.; Abshiru N.; Perry R.; Wu J.; Yang C.; Zheng Y. G.; Speicher D. W.; Thibault P.; Verreault A.; Johnson F. B.; Berger S. L.; Sternglanz R.; McMahon S. B.; Côté J.; Marmorstein R. (2012) MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 31, 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D.; Rivera M.; Zor T.; Henrion-Caude A.; Radhakrishnan I.; Kumar A.; Shapiro L. H.; Wright P. E.; Montminy M.; Brindle P. K. (1999) Role of secondary structure in discrimination between constitutive and inducible activators. Mol. Cell. Biol. 19, 5601–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. S.; Green E. W.; Zhao Y.; van Ooijen G.; Olmedo M.; Qin X.; Xu Y.; Pan M.; Valekunja U. K.; Feeney K. A.; Maywood E. S.; Hastings M. H.; Baliga N. S.; Merrow M.; Millar A. J.; Johnson C. H.; Kyriacou C. P.; O’Neill J. S.; Reddy A. B. (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J. S.; Reddy A. B. (2011) Circadian clocks in human red blood cells. Nature 469, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]