Abstract
Circadian clocks are self-sustaining timekeepers found in almost all organisms on earth. The filamentous fungus Neurospora crassa is a preeminent model for eukaryotic circadian clocks. Investigations of the Neurospora circadian clock system have led to elucidation of circadian clock regulatory mechanisms that are common to all eukaryotes. In this work, we will focus on the Neurospora circadian oscillator mechanism with an emphasis on the regulation of the core clock component FREQUENCY.
Eukaryotic circadian oscillators are comprised of interlocked autoregulatory feedback loops that control gene expression at the levels of transcription and translation.1,2 Despite the evolutionary distance, the molecular mechanism of the circadian oscillator of the filamentous fungus Neurospora crassa is remarkably similar to those of higher eukaryotes.3−5 The asexual spore formation of Neurospora is governed by the circadian clock; therefore, Neurospora circadian rhythms are readily monitored.6 Recently, a bioluminescence reporter has been established to monitor circadian behavior of various promoters.7 In addition, the availability of a whole genome knockout collection of Neurospora has made this fungus as an outstanding model organism for research on the molecular architecture of eukaryotic circadian clocks.8
FREQUENCY (FRQ) is a central component of the Neurospora circadian oscillator like PERIOD (PER) in animals. FRQ forms FFC (FRQ-FRH complex) with its partner FRQ-interacting RNA helicase (FRH) and functions as the negative limb in the core circadian negative feedback loop.9,10 The transcription of the frq gene is activated by the positive element, WHITE COLLAR complex (WCC), which is formed by the interaction of two PER-ARNT-SIM (PAS) domain-containing transcription factors WC-1 and WC-2, like CLOCK and BMAL1 in animals. WCC binds rhythmically to the promoter of frq to induce transcription.11−15 To close the negative feedback loop, FFC inhibits the activity of WCC. After FRQ is synthesized, it is subjected to progressive post-translational modifications by phosphorylations that lead to its degradation by the ubiquitin/proteasome pathway.16−22 When the amount of FRQ drops below a certain level, WCC is reactivated and a new cycle begins. As a result of this negative feedback loop, frq mRNA and FRQ protein exhibit robust rhythms with a period close to 24 h. The core negative feedback loop is interlocked with positive feedback loops to maintain robust rhythmicity.23,24 This review will focus on the recent progress in understanding the molecular network governing the expression and activity of FRQ.
Molecular Architecture of the Neurospora Circadian Clock
Identification of clock components has been central to our understanding of the clock mechanisms. Since the cloning of the frq gene,25 more than 20 Neurospora clock components that regulate FRQ expression, function, and stability at various levels have been identified (Table 1). These components form the network that is important for the control of the clock (Figure 1).
Table 1. Components of the Neurospora Circadian Clock.
| protein (complex) | functions |
|---|---|
| FRQ (component of core loop) | forms a complex with FRH, recruits CKs to phosphorylate itself and WCC |
| FRH (component of core loop) | interacts with FRQ to form FFC, which stabilizes FRQ protein and recruits exosome to regulate frq mRNA stability |
| WC-1 (component of core loop) | forms WCC with WC-2; WCC activates frq and clock-controlled genes |
| WC-2 (component of core loop) | forms WCC with WC-1; WCC activates frq and clock-controlled genes |
| CKI (CK-1a) | phosphorylates FRQ and WCC |
| CKII | phosphorylates FRQ and WCC |
| PKA | phosphorylates FRQ and WCC |
| CAMK-1 | phosphorylates FRQ |
| CHK2 | phosphorylates FRQ |
| PP1 | dephosphorylates (to stabilize) FRQ |
| PP2A | dephosphorylates WCC |
| PP4 | dephosphorylates (to stabilize) FRQ, dephosphorylates WCC |
| FWD-1 | part of SCFFWD-1, the E3 ligase responsible for FRQ ubiquitination |
| COP-9 signalosome | regulates the stability of SCFFWD-1 |
| CSW-1 | tegulates chromatin structure, changes DNase sensitivity of the frq promoter |
| CHD1 | enhances DNA methylation on frq locus and regulates frq transcription |
| CATP | regulates histone occupancy of the frq locus and enhances WCC binding |
| Not1–Ccr4 complex | stabilizes WCC |
| RCO-1 | part of a transcription repressor complex that suppresses WC-independent transcription of frq |
| SET1 | histone H3K4 methyltransferase; regulates frq transcription |
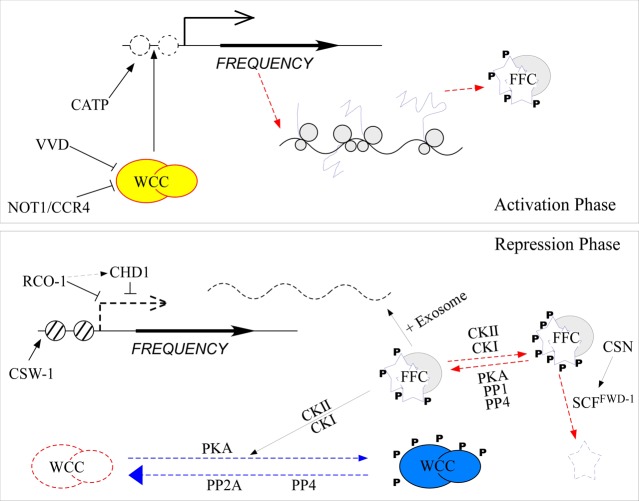
Figure 1.
Model describing the core circadian negative feedback loop of Neurospora. In the activation phase, CATP reduces the histone occupancy on the frq promoter, which promotes WCC binding and activation of frq transcription. After transcription of frq mRNA, the nonoptimal codons in frq mRNA regulate translation speed to allow proper cotranslational folding of FRQ. The resulting FRQ protein is stabilized by its interaction with FRH to form the FFC complex. In the repression phase, FFC recruits casein kinases to phosphorylate PKA-primed WCs and promotes degradation of frq mRNA by the exosome. Phosphorylation of WCC inhibits its DNA binding activity and sequesters it in the cytoplasm. FRQ is progressively phosphorylated by casein kinases and degraded by the ubiquitin/proteasome system, a process that is counterbalanced by the actions of PKA, PP1, and PP4. CSW-1 relocates nucleosomes to suppress frq activation, whereas RCO-1 and CHD1 suppress WC-independent frq transcription to permit WCC-regulated frq transcription. Dephosphorylation of WCC by PP2A and PP4 reactivates WCC to allow reactivation of frq transcription.
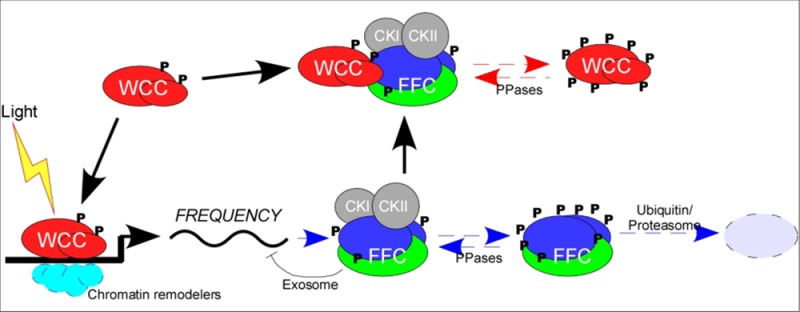
During the early subjective day in the constant darkness, WCC activates frq transcription by binding to the clock box (c-box) in the frq promoter; this binding is essential for the circadian transcription of frq as well as other clock-controlled genes.11,14,23 Transcriptional activation is followed by an increase in levels of FRQ protein, which forms a homodimer that interacts with FRH to form FFC.26−28 The FRQ–FRH interaction is important for maintaining the steady-state levels and proper structure of FRQ.27−31 In the absence of FRH, FRQ is unstable. FFC interacts with WCC and promotes phosphorylation of WCs. This phosphorylation inhibits WCC activity, resulting in a decrease in the level of frq transcription.21,32−34 In addition, there is WC-independent transcription of frq that is suppressed by a mechanism that involves the RCO-1 repressor complex.35 In addition, FFC recruits the RNA exosome complex to the frq mRNA to regulate its half-life.29
After its synthesis, FRQ is progressively phosphorylated by casein kinases (CKI and CKII) and other kinases through the subjective day and evening, leading to its degradation by the ubiquitin/proteasome pathway.17−21,36−39 FWD-1 is the E3 ubiquitin ligase responsible for FRQ ubiquitination, and the COP9 signalosome regulates FRQ stability by modulating the activity and stability of the SCFFWD-1 complex that is critical for FRQ degradation. In contrast to the role of casein kinases in promoting the degradation of FRQ, protein kinase A (PKA) phosphorylates and stabilizes FRQ. The roles of these kinases are countered by multiple protein phosphatases, including PP1, PP2A, and PP4.32,40,41 Thus, the phosphorylation of FRQ is fine-tuned by a series of regulators to determine its stability and the period length of the clock.22,42 Degradation of FRQ to a certain level releases the inhibition of FRQ on WCC and results in its dephosphorylation and reactivation, a process that involves PP2A and PP4.32,41,43 The reactivated WCC is transported to the nucleus, binds to the frq c-box with the help of chromatin modulating factors, and drives the next cycle of frq transcription.15,41,44,45 The resulting rhythms of the abundance of frq mRNA and FRQ protein and the phosphorylation profile of FRQ are the hallmarks of the Neurospora circadian oscillation.
In addition to its role in inhibiting WCC activity, FRQ also positively regulates the expression of both WC-1 and WC-2.23,24,46,47 The detailed mechanisms are still unknown, but FRQ regulates WC-1 expression at the post-transcriptional level, in a manner independent of WC-2. FRQ also enhances wc-2 transcription. These positive feedback loops are interlocked with the core negative feedback loop, a feature that is shared in animal clocks.48,49
Transcriptional Regulation of frq
Post-translational Regulations of the WCC: Stability, Activity, and Localization
Transcriptional regulation of frq is mainly achieved by post-translational regulation of its activator, WCC. Because FRQ represses its own expression, it has been proposed that FRQ can regulate WCC activity by physical interactions alone.26,47,50 However, the concentration of nuclear FFC is much lower than that of the nuclear WCC. Thus, the amount of nuclear FRQ is not sufficient to inhibit nuclear WCC through a direct interaction alone,23,27,33,50−52 suggesting that FFC may behave like an enzyme to inhibit the WCC. Both biochemical purification and genetic analyses identified CKI and CKII as FRQ-interacting kinases that inhibit WCC through phosphorylation.21 FFC acts as a substrate-recruiting subunit of CKs and mediates phosphorylation by a physical interaction with WCC. Phosphorylation of WCC occurs sequentially at multiple residues: first by PKA, which functions in a manner independent of FRQ, and then by the FRQ-recruited CKs.21,34,40 The phosphorylation of WCs can be reversed by phosphatases PP2A and PP4 to counterbalance the actions of kinases.32,41,43
Phosphorylation of clock proteins is also critical to the negative feedback mechanisms in the animal circadian systems. PER-dependent rhythms of CLK phosphorylation and E-box binding were shown in Drosophila, and PER-DBT (doubletime kinase) interaction is required for the transcriptional repression process.53,54 On the other hand, it was proposed that the 1:1 PER–CLK interaction in Drosophila can sequester CLK from DNA to mediate the feedback process,55 but it is not clear whether the interaction alone is sufficient. In mammals, CK2α was shown to phosphorylate BMAL1 in vitro and in vivo to regulate its nuclear accumulation and clock function, and furthermore, hyperphosphorylated CLOCK in the repressed phase displayed less binding to E-box.56,57 These findings suggest the conservation of the negative feedback mechanisms in eukaryotic clocks, although the exact mechanism in each system may differ.
On the basis of the finding that the frq-null strain as well as FRQ kinase mutants shows low levels of WCC, it has been proposed that the phosphorylation of WCC stabilizes the WC proteins.21,23,24,34,40,46 Consistent with this notion, VIVID (VVD), a small photoreceptor protein, can physically interact with the light-activated WCC, inhibiting its activity and degradation.58,59 Therefore, FFC and VVD independently and cooperatively regulate the stability of WCC by distinct mechanisms. In addition, the CCR4–Not complex, conserved among all eukaryotes, interacts with WC-1 and stabilizes it, probably by affecting its phosphorylation status.60 These findings suggest that the phosphorylation of WCC plays a role in the stability of WCs and ensures the timely activation of the complex.
Biochemical analyses also showed that dephosphorylation of WCC enhances its DNA binding activity in vitro. In mutants in which the level of FFC-mediated WCC phosphorylation is reduced, higher WCC occupancy on the frq promoter is observed.21,33,40,61 In contrast, hyperphosphorylated WCC and a reduced level of binding of WCC to the c-box of frq promoter were observed in the phosphatase mutants.32,41,43 Therefore, the PKA-primed and FFC-mediated sequential phosphorylations of WCC can remove the complex from the frq promoter, resulting in the repression of frq transcription and, at the same time, stabilization of WCC.
Furthermore, the FFC-mediated phosphorylation of WCC also sequesters the WCs in the cytoplasm.41,43,62 The ratio of nuclear to cytoplasmic WCC changes in a circadian manner: when WCC is hyperphosphorylated, more WCC is present in the cytoplasm; the nuclear level increases when the complex is hypophosphorylated. In strains with a defective PP2A or disrupted pp4 gene, significantly less WCC is found in accompanying nuclei, and there are decreased levels of WC DNA binding and reduced frq transcription relative to wild-type levels. These studies suggest that there are multiple regulators of WC phosphorylation that control its activity, stability, and subcellular localization.
Chromatin Modulations at the frq Locus
The nucleosome is the basic unit of the eukaryotic chromosome. Nucleosomes are assembled from histone octamers bound to DNA strands. Post-translational modifications of histones, including H3 and H4 acetylation and methylation, are known to be the basis of epigenetic regulation that control transcription.63 Nucleosome depletion or exchange at the promoter can modulate accessibility of DNA to transcription factors. It has been shown that many clock-controlled and core clock genes exhibit rhythmic histone modifications, which oscillate in accordance with RNA polymerase II occupancies at those loci.64−68
In Neurospora, rhythmic nucleosome occupancy and DNase hypersensitive regions are observed on the frq promoter.15,44,45 Two homologues of ATP-dependent chromatin-remodeling factors, CLOCKSWITCH (CSW-1) and chromodomain helicase DNA-binding-1 (CHD1), as well as a homologue of a bromodomain-containing ATPase called Clock ATPase (CAPT) are involved in clock function through regulation of frq transcription. These proteins function by regulating the chromatin status of the frq locus through distinct mechanisms. CSW-1 and CATP affect WCC occupancy on the frq promoter in opposite ways: CSW-1 appears to promote nucleosome compaction because the csw-1-null mutant has a relatively open chromatin structure. In contrast, CATP may be involved in the histone eviction process, and the catp mutants have a histone occupancy on the frq promoter higher than that of wild-type cells. Consistent with this notion, homologues of CATP interact with histones to activate transcription.69−72 Furthermore, rhythmic changes in H3K14 acetylation on the frq promoter are lost in the catp-null strain, suggesting that additional histone modifiers, including acetyltransferases and deacetylases, are also involved in the control of frq transcription.
The helicase CHD1 probably regulates frq transcription by modulating DNA structure at the frq locus. The chd1-null strain exhibits hypermethylation at the frq locus and dampened FRQ oscillation. The molecular mechanism through which DNA methylation regulates frq transcription is still not clear. A histone H3K4 methyltransferase SET1 is also required for normal circadian rhythms and normal WCC binding at the frq promoter.73 Together, these findings emphasize the importance of epigenetic regulations in the function of the Neurospora circadian clock.
WC-Independent Transcription of frq
The rhythmic transcriptional activation by WCC is essential for rhythmic frq expression. Because the levels of frq mRNA are extremely low in wc-1 and wc-2 mutants,11 WCC was thought to be the only transcriptional regulator of frq transcription. Recently, however, transcriptional repressor RCO-1 was shown to be an essential clock component that represses WC-independent frq transcription.35 RCO-1 is a homologue of yeast Tup1p. RCO-1 interacts with Csp1 to regulate the expression of clock-controlled genes implicated in metabolism.74,75 In the rco-1-null strain, frq mRNA and FRQ protein levels are constantly high, and conidiation and molecular rhythms are abolished. More interestingly, in the wc rco-1 double mutants, near wild-type amounts of frq mRNA are observed, indicating that the suppression of WC-independent transcription of frq is essential for rhythmicity. Such an elevation of the level of frq transcription is also observed in the wc-1 chd1 double mutant, suggesting that CHD1 mediates the repression of WC-independent frq transcription by regulating chromatin structure. The conserved RCO-1 complex is also known to participate in clock regulation in plants. In plants, the TOPLESS/TOPLESS RELATED (TPL/TPR) complex cooperates with pseudoresponse regulators, the major players of the plant circadian feedback loops, to repress the transcription of clock genes.76
Role of frq Codon Usage in the Control of FRQ Expression and Function
There are 61 genetic codes for the 20 different translated amino acids, and thus, most amino acids (except for methionine and tryptophan) are coded by two to six synonymous codons. Different genomes have preferences for certain synonymous codons over others; this phenomenon is called codon usage bias. Codon usage bias impacts protein expression in prokaryotes and eukaryotes.7,77−79 Codon optimization is used in research and industrial laboratories to achieve optimal protein expression.80 Recently, codon usage has been proposed to play a role in regulating protein folding and activity;81−83 however, its in vivo relevance has not been demonstrated.
Selection for codons that ensure efficient translation is thought to be the major cause of codon usage bias in different organisms. Highly expressed genes are encoded predominantly by codons that correspond to highly abundant tRNAs. This codon usage bias is thought to allow highly expressed genes to be rapidly translated with high fidelity.84,85 In contrast, the core clock genes such as frq in Neurospora and kaiBC in cyanobacteria have nonoptimal codons in their open reading frames.86,87
We showed recently that N-terminal codon optimization of frq in Neurospora not only results in higher FRQ expression levels but also abolishes circadian clock function.86 Several lines of biochemical evidence suggested that codon optimization impairs FRQ function in both circadian negative and positive feedback loops and alters FRQ structure. The effect of nonoptimal codon usage was partially reversed by growing Neurospora at low temperatures, suggesting that the codon optimization accelerates translation and affects the cotranslational folding of the FRQ protein. Furthermore, codon optimization of the central region of FRQ, containing the CKI-interacting domains, alters the FRQ phosphorylation profile and stabilizes the protein. These results suggest that the nonoptimal codons in frq are evolutionarily selected to ensure that the pace of translation in these regions allows proper folding.
Nonoptimal codon usage of clock components also plays an important role in cyanobacteria. The nonoptimal codon usage in the kaiBC gene in cyanobacteria enhances organismal fitness at cool temperatures.87 This is an example of how natural selection resulted in the use of nonoptimal codons so that the organism can properly grow under different environmental conditions. These in vivo studies in Neurospora and cyanobacteria suggest that codon usage is an important mechanism that can regulate both protein expression and protein structures, previously thought to be controlled mainly by transcriptional and post-translational mechanisms, respectively.
Post-translational Regulation of FRQ
Similar to its animal homologue PER, FRQ is progressively phosphorylated as soon as it is made and becomes extensively phosphorylated before degradation.16 Mass spectrometry (MS) analyses of recombinant peptides phosphorylated in vitro by the casein kinases and of FRQ protein purified from Neurospora led to the identification of more than 100 phosphorylation sites in the 989-amino acid protein.22,42 The comparison of in vitro and in vivo sites confirmed that CKI and CKII are responsible for most of the phosphorylation events. FRQ phosphorylation is known to promote its degradation through the ubiquitin/proteasome pathway mediated by the ubiquitin E3 ligase-containing complex SCFFWD-1.19,37,38 FWD-1 acts as the substrate-recruiting subunit of the E3 ligase that recognizes and binds phosphorylated FRQ through its WD-40 domain. It is known that the mammalian FWD-1 homologue β-TRCP recognizes a DpSGϕXpS (ϕ is any hydrophobic residue) motif in the substrates.88 The substrate-recognizing domain of β-TRCP is conserved in FWD-1, but the target motif is not found in any of the phosphorylated regions of FRQ. This suggests that FWD-1 may not recognize phosphorylated FRQ through a single high-affinity binding site. Instead, it may sense a conformational change of FRQ that results from extensive phosphorylations. The presence of multiple low-affinity binding sites in FRQ may result in high-affinity binding to FWD-1. This model predicts that multiple FRQ phosphorylation events contribute to FRQ stability. Indeed, systematic mutagenesis of identified phosphorylation sites showed that mutation of many phosphorylation sites affects FRQ stability.22,42 In addition, although most FRQ phosphorylation events promote its degradation, phosphorylation of the C-terminal region of FRQ results in its stabilization, suggesting the importance of interdomain interactions to FRQ stability.
Unlike most kinase substrates, FRQ forms a stable complex with CK-1a through two separate motifs on FRQ.18,21,89 Both motifs are required for the FRQ–CK1-a interaction and CK-1a-dependent phosphorylation of FRQ and WCs. Biochemical analyses suggest that these two motifs may interact with each other and mediate the CK-1a interaction and the subsequent phosphorylation may trigger a change in FRQ conformation that facilitates the degradation of FRQ by the ubiquitin/proteasome pathway.89
Although FRQ nuclear localization is essential for its function in the circadian clock, most FRQ is found in the cytoplasm.23,27,33,51 An SV-40-like nuclear localization signal located downstream of the coiled-coil domain is required and sufficient for the nuclear localization of FRQ, suggesting that the FRQ cellular distribution profile is regulated by an active nuclear export process. A study using a FRQ–FRB fusion protein showed that there is rapid nuclear–cytoplasmic shuttling of FRQ, suggesting that the phosphorylation of FRQ inhibits its nuclear import.90 However, we showed in a later analysis that the nuclear to cytoplasmic ratio of the FRQ protein does not significantly change during the circadian cycle despite differences in phosphorylation status.91 In addition, mutations in FRQ kinases, phosphatases, and FWD-1, which all have severe effects on FRQ phosphorylation profiles, do not significantly alter the nuclear to cytoplasmic ratios of FRQ relative to that of the wild-type strain. Furthermore, period-changing mutations of FRQ-phosphorylated residues do not significantly alter the subcellular localization. These results argue against the role of FRQ phosphorylation in the cellular localization of FRQ.
What then is the function of FRQ phosphorylation in addition to its effect on FRQ stability? Biochemical analyses suggest that the ability of FRQ to interact with its partners changes during a circadian cycle.42,91 When FRQ is hypophosphorylated in the subjective early day, it interacts with WCs and CK-1a. When FRQ becomes hyperphosphorylated in the subjective evening, however, its interaction with WC-2 is weakened. High levels of hyperphosphorylated FRQ accumulate in the fwd-1-null strain because of the inability of the phosphorylated FRQ to be ubiquitinated and degraded through the proteasome pathway. This hyperphosphorylated FRQ is not functional in the negative feedback loop: It does not repress the transcription of frq.91 Furthermore, the levels of FRQ–WC and FRQ–CK-1a interaction are dramatically reduced in the fwd-1-null mutant. These results indicate that the phosphorylation of FRQ regulates FRQ activity in the negative feedback loop by affecting its ability to interact with WCs and CK-1a. Thus, the rhythmic FRQ phosphorylation profile alone can lead to rhythmic FRQ activity, so rhythm generation may not be totally dependent on the amount of FRQ.
Conclusion
During the past two decades, Neurospora has been an excellent model organism for studying the molecular mechanisms of eukaryotic circadian clocks. Studies of Neurospora have uncovered mechanisms that are critical for circadian clock functions. These mechanisms operate at the transcriptional, post-transcriptional, cotranslational, and post-translational levels to regulate frq expression, FRQ activity, and stability. These analyses not only established the FRQ-based negative feedback loop as the core of the circadian machinery but also provided an explanation of how circadian period length and clock entrainment are determined at the molecular level.
This work was supported by grants from the National Institutes of Health and the Welch Foundation (I-1560) to Y.L.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Dunlap J. C. (1999) Molecular bases for circadian clocks. Cell 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Young M. W.; Kay S. A. (2001) Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet. 2, 702–715. [DOI] [PubMed] [Google Scholar]
- Heintzen C.; Liu Y. (2007) The Neurospora crassa circadian clock. Adv. Genet. 58, 25–66. [DOI] [PubMed] [Google Scholar]
- Cha J.; Huang G.; Guo J.; Liu Y. (2007) Posttranslational control of the Neurospora circadian clock. Cold Spring Harbor Symp. Quant. Biol. 72, 185–191. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas P. L.; Bell-Pedersen D.; Brody S. (2011) The genetics of circadian rhythms in Neurospora. Adv. Genet. 74, 55–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent M. L.; Briggs W. R.; Woodward D. O. (1966) Circadian nature of a rhythm expressed by an invertaseless strain of Neurospora crassa. Plant Physiol. 41, 1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooch V. D.; Mehra A.; Larrondo L. F.; Fox J.; Touroutoutoudis M.; Loros J. J.; Dunlap J. C. (2008) Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryotic Cell 7, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H. V.; Park G.; Turner G. E.; Ringelberg C.; Crew C. M.; Litvinkova L.; Weiss R. L.; Borkovich K. A.; Dunlap J. C. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U.S.A. 103, 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson B. D.; Johnson K. A.; Loros J. J.; Dunlap J. C. (1994) Negative feedback defining a circadian clock: Autoregulation of the clock gene frequency. Science 263, 1578–1584. [DOI] [PubMed] [Google Scholar]
- Cheng P.; He Q.; Wang L.; Liu Y. (2005) Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 19, 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite S. K.; Dunlap J. C.; Loros J. J. (1997) Neurospora wc-1 and wc-2: Transcription, photoresponses, and the origins of circadian rhythmicity. Science 276, 763–769. [DOI] [PubMed] [Google Scholar]
- Cheng P.; Yang Y.; Gardner K. H.; Liu Y. (2002) PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady-state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol. Cell. Biol. 22, 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P.; Yang Y.; Wang L.; He Q.; Liu Y. (2003) WHITE COLLAR-1, a multifunctional neurospora protein involved in the circadian feedback loops, light sensing, and transcription repression of wc-2. J. Biol. Chem. 278, 3801–3808. [DOI] [PubMed] [Google Scholar]
- Froehlich A. C.; Loros J. J.; Dunlap J. C. (2003) Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc. Natl. Acad. Sci. U.S.A. 100, 5914–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden W. J.; Loros J. J.; Dunlap J. C. (2007) Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol. Cell 25, 587–600. [DOI] [PubMed] [Google Scholar]
- Garceau N. Y.; Liu Y.; Loros J. J.; Dunlap J. C. (1997) Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89, 469–476. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Loros J.; Dunlap J. C. (2000) Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 97, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorl M.; Merrow M.; Huttner B.; Johnson J.; Roenneberg T.; Brunner M. (2001) A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J. 20, 7074–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q.; Cheng P.; Yang Y.; He Q.; Yu H.; Liu Y. (2003) FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. EMBO J. 22, 4421–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Cheng P.; He Q.; Wang L.; Liu Y. (2003) Phosphorylation of FREQUENCY protein by casein kinase II is necessary for the function of the Neurospora circadian clock. Mol. Cell. Biol. 23, 6221–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q.; Cha J.; Lee H. C.; Yang Y.; Liu Y. (2006) CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 20, 2552–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C. T.; Li S.; Long C.; Cha J.; Huang G.; Li L.; Chen S.; Liu Y. (2009) Setting the pace of the Neurospora circadian clock by multiple independent FRQ phosphorylation events. Proc. Natl. Acad. Sci. U.S.A. 106, 10722–10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P.; Yang Y.; Liu Y. (2001) Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc. Natl. Acad. Sci. U.S.A. 98, 7408–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeier T.; Kaldi K.; Diernfellner A.; Mohr C.; Brunner M. (2006) Phosphorylation-dependent maturation of Neurospora circadian clock protein from a nuclear repressor toward a cytoplasmic activator. Genes Dev. 20, 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C. R.; Fox B. A.; Dunlap J. C. (1989) The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period. Nature 339, 558–562. [DOI] [PubMed] [Google Scholar]
- Cheng P.; Yang Y.; Heintzen C.; Liu Y. (2001) Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J. 20, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P.; He Q.; He Q.; Wang L.; Liu Y. (2005) Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 19, 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.; Cheng P.; Liu Y. (2010) Functional significance of FRH in regulating the phosphorylation and stability of Neurospora circadian clock protein FRQ. J. Biol. Chem. 285, 11508–11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.; Cheng P.; Yuan H.; Liu Y. (2009) The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell 138, 1236–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M.; Collett M.; Loros J. J.; Dunlap J. C. (2010) FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock. Genetics 184, 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. M.; Larrondo L. F.; Loros J. J.; Dunlap J. C. (2013) Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered Neurospora clock protein FRQ. Mol. Cell 52, 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; He Q.; Cheng P.; Wrage P.; Yarden O.; Liu Y. (2004) Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes Dev. 18, 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeier T.; Haase A.; Kaldi K.; Scholz J.; Fuchs M.; Brunner M. (2005) Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell 122, 235–246. [DOI] [PubMed] [Google Scholar]
- He Q.; Shu H.; Cheng P.; Chen S.; Wang L.; Liu Y. (2005) Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. J. Biol. Chem. 280, 17526–17532. [DOI] [PubMed] [Google Scholar]
- Zhou Z.; Liu X.; Hu Q.; Zhang N.; Sun G.; Cha J.; Wang Y.; Liu Y.; He Q. (2013) Suppression of WC-independent frequency transcription by RCO-1 is essential for Neurospora circadian clock. Proc. Natl. Acad. Sci. U.S.A. 110, E4867–E4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Cheng P.; Zhi G.; Liu Y. (2001) Identification of a calcium/calmodulin-dependent protein kinase that phosphorylates the Neurospora circadian clock protein FREQUENCY. J. Biol. Chem. 276, 41064–41072. [DOI] [PubMed] [Google Scholar]
- He Q.; Cheng P.; He Q.; Liu Y. (2005) The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 19, 1518–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q.; Liu Y. (2005) Degradation of the Neurospora circadian clock protein FREQUENCY through the ubiquitin-proteasome pathway. Biochem. Soc. Trans. 33, 953–956. [DOI] [PubMed] [Google Scholar]
- Pregueiro A. M.; Liu Q.; Baker C. L.; Dunlap J. C.; Loros J. J. (2006) The Neurospora checkpoint kinase 2: A regulatory link between the circadian and cell cycles. Science 313, 644–649. [DOI] [PubMed] [Google Scholar]
- Huang G.; Chen S.; Li S.; Cha J.; Long C.; Li L.; He Q.; Liu Y. (2007) Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev. 21, 3283–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J.; Chang S. S.; Huang G.; Cheng P.; Liu Y. (2008) Control of WHITE COLLAR localization by phosphorylation is a critical step in the circadian negative feedback process. EMBO J. 27, 3246–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L.; Kettenbach A. N.; Loros J. J.; Gerber S. A.; Dunlap J. C. (2009) Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol. Cell 34, 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeier T.; Diernfellner A.; Schafer A.; Dintsis O.; Neiss A.; Brunner M. (2008) Circadian activity and abundance rhythms of the Neurospora clock transcription factor WCC associated with rapid nucleo-cytoplasmic shuttling. Genes Dev. 22, 3397–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden W. J.; Lewis Z. A.; Selker E. U.; Loros J. J.; Dunlap J. C. (2011) CHD1 remodels chromatin and influences transient DNA methylation at the clock gene frequency. PLoS Genet. 7, e1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J.; Zhou M.; Liu Y. (2013) CATP is a critical component of the Neurospora circadian clock by regulating the nucleosome occupancy rhythm at the frequency locus. EMBO Rep. 14, 923–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.; Loros J. J.; Dunlap J. C. (2000) Interconnected feedback loops in the Neurospora circadian system. Science 289, 107–110. [DOI] [PubMed] [Google Scholar]
- Merrow M.; Franchi L.; Dragovic Z.; Gorl M.; Johnson J.; Brunner M.; Macino G.; Roenneberg T. (2001) Circadian regulation of the light input pathway in Neurospora crassa. EMBO J. 20, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glossop N. R.; Lyons L. C.; Hardin P. E. (1999) Interlocked feedback loops within the Drosophila circadian oscillator. Science 286, 766–768. [DOI] [PubMed] [Google Scholar]
- Shearman L. P.; Sriram S.; Weaver D. R.; Maywood E. S.; Chaves I.; Zheng B.; Kume K.; Lee C. C.; van der Horst G. T.; Hastings M. H.; Reppert S. M. (2000) Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019. [DOI] [PubMed] [Google Scholar]
- Denault D. L.; Loros J. J.; Dunlap J. C. (2001) WC-2 mediates WC-1-FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J. 20, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C.; Loros J. J.; Dunlap J. C. (1998) Nuclear localization is required for function of the essential clock protein FRQ. EMBO J. 17, 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q.; Cheng P.; Yang Y.; Wang L.; Gardner K. H.; Liu Y. (2002) White collar-1, a DNA binding transcription factor and a light sensor. Science 297, 840–843. [DOI] [PubMed] [Google Scholar]
- Yu W.; Zheng H.; Houl J. H.; Dauwalder B.; Hardin P. E. (2006) PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 20, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. Y.; Ko H. W.; Yu W.; Hardin P. E.; Edery I. (2007) A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol. Cell. Biol. 27, 5014–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet J. S.; Abruzzi K. C.; Desrochers J.; Rodriguez J.; Rosbash M. (2010) Dynamic PER repression mechanisms in the Drosophila circadian clock: From on-DNA to off-DNA. Genes Dev. 24, 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitane H.; Takao T.; Satomi Y.; Du N. H.; Okano T.; Fukada Y. (2009) Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol. Cell. Biol. 29, 3675–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru T.; Hirayama J.; Isojima Y.; Nagai K.; Norioka S.; Takamatsu K.; Sassone-Corsi P. (2009) CK2α phosphorylates BMAL1 to regulate the mammalian clock. Nat. Struct. Mol. Biol. 16, 446–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S. M.; Thompson S.; Elvin M.; Heintzen C. (2010) VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. Proc. Natl. Acad. Sci. U.S.A. 107, 16709–16714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H.; DeMay B. S.; Gladfelter A. S.; Dunlap J. C.; Loros J. J. (2010) Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc. Natl. Acad. Sci. U.S.A. 107, 16715–16720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.; He Q.; Guo J.; Cha J.; Liu Y. (2013) The Ccr4-not protein complex regulates the phase of the Neurospora circadian clock by controlling white collar protein stability and activity. J. Biol. Chem. 288, 31002–31009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q.; Liu Y. (2005) Molecular mechanism of light responses in Neurospora: From light-induced transcription to photoadaptation. Genes Dev. 19, 2888–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C. I.; Ruoff P.; Loros J. J.; Dunlap J. C. (2008) Closing the circadian negative feedback loop: FRQ-dependent clearance of WC-1 from the nucleus. Genes Dev. 22, 3196–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S. L. (2007) The complex language of chromatin regulation during transcription. Nature 447, 407–412. [DOI] [PubMed] [Google Scholar]
- Etchegaray J. P.; Lee C.; Wade P. A.; Reppert S. M. (2003) Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421, 177–182. [DOI] [PubMed] [Google Scholar]
- Ripperger J. A.; Schibler U. (2006) Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38, 369–374. [DOI] [PubMed] [Google Scholar]
- Katada S.; Sassone-Corsi P. (2010) The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol. 17, 1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTacchio L.; Le H. D.; Vollmers C.; Hatori M.; Witcher M.; Secombe J.; Panda S. (2011) Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 333, 1881–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N.; Yoo S. H.; Huang H. C.; Kumar V.; Lee C.; Kim T. K.; Takahashi J. S. (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J. X.; Revenko A. S.; Li L. B.; Gemo A. T.; Chen H. W. (2007) ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERα, is required for coregulator occupancy and chromatin modification. Proc. Natl. Acad. Sci. U.S.A. 104, 18067–18072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradolatto A.; Smart S. K.; Byrum S.; Blair L. P.; Rogers R. S.; Kolar E. A.; Lavender H.; Larson S. K.; Aitchison J. D.; Taverna S. D.; Tackett A. J. (2009) A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity. Mol. Cell. Biol. 29, 4604–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenko A. S.; Kalashnikova E. V.; Gemo A. T.; Zou J. X.; Chen H. W. (2010) Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol. Cell. Biol. 30, 5260–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L. M.; Ellahi A.; Rine J. (2011) Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc. Natl. Acad. Sci. U.S.A. 108, E1302–E1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raduwan H.; Isola A. L.; Belden W. J. (2013) Methylation of histone H3 on lysine 4 by the lysine methyltransferase SET1 protein is needed for normal clock gene expression. J. Biol. Chem. 288, 8380–8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowicz P.; Pawlak R.; Correa A.; Bell-Pedersen D.; Ebbole D. J. (2002) The Neurospora crassa pheromone precursor genes are regulated by the mating type locus and the circadian clock. Mol. Microbiol. 45, 795–804. [DOI] [PubMed] [Google Scholar]
- Sancar G.; Sancar C.; Brugger B.; Ha N.; Sachsenheimer T.; Gin E.; Wdowik S.; Lohmann I.; Wieland F.; Hofer T.; Diernfellner A.; Brunner M. (2011) A global circadian repressor controls antiphasic expression of metabolic genes in Neurospora. Mol. Cell 44, 687–697. [DOI] [PubMed] [Google Scholar]
- Wang L.; Kim J.; Somers D. E. (2013) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U.S.A. 110, 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M.; Tuohy T. M. F.; Mosurski K. R. (1986) Codon usage in yeast: Cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 14, 5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M. A.; Kurland C. G.; Pedersen S. (1989) Codon usage determines translation rate in Escherichia coli. J. Mol. Biol. 207, 365–377. [DOI] [PubMed] [Google Scholar]
- Lavner Y.; Kotlar D. (2005) Codon bias as a factor in regulating expression via translation rate in the human genome. Gene 345, 127–138. [DOI] [PubMed] [Google Scholar]
- Brocca S.; Schmidt-Dannert C.; Schmid R. D.; Lotti M.; Alberghina L. (1998) Design, total synthesis, and functional overexpression of the Candida rugosa lipl gene coding for a major industrial lipase. Protein Sci. 7, 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.; Hubalewska M.; Ignatova Z. (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 16, 274–280. [DOI] [PubMed] [Google Scholar]
- Siller E.; DeZwaan D. C.; Anderson J. F.; Freeman B. C.; Barral J. M. (2010) Slowing Bacterial Translation Speed Enhances Eukaryotic Protein Folding Efficiency. J. Mol. Biol. 396, 1310–1318. [DOI] [PubMed] [Google Scholar]
- Spencer P. S.; Siller E.; Anderson J. F.; Barral J. M. (2012) Silent Substitutions Predictably Alter Translation Elongation Rates and Protein Folding Efficiencies. J. Mol. Biol. 422, 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H. (1994) Synonymous codon usage in Drosophila melanogaster: Natural selection and translational accuracy. Genetics 136, 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D. A.; Wilke C. O. (2008) Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M.; Guo J.; Cha J.; Chae M.; Chen S.; Barral J. M.; Sachs M. S.; Liu Y. (2013) Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495, 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Ma P.; Shah P.; Rokas A.; Liu Y.; Johnson C. H. (2013) Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495, 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.; Xu G.; Schulman B. A.; Jeffrey P. D.; Harper J. W.; Pavletich N. P. (2003) Structure of a β-TrCP1-Skp1-β-catenin complex: Destruction motif binding and lysine specificity of the SCF(β-TrCP1) ubiquitin ligase. Mol. Cell 11, 1445–1456. [DOI] [PubMed] [Google Scholar]
- Querfurth C.; Diernfellner A. C.; Gin E.; Malzahn E.; Hofer T.; Brunner M. (2011) Circadian conformational change of the Neurospora clock protein FREQUENCY triggered by clustered hyperphosphorylation of a basic domain. Mol. Cell 43, 713–722. [DOI] [PubMed] [Google Scholar]
- Diernfellner A. C.; Querfurth C.; Salazar C.; Hofer T.; Brunner M. (2009) Phosphorylation modulates rapid nucleocytoplasmic shuttling and cytoplasmic accumulation of Neurospora clock protein FRQ on a circadian time scale. Genes Dev. 23, 2192–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J.; Yuan H.; Liu Y. (2011) Regulation of the activity and cellular localization of the circadian clock protein FRQ. J. Biol. Chem. 286, 11469–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]