Abstract
Bacillithiol is produced by many Gram-positive bacteria via a pathway utilizing the enzymes BshA, BshB, and BshC. Here we report the 1.77 Å resolution crystal structure of BshC, the putative cysteine ligase in bacillithiol production. The structure reveals that BshC contains a core Rossmann fold with connecting peptide motifs (CP1 and CP2) and a unique α-helical coiled-coil domain that facilitates dimerization. The model contains citrate and glycerol in the canonical active site and ADP in a second binding pocket. The overall structure and bound ligands give insight into the function of this unique enzyme.
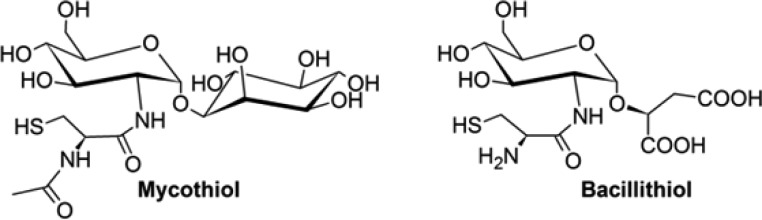
Low-molecular weight thiols are involved in the maintenance of cellular redox balance and detoxification of xenobiotic agents.1 Eukaryotes and Gram-negative bacteria utilize glutathione in this capacity, whereas mycobacteria use mycothiol (MSH) (Scheme 1).2 Gram-positive bacteria such as Staphylococcus aureus, Bacillus anthracis, and other Firmicutes utilize bacillithiol (BSH),3−6 which is the preferred cosubstrate for FosB-type fosfomycin resistance enzymes.7−9 BSH knockout bacterial strains are more sensitive to extremes in salinity, pH, and the presence of electrophilic compounds, making the enzymes responsible for BSH biosynthesis attractive targets for combating resistance to fosfomycin.
Scheme 1. Mycothiol and Bacillithiol.
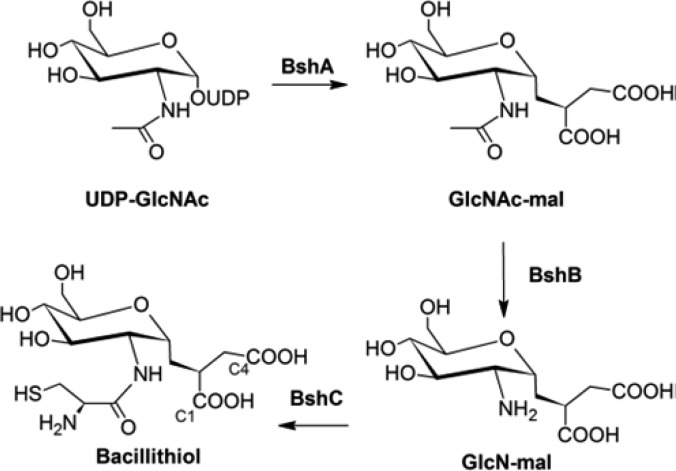
Studies suggest that BSH is produced from UDP-N-acetylglucosamine in three steps conducted by the glycosyltransferase BshA (YpjH), the deacetylase BshB (YpjG), and the putative cysteine ligase BshC (YllA) (Scheme 2).3 The activities of the BshA and BshB enzymes have been confirmed via in vitro assays, but BshC enzymatic activity has not been successfully demonstrated. Cysteine-tRNA ligases and the MSH cysteine ligase MshC add cysteine to an acceptor molecule via a process involving the activation of cysteine by ATP and subsequent attachment of cysteine to the acceptor.10 Given the similarities between MSH and BSH, cysteine ligation in the BSH biosynthesis pathway might occur in a congruent manner. However, ATP-dependent cysteine ligase activity has not been demonstrated with BshC even though glucosaminyl-malate (GlcN-mal) accumulates in YllA knockouts and they do not produce BSH.3 The lack of activity may be due to the absence of a cofactor, substrate, or additional protein. The BshC amino acid sequences from various Firmicutes are related to each other but share very little sequence identity with MshC or cysteine-tRNA ligases. These differences suggest that BSH cysteine ligation may operate via a novel mechanism.
Scheme 2. Proposed BSH Biosynthesis Pathway.
To characterize BshC and help elucidate its role in BSH biosynthesis, we embarked on an X-ray crystallographic study of it. Here we report the 1.77 Å resolution crystal structure of BshC from Bacillus subtilis 168 refined to an overall R factor of 17.0% (Tables S1 and S2 of the Supporting Information, Protein Data Bank entry 4WBD).
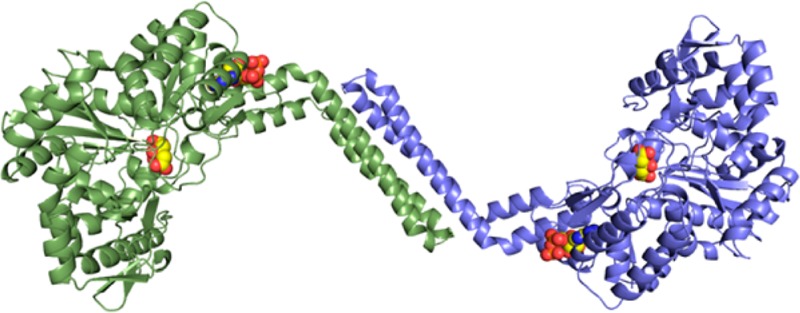
BshC consists of 539 amino acid residues with a core Rossmann fold domain, two connecting peptide motifs (CP1 and CP2), and an α-helical coiled-coil domain (Figure 1 and Figure S1 of the Supporting Information). Our gel filtration experiments suggest that BshC is a dimer in solution, and examination of symmetry-equivalent polypeptides in the unit cell reveals a striking interaction in which the coiled-coil domain of each subunit forms a four-helix bundle (Figure 2A). Several conserved nonpolar residues are buried at the interface between the two subunits (Figure 2B and Figure S5 of the Supporting Information), and the online tool PDBePISA11 estimated a ΔG of −30 kcal/mol for the formation of this dimer. Taken together, these analyses strongly suggest that BshC forms a dimer via its coiled-coil domain. The sequences of BshC from several other Firmicutes show the presence of this coiled-coil domain (Figure S4 of the Supporting Information), suggesting that the formation of a dimer in this manner is a feature common to all BshC enzymes. The active sites within the BshC dimer are open to solvent (Figure 2A), and enough space is present to allow interaction with an additional protein. If such a protein is indeed critical for BshC function, then its absence in our study could explain the lack of observable enzymatic activity.
Figure 1.
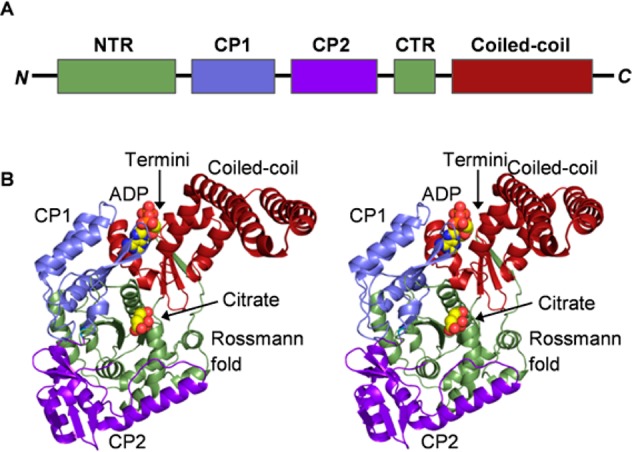
Overall structure of BshC from B. subtilis. (A) Schematic of the BshC domains. The N-terminal portion of the Rossmann fold (NTR) is colored green. The CP1 and CP2 domains are colored light blue and purple, respectively. The C-terminal portion of the Rossmann fold (CTR) is colored green. The α-helical coiled-coil domain is colored red. (B) Stereo representation of the BshC monomer with the domains colored as they are in panel A. The ligands are shown as spheres, with citrate situated within the canonical active site in front of the Rossmann fold parallel β-sheet and ADP bound between the CP1 and coiled-coil domains. All macromolecular graphics were prepared with PyMOL.12
Figure 2.
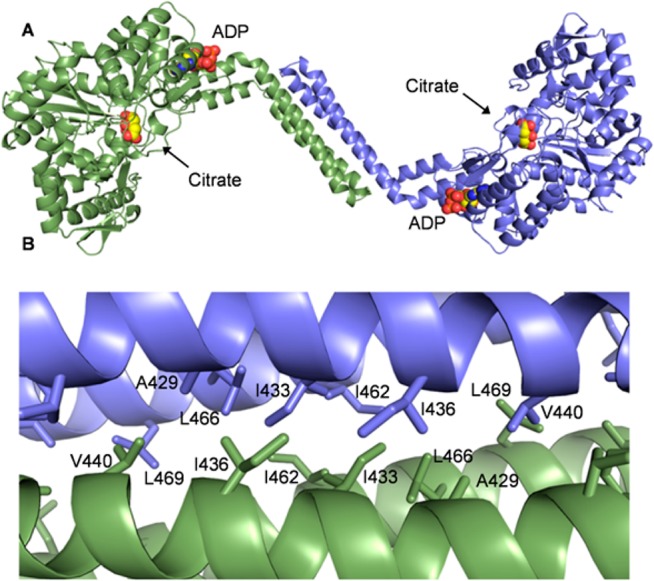
Quaternary structure of BshC. (A) Cartoon representation of the BshC dimer colored by chain. (B) Close-up view of the four-helix bundle located at the dimer interface. Nonpolar side chains between the coiled-coil domains are shown as sticks.
The BshC crystals used in this study were grown in the presence of 200 mM citrate, and electron density corresponding to a citrate molecule was observed within the canonical Rossmann fold active site (Figure S2 of the Supporting Information). The 5- and 6-carboxylate groups of citrate likely mimic the 4- and 1-carboxylate groups, respectively, of the malyl moiety of glucosaminyl-malate and BSH. Thus, this citrate molecule provides clues about how BshC accommodates that portion of its substrate and product (Figure 3A). The 5- and 6-carboxylate groups of citrate interact with the guanidinium groups of Arg 377 and Arg 504 and main chain amide groups of Gly 352 and Glu 353, respectively. The remaining citrate carboxylate group is less ordered and makes no direct interactions with the enzyme. It extends into a solvent-filled cavity, which has enough space to accommodate larger ligands. Electron density corresponding to a glycerol molecule was observed near the citrate (Figure S2 of the Supporting Information). This glycerol molecule interacts with the side chain of Glu 353 and an ordered water molecule (Figure 3A).
Figure 3.
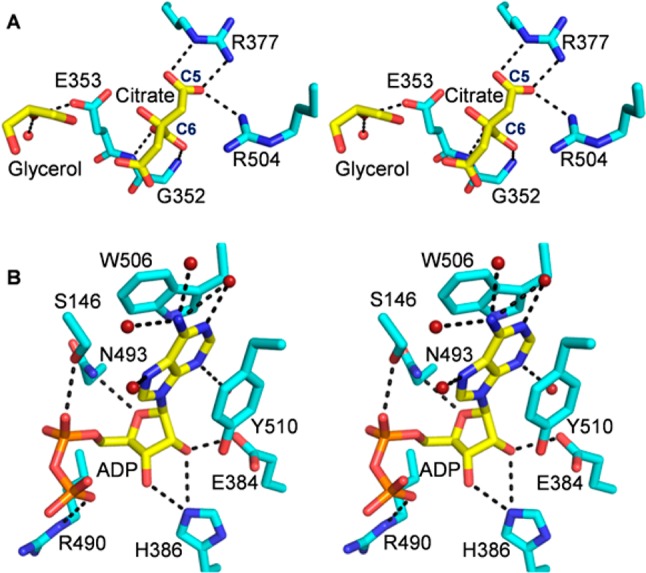
Stereoviews of BshC ligands. Potential hydrogen bonding interactions within 3.3 Å of the ligands are indicated by dashed lines. (A) Close-up view of the canonical Rossmann fold active site with glycerol and citrate. (B) Close-up view of the second ligand-binding site containing an ADP molecule.
An ADP molecule is bound within a pocket distinct from the canonical active site. The electron densities corresponding to the adenine, ribose, and α-phosphate portions of ADP are well-defined (Figure S2 of the Supporting Information). The electron density for the β-phosphoryl group is somewhat ambiguous, and it is possible that a group other than phosphate is present at this position. However, peaks of electron density at the α- and β-phosphorus positions remain in the Fo – Fc omit map when contoured above 8σ, suggesting that electron-rich groups occupy these locations. It is possible that an additional group is attached to the β-phosphate but is too disordered to be observed. Interestingly, no nucleotides were added during purification, dialysis, or crystallization, and thus, this ligand came along with the protein when it was expressed in Escherichia coli. Mass spectroscopic analysis of the purified protein suggests the presence of an adenosine nucleoside (Figure S3 of the Supporting Information), but unequivocal identification of the ligand was not possible via this technique. We have chosen to model the ligand as ADP on the basis of the electron density map, but we acknowledge that an additional group may be attached to it at the β-position.
The ADP molecule sits in a pocket separated from the canonical active site by a portion of α-helix 19. This pocket comprises residues from α-helices 14, 18, and 19 and β-strand 4, which are parts of the CP1 and coiled-coil domains. The adenine base of ADP is involved in π-stacking interactions with the Trp 506 and Tyr 510 side chains (Figure 3B). All of the potential hydrogen bonding interactions of the adenine base are met through interactions with ordered water molecules. The ribose polar groups interact with the Glu 384, His 386, and Asn 493 side chains. The α- and β-phosphate groups interact with the side chains of Ser 146 and Arg 490, respectively.
The ADP-binding pocket is not present in MshC or cysteine-tRNA ligases. Strikingly, although the CP1 and coiled-coil domains are present among various BshC orthologs, the residues that interact with ADP in the B. subtilis enzyme are not highly conserved across these species (Figures S4 and S5 of the Supporting Information). The biological significance of the ADP molecule and the binding site that it occupies is an intriguing mystery. The binding pocket may function in regulation, or perhaps it serves as a second catalytic site. It may simply be an evolutionary vestige. Regardless, structural analysis of BshC orthologs with various ligands is presently in progress to further characterize this binding site.
BshC and MshC from Mycobacterium smegmatis(13) superimpose with a root-mean-square deviation of 3.4 Å for 207 structurally equivalent α-carbons (of 539 total amino acid residues). This superposition provides insight into the function of BshC (Figure 4A). MshC has a shorter CP domain, lacks the coiled-coil domain entirely, and retains a portion of the anticodon binding domain found in cysteine-tRNA ligases. The BshC and MshC Rossmann fold β-sheet and α-helices superimpose well (Figure 4B). The HIGH and MKSKS sequence motifs present in cysteine-tRNA ligases and the ERGGDP sequence motif found in MshC are not present in BshC. However, BshC orthologs contain strictly conserved V331VRTP, G352PGEXXYW, and Q502ER sequences (Figure S4 of the Supporting Information) that are within the canonical active site.
Figure 4.
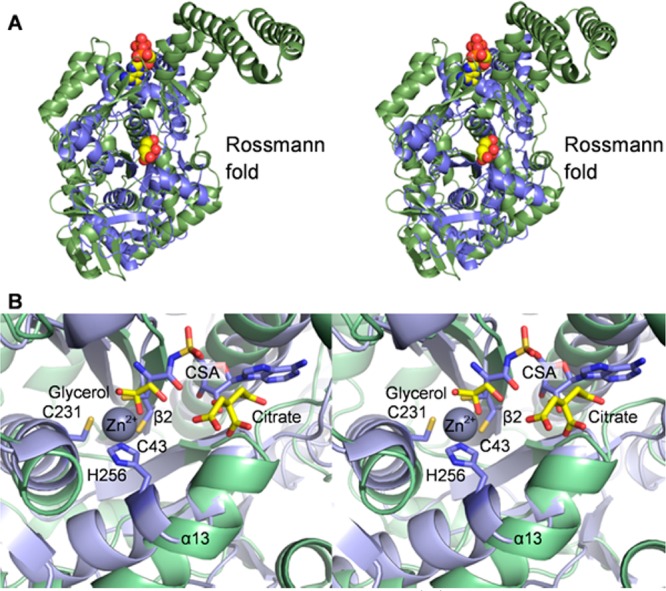
Comparison of BshC and MshC. (A) Stereoview of the superposition of BshC (green) and MshC (blue) structures. (B) Close-up stereoview of the superimposed BshC and MshC active sites. BshC ligands are shown with yellow carbon atoms, whereas MshC ligands are shown with blue carbon atoms.
The structure of MshC was determined in the presence of the cysteinyl-AMP analogue 5′-O-[N-(l-cysteinyl)-sulfamonyl]-adenosine (CSA). In the superposition shown in Figure 4B, CSA is bound near the citrate and glycerol molecules in BshC. The glycerol molecule bound in the BshC active site occupies the same position as the cysteinyl portion of CSA in MshC and amino acids in type 1 aminoacyl-tRNA ligases.14 Thus, the glycerol molecule serves as a mimic for an amino acid and indicates where cysteine binds in BshC. Glucosaminyl-malate, if modeled into this binding site, can be oriented in such a way that its amino group points toward the glycerol molecule (not shown), as would be expected to occur during cysteine ligation.
MshC and the cysteine-tRNA ligases utilize a zinc ion to allow differentiation between cysteine and serine. In the structure of MshC, the zinc ion is held in the active site via interactions with the Cys 43, Cys 231, and His 256 side chains (Figure 4B). These residues, conserved among MshC and cysteine-tRNA ligases, are not found in BshC. In fact, α-helix 13 and β-strand 2, which occupy positions in BshC near the zinc-binding residues in MshC, are shifted significantly, and the BshC active site appears to be devoid of a constellation of zinc-biding residues. If BshC does not bind a zinc ion, then it must discriminate between cysteine and serine via a mechanism distinct from that of MshC. Our efforts to soak or cocrystallize BshC crystals with zinc, BSH, and other ligands have not yet met with success.
The proliferation of multi-drug-resistant microorganisms, such as the Gram-positive methicillin-resistant S. aureus (MRSA), represents a growing threat to human health and quality of life. A thorough characterization of the BSH biosynthesis pathway, including BshC, will provide insight into new therapeutic targets to combat resistance to the antibiotic fosfomycin. The structure of BshC presented here provides a foundation for further structural and functional studies into this unique and intriguing enzyme.
Acknowledgments
Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract DE-AC02-06CH11357. Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corp. and the Michigan Technology Tri-Corridor (Grant 085P1000817). We gratefully acknowledge the assistance of Dr. David Smith and Dr. Zdzislaw Wawrzak at Argonne National Laboratory for assistance with the collection of X-ray data. We thank Dr. Richard N. Armstrong for supporting the initial stages of this investigation. Finally, we thank Dr. Hazel M. Holden, Dr. David A. Leonard, and other colleagues for helpful discussions.
Supporting Information Available
Detailed experimental procedures, crystallographic and refinement statistics, and additional figures, including ligand electron density. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
Coordinates have been deposited in the Protein Data Bank as entry 4WBD.
This research was supported in part by National Institutes of Health Grants F32GM093507 (to P.D.C.) and R01GM030910 (to Dr. Richard Armstrong, Vanderbilt University School of Medicine, Nashville, TN).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Zuber P. (2009) Management of oxidative stress in Bacillus. Annu. Rev. Microbiol. 63, 575–597. [DOI] [PubMed] [Google Scholar]
- Fan F.; Vetting M. W.; Frantom P. A.; Blanchard J. S. (2009) Structures and mechanisms of the mycothiol biosynthetic enzymes. Curr. Opin. Chem. Biol. 13, 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A.; Newton G. L.; Antelmann H.; Parsonage D.; Upton H.; Rawat M.; Claiborne A.; Fahey R. C.; Helmann J. D. (2010) Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc. Natl. Acad. Sci. U.S.A. 107, 6482–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D. (2011) Bacillithiol, a new player in bacterial redox homeostasis. Antioxid. Redox Signaling 15, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton G. L.; Rawat M.; La Clair J. J.; Jothivasan V. K.; Budiarto T.; Hamilton C. J.; Claiborne A.; Helmann J. D.; Fahey R. C. (2009) Bacillithiol is an antioxidant thiol produced in Bacilli. Nat. Chem. Biol. 5, 625–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D. J. (2014) Bacillithiol, a new role in buffering intracellular zinc. Mol. Microbiol. 94, 743–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers A. P.; Keithly M. E.; Kim K.; Cook P. D.; Stec D. F.; Hines K. M.; Sulikowski G. A.; Armstrong R. N. (2012) Synthesis of bacillithiol and the catalytic selectivity of FosB-type fosfomycin resistance proteins. Org. Lett. 14, 5207–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. V.; Jothivasan V. K.; Newton G. L.; Upton H.; Wakabayashi J. I.; Kane M. G.; Roberts A. A.; Rawat M.; La Clair J. J.; Hamilton C. J. (2011) Chemical and chemoenzymatic syntheses of bacillithiol: A unique low-molecular-weight thiol amongst low G + C Gram-positive bacteria. Angew. Chem., Int. Ed. 50, 7101–7104. [DOI] [PubMed] [Google Scholar]
- Thompson M. K.; Keithly M. E.; Goodman M. C.; Hammer N. D.; Cook P. D.; Jagessar K. L.; Harp J.; Skaar E. P.; Armstrong R. N. (2014) Structure and function of the genomically encoded fosfomycin resistance enzyme, FosB, from Staphylococcus aureus. Biochemistry 53, 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D.; Steffek M.; Newton G. L.; Fahey R. C. (2002) ATP-dependent l-cysteine:1d-myo-inosityl 2-amino-2-deoxy-α-d-glucopyranoside ligase, mycothiol biosynthesis enzyme MshC, is related to class I cysteinyl-tRNA synthetases. Biochemistry 41, 6885–6890. [DOI] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797. [DOI] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, version 1.5.0.4 (2012) Schrödinger, LLC, Portland, OR. [Google Scholar]
- Tremblay L. W.; Fan F.; Vetting M. W.; Blanchard J. S. (2008) The 1.6 Å crystal structure of Mycobacterium smegmatis MshC: The penultimate enzyme in the mycothiol biosynthetic pathway. Biochemistry 47, 13326–13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue P.; Luthey-Schulten Z. (2003) On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol. Mol. Biol. Rev. 67, 550–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.