Abstract
Sequence-specific recognition of DNA by small organic molecules offers a potentially effective approach for the external regulation of gene expression and is an important goal in cell biochemistry. Rational design of compounds from established modules can potentially yield compounds that bind strongly and selectively with specific DNA sequences. An initial approach is to start with common A·T bp recognition molecules and build in G·C recognition units. Here we report on the DNA interaction of a synthetic compound that specifically binds to a G·C bp in the minor groove of DNA by using an azabenzimidazole moiety. The detailed interactions were evaluated with biosensor-surface plasmon resonance (SPR), isothermal calorimetric (ITC), and mass spectrometry (ESI-MS) methods. The compound, DB2277, binds with single G·C bp containing sequences with sub-nanomolar potency and displays slow dissociation kinetics and high selectivity. A detailed thermodynamic and kinetic study at different experimental salt concentrations and temperatures shows that the binding free energy is salt concentration dependent but essentially temperature independent under our experimental conditions, and binding enthalpy is temperature dependent but salt concentration independent. The results show that in the proper compound structural context novel heterocyclic cations can be designed to strongly recognize complex DNA sequences.
Heterocyclic diamidine minor groove binders have been successful in therapeutic targeting of DNA structures in various types of cells and particularly parasitic microorganisms.1−11 Selective targeting cellular DNA has been shown with compounds that have intrinsic fluorescence in cells2,12−14 and which have been used in human clinical trials.1,11−17 Given the limited number of new antiparasitic drugs, the success of amidines against those diseases is attractive for continued compound development.5,7,10,13,17−20
In considering methods to increase heterocyclic diamidine success in selective targeting of cellular DNAs, we have focused on two A·T sequences separated by a single G·C base pair (bp). In the sequence of kinetoplast DNA, for example, a large number of A·T bp sites of 3 to 4 bp or larger, are commonly separated by one or two G·C bps.2,5,21−23 These sequence motifs suggest that including new heterocycles with H-bond accepting units at the appropriate position in the diamidine derivatives could provide G·C bp recognition capability in the AT sequence context. In addition, the increased sequence recognition capability could provide a very productive approach to enhance the design of new minor groove targeting drugs against a range of diseases.13 A similar and complementary approach has been very successful with design of mixed sequence recognizing polyamides.24−30 These compounds, however, have not been as successful in animal studies and have not gone into human trials. Our rationally designed compounds, with mixed DNA sequence recognition capability, would mark a much needed breakthrough in expanding the DNA targeting field.
Unfortunately, our understanding of G·C bp recognition by small molecule minor groove binders is very limited, and this lack severely restricts the use of the DNA minor groove for new drug design and development. For the design of mixed binding site compounds, incorporation of modules with H-bond accepting groups for G·C bps is essential. As an initial design, compounds with azabenzimidazole H-bond acceptors in quite different types of structures have been prepared (Figure 1).31 We have been able to achieve G·C bp selective binding in this new series and the ability to bind selectively to a G·C bp was found to be quite compound structure and DNA sequence dependent. This result shows that, although there is still much that we do not know about mixed bp recognition in the DNA minor groove, the new series with G·C recognition offers a critically needed new lead. Such compounds are essential for the development of diamidine derivative pairing rules for a novel DNA minor groove binding language. To address the basis for the new recognition module, we report a detailed thermodynamic and kinetic study at different salt concentrations and temperatures. The results reveal the molecular interaction details between mixed A·T/G·C DNA sequences and heterocyclic azabenzimidazole diamidine molecules. The binding free energies have been determined from biosensor surface plasmon resonance (SPR), while binding enthalpies and heat capacities are from isothermal calorimetric titration (ITC).
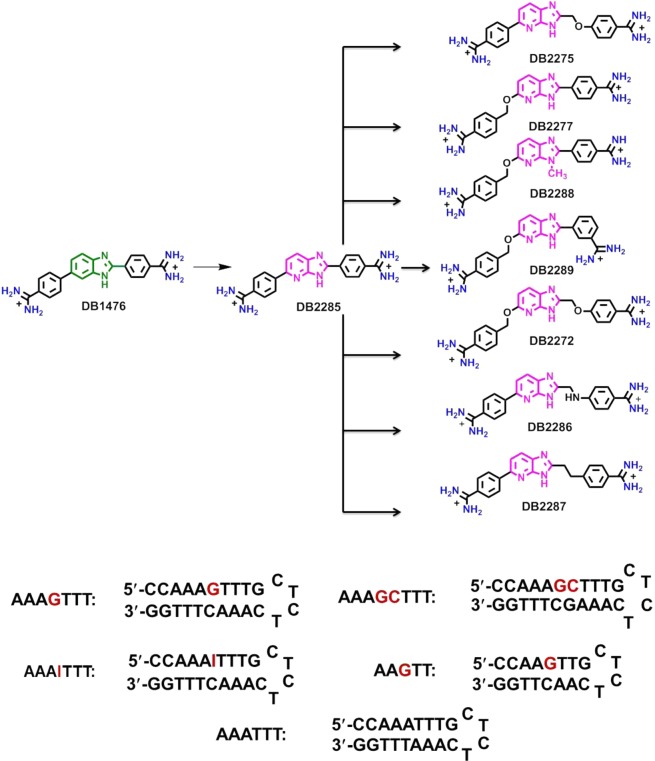
Figure 1.
Structure of compounds and DNA sequences used in this study. For SPR experiments, 5′-biotin labeled DNA sequences are used.
Compound Design for G·C Base Pair Recognition
The benzimidazole-based diamidine, DB1476 (Figure 1),32 is a very strong A·T specific minor groove binder with poor G·C sequence recognition. An initial test to determine whether DB1476 could be converted to a G·C recognition compound with a benzimidazole to an azabenzimidazole conversion yielded DB2285. DB2285 showed a large reduction in A·T binding with a slight enhancement of G·C binding strength such that it had a slight G·C selectivity.27 With this encouraging result we considered various compound design features to enhance the G·C selectivity. Based on our previous results with alkyl–aryl compounds,33 we felt that some flexibility in the system might enhance G·C bp recognition in DB2285. This is possible since specific H-bonding with the G-NH2 group is critically dependent on the correct distance and angle for a strong interaction. With our limited knowledge of the requirements in minor groove binders, in general for G·C recognition, a number of modifications of DB2285 were prepared and their interactions with different DNA sequences have been evaluated. By introducing mono −CH2O– or −OCH2– substituted flexible isomers, DB2275 and DB2277 were prepared along with the disubstituted compound, DB2272. Several analogues of these compounds were also prepared (Figure 1) with the goal of finding the requirements for strong and specific G·C recognition in this series. This compound set illustrates the need for exploring wide chemical and structural space in a series of compounds when searching for G·C bp specific recognition. In the entire series, only DB2277, and not, for example, the isomer DB2275 gave strong G·C bp specific binding.31 The strongest binding of DB2277 was found with A-tract flanking sequences of the type, AnGTn. To help understand the molecular basis for this result, detailed binding studies were conducted for these compounds and an A-tract mixed sequence, and the results are presented along with a discussion of why we think the ability to recognize a G·C bp is quite different and structure-dependent in this series of derivatives.
Materials and Methods
DNA Oligonucleotides
For the ITC experiments, the hairpin DNA oligomers used were AAGTT [5′-CCAAGTTGCTCTCAACTTGG-3′], AAAGTTT [5′-CCAAAGTTTGCTCTCAAACTTTGG-3′], and AAATTT [5′-CCAAATTTGCCTCTGCAAATTTGG-3′], with the hairpin loop sequences underlined. Lyophilized DNA oligomers were purchased from Integrated DNA Technologies, Inc. (IDT, Coralville, IA) with HPLC purification. Doubly distilled water was added to the solid DNAs to bring the concentration to approximately 1.0 mM, based on the reported amount of DNA from IDT. The molar concentrations of these hairpin DNAs were then determined using a Cary 300 UV–vis spectrophotometer (Varian, Walnut Creek, CA) at 260 nm based on the molar extinction coefficients (ε260) calculated by the nearest-neighbor method.
Isothermal Titration Calorimetry (ITC)
ITC experiments were performed using a MicroCal VP-ITC (MicroCal Inc., Northampton, MA) interfaced with a computer equipped with VP-2000 software for instrument control and Origin 7.0 for data analysis. The sample cell was filled with 10 μM hairpin DNA in 50 mM Tris-HCl buffer (50 mM Tris-HCl pH 7.4 having 1 mM EDTA, and 50–600 mM NaCl concentration), and 29 injections of 10 μL of the compound solution were performed incrementally at 20–45 °C. A delay of 300 s was used between each injection to ensure the equilibration of the baseline. The heat for each injection was obtained by integration of the peak area as a function of time. The heats of dilution, determined by injecting the compound into the sample cell containing only buffer, were subtracted from those in compound/DNA titrations to present the corrected binding induced enthalpy changes. Because all the ligands bind quite strongly to the sequences in this work, the heat/mol of added compound is essentially constant in the initial titration region where all added compound is bound to DNA. The ΔHb can be determined by a linear fit as shown in Figures 5 and 6. A range of compound concentrations and temperatures were used in these experiments to optimize conditions for data collection.
Figure 5.
ITC data for the titration of (A) DB2277-AAAGTTT, (B) DB2277-AAGTT, and (C) DB2275-AAAGTTT. Injections of 10 μL aliquots of 50 μM ligand into 10 μM hairpin duplex DNA at 25 °C in 50 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA buffer, pH 7.4. The ITC raw data, located in the top panel, is the power output per injection as a function of time. The bottom panel is the peak integration of the data that shows the heat produced per injection as a function of the hairpin/ligand molar ratio.
Figure 6.
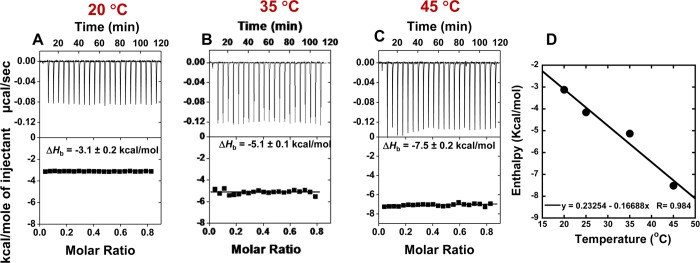
(A–C) ITC data for the titration of DB2277 and AAAGTTT DNA at different experimental temperatures. (D) Plot of ΔHb versus temperature for DB2277 with AAAGTTT DNA, and the linear fit yields a ΔCp of −172 ± 5 cal M–1 K–1.
Biosensor Surface Plasmon Resonance (SPR)
SPR measurements were performed with four-channel Biacore T200 optical biosensor systems (GE Healthcare, Inc., Piscataway, NJ). A streptavidin-derivatized (SA) sensor chip was prepared for use by conditioning with a series of 60 s injections of 1 M NaCl in 50 mM NaOH (activation buffer) followed by extensive washing with HBS buffer [10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.05% P20 (pH 7.4)]. Biotinylated DNA samples [AAATTT, AAAGTTT, AAAGCTTT, and AAAITTT hairpins] (25–50 nM) were prepared in HBS buffer and immobilized on the flow cell surface by noncovalent capture as previously described.34,35 Flow cell 1 was left blank as a reference, while flow cells 2–4 were immobilized with DNA by manual injection of DNA stock solutions (flow rate of 1 μL/min) until the desired amount of DNA response units (RU) was obtained (320–330 RU). Ligand solutions were prepared with degassed and filtered 50 mM Tris-HCl buffer (at pH 7.42) by serial dilutions from a concentrated stock solution. Typically, a series of different ligand concentrations (from 1 nM to 1 μM) were injected over the DNA sensor chip at a flow rate of 100 μL/min until a constant steady-state response was obtained (3 min), and this was followed by buffer flow for ligand dissociation (10–20 min). After each cycle, the sensor chip surface was regenerated with a 10 mM glycine solution at pH 2.5 for 30 s followed by multiple buffer injections to yield a stable baseline for the following cycles. RUobs was plotted as a function of free ligand concentration (Cfree), and the equilibrium binding constants (KA) were determined either with a one-site binding model (K2 = 0) or with a two-site model, where r represents the moles of bound compound per mole of DNA hairpin duplex and K1 and K2 are macroscopic binding constants.
| 1 |
RUmax in the equation was used as a fitting parameter, and the obtained value was compared to the predicted maximal response per bound ligand to independently evaluate the stoichiometry.35 Kinetic analyses were performed by globally fitting the binding results for the entire concentration series using a standard 1:1 kinetic model with integrated mass transport-limited binding parameters as described previously.33
Mass Spectrometry
Mass spectrometry (MS) analyses were performed on a Waters Q-TOF spectrometer (Waters Corporate, Milford, MA) equipped with an electrospray ionization source (ESI) in negative ion mode. Samples of compound/DNA (1:1 and 2:1 ratios) were prepared in 100 mM ammonium acetate with 5% v/v methanol at pH 7.4 and introduced into the ion source through direct infusion at 5 μL/min flow rate. The instrument parameters were as follows: capillary voltage of 2200 V, sample cone voltage of 30 V, extraction cone voltage of 2.5 V, desolvation temperature of 70 °C, and source temperature of 100 °C. Nitrogen was used as nebulizing and drying gas. Spectra were collected for the mass/charge region of 300–3000 for 10 min, and the last 2 min of the scan were used for analyses. Analyses and interpretation of the deconvoluted spectra were performed using MassLynx 4.1 software.
Results
Biosensor-SPR: Binding Affinity and Kinetics
The biosensor-SPR method provides an excellent way to quantitatively evaluate the interaction of a set of synthetic small molecules with immobilized biomolecules.33−36 This technique provides sensitive real time progress of interaction rates as well as the equilibrium binding affinities of biomolecular interactions. In our previous study31 we observed that azabenzimidazole substituted diamidines (Figure 1) prefer to bind to an A-tract sequence rather than to the wider minor groove of alternating A·T (e.g., ATAGTAT) sequences. Hence, in this work, detailed SPR experiments were conducted with immobilized DNA hairpin duplex sequences containing either a pure A-tract sequence (AAATTT) or mixed DNA sequences containing a pure A-tract flanking sequence with one or two G·C bp, such as AAAGTTT and AAAGCTTT (Figure 1).
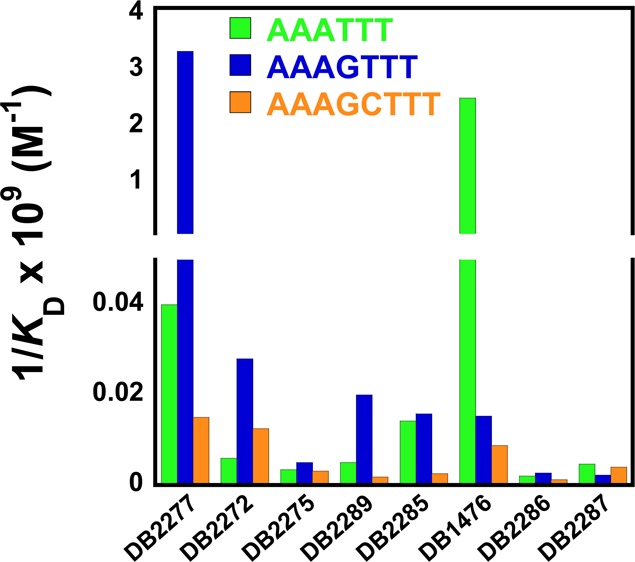
As previously reported DB1476,32 a benzimidazole core diamidine, is a strong binder with the pure A·T sequence, AAATTT [KD = 0.4 nM (Table 1)]. DB2285, the azabenzimidazole substituted analogue of DB1476, gave a 175-fold reduction in binding affinity to AAATTT (KD = 70 nM) relative to DB1476, and it binds with almost the same affinity with mixed sequences that have a G·C bp (AAAGTTT, KD = 63 nM). While DB2285 has improved G·C specificity, it is clearly not well optimized for mixed sequence recognition. In an effort to enhance the DNA minor groove binding affinity and selectivity, an −OCH2– group was inserted as a linker between the azabenzimadole and phenyl group to obtain DB2277. A striking result is the strong interaction of DB2277 with AAAGTTT and particularly noteworthy is the very slow dissociation of the complex (Figure 2). Even with a quite long experimental dissociation time (∼20 min), total dissociation of DB2277 from the complex is not complete. Global kinetics fitting yielded a single binding site and an approximate KD of 0.3 nM for DB2277 with AAAGTTT. DB2277 shows very different affinity toward the narrower minor groove of pure A·T sequences with 80-fold (KD = 24 nM) weaker binding with AAATTT than with the single G·C bp sequence. The sensorgram of AAATTT shows an off-rate that is much faster and complete dissociation from the complex occurs within the first few minutes of the dissociation phase (Figure 2C). With the wider minor groove sequence AAAGCTTT, DB2277 shows 200-fold weaker binding affinity under the same experimental conditions. When “G” (AAAGTTT) is substituted by “I” (inosine) (AAAITTT) at the minor groove binding site, the binding affinity decreases by 30-fold (KD = 10 nM) as would be expected for disruption of hydrogen bonding between the azabenzimidazole and G-NH2 (Figure 2B). These results suggest that the binding aptitude of DB2277 depends on the structure, width and chemistry of the minor groove of duplex DNA and for this reason this molecule has excellent sequence selectivity.
Table 1. Biosensor-SPR Equilibrium Dissociation Constants (KD, nM) of DB2277 and Analogues with Pure A·T and Mixed DNA Sequencesa.

The listed binding affinities are an average of two independent experiments carried out with two different sensor chips and the values are reproducible within 10% experimental errors. The experiments were conducted in Tris-HCl buffer (50 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, pH 7.4) at 25 °C.
Figure 2.
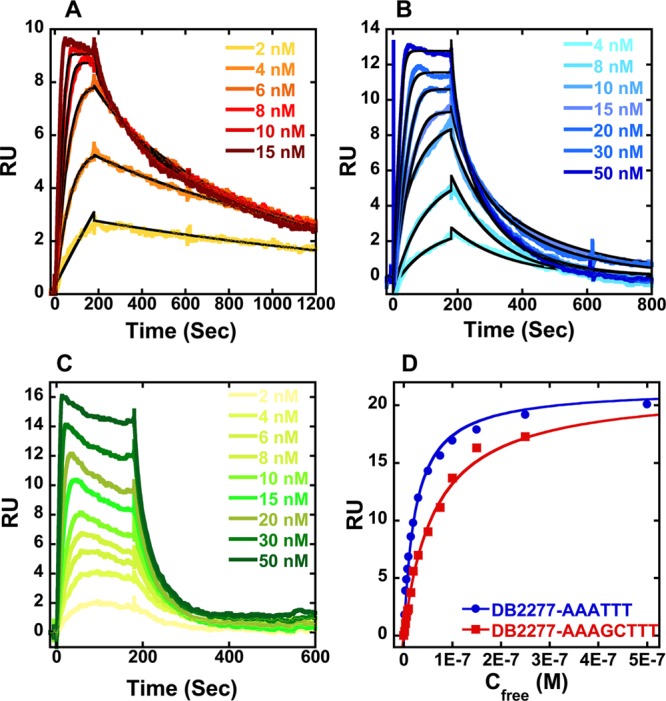
Representative SPR sensorgrams for DB2277 in the presence of (A) AAAGTTT, (B) AAAITTT, and (C) AAATTT hairpin DNAs. (D) Comparison of steady-state binding plots for AAATTT and AAAGCTTT with DB2277. The data are fitted to a steady state binding function using a 1:1 model to determine equilibrium binding constants. In (A) and (B), the solid black lines are best fit values for global kinetic fitting of the results with a single site function.
The para to meta positional change of the amidine on the phenyl group in DB2277, DB2289, results in a 150-fold decrease in binding affinity with the single G·C bp sequence, and only a 5-fold sequence selectivity is observed. The differences in binding affinity and sequence selectivity for these two compounds, which differ only at the amidine group, are very striking. DB2275, the structural isomer of DB2277, binds with AAAGTTT as a much weaker monomer (KD = 185 nM) and has lower binding affinities with all of the DNA sequences (Table 1 and Supporting Information Figure S1). Two analogues of DB2275, DB2286 and DB2287, show similar results. DB2272 has the −OCH2– and −CH2O– linkers at both sides, and it recognizes the single G·C bp sequence about 100-fold more weakly than DB2277 (KD = 35 nM). The combined SPR results of all the compounds clearly highlight both the difficulties and significant successes in the development of new ligands specific for G·C bp containing sequences.
The Effects of Salt Concentration on DB2277 Binding to AAAGTTT
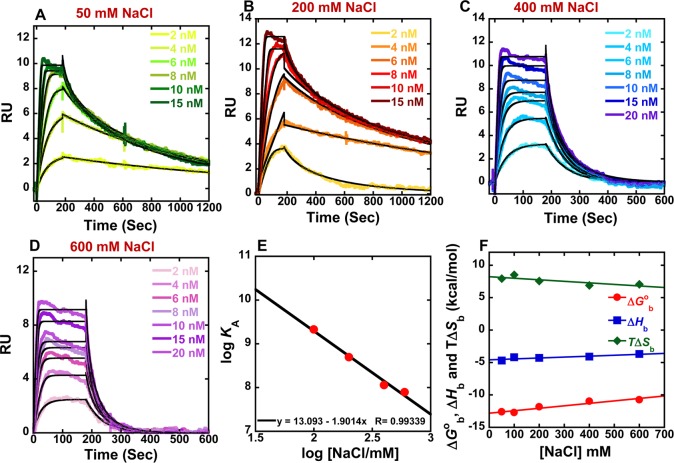
The SPR binding results indicate that DB2277 has an optimized size and curvature for strong and selective recognition of a single G·C bp in an A-tract sequence. To understand these significant findings in more detail, it is essential to evaluate the thermodynamics of this compound with the single G·C bp sequence. To compare the effects of salt concentration on binding of DB2277 with the single G·C bp sequence, SPR experiments were carried out at 25 °C from 50 to 600 mM NaCl concentrations (Figure 3). The equilibrium binding constants obtained by both global kinetic fits and steady state fits are in excellent agreement and are collected in Table 2 and Supporting Information Figure S2A. According to the counterion condensation theory,37−40 the logarithm of the equilibrium binding constants KA (=1/KD) (from global kinetic fits) should be a linear function of the logarithm of salt concentration as shown in Figure 3E. The KA values decrease significantly as the salt concentration increases as is typical for DNA–cation complexes.40 The slopes of the linear fits are ∼1.5 which is reasonable for a dication on DNA complex formation.40 The number of phosphate contacts (Z) between DB2277 and hairpin duplex DNA can be obtained by slope/Ψ (Ψ = fraction of phosphate shielded by condensed counterions and is 0.88 for double stranded B-DNA),41,42 and this gives a Z of 1.70 ± 0.2. Thus, there are about two phosphate contacts between DB2277 and DNA which is a very realistic value for this dicationic molecule.
Figure 3.
(A–D) SPR sensorgrams (color) and global kinetic fits (black overlays) for DB2277 with the AAAGTTT DNA sequence at different salt concentrations. (E) Salt dependence of KA for DB2277 binding as determined by SPR. The KA values were obtained by both global kinetic and steady state fits at the two higher salt concentrations. (F) Plot of ΔGb°, ΔHb, and TΔSb versus salt concentrations for DB2277 with the AAAGTTT sequence at 25 °C.
Table 2. Kinetics and Steady-State Analysis of DB2277 with the AAAGTTT Sequence at Different Salt Concentrations in 50 mM Tris-HCl, 1 mM EDTA Buffer, pH 7.4 at 25 °Ca.
|
KD (nM) |
|||||
|---|---|---|---|---|---|
| [NaCl] mM | kinetic fit | steady state | ΔGb° (kcal/mol) | ΔHb (kcal/mol) | TΔSb (kcal/mol) |
| 50d | 0.2 ± 0.4 | –13.2d | –4.7 ± 0.2 | 8.5 | |
| 100 | 0.3 ± 0.2 | 0.2 ± 0.1 | –13.0b | –4.2 ± 0.3 | 8.8 |
| 200 | 2.1 ± 0.6 | 4.3 ± 0.8 | –11.8b | –4.3 ± 0.1 | 7.5 |
| 400 | 8.8 ± 0.9 | 9.5 ± 2.0 | –11.0c | –4.1 ± 0.2 | 6.9 |
| 600 | 12.8 ± 2.0 | 13.2 ± 1.0 | –10.7c | –3.7 ± 0.4 | 7.0 |
Kinetic analysis was performed by global fitting with a 1:1 binding model.
Data obtained from kinetic fit values.
Data obtained from steady-state fit values; ΔHb was determined in ITC experiments; TΔSb = −ΔGb° + ΔHb.
Results are calculated from extrapolation of log KA vs log [NaCl/mM] (Figure 3E).
The Effects of Temperature on the DNA Binding Affinity of DB2277
SPR experiments of DB2277 and AAAGTTT were also conducted at different temperatures at 200 mM salt concentration (Supporting Information Figures S2A and S3), and it is visually apparent from the sensorgrams that the temperature has a significant effect on the on/off rate of ligand binding (Table 3). However, temperature changes have a small effect on the ΔGb° (Figure 4 and Table 3) as has frequently been seen with DNA complexes near 25 °C.40
Table 3. Kinetics and Thermodynamic Results for DB2277 with the AAAGTTT Sequence at Different Experimental Temperatures at 50 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA Buffer at pH 7.4a.
|
KD (nM) |
|||||
|---|---|---|---|---|---|
| T (°C) | kinetic fit | steady state | ΔGb° (kcal/mol) | ΔHb (kcal/mol) | TΔSb (kcal/mol) |
| 20 | 0.8 ± 0.4 | 0.5 ± 0.3 | –12.4b | –3.1 ± 0.2 | 9.3 |
| 25 | 2.1 ± 0.6 | 4.3 ± 0.8 | –11.8b | –4.3 ± 0.1 | 7.5 |
| 35 | 4.2 ± 1.3 | 6.1 ± 0.7 | –11.6c | –5.1 ± 0.1 | 6.5 |
| 45 | 5.0 ± 0.8 | 8.2 ± 2.0 | –11.8c | –7.5 ± 0.2 | 4.3 |
Kinetic analysis was performed by global fitting with a 1:1 binding model.
Data obtained from kinetic fit values.
Data obtained from steady-state fit values; ΔHb was determined in ITC experiments; TΔSb = −ΔGb° + ΔHb
Figure 4.
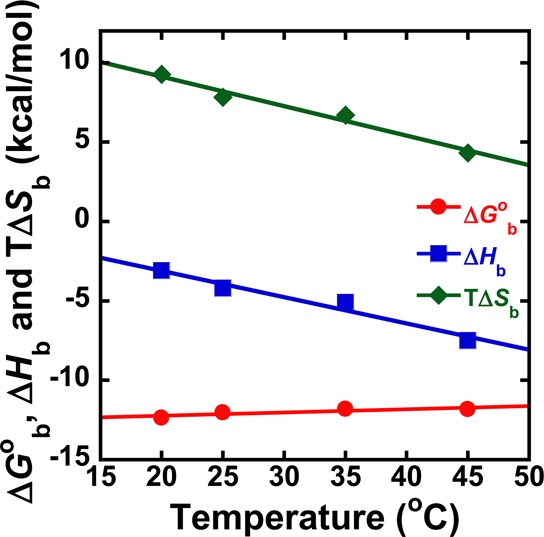
Thermodynamic results, from Table 3 for binding of DB2277 to the AAAGTTT site at different temperatures.
Isothermal Titration Calorimetry (ITC) of Complex formation
ITC experiments were conducted to develop a detailed understanding of the energetic basis for the strong and weak binding of DB2277 and DB2275 with the AAAGTTT site DNA. ITC provides a direct determination of the enthalpy, ΔHb, and allows calculation of the entropy of binding, ΔSb. In the experiments reported here, excess DNA is used in the calorimeter cell such that all added compound is fully bound to DNA and ΔHb can be directly obtained from the average binding heat per mol without any specific model for fitting. Subtracting the integrated peak areas for ligand/buffer titration from the ligand/DNA titration allows a direct determination of ΔHb (Table 3). Figure 5 shows a titration of DB2277 into AAAGTTT and AAGTT with the buffer blank correction and the ΔHb values are −4.2 ± 0.3 and −5.4 ± 0.1 kcal/mol, respectively, at 25 °C at 100 mM salt concentration. The same experimental conditions with DB2275, however, give a less exothermic enthalpy change (−1.9 ± 0.1 kcal/mol). Thus, changing the linker position at DB2275, to give DB2277, results in a significant strong and selective exothermic enthalpy that accounts for highly favorable ΔGb° for DB2277. On the basis of SPR binding free energy value, ΔGb = −RT ln K (R = 1.987 and T = 298 K), and from the ITC enthalpy value the TΔSb was calculated from TΔSb = ΔGb° + ΔHb. The binding TΔSb is highly favorable for the DB2277-DNA complex (8.8 kcal/mol). However, the DB2275–AAAGTTT complex shows a reduction in TΔSb (7.2 kcal/mol).
Determination of the Heat Capacity of the DB2277–AAAGTTT Complex
The ITC experiments of DB2277 with AAAGTTT were also carried out at different temperatures (20–45 °C) with constant 200 mM NaCl concentration (Figure 6) and the titration profile clearly indicated that enthalpy of binding strongly depends on the experimental temperature (Table 3). The heat capacity (ΔCp) of DB2277 was calculated from the slope of the linear fit of ΔHb versus temperature plot (Figure 6D) and the linear fit yields a ΔCp of −172 ± 5 cal M–1 K–1.
Effects of Salt Concentrations on Ligand-DNA Binding Enthalpy
To evaluate the relationship between experimental salt concentrations and the binding enthalpy and entropy, ITC experiments were carried out at several salt concentrations and Figure S4 shows the titration profile of DB2277 with AAAGTTT. The results show that salt concentration has a much smaller effect on ΔHb than on ΔGb° (Figure 3 and Table 2).
Mass Spectrometry: Stoichiometry, and Relative Affinity
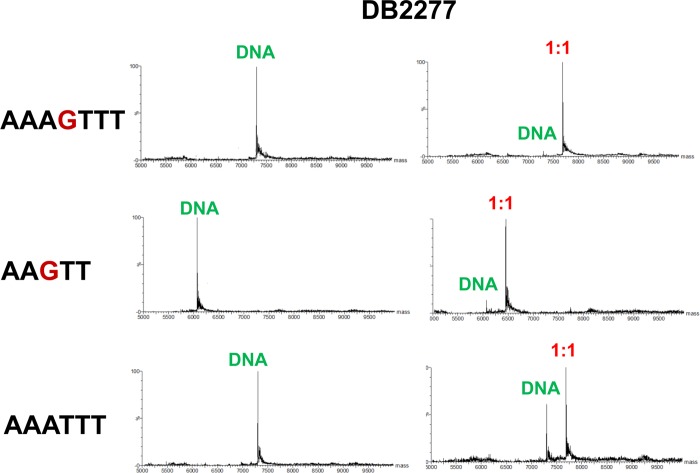
Electrospray ionization mass spectrometry (ESI-MS) experiments allow the resolution of complex mixtures and determination of stoichiometries and qualitative affinity for complexes that are present simultaneously in an injected sample.43,44 Figure 7 shows mass spectral comparisons of selected ligand–DNA complexes. DB2277 shows a very high intensity (1:1) peak for both the AAAGTTT and AAGTT sequences. However, DB2277 shows a (1:1) complex peak also for AAATTT, but up to (2:1) (results not shown) compound to DNA ratios of the original DNA peak is present, which is not observed for single G·C bp containing sequences. These results provide excellent support for the strong monomer binding of DB2277 with single G·C containing sequences.
Figure 7.
Mass spectra of DB2277 with various hairpin DNA sequences. Samples containing (1:1) (10 μM of each oligonucleotide) compound to DNA ratio at 150 mM ammonium acetate/5% methanol (v/v) buffer at pH 6.8.
Discussion
The sequence selective recognition of G·C containing mixed base pair DNA sequences, which are widely found in many critical biological sequences, by rationally designed small molecules is one of the most challenging areas of research in molecular recognition. DB2277 is the first designed, nonpolyamide minor groove binder that can selectively bind to a mixed A·T and G·C bps DNA sequence.31 Initial results showed that DB2277 has excellent sequence selectivity and strong binding affinity with sequences such as AAAGTTT. This important finding shows that rational design of compounds to recognize mixed bp sequences is quite feasible. To better understand this unique DNA–compound complex, more detailed biophysical and thermodynamic analyses were conducted. Although there are many studies of A·T specific minor groove binding, our knowledge of minor groove binders with mixed A·T/G·C bp sequences is mostly limited to polyamides.45 Detailed studies of the salt and temperature dependent thermodynamics of DB2277 binding can help fill a missing piece of fundamental information about compounds that target the minor groove of DNA. DB2277 is a paradigm for design of a new class of sequence specific DNA binding agents.
DB1476 is a control benzimidazole-diamidine compound with strong A·T selective minor-groove binding. This type of recognition is understandable given its shape match to the minor groove and its strong hydrogen bond donating ability with A·T bp through the benzimidazole and amidine moieties. Changing the benzimidazole to an azabenzimidazole core in DB2285 resulted in a decrease in A·T sequence specific binding by a factor of 175. Given the structural similarities between benzimidazole and the aza analogue, structural differences cannot explain the large decrease. The aza −N, however, can hydrogen bond with water when the compound is not bound, but the water and H-bonds are lost on binding. The aza interaction with an A·T bp is not favorable and all of these features result in a large binding decrease. At the same time the binding affinity with a single G·C bp containing sequence remains the same or slightly higher than DB1476, which is a very key observation toward the aim of selective mixed DNA sequence recognition. The azabenzimidazole group thus binds well to a G·C bp with adjacent A·T bps but it binds poorly in a pure A·T sequence. DB2285, however, is clearly not an optimum shape to target mixed sequences and we initiated synthetic efforts to prepare an optimized compound.
A significant breakthrough in DNA binding affinity and selectivity was observed on incorporation of a single −OCH2– linker between the azabenzimidazole and phenyl in DB2285 to give DB2277 (Figure 1). The flexibility, curvature of the compound, and the minor groove shape of G·C bp containing sequences allow close proximity between the azabenzimidazole-N (Ar-N) and G-NH2. DB2277 binds with AAAGTTT and AAGTT sequences as a monomer in the sub-nanomolar KD (0.3 nM) range and, very importantly, gives 80 fold selectivity over the narrower minor groove of pure A·T sequences. On changing the nucleobase from G to I, the curvature and the width of the minor groove remain almost the same46 but the absence of the G-NH2 group, which plays a very important role in the drug–DNA hydrogen bond interaction,45 causes a 30-fold drop in binding affinity for the DB2277–AAAITTT complex. Reduction in binding affinity of this complex clearly supports a model with the Ar-N of DB2277 taking part in strong H-bonding with the G-NH2 group in sequences such as AAAGTTT.
It is quite informative that a structural isomer of DB2277, DB2275, shows very poor DNA binding ability. Placing −CH2O– and −OCH2– linkers at both positions in DB2272 also results in a loss (∼100-fold) in complex stability. The optimized geometry results of these molecules (Supporting Information Figure S5) (B3LYP/6-31G* level) revealed important features about their conformation which can help explain the different DNA binding affinity. By introducing the −OCH2– flexible linker between the phenylamidine and the azabenzimidazole, DB2277, the proper curvature and shape of the compound for the minor groove recognition is obtained. The flexibility of DB2277 helps to orient the azabenzimidazole moiety close to the G-NH2 group to allow a hydrogen bond between the Ar–N of the compound and G-NH2 (Figure 8). The adjacent −O– and aza −N– may also enhance the interaction with the G-NH2. The optimized geometry of DB2275 shows that insertion of a −CH2O– linker at the 2-position of the azabenzimidazole core causes a low energy out-of-plane geometry of the phenylamidine group (Supporting Information Figure S5) This conformation lowers the DNA binding ability due to the energy cost of converting DB2275 to a more planar conformation to fit the minor groove width. In DB2272, the presence of both the −OCH2– and −CH2O– linkers at two sites of the azabenzimidazole moiety adopted the mixed conformation of DB2277 and DB2275 which explains the in-between DNA binding affinity of the molecule. Therefore, in DB2277, we have optimized the flexibility, linker position, and the curvature for selective recognition of single G·C bp containing mixed DNA sequences. Moving one terminal amidine to a meta position (DB2289) results in a dramatic decrease in binding affinity and sequence selectivity, and this indicates that not only the shape of the main scaffold but also the position, molecular curvature, and the distances between two positively charged amidine groups play crucial roles in ligand–DNA interaction.
Figure 8.
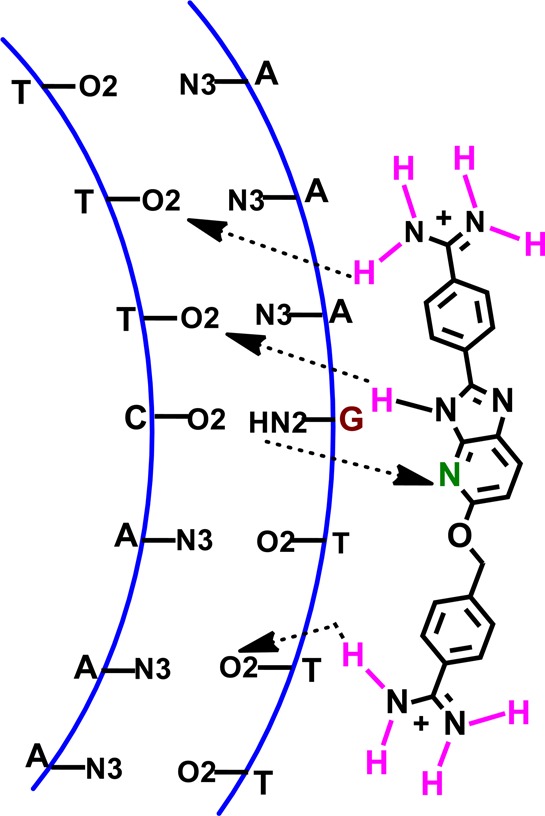
Schematic model of the A·T/G·C containing mixed DNA sequence (AAAGTTT) and azabenzimidazole diamidine (DB2277) interaction.
A very rapid association and very slow dissociation has been observed from SPR experiments for DB2277-AAAGTTT complex formation. The rapid association is in agreement with a well optimized geometry of DB2277 for a slightly wider minor groove of a single G·C bp containing sequence. The on rate in SPR for the second order reaction of DB2277 with DNA at 6 nM, a concentration significantly above the KD, for example, has an apparent half-life of 32 s while the half-life for the first order dissociation reaction is more than 230 s. This gives exceptionally strong binding with a KD of approximately 0.3 nM.
The binding affinity between small molecules and DNA depends on changes in enthalpy and entropy during the binding processes. Previously, we have determined detailed thermodynamic results mainly for heterocyclic dications which selectively bind A·T sequences.47 All of these molecules give binding enthalpies ΔHb = ∼−5 kcal/mol or less and a significant entropy component at 25 °C with an AATT sequence which has a narrow, A-tract type, minor groove geometry.47
The full thermodynamic data for binding of DB2277 and a mixed A·T/G·C bp containing sequence, AAAGTTT, are summarized in Figure 4 and Tables 2 and 3. The negative enthalpy indicates a favorable contribution to complex formation for DB2277 with AAAGTTT. The very slow dissociation and negative enthalpy change for the DB2277–AAAGTTT complex agree with strong hydrogen bond formation between the Ar-N and G-NH2. The calculated high TΔSb value (8.8 kcal/mol) indicates displacement of structured bound water molecules from the minor groove of duplex DNA during the DB2277–AAAGTTT complex formation. This is expected for the A-tract regions flanking the G·C bp.40 The complex thus has very favorable enthalpy and entropy components in binding.
Due to the dicationic nature of DB2277, the effects of electrostatic contribution or ionic strength of the medium play a crucial role on DNA–ligand complex formation. It is clear from the shapes of the binding curves and the fitting of the SPR results (Supporting Information Figure S1) that DB2277 has varying on and off rates when changing the salt concentration. The rate of association decreases and rate of dissociation increases with increasing the salt concentration. A linear dependence has been observed between the logarithm of the binding constant and the logarithm of salt concentrations (Figure 3E) and it shows that the binding affinity decreases with an increase in the ionic strength or salt concentration, as expected.40 The enthalpy of complex formation, which is primarily the result of interactions like hydrogen bond formation and van der Waals interactions, is essentially independent of salt concentration, again as expected. On the other hand, the electrostatic component of binding, which is mainly responsible for entropic change of the complex formation, changes significantly on changing the experimental salt concentration. The Gibbs free energy, the difference of sum of the binding enthalpy and the entropy (TΔSb), also changes with salt concentrations.
Changes in the experimental temperatures from 20 to 45 °C result in the binding enthalpy becoming more negative which indicates a more favorable contribution to complex formation as expected from the water loss in A-tract regions. On the contrary, the binding entropy becomes less positive with increasing temperature. With increasing temperature the highly ordered tight bound water molecules become more mobile which gives less entropy changes after these are displaced by DB2277 from the minor groove. A negative ΔCp of −172 cal M–1 K–1 has been calculated from the slope of ΔHb vs temperature plot and is in agreement with results for other similar minor groove binding dications of a similar size.40 However, the Gibbs free energy depends on the difference of the sum of the binding enthalpy and the entropy of complex formation and changes very little with temperatures.
In summary, a library of azabenzimidazole substituted molecules, based on the parent pure A·T specific ligand DB1476, has been designed with the goal of recognizing mixed DNA sequences with A·T and G·C bp. The combined experimental data established how a sequence specific minor groove binding ligand can be obtained with an azabenzimidazole core and the correct shape. DB2277 represents a major breakthrough in this process. The interactions, which give a high association and slow dissociation rate constant with an AAAGTTT sequence, make DB2277 a strong sequence specific ligand. On the other hand, the binding decreases after replacement of G by I in the minor groove of the DNA sequence and is a clear indication of a strong H-bond interaction between G-NH2 and the central azabenzimidazole moiety. The detailed thermodynamic and kinetic analyses reveal that, like other DNA minor groove binding organic small molecules, the binding enthalpy, entropy, and free energy of the DB2277–AAAGTTT complex highly depend on the experimental salt concentrations and the temperatures. Importantly this study opens a new horizon of DNA recognition by rationally designed organic small molecules. To get more detailed DNA–ligand structural information extensive 2D NMR studies for DB2277, and single G·C bp containing DNA sequences are ongoing in our laboratory and will be reported in due course. Encouraged by the excellent affinity and sequence specificity of DB2277, our research is focusing on the design of other classes of ligands which will bind more complex sequences with high specificity.
Acknowledgments
The authors thank Carol Wilson for manuscript assistance.
Glossary
Abbreviations
- bp
base pair
- SPR
surface plasmon resonance
- RU
response unit of SPR instruments
- ITC
isothermal titration calorimetry
- ESI-MS
electrospray ionization mass spectrometry
- B3LYP
Becke 3-parameter hybrid functional using Lee, Yang and Parr correlation functional
- DFT
density functional theory
Supporting Information Available
Figures S1 and S2 show SPR plots for several compounds with different DNAs and different conditions. Figure S3 shows SPR sensorgrams and global kinetic fits for DB2277 with the AAAGTTT DNA sequence at different temperatures. Figure S4 shows ITC titrations of DB2277 with the AAAGTTT hairpin DNA at different salt concentrations. Figure S5 shows DFT ab initio calculations for DB2277, 2275 and 2272. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
† Y.C.: Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100050, China.
The work at Georgia State University was supported by National Institutes of Health (NIH) grants AI064200 and GM111749 awarded to W.D.W. and D.W.B.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Tidwell R. R., and Boykin D. W. (2003) In DNA and RNA Binders: From Small Molecules to Drugs (Demeunynck M.; Bailly C.; Wilson W. D., Eds.), Vol. 2, pp 414–460, Wiley-VCH, Weinheim. [Google Scholar]
- Wilson W. D.; Tanious F. A.; Mathis A.; Tevis D.; Hall J. E.; Boykin D. W. (2008) Antiparasitic compounds that target DNA. Biochimie 90, 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouteille B.; Oukem O.; Bisser S.; Dumas M. (2003) Treatment perspectives for human African trypanosomiasis. Fundam. Clin. Pharmacol. 17, 171–181. [DOI] [PubMed] [Google Scholar]
- Lombardy R. L.; Tanious F. A.; Ramachandran K.; Tidwell R. R.; Wilson W. D. (1996) Synthesis and DNA interactions of benzimidazole dications which have activity against opportunistic infections. J. Med. Chem. 39, 1452–1462. [DOI] [PubMed] [Google Scholar]
- Brendle J. J.; Outlaw A.; Kumar A.; Boykin D. W.; Patrick D. A.; Tidwell R. R.; Werbovetz K. A. (2002) Antileishmanial activities of several classes of aromatic dications. Antimicrob. Agents Chemother. 46, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakunova S. M.; Bakunov S. A.; Patrick D. A.; Kumar S. E. V. K.; Bridges K. A.; Wenzler T.; Barszcz T.; Jones S. K.; Werbovetz K. A.; Brun R.; Tidwell R. R. (2009) Structure–activity study of pentamidine analogues as antiprotozoal agents. J. Med. Chem. 52, 2016–2035. [DOI] [PubMed] [Google Scholar]
- Thuita J. K.; Wang M. Z.; Kagira J. M.; Denton C. L.; Paine M. F.; Mdachi R. E.; Murilla G. A.; Ching S.; Boykin D. W.; Tidwell R. R.; Hall J. E.; Brun R. (2012) Pharmacology of DB844, an orally active aza analogue of pafuramidine, in a monkey model of second stage Human African Trypanosomiasis. PLoS Neglected Trop. Dis. 6(e1734), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose S.; Muleme H. M.; Onyilagha C.; Singh R.; Jia P.; Uzonna J. E. (2012) Diminazene aceturate (Berenil) modulates the host cellular and inflammatory responses to Trypanosoma congolense Infection. PLoS One 7(e48696), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W. D.; Nguyen B.; Tanious F. A.; Mathis A.; Hall J. E.; Stephens C. E.; Boykin D. W. (2005) Dications that target the DNA minor groove: Compound design and preparation, DNA interactions, cellular distribution and biological activity. Curr. Med. Chem.: Anti-Cancer Agents 5, 389–408. [DOI] [PubMed] [Google Scholar]
- Paine M. F.; Wang M. Z.; Generaux C. N.; Boykin D. W.; Wilson W. D.; De Koning H. P.; Olson C. A.; Pohlig G.; Burri C.; Brun R.; Murilla G. A.; Thuita J. K.; Barrett M. P. (2010) Diamidines for Human African Trypanosomiasis. Curr. Opin. Invest. Drugs 11, 876–883. [PubMed] [Google Scholar]
- Boykin D. W.; Kumar A.; Xiao G.; Wilson W. D.; Bender B. C.; McCurdy D. R.; Hall J. E.; Tidwell R. R. (1998) 2,5-Bis[4-(N-alkylamidino) phenyl]furans as anti-Pneumocystis carinii agents. J. Med. Chem. 41, 124–129. [DOI] [PubMed] [Google Scholar]
- Giordani F.; Munde M.; Wilson W. D.; Ismail M. A.; Kumar A.; Boykin D. W.; Barrett M. P. (2014) Green fluorescent diamidines as diagnostic probes for trypanosomes. Antimicrob. Agents Chemother. 58, 1793–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munde M.; Wang S.; Kumar A.; Stephens C. E.; Farahat A. A.; Boykin D. W.; Wilson W. D.; Poon G. M. (2014) Structure-dependent inhibition of the ETS-family transcription factor PU.1 by novel heterocyclic diamidines. Nucleic Acids Res. 42, 1379–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjunda R., and Wilson W. D. (2012) Binding to the DNA Minor Groove by Heterocyclic Dications: From AT-Specific Monomers to GC Recognition with Dimers. In Current Protocols in Nucleic Acid Chemistry, Chapter 8, Unit 8.8, Wiley, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory M.; Tidwell R. R.; Fairley T. A. (1992) Structure and DNA binding activity of analogues of 1,5-bis(4-amidinop henoxy pentane (pentamidine). J. Med. Chem. 35, 431–438. [DOI] [PubMed] [Google Scholar]
- Bakshi R. P.; Shapiro T. A. (2003) DNA topoisomerases as targets for antiprotozoal therapy. Mini Rev. Med. Chem. 3, 597–608. [DOI] [PubMed] [Google Scholar]
- Bell C. A.; Hall J. E.; Kyle D. E.; Grogl M.; Ohemeng K. A.; Allen M. A.; Tidwell R. R. (1990) Structure-activity relationships of analogs of pentamidine against Plasmodium falciparum and Leishmania mexicana amazonensis. Antimicrob. Agents Chemother. 34, 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matovu E.; Stewart M. L.; Geiser F.; Brun R.; Maser P.; Wallace L. J.; Burchmore R. J.; Enyaru J. C.; Barrett M. P.; Kaminsky R.; Seebeck T.; De Koning H. P. (2003) Mechanisms of arsenical and diamidine uptake and resistance in Trypanosoma brucei. Eukaryotic Cell 2, 1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boykin D. W.; Kumar A.; Spychala J.; Zhou M.; Lombardy R. J.; Wilson W. D.; Dykstra C. C.; Jones S. K.; Hall J. E.; Tidwell R. R.; Laughton C.; Nunn C. M.; Neidles S. (1995) Dicationic diarylfurans as anti-Pneumocystis carinii agents. J. Med. Chem. 38, 912–916. [DOI] [PubMed] [Google Scholar]
- Wenzler T.; Boykin D. W.; Ismail M. A.; Hall J. E.; Tidwell R. R.; Brun R. (2009) New treatment option for second-stage African sleeping sickness: In vitro and in vivo efficacy of aza analogs of DB289. Antimicrob. Agents Chemother. 53, 4185–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingbeil M. M.; Drew M. E.; Liu Y.; Morris J. C.; Motyka S. A.; Saxowsky T. T.; Wang Z.; Englund P. T. (2001) Unlocking the secrets of trypanosome kinetoplast DNA network replication. Protist 152, 255–262. [DOI] [PubMed] [Google Scholar]
- Roy Chowdhury A.; Bakshi R.; Wang J.; Yildirir G.; Liu B.; Pappas-Brown V.; Tolun G.; Griffith J. D.; Shapiro T. A.; Jensen R. E.; Englund P. T. (2010) The killing of African trypanosomes by ethidium bromide. PLoS Pathog. 6(e1001226), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro T. A.; Englund P. T. (1995) The structure and replication of kinetoplast DNA. Annu. Rev. Microbiol. 49, 117–143. [DOI] [PubMed] [Google Scholar]
- Meier J. L.; Yu A. S.; Korf I.; Segal D. J.; Dervan P. B. (2012) Guiding the design of synthetic DNA-binding molecules with massively parallel sequencing. J. Am. Chem. Soc. 134, 17814–17822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F.; Nickols N. G.; Li B. C.; Marinov G. K.; Said J. W.; Dervan P. B. (2013) Antitumor activity of a pyrrole-imidazole polyamide. Proc. Natl. Acad. Sci. U. S. A. 10, 1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavda S.; Liu Y.; Babu B.; Davis R.; Sielaff A.; Ruprich J.; Westrate L.; Tronrud C.; Ferguson A.; Franks A.; Tzou S.; Adkins C.; Rice T.; Mackay H.; Kluza J.; Tahir S. A.; Lin S.; Kiakos K.; Bruce C. D.; Wilson W. D.; Hartley J. A.; Lee M. (2011) Hx, a novel fluorescent, minor groove and sequence specific recognition element: design, synthesis, and DNA binding properties of p-anisylbenzimidazole-imidazole/pyrrole-containing polyamides. Biochemistry 19, 3127–3136. [DOI] [PubMed] [Google Scholar]
- He G.; Vasilieva E.; Davis G. Jr.; Koeller K. J.; Bashkin J. K.; Dupureur C. M. (2014) Binding studies of a large antiviral polyamide to a natural HPV sequence. Biochimie 102, 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Nanjunda R.; Aston K.; Bashkin J. K.; Wilson W. D. (2012) Correlation of local effects of DNA sequence and position of β-alanine inserts with polyamide-DNA complex binding affinities and kinetics. Biochemistry 51, 9796–9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando T.; Sugiyama H. (2006) Synthesis and biological properties of sequence-specific DNA-alkylating pyrrole-imidazole polyamides. Acc. Chem. Res. 39, 935–944. [DOI] [PubMed] [Google Scholar]
- Wang X.; Nagase H.; Watanabe T.; Nobusue H.; Suzuki T.; Asami Y.; Shinojima Y.; Kawashima H.; Takagi K.; Mishra R.; Igarashi J.; Kimura M.; Takayama T.; Fukuda N.; Sugiyama H. (2010) Inhibition of MMP-9 transcription and suppression of tumor metastasis by pyrrole-imidazole polyamide. Cancer Sci. 101, 759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y.; Paul A.; Rettig M.; Wilson W. D.; Boykin D. W. (2014) Design and synthesis of heterocyclic cations for specific DNA recognition: from AT-rich to mixed-base-pair DNA sequences. J. Org. Chem. 79, 852–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farahat A. A.; Paliakova E.; Kumar A.; Barghashb A-E. M.; Goda F. E.; Eisa H. M.; Wenzler T.; Brun R.; Liu Y.; Wilson W. D.; Boykin D. W. (2011) Exploration of larger central ring linkers in furamidine analogues: synthesis and evaluation of their DNA binding, antiparasitic and fluorescence properties. Bioorg. Med. Chem. 19, 2156–2167. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Chai Y.; Kumar A.; Tidwell R. R.; Boykin D. W.; Wilson W. D. (2012) Designed compounds for recognition of 10 base pairs of DNA with two AT binding sites. J. Am. Chem. Soc. 134, 5290–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen B.; Tanious F. A.; Wilson W. D. (2007) Biosensor-surface plasmon resonance: Quantitative analysis of small molecule–nucleic acid interactions. Methods 42, 150–161. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Wilson W. D. (2012) Quantitative analysis of small molecule-nucleic acid interactions with a biosensor surface and surface plasmon resonance detection. Methods Mol. Biol. 613, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjunda R., Munde M., Liu Y., and Wilson W. D. (2011) In Methods for Studying DNA/drug Interactions (Wanunu M., and Tor Y., Eds.), Chapter 4, CRC Press-Taylor & Francis Group: Boca Raton, FL. [Google Scholar]
- DeHaseth P. L.; Lohman T. M.; Record M. T. Jr. (1977) Nonspecific interaction of iac repressor with DNA: An association reaction driven by counterion release. Biochemistry 16, 4783–4790. [DOI] [PubMed] [Google Scholar]
- Wilson W. D.; Krishnamoorthy C. R.; Wang Y. H.; Smith J. C. (1985) Mechanism of intercalation: Ion effects on the equilibrium and kinetic constants for the interaction of propidium and ethidium with DNA. Biopolymers 24, 1941–1961. [DOI] [PubMed] [Google Scholar]
- Lohman T. M.; deHaseth P. L.; Record M. T. Jr. (1978) Analysis of ion concentration effects of the kinetics of protein-nucleic acid interactions. Application to lac repressor-operator interactions. Biophys. Chem. 8, 281–294. [DOI] [PubMed] [Google Scholar]
- Wang S.; Kumar A.; Aston K.; Nguyen B.; Bashkin J. K.; Boykin D. W.; Wilson W. D. (2013) Different thermodynamic signatures for DNA minor groove binding with changes in salt concentration and temperature. Chem. Commun. 49, 8543–8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim N.; Lamichhane R.; Zhao R.; Banerjee T.; Philip J.; Rueda D.; Feig A. L. (2012) Thermodynamic and kinetic analysis of an RNA kissing interaction and its resolution into an extended duplex. Biophys. J. 102, 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalov P. L.; Dragan A. I.; Crane-Robinson C. (2011) Interpreting protein/DNA interactions: Distinguishing specific from non-specific and electrostatic from non-electrostatic components. Nucleic Acids Res. 39, 2483–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu F.; De Pauw E.; Gabelica V. (2008) Electrospray mass spectrometry to study drug-nucleic acids interactions. Biochimie 90, 1074–1087. [DOI] [PubMed] [Google Scholar]
- Lombardo C. M.; Martínez I. S.; Haider S.; Gabelica S. V.; Pauw E. D.; Moses J. E.; Neidle S. (2010) Structure-based design of selective high-affinity telomeric quadruplex-binding ligands. Chem. Commun. 46, 9116–9118. [DOI] [PubMed] [Google Scholar]
- Dervan P. B.; Poulin-kerstien A. T.; Fechter E.; Edelson B. S. (2005) Regulation of gene expression by synthetic DNA-binding ligands. Top Curr. Chem. 253, 1–31. [Google Scholar]
- Xuan J.-C.; Weber I. T. (1992) Crystal structure of a B-DNA dodecamer containing inosine, d(CGCIAATTCGCG), at 2.4 Å resolution and its comparison with other B-DNA dodecamers. Nucleic Acids Res. 20, 5457–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munde M.; Lee M.; Neidle S.; Arafa R.; Boykin D. W.; Liu Y.; Bailly C.; Wilson W. D. (2007) Induced fit conformational changes of a “reversed amidine” heterocycle: optimized interactions in a DNA minor groove complex. J. Am. Chem. Soc. 129, 5688–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.