Abstract
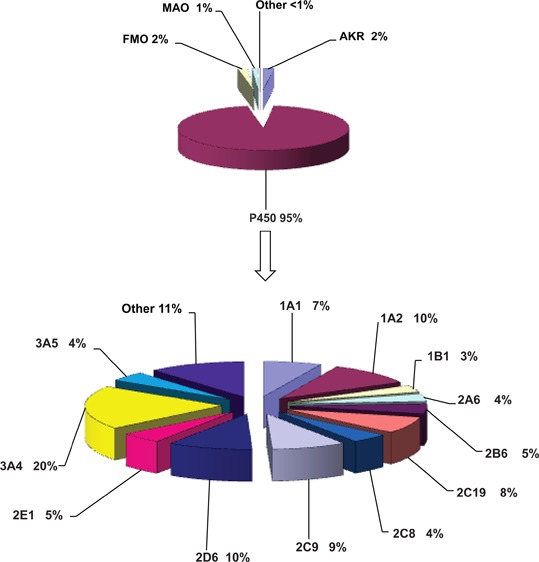
Analyzing the literature resources used in our previous reports, we calculated the fractions of the oxidoreductase enzymes FMO (microsomal flavin-containing monooxygenase), AKR (aldo-keto reductase), MAO (monoamine oxidase), and cytochrome P450 participating in metabolic reactions. The calculations show that the fractions of P450s involved in the metabolism of all chemicals (general chemicals, natural, and physiological compounds, and drugs) are rather consistent in the findings that >90% of enzymatic reactions are catalyzed by P450s. Regarding drug metabolism, three-fourths of the human P450 reactions can be accounted for by a set of five P450s: 1A2, 2C9, 2C19, 2D6, and 3A4, and the largest fraction of the P450 reactions is catalyzed by P450 3A enzymes. P450 3A4 participation in metabolic reactions of drugs varied from 13% for general chemicals to 27% for drugs.
1. Introduction
In our previous report,1 we analyzed the literature reporting the metabolism of carcinogens divided into the groups of general chemicals (environmental/industrial), drugs, and natural/physiological compounds. The results showed a dominant role for cytochrome P450 (P450) in the activation of chemicals, especially the three Family 1 P450 enzymes (1A1, 1A2, and 1B1) and P450s 2A6, 2E1, and 3A4. The aldo-keto reductase (AKR) enzymes were also highly represented. In the metabolic activation of the potential carcinogens, six P450s, 1A1, 1A2, 1B1, 2A6, 2E1, and 3A4, account for 77% of the reported activations. In the present review, analyzing the literature resources used in our previous reports,1−3 we calculated the participation and the fractions of the oxidoreductase enzymes FMO (microsomal flavin-containing monooxygenase), AKR (aldo-keto reductase), and MAO (monoamine oxidase), and P450s involved in metabolic reactions, as reported in the literature. Typical reactions catalyzed by these enzymes are
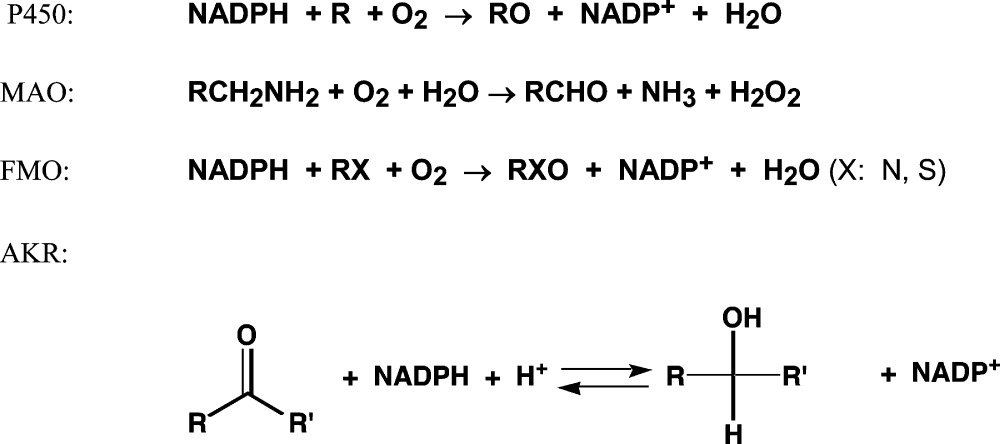 |
where R denotes part of the molecule.
We compared the results with those obtained when fractions of the enzymes participating in metabolic activation was considered.1
To our knowledge, there has not been an effort to categorize in detail the human P450 enzymes involved in the metabolism of all chemicals, to compare the results with the fraction of the enzymes involved in the metabolism of the chemicals as divided into subgroups such as general chemicals (environmental/industrial chemicals), natural and physiological compounds, and drugs (as divided into marketed drugs and new chemical entities or drug candidates), taking into account also the major and minor metabolic reactions of each particular chemical or drug. We thought that this would be a useful exercise in light of continuing scientific interest in the metabolism of chemicals, drugs, and natural products in relation to drug–general chemical, drug–natural compound, and general chemicals–natural compound interactions and the possible clinical consequences of such interactions.
2. Approach
The analysis of the literature data was performed by calculating the number of reactions in which a particular chemical interacted with a specific enzyme. Furthermore, the reactions were additionally divided into major and minor reactions based on the kinetic values and/or description of the role of the reaction in the overall metabolism of a particular chemical as reported in the literature resources.1−3 In addition to the reactions in which parent compounds participated as substrates, reactions of metabolites were also counted when they participate as substrates in particular reactions. Searches were done using the PubMed database, accessing the MEDLINE database of references and abstracts. In the latter stages, the existing literature and the original papers were systematically analyzed, extracting those data related to the metabolism of chemicals by subdividing the compounds into three groups: “general” chemicals, drugs, and physiological (i.e., natural) compounds. This is a rather qualitative evaluation, and the reader is referred to a more comprehensive list in the Supporting Information. The literature sources used were original, as well as review papers covering the publishing period from the years 1976–2008. In addition to the cited references, the literature sources, as well as the list of compounds and the reactions taken into account for calculations, are presented in the (Supporting Information) or can be searched by using the Web searchable ADME database.4
The following limits were used when numerical values of the kinetic parameters from the literature were interpreted as “low” for minor reactions: kcat < 1 min–1, Km > 2 μM, and catalytic efficiency (kcat/Km) < 0.02 min–1 μM–1.3
3. FMO, AKR, and MAO Enzymes
Participation of human oxidoreductases and cytochrome P450 enzymes in the metabolism of chemicals as a general group and divided into separate subgroups (i.e., drugs, physiological compounds, and general chemicals, environmental/industrial chemicals) is presented in Figures 1–4 or Table 1. As anticipated, the results (Figures 1–4) show that 92 to 96% of metabolic oxidation–reduction reactions of all chemicals are catalyzed by P450s. When the groups of chemicals were analyzed, the results showed the highest value for participation of P450 enzymes (∼96%, Figure 3A) in the metabolism of drugs, and the lowest value (∼92%, Figure 2A) was calculated for general chemicals as substrates. An intermediate value (∼94%) for participation of the P450 enzymes was calculated for natural and physiological compounds as substrates (Figure 4A). FMO, AKR, and MAO enzymes collectively participate in the metabolism of all chemicals to the extent of ∼5%. When general chemicals as a separate group are taken as substrates, FMO and AKR enzymes participate at equally low extents (∼3%, Figure 2A). However, the representation of FMO enzymes is higher when drugs were analyzed as substrates (∼2%, Figure 3A). Taking natural and physiological compounds into calculation (Figure 4A), AKR enzymes predominate (∼4%) over FMO and MAO enzymes (∼1% each). However, it should be emphasized that the numbers presented in this work are estimates and cannot be precise, in that they are influenced by a number of different factors such as quality of the research done, conclusions and interpretations by authors (of the articles in the database), and by the subjective approach of the collector and interpreters of the published results. Therefore, the small differences in percentages, although minor, may exist, but our reuslts are supported by the data used for the calculations.
Figure 1.
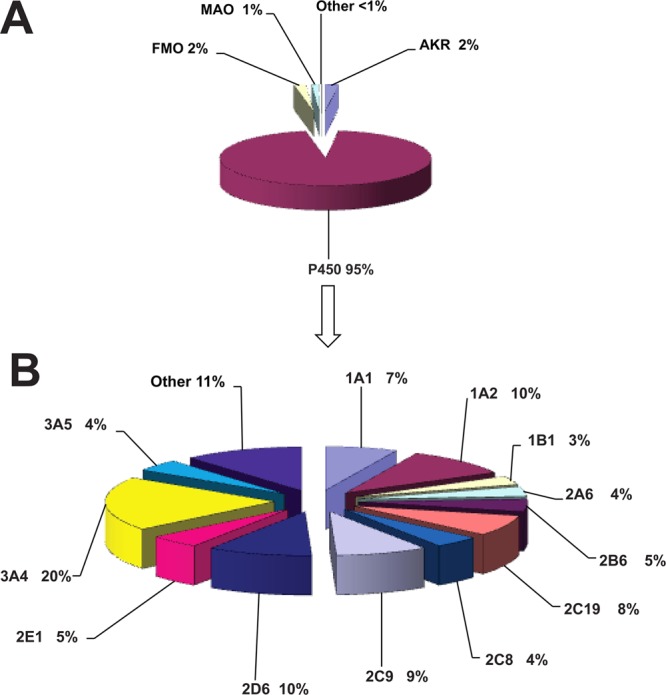
(A) Human oxidoreductases participating in the metabolism of all chemicals. n = 8320 reactions; 1829 compounds used in calculations. (B) Human P450s participating in the metabolism of all chemicals (drugs, physiological compounds, and general chemicals), n = 7906 data reactions; 1829 compounds used in calculations.
Figure 4.
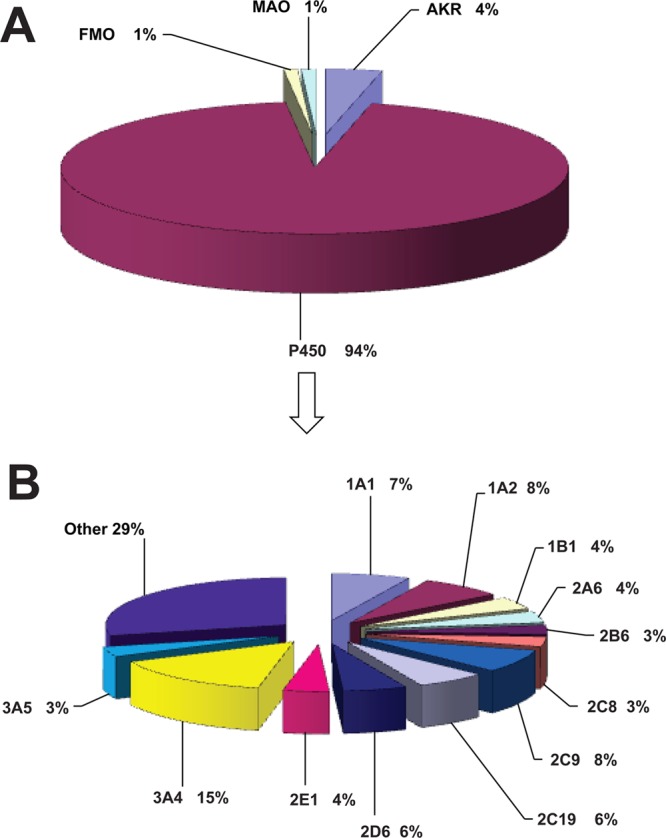
(A) Human oxidoreductases participating in the metabolism of natural and physiological compounds. n = 1693 reactions; 331 compounds used in calculations. (B) Human P450s participating in the metabolism of natural/physiological compounds. n = 1601 reactions; 331 compounds used in calculations.
Table 1. Summary of Percentages of Metabolism Attributed to Enzymesa.
| enzymes | all chemicals | general chemicals | natural and physio- logical chemicals | drugs | marketed drugs | drugs in development |
|---|---|---|---|---|---|---|
| FMO | 2 | 3 | 1 | 2 | ||
| MAO | 1 | 2 | 1 | 1 | ||
| AKR | 2 | 3 | 4 | 1 | ||
| other (non-P450) | <1 | <1 | <1 | <1 | ||
| P450s | ||||||
| 1A1 | 7 | 11 | 7 | 5 | 4 | 4 |
| 1A2 | 10 | 15 | 8 | 9 | 8 | 8 |
| 1B1 | 3 | 6 | 4 | 1 | 1 | 1 |
| 2A6 | 4 | 5 | 4 | 2 | 3 | 2 |
| 2B6 | 5 | 6 | 3 | 4 | 5 | 4 |
| 2C8 | 8 | 3 | 3 | 5 | 6 | 5 |
| 2C9 | 4 | 7 | 8 | 10 | 10 | 10 |
| 2C19 | 9 | 6 | 6 | 9 | 9 | 8 |
| 2D6 | 10 | 8 | 6 | 13 | 14 | 12 |
| 2E1 | 5 | 8 | 4 | 3 | 4 | 3 |
| 3A4 | 20 | 13 | 15 | 27 | 22 | 29 |
| 3A5 | 4 | 2 | 3 | 6 | 6 | 6 |
| other P450s | 11 | 10 | 29 | 6 | 8 | 8 |
Figure 3.
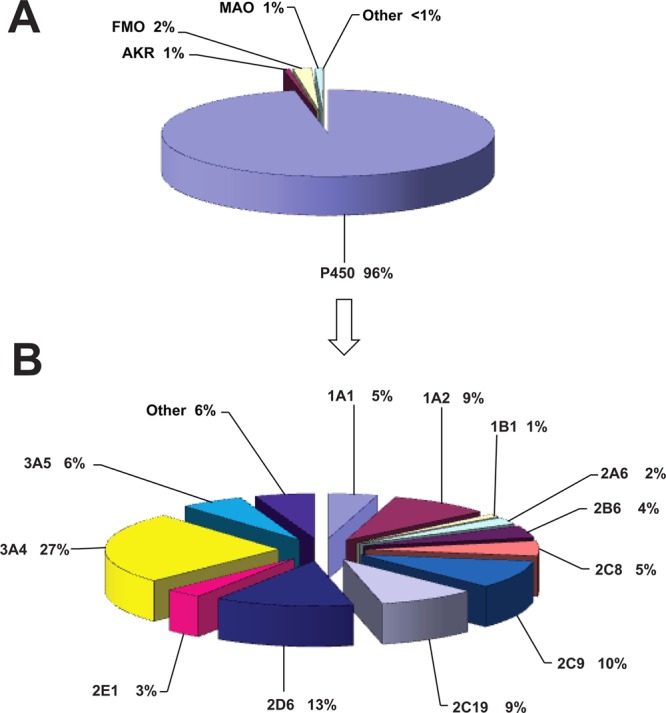
(A) Human oxidoreductases participating in the metabolism of drugs (calculation for drugs under development and marketed drugs). n = 4192 reactions; 860 drugs used in calculations. (B) Human P450 enzymes in the metabolism of drugs (data calculated for minor and major reactions, drugs under development, and marketed drugs). n = 4058 reactions; 860 drugs used in calculations.
Figure 2.
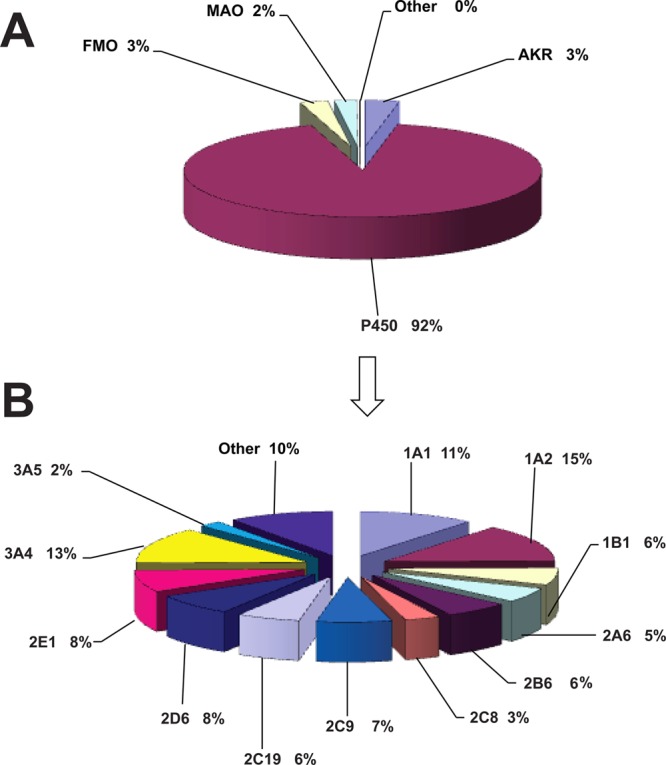
(A) Human oxidoreductases participating in metabolism of general chemicals. n = 2437 reactions; 638 compounds used in calculations. (B) Human P450s participating in metabolism of general chemicals. n = 2248 reactions; 638 compounds used in calculations.
4. P450s
Two prominent analyses of the literature5,6 show that fractions of P450s involved in the metabolic clearance of the 200 most prescribed drugs are rather consistent in the findings that (i) ∼75% of drugs are metabolically cleared mainly by P450s, (ii) ∼90% of the P450 reactions can be accounted for by a set of five P450s, 1A2, 2C9, 2C19, 2D6, and 3A4, and that (iii) the largest fraction (∼46%) of the P450 reactions is catalyzed by P450 3A enzymes.
Our analysis showed the following: participation of P450s in metabolism when data for all chemicals (drugs, physiological compounds, and general chemicals) were analyzed showed, as expected, that P450 3A4 is a major enzyme (20%), followed by P450 2D6 and P450 1A2 (10% each), P450 2C9 (9%), P450 2C19 (8%), and P450 1A1 (7%) (Figure 1B and Table 1). Minor contributions were seen for P450 2E1 (5%), P450s 3A5, 2A6, and 2C8 (4% each), and P450 1B1 (3%), while others of the set of 57 human P450s contributed an additional 11% of the metabolic reactions. Interestingly, the enzymes that are responsible for activation of chemicals to toxic products, such as P450s 1B1, 2A6, and 2E1,1 contributed to the metabolism of all compounds only to a minor extent.
Analysis of P450s in the metabolism of general chemicals (environmental/industrial) revealed that the major participating P450 enzymes were 1A2 (15%), 3A4 (13%), P450 1A1 (11%), 2D6 and 2E1 (8% each), 2C9 (7%), 2C19, and 1B1 and P450 2B6 (6% each) (Figure 2B and Table 1). Minor contributions were calculated for P450s 2A6 (5%), 2C8 (3%), and 3A5 (2%). Other P450 enzymes contributed a total of 10% to the metabolic reactions. These results are in good agreement with the results on the participation of the major enzymes in activation of potential toxicants (i.e., P450s 1A1, 1A2, 1B1, 2A6, 2E1, and 3A4).1
Participation of P450s in metabolism, using the data for drugs under development and marketed drugs, showed that the major enzymes are P450s 3A4 (27%), 2D6 (13%), 2C9 (10%), 2C19 (9%), and 1A2 (9%) (Figure 3B and Table 1). Minor contributions were seen for P450s 3A5 (6%), 1A1 and 2C8 (5% each), 2B6 (4%), 2E1 (3%), 2A6 (2%), and 1B1 (1%). All other human P450s contributed a total of 6% to the metabolic reactions.
Analysis of P450s in metabolism for natural and physiological compounds gave the following results: the major enzymes were P450s 3A4 (15%), 2C9 (8%), 1A2 (8%), 1A1 (7%), and 2C19 and 2D6 (6% each) (Figure 4B and Table 1). Minor contributions were seen for P450s 1B1, 2E1, and 2A6 (4% each), and 2B6, 2C8, and 3A5 (3% each). Other P450s contributed a total of 29% to the metabolic reactions. Also in this case, the percentages we calculated might be influenced by factors mentioned before. In addition (and somewhat surprisingly), drugs are not metabolized to a great extent by AKR enzymes compared to that of the other groups of compounds investigated.
To further analyze the participation of P450 enzymes in the metabolism of drugs, calculations were performed using data only for drugs which are on the market and data for both the drugs on the market and drugs under development, selecting also for the data for major reactions (Table 1). The results were very similar to those presented in Figure 3B, with minor differences. Calculated data show again that the major participating enzymes are P450s 3A4 (22 and 29%, marketed drugs and drugs under development, respectively), 2D6 (14% and 12%, marketed drugs and drugs under development, respectively), 2C9 (10%, both marketed drugs and drugs under development,), 2C19 (9% and 8%, marketed drugs and drugs under development, respectively), and 1A2 (8%, marketed drugs and drugs under development) (Table 1). These data correspond well to those presented in Figure 3B and show that the method of calculation did not influence the results in a major way.
5. Discussion
This is an effort to add to our work on chemical carcinogens in 2012.1 The most commonly cited references on the topic are papers by Williams et al.5 and Wienkers et al.,6 both of which are from pharmaceutical industry authors and deal with drugs. Ten years later, we note that the general conclusions are still valid, with some possible changes. We have expanded the effort to include other chemicals.
At the outset, we should discuss some caveats about interpretation of the results. First, we emphasize that this analysis is restricted to oxidation and reduction (Figures 1–4). In previous analysis with drugs,5 UDP-glucuronosyl transferases and esterases were the major non-450 enzymes involved in metabolism, together accounting for most of the 25% of drug metabolism outside of the P450s.
The second major caveat is that the percentages in the graphs (Figures 1–4) and Table 1 reflect the groups of compounds that have been studied the most. As we pointed out in our work with chemical carcinogens,1 the fact that many polycyclic aromatic hydrocarbons have been studied extensively skews the charts in favor of Family 1 P450s. Likewise, the same point can be made about some P450s oxidizing members of closely related families of drugs or natural products.
The third caveat is that including a report of the involvement of a particular enzyme in the oxidation of a compound does not necessarily reflect the extent of the contribution of that enzyme to the reaction/metabolism. For example, if one enzyme contributed 20% to the clearance of a chemical and another 60%, both might be included. In many cases, the exact contributions are not known, and there is no good way to circumvent this issue in arriving at simple conclusions.
The fourth caveat deals with enzyme overlap. The most apparent case is with P450s 3A4 and 3A5. Many reactions catalyzed by P450 3A4 are also catalyzed by P450 3A5, although generally (but not always) at lower rates. Therefore, the contributions of P450 3A5 presented here may be misleading in terms of the importance of that enzyme.
Finally, the fractions in Figures 1–4 may change as orphan enzymes (P450s)7 are characterized. We cannot yet approximate how much of an effect there will be.
Comparisons may be made with previous percentages, at least for drugs.5,6 The main five P450s are still 1A2, 2C9, 2C19, 2D6, and 3A4. Collectively, these five P450s accounted for 74% of the P450 metabolism of drugs (when P450 3A5 is included along with 3A4). It is somewhat surprising that P450s 2C19 and 2D6 still account for such large fractions of drug metabolism (8 and 12%, respectively, Figure 3B and Table 1), in spite of bias against these in development (due to their extensive polymorphism).
Several reports indicate that several steroid metabolizing P450s can bind to and oxidize drug molecules.8−12 These results are unexpected, but the fraction of all drugs metabolized by “other P450s” is still relatively low (8%, Table 1).
In this and our recent review of enzymes involved in carcinogen metabolism,1 we have tried to catalog which enzymes have major roles in chemical oxidations. We conclude those P450s are the major ones for all chemicals. All of the P450s are of interest for a variety of reasons. We present this information as a guide to promote interest in these and to consider priorities in terms of analysis of the enzymes involved in metabolism of chemicals.
Glossary
Abbreviations
- AKR
aldo-keto redctuase
- FMO
microsomal flavin-containing monooxygenase
- MAO
monoamine oxidase
Supporting Information Available
MS Word and Excel databases of chemicals analyzed in this review (searchable). This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported in part by National Institutes of Health Grants R37 CA090426, R01 ES010546, and R01 GM103937 (to F.P.G.).
The authors declare no competing financial interest.
Dedication
This review is dedicated to the memory of Vjeka Rendic who passed away recently. Vjeka, the wife of one of the coauthors of this review (Professor S. Rendic), was of great support to him during his long term academic, research, and publishing activity in the field of P450s.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Rendic S.; Guengerich F. P. (2012) Contributions of human enzymes in carcinogen metabolism. Chem. Res. Toxicol. 25, 1316–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendic S. (2002) Summary of information on human CYP enzymes: Human P450 metabolism data. Drug Metab. Rev. 34, 83–448. [DOI] [PubMed] [Google Scholar]
- Rendic S.; Di Carlo F. J. (1997) Human cytochrome P450 enzymes: a status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab. Rev. 29, 413–580. [DOI] [PubMed] [Google Scholar]
- ADME Database: WEB Searchable ADME Database of Substrates, Inhibitors, Inducers and Activators of Cytochrome P450, Other Enzymes and Drug Transporters, Fujitsu Kyushu System Ltd., Fukuoka, Japan, http://jp.fujitsu.com/group/kyushu/en/services/admedatabase/. [Google Scholar]
- Williams J. A.; Hyland R.; Jones B. C.; Smith D. A.; Hurst S.; Goosen T. C.; Peterkin V.; Koup J. R.; Ball S. E. (2004) Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab. Dispos. 32, 1201–1208. [DOI] [PubMed] [Google Scholar]
- Wienkers L. C.; Heath T. G. (2005) Predicting in vivo drug interactions from in vitro drug discovery data. Nat. Rev. Drug Discovery 4, 825–833. [DOI] [PubMed] [Google Scholar]
- Guengerich F. P.; Cheng Q. (2011) Orphans in the human cytochrome P450 superfamily: Approaches to discovering functions and relevance in pharmacology. Pharmacol. Rev. 63, 684–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.; Flint O.; Wang L.; Gupta A.; Westhouse R. A.; Zhao W.; Raghavan N.; Caceres-Cortes J.; Marathe P.; Shen G.; Zhang Y.; Allentoff A.; Josephs J.; Gan J.; Borzilleri R.; Humphreys W. G. (2012) Cytochrome P450 11A1 bioactivation of a kinase inhibitor in rats: use of radioprofiling, modulation of metabolism, and adrenocortical cell lines to evaluate adrenal toxicity. Chem. Res. Toxicol. 25, 556–571. [DOI] [PubMed] [Google Scholar]
- Mast N.; Li Y.; Linger M.; Clark M.; Wiseman J.; Pikuleva I. A. (2014) Pharmacologic stimulation of cytochrome P450 46A1 and cerebral cholesterol turnover in mice. J. Biol. Chem. 289, 3529–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafaati M.; Mast N.; Beck O.; Nayef R.; Heo G. Y.; Björkhem-Bergman L.; Lutjohann D.; Björkhem I.; Pikuleva I. A. (2010) The antifungal drug voriconazole is an efficient inhibitor of brain cholesterol 24S-hydroxylase in vitro and in vivo. J. Lipid Res. 51, 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast N.; Zheng W.; Stout C. D.; Pikuleva I. A. (2013) Binding of a cyano- and fluoro-containing drug bicalutamide to cytochrome P450 46A1: Unusual features and spectral response. J. Biol. Chem. 288, 4613–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast N.; Norcross R.; Andersson U.; Shou M.; Nakayama K.; Björkhem I.; Pikuleva I. A. (2003) Broad substrate specificity of human cytochrome P450 46A1 which initiates cholesterol degradation in the brain. Biochemistry 42, 14284–14292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


