Correlative gene expression analysis of two putative mouse “immunogenicity” genes, ZG16 and HORMAD1, was used to assay their human homologous expression levels in human pluripotent stem cells and their derivatives. We found that ZG16 expression is heterogeneous across multiple human embryonic stem cell and human induced pluripotent stem cell-derived cell types. Neither of the previous immunogenicity-associated genes in the mouse currently appears to be relevant in a human context.
Keywords: Human induced pluripotent stem cell, Human embryonic stem cell, Immunogenicity, Peripheral blood mononuclear cells, HORMAD1, ZG16
Abstract
Autologous human induced pluripotent stem cells (hiPSCs) should allow cellular therapeutics without an associated immune response. This concept has been controversial since the original report that syngeneic mouse iPSCs elicited an immune response after transplantation. However, an investigative analysis of any potential acute immune responses in hiPSCs and their derivatives has yet to be conducted. In the present study, we used correlative gene expression analysis of two putative mouse “immunogenicity” genes, ZG16 and HORMAD1, to assay their human homologous expression levels in human pluripotent stem cells and their derivatives. We found that ZG16 expression is heterogeneous across multiple human embryonic stem cell and hiPSC-derived cell types. Additionally, ectopic expression of ZG16 in antigen-presenting cells is insufficient to trigger a detectable response in a peripheral blood mononuclear cell coculture assay. Neither of the previous immunogenicity-associated genes in the mouse currently appears to be relevant in a human context.
Introduction
Human induced pluripotent stem cells (hiPSCs) provide a viable alternative to human embryonic stem cells (hESCs) as an autologous stem cell source, ideally eliciting no detectable immune response in a transplant setting [1]. Multiple groups, however, have provided evidence of differences manifested in the transcriptome, epigenome, genomic imprinting, somatic mutations, and differentiation efficiencies between ESCs and iPSCs [2–12]. Furthermore, it was reported that syngeneic transplanted mouse iPSCs, but not ESCs, were capable of eliciting an immune response on teratoma formation [13]. Follow-up studies in the mouse have produced variable and contradictory results [14, 15]. Only one group has investigated the immunogenicity of hPSCs, although differences in HORMAD1 and ZG16 between lines have suggested no significant variation that would manifest as a possible immunogenic difference between hiPSCs and hESCs [16]. Expanding on that study, we present a comprehensive analysis of the expression of the human homologs of mouse HORMAD1 and ZG16 in both undifferentiated hPSCs and varying degrees of differentiated derivatives. We also used a modified peripheral blood mononuclear cell (PBMC) coculture assay to test for an in vitro-mediated acute immune response.
Materials and Methods
Ethics Statement
Written approval and informed consent regarding human skin biopsy procedures and human fibroblast derivation, culture, and experimental use are detailed elsewhere [17].
Tissue Culture Maintenance of Primary Human Skin Cells
The human skin-derived primary cell line used in our study was derived and cultured as previously described [17]. Additionally, two other fibroblast lines, MGM2 and LAVIV (azficel-T, part no. DR01; Fibrocell Science, Exton, PA, http://www.fibrocellscience.com), used in the present study are detailed as previously described [17]. LAVIV adult human skin-derived dermal fibroblasts were obtained from a 4-mm skin punch biopsy, as described in the Isolagen Standardized Manufacturing Process EX-GTR-110, version 00 (Fibrocell Science). All three fibroblast lines were cultured in standard fibroblast media conditions, as detailed previously [17]. In brief, the fibroblast lines were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) nutrient mixture/F-12 (DMEM/F-12) supplemented with fetal bovine serum (FBS), 1× nonessential amino acids, 1× GlutaMAX, and 100 IU/ml penicillin-streptomycin (Invitrogen/Gibco, Carlsbad, CA, http://www.invitrogen.com) and maintained in a 37°C in a 5% CO2 incubator. Regular passaging with 0.05% trypsin (Invitrogen) and banking was done in standard fibroblast medium supplemented with 10% dimethyl sulfoxide (Fisher Scientific, Fair Lawn, NJ, http://www.thermofisher.com).
In Vitro Culture of Stem Cell Lines
Human embryonic stem cell (hESC) lines 1 and 9 were procured from WiCell Research Institute (Madison, WI, http://www.wicell.org). UCLA embryonic stem cell lines 2, 3, and 6 were procured from the Eli and Edythe Broad Stem Cell Research Center, Stem Cell Core, University of California, Los Angeles (UCLA) (Los Angeles, CA, http://www.stemcell.ucla.edu). hESC lines 1, 2, 3, 6, and 9 are hereafter referred to as ES1 through ES5, respectively. Multiple integration iPSCs were derived as previously reported [18]. mRNA, adult pre- and postexcision hiPSCs, and MGM 2.19, 6.7, and 13.1.0 hiPSCs were derived from patient-derived fibroblasts using standard skin biopsy procedures. hiPSCs were derived using the stem cell cassette, lentiviral-based reprogramming method [17, 19, 20]. The pre- and postexcised hiPSCs (genetically identical lines) are hereafter referred to as iPS1 and iPS2, respectively. The mRNA-derived line is hereafter referred to as iPS3. MGM 2.19, 6.7, and 13.1.0 are hereafter referred to as iPS4, iPS5, and iPS6, respectively. The multiple integration line is hereafter referred to as iPS7. All hESC lines were originally plated on mouse embryonic fibroblasts and maintained in hESC media as previously described [17]. The colonies were subsequently passaged into feeder-free conditions using an 18-gauge needle (Fisher Scientific) onto reduced growth factor Matrigel (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com). All further stem cell culture in feeder-free conditions was performed as previously published for all hESC and hiPSC lines [17]. In brief, all stem cells, once converted to feeder-free conditions consisting of Matrigel as a substrate, used a 50:50 blend of Nutristem (Stemgent, San Diego, CA, http://www.stemgent.com) and mTeSR1 medium (STEMCELL Technologies Inc., Vancouver, BC, Canada; http://stemcell.com). The cells were regularly passaged with either an 18-gauge needle or a StemPro EZPassage Tool (Life Technologies, Carlsbad, CA, http://www.lifetechnologies.com) every 4–5 days.
Teratoma Formation
One 10-cm dish of each individual stem cell line was grown to 95% confluence, the cells were removed in clumps with a 25-ml serological pipette, and the plate was rinsed with DMEM/F-12 (Invitrogen and Gibco). The cells were spun down at 200g for 5 minutes and resuspended in ice-cold Matrigel diluted at 1:2 in DMEM to a total volume of 50 µl. Each 10-cm dish was split into two (e.g., 7.5 million cells per injection site). For the testicular injections, both testes in a severe combined immunodeficient (SCID) adult male beige mouse were injected with 50 µl of the cell/Matrigel slurry. For subcutaneous injections, 7.5 million cells were injected into the subcutaneous space in each hind leg of the SCID beige mice. For both testicular and subcutaneous injections, the mice were anesthetized; this was used for the nonsurgical subcutaneous injections to ensure the cells were not immediately dispersed on movement and an adequate interval for Matrigel solidification could occur. The teratomas were harvested at 7 weeks for both testicular and subcutaneous teratomas by surgery. Immediately, one half of the teratoma was sectioned with a scalpel into 10 pieces and placed into RNAlater buffer (Qiagen, Valencia, CA, http://www.qiagen.com). The other half of the teratomas were fixed in 4% formaldehyde, and the sections were embedded in paraffin and stained with hematoxylin and eosin for histological analysis at the UCLA Translational Pathology Laboratory. All animal experiments were performed in accordance with the UCLA Animal Research Committee and Division of Laboratory Animal Medicine.
Nondirected Embryoid Body Differentiation
Embryoid bodies (EBs) were made by taking 95% confluent 10-cm dishes of hESCs or hiPSCs and washing them once with 1× phosphate-buffered saline (PBS) (Invitrogen). This was followed by incubation for 5 minutes with StemPro Accutase (Life Technologies) to form a single cell suspension. The plate was rinsed two times with nonsupplemented DMEM/F-12 and spun down at 300g for 5 minutes. This pellet was then resuspended in AggreWell Medium (STEMCELL Technologies) with rho-associated protein kinase inhibitor. Different EB sizes were created by changing the number of input hES or hiPS single cells from 100 cells per EB, 1,000 cells per EB, and 10,000 cells per EB into AggreWell 400 or AggreWell 800 plates (STEMCELL Technologies). The EBs were placed in ultra-low attachment multiwall plates (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) for 24 hours and then underwent one media change with AggreWell medium (STEMCELL Technologies) for 24 hours and then plated onto 0.2%-coated gelatin wells in a 6-well plate in standard fibroblast-containing media until harvested at the designated points. The media were changed every 2 days for the duration of the experiment.
Directed Trilineage Differentiation
Directed differentiation into mesoderm was performed as previously published [21]. In brief, hESCs and hiPSCs were routinely passaged at high confluence onto Matrigel (BD Biosciences) with daily media changes. After 48 hours, the stem cell media were replaced with basal differentiation media (STEMdiff APEL, STEMCELL Technologies) supplemented with 5-µM GSKi (CHIR99021; Stemgent) for 24 hours and further differentiated in APEL media supplemented with 25 ng/ml human recombinant bone morphogenetic protein 4 (PeproTech, Rocky Hill, NJ, http://www.peprotech.com) for 24 hours. The hESCs and hiPSCs were differentiated into ectoderm by following the manufacture’s protocol (STEMCELL Technologies). In brief, the stem cell colonies were made into a single cell suspension as detailed previously using StemPro Accutase (Life Technologies). The cells were plated onto Matrigel-coated plates or glass cover slips (BD Biosciences) overnight and then rinsed with DMEM/F-12. Appropriate volumes of STEMdiff Neural Induction Media (STEMCELL Technologies) were placed onto the cells for a 10-day period with daily media changes. The hESCs and hiPSCs were differentiated into endoderm by following the manufacture’s protocol (STEMCELL Technologies). In brief, the stem cell colonies were made into a single cell suspension as detailed and plated onto Matrigel-coated wells or glass cover slips (BD Biosciences) overnight and then rinsed with DMEM/F-12. The cells were then incubated with the specific media and supplements as indicated by the protocol for a 5-day period with daily media changes.
Directed Differentiation Into Cardiomyocytes, Oligodendrocyte Progenitor Cells, and Hepatocytes
For cardiomyocyte differentiation, the hESCs and hiPSCs were differentiated as previously published [22, 23]. Specifically, 1 million cells originally plated onto Matrigel-coated wells (BD Biosciences) were found to be the optimal cell density for the hESC and hiPSC lines. The oligodendrocyte progenitor cells (OPCs) were differentiated as previously published [24]. Specifically, EBs were made with 5,000 cells per EB, because it has been shown that larger EBs tend to result in better neural lineages [25] Hepatocytes were differentiated as previously published [17, 26]. Human fetal cardiomyocytes, hepatocytes, oligodendrocyte progenitor cells, and adult cardiomyocytes were purchased and used as controls (ScienCell Research Laboratories, Carlsbad, CA, http://www.sciencellonline.com). Adult hepatocytes were also used as a control (Triangle Research Laboratories, Charlottesville, VA, http://triangleresearchlabs.net).
RNA Isolation and Reverse Transcription Polymerase Chain Reaction
Total RNA was isolated using a High Pure RNA Isolation Kit as per the manufacturer’s recommendations (Roche Diagnostics USA, Indianapolis, IN, http://www.roche-diagnostics.us). cDNA was synthesized using 1,000 ng/µl total RNA using the Transcriptor First Strand cDNA Synthesis Kit and both anchored-oligo(dT)18 and random hexamer primers (Roche Diagnostics USA) in accordance with the manufacturer’s recommendations. Reverse transcription polymerase chain reactions (PCRs) were performed as previously described [17]. The primer sequences were as follows: oligodendrocyte transcription factor 2, forward, 5′-CTCCTCAAATCGCATCCAG-3′, reverse, 5′-ACTTCTCGCTTTTGGTGAGG-3′; platelet-derived growth factor receptor-α, forward, 5′-CCTGAAAAGGGTCAGAAGGA-3′, reverse, 5′-GTGGTTGAATGCCAACCCTG-3′; NKX homeobox 2, forward, 5′-TGGCCATGTAAACGTTCTGA-3′, reverse, 5′-GCCGAATAGCTGAGCTCCAA-3′; myelin-associated glycoprotein, forward, 5′-TATGATTTCAGGTAACGGCTGAC-3′, reverse, 5′-AAGTACCAGACACCATGCAC-3′; albumin, forward, 5′-ACCCCAAGTGTCAACTCCAA-3′, reverse, 5′-CTGAAAAGCATGGTCGCCTG-3′; α1-antitrypsin, forward, 5′-AAGGACACCGAGGAAGAGGA-3′, reverse, 5′-CTTGGAGAGCTTCAGGGGTG-3′; tryptophan 2,3 dioxygenase, forward, 5′- TGGAGACGATGACAGCCTTG-3′, reverse, 5′-TCCAGAAGTGTCTTTTCCTGCT-3′; cytochrome P450, forward, 5′-GGTGGTGAATGAAACGCTCAG-3′, reverse, 5′-GGTCCACTTCCAAAGGGTGT-3′; GATA binding protein, forward, 5′-TCCAAACCAGAAAACGGAAG-3′; reverse, 5′-AAGACCAGGCTGTTCCAAGA-3′; T-box2, forward, 5′-AGTGGATGGCTAAGCCTGTG-3′, reverse, 5′-ACGGGTTGTTGTCGATCTTC-3′; Wilms tumor 1, forward, 5′-GGGCAGAGCAACCACAGCACA-3′, reverse, 5′-GCCACCGACAGCTGAAGGGC-3′; phospholamban, forward, 5′-ACAGCTGCCAAGGCTACCTA-3′, reverse, 5′-GCTTTTGACGTGCTTGTTGA-3′.
Quantitative PCR
All PCR reactions were conducted in triplicate and used two housekeeping genes, HPRT1 and GAPDH, as normalization controls. Equal amounts of cDNA (10 ng), 2× LightCycler 480 Probes Master, specific Universal Probe Library probe, PCR grade water (Life Science, Roche Diagnostics), and primers (ValueGene, San Diego, CA, http://www.valuegene.com) were used per reaction. The reactions were loaded onto a LightCycler 480 Multiwell Plate 96 (Roche Diagnostics) and sealed with sealing foil. The total volume per well was 20 µl and amplified on a LightCycler 480 Real-Time PCR System. All data analyzed with LightCycler 480 software, release 1.5.0, using the all-to-mean pairing rule for advanced relative quantification. A list of the primers and probes used are listed in supplemental online Table 1.
Immunocytochemistry
All cells for staining were plated onto glass cover slips and fixed for 15 minutes in 4% paraformaldehyde. The cells were washed two times with 1× PBS supplemented with 100 mM glycine for 5 minutes and permeabilized, when needed, with 0.5% Triton X-100 (Sigma-Aldrich) in 1× PBS for 60 minutes at room temperature. Blocking used 10% goat or donkey serum in blocker casein in PBS (Thermo Scientific, Rockford, IL, http://www.thermoscientific.com) for 60 minutes at room temperature. The cells were washed with 1× PBS after primary staining and each subsequent step. After primary antibody incubation, appropriate secondary Alexa Fluor antibodies (Invitrogen, Life Technologies) were incubated for 1 hour at room temperature in the dark and mounted on Prolong Gold with 4′,6-diamidino-2-phenylindole (Invitrogen, Life Technologies). The cover slips were visualized with an AxioCam MR Monocolor Camera and AxioVision Digital Image Processing Software (Axio Observer Inverted Microscope, Carl Zeiss, Jena, Germany, http://www.zeiss.com). A list of the primary antibodies used is provided in supplemental online Table 2.
DNA Extraction and Human Leukocyte Antigen Typing
DNA extraction from peripheral blood mononuclear cells (PBMCs) was performed using the PureLink Genomic DNA Mini Kit (Invitrogen, Life Technologies) in accordance with the manufacturer’s instructions. Extracted DNA was sent for human leukocyte antigen (HLA)-A, -B, and -DRB typing at the UCLA Immunogenetics Center (Los Angeles, CA).
PBMC Extraction, Culture, and Mixed Lymphocyte Reaction
PBMC Preparation
Blood was drawn using a 25-guage butterfly needle (BD Biosciences) to heparin-coated Vacutainers (BD Biosciences). Whole blood was diluted 1:3 with Dulbecco’s PBS (DPBS) and layered onto Ficoll-Paque PLUS (GE Healthcare, Pittsburgh, PA, http://www.gelifesciences.com). PBMCs were separated from whole blood by spinning at 400g for 30 minutes with no brake. The buffy coat was collected and washed three times with DPBS, enumerated, and frozen in 10% dimethyl sulfoxide/90% FBS.
Coculture
Cryopreserved cells were thawed and enumerated. The cells were resuspended at the appropriate density (i.e., 3 × 105 or 1.5 × 105 per milliliter) in RPMI plus 10% FBS or serum-free medium, as indicated. Next, 1 ml of PBMC suspension was preincubated in each well, followed by the addition of 1 ml of PBMC suspension that had been transduced with the appropriate vector as detailed below. The cells were incubated for 5 and 15 days. On days 5 and 15, 250 µl of medium was collected for enzyme-linked immunosorbent assay (ELISA), and 250 µl of fresh medium was added back to the culture.
Enzyme-Linked Immunosorbent Assay
Evaluation of interferon-γ (IFN-γ) release was determined using the Human IFN-γ ELISA Ready-SET-Go! kit per the manufacturer’s instructions (Affymetrix; eBioscience, San Diego, CA, http://www.ebioscience.com). The plates were read on a Tecan Infinite M1000 (Tecan, San Jose, CA, http://www.tecan.com). The 450-nm optical density (OD) was measured using the Infinite 200 multimode microplate reader provided with the Tecan-i-Control Plate reader analysis software (Tecan). A separate analysis was performed to normalize the result to a total protein quantity that was measured using the Bio-Rad protein assay kit I (no. 500-0001; Bio-Rad Laboratories, Hercules, CA, http://www.bio-rad.com), according to the manufacturer’s instructions. The OD595 was measured using the Infinite 200 multimode microplate reader provided with the Tecan-i-Control Plate reader analysis software (Tecan).
HORMAD1 and ZG16 Lentiviral Production and Transduction Into Human Fibroblasts
Lentiviral Vector Construction and Production
ZG16 was cloned as a PCR product from human hepatocyte cDNA and inserted into a third-generation lentiviral vector with a murine stem cell virus promoter for PBMC transduction. Vesicular stomatitis virus G pseudotyped lentivirus preparations were produced by cotransfection of packaging plasmids pMD2.G, pMDLg/pRRE, pRSV-Rev, and transfer vector into 293T cells (American Type Culture Collection, Manassas, VA, http://www.atcc.org) with jetPRIME reagent (Polyplus Transfection, New York, NY, http://www.polypus-transfection). Forty-eight hours later, virus-containing supernatant was collected, filtered, and concentrated by ultracentrifugation. The virus titers were determined by p24 ELISA, performed by the UCLA Virology Core (Los Angeles, CA), and quantitative PCR of transduced genomic DNA.
PBMC Transduction
The cells were thawed and stimulated for 72 hours with Dynabeads Human T-Activator CD3/CD28 (Life Technologies) per the manufacturer’s instructions. The cells were isolated from beads and transduced with viruses, as described, at a vector concentration of 1 × 106 transduction units per milliliter for 48 hours. The cells were subsequently cocultured at a 1:1 ratio with freshly thawed PBMCs for 5 and 15 days before the medium was collected for ELISA (described above).
Western Blot
Jurkat cells and PBMCs were transduced as described above. At 1 week after transduction, whole cell lysate was generated using RIPA buffer (Thermo Fisher Scientific) and HALT Protease Inhibitor (Thermo Fisher Scientific). Next, 12 μg of lysate was loaded and run on a 4%–12% bis-tris gradient gel (Invitrogen, Life Technologies). The separated gels were loaded onto polyvinyldine difluoride membranes (Invitrogen, Life Technologies), and blocked with 5% milk in PBS with Tween (PBST) (0.05%) for 1 hour at RT. The blots were probed using ZG16 antibody (Proteintech Group, Inc., Chicago, IL, http://www.ptglab.com) at 1:500 in blocking buffer and α-tubulin antibody (Sigma-Aldrich) at 1:1,000 in blocking buffer overnight at 4°C. The membranes were washed three times with PBST, probed with secondary anti-mouse and anti-rabbit peroxidase (Sigma-Aldrich) at 1:5,000 in blocking buffer for 1 hour at room temperature. The membranes were washed three times with PBST. Peroxidase activity was detected using ECL-PLUS (Thermo Fisher Scientific) and Typhoon FLA 9000 (GE Healthcare).
Statistical Analysis
The results are presented as the mean ± SD and mean ± SE. The statistical significance of the differences for all gene expression analyses was evaluated using SPSS, version 21 (IBM Corp., Armonk, NY, http://www-01.ibm.com/software/analytics/spss/). The results from analysis of variance, the t test for independent samples, Levene’s homogeneity of variance test, and the Mann-Whitney and Kruskal-Wallis nonparametrical one-way analysis of variance tests were considered statistically significant at p < .05.
Results
Teratoma-Based Immunogenicity Gene Expression Profiling
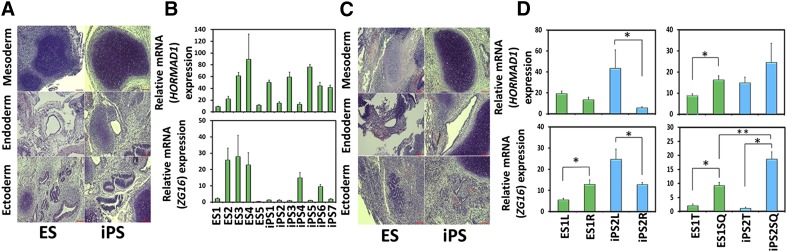
To duplicate the teratoma-based iPSC-specific immune response assay, teratomas were created from a variety of hiPSCs and hESCs that encompass a spectrum of hPSCs and therefore should represent the inherent variability among stem cell lines and any putative differences in hESCs and hiPSCs. All tested hESC and hiPSC lines formed intratesticular teratomas consisting of all three germ layers (Fig. 1A; supplemental online Fig. 1A). Because of their rich tissue diversity, teratomas were sectioned into 10 pieces and analyzed via quantitative PCR (QPCR) for both HORMAD1 and ZG16. After averaging the 10 pieces, HORMAD1 expression was found to be very low, although heterogeneous, with high QPCR-based cycle threshold values present across all hPSC lines analyzed (Fig. 1B, top). Concordantly, although ZG16 had much higher expression, it too exhibited heterogeneous expression across all hPSC lines (Fig. 1B, bottom). Statistically significant gene expression differences were observed between the hESC lines and hiPSC lines. We classified hESC and hiPSC differences between specific hESC and hiPSC lines as “interexperimental variation.” Statistically significant differences were also observed when comparing individual hiPSC lines, or individual hESC lines, with other hiPSC, or hESC, lines, respectively. We classified these differences as “intraexperimental variation.” The immunogenicity gene expression variation previously observed proved inconsistent between the hiPSC and hESC inter- and intraexperimental lines. This inconsistent variation provides evidence that interline variation (i.e., between hiPSC and hESC lines) does not provide any more variation than the intraline differences observed in hESCs or hiPSC lines.
Figure 1.
Teratomas from human embryonic stem cell (hESC) and human induced pluripotent stem cell (hiPSC) differentiation showed variable HORMAD1 and ZG16 expression levels in vivo. (A): Representative hematoxylin and eosin staining (H&E) from hESC and hiPSC testicular-derived teratomas showing representatives of the germ layers: cartilage (mesoderm), gut epithelium (endoderm), and neural tube (ectoderm) (supplemental online Fig. 1A). Scale bars = 100 µm. (B): Total RNA from whole testicular-derived teratoma tumors was used in quantitative polymerase chain reaction (QPCR) for HORMAD1 and ZG16 (top and bottom, respectively). Overall, low and variable HORMAD1 expression was observed (top), but substantially higher and heterogeneous expression of ZG16 was seen across all hESC and hiPSC lines (bottom). The results were normalized to HPRT1 and GAPDH. Data are presented as mean ± SEM of triplicates of 10 different pieces quantified. ZG16 y-values are in units of hundreds. Scale bars = 100 µm. (C): Representative H&E staining from hESC and hiPSC subcutaneously derived teratomas showing representatives of the germ layers as detailed in (A) (supplemental online Fig. 1A). Scale bars = 100 µm. (D): Total RNA from whole subcutaneously derived teratoma was used in QPCR that yielded low HORMAD1 expression (top). In contrast, higher ZG16 expression was identified, with significant differences identified. Results were normalized to HPRT1 and GAPDH. Data are presented as mean ± SEM of triplicates of 10 different pieces quantified. ZG16 y-values are in units of hundreds. ∗, p < .05. Abbreviations: ES, human embryonic stem cell line; iPS, human induced pluripotent stem cell line.
We hypothesized that the gene expression differences in testicular teratomas had resulted from intrinsic differences between the different hESC and hiPSC lines, manifesting in differential responses to niche differentiation factors. We tested this potential inherent variability of teratoma formation by injecting cells into the same mouse; we transplanted ES1 or iPS2 cells into the left and right hind legs of an immunocompromised mouse (Fig. 1C) and again found low overall expression of HORMAD1. In addition, only the iPS2 cells showed a significant difference between the left and right hind leg teratomas (Fig. 1D, top left). We again noted that ZG16 expression was higher than HORMAD1 expression. Both lines analyzed displayed a significant difference between the left and right hind leg teratomas, indicating induced differences despite forming in the same mouse and injection site location (Fig. 1D, bottom left). When the subcutaneous and testicular teratomas from the ES1 and iPS2 lines were compared, a significant difference was found in relation to HORMAD1 expression between the ES1 teratomas, but not the iPS2 teratomas (Fig. 1D, top right). However, not only was ZG16 found to differ between the ES1 and iPS2 subcutaneous- and testicular-derived teratomas, but a difference was also exhibited between ES1 and iPS2 subcutaneous teratomas (Fig. 1D, bottom right). Notwithstanding, because of the variation previously observed between the left and right hind leg teratomas, we have concluded that the differences between the subcutaneous and testicular teratomas in both lines for HORMAD1 and ZG16 display variable expression, likely owing to line-to-line and clonal variation.
Putative Immunogenicity Gene Expression Across hPSC Derivatives
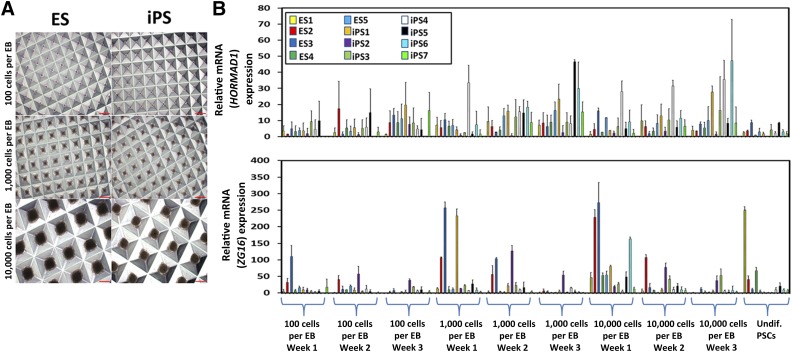
To maintain clinical applicability, our investigation focused on the expression patterns of HORMAD1 and ZG16 across varying levels of nondirected, partially directed, and fully directed differentiation of hPSCs. We hypothesized that, owing to the gene expression heterogeneity observed in teratomas, we would again see broad expression variance owing to hPSC line variation and differential amenability to differentiation protocols. Therefore, the hESC and hiPSC lines were first differentiated in a nondirected manner by EB formation (Fig. 2A; supplemental online Fig. 1B). EBs are three-dimensional multicellular aggregates that not only mimic early embryogenesis (e.g., can differentiate into mesoderm, endoderm, and ectoderm), but also differentiate spontaneously in suspension culture and in basic serum-containing media [27, 28]. Additionally, EB size and measurement points can bias EBs and cause different and/or multiple differentiation trajectories. We chose to make EBs with 100, 1,000, and 10,000 cells per EB and then transferred these EBs onto gelatin-coated plates for 1, 2, and 3 weeks of growth before harvesting [25, 29]. HORMAD1 expression was again extremely low, with expression levels that varied across the hESC and hiPSC lines (Fig. 2B, top). We observed high ZG16 expression compared with HORMAD1, although still at relatively low levels compared with teratoma expression, which varied significantly between the inter- and intraexperimental hES and hiPS lines (Fig. 2B, bottom). Statistically significant variance was observed for both HORMAD1 and ZG16, although the levels were inconsistent and no specific correlation between the different cell numbers and time points across all lines tested was observed.
Figure 2.
Differentiation into EBs yields heterogeneous HORMAD1 and ZG16 expression levels. (A): Representative human embryonic stem cell (hESC)- and human induced pluripotent stem cell (hiPSC)-derived EBs at three different sizes using a standardized EB formation microwell plate (supplemental online Fig. 1B). Scale bars = 200 µm. (B): EBs made at different sizes (100, 1,000, and 10,000 cells per EB) were plated onto gelatin-coated plates and harvested at different points (1, 2, or 3 weeks after plating) for total RNA. Quantitative polymerase chain reaction (PCR) was performed. Overall low and heterogeneous expression of HORMAD1 was observed with no significant trend seen between the size or time samples (top). ZG16 expression was higher, and some lines at specific points had significant increases or decreases, although no significant trend was observed across the size or measurement points (bottom). Error bars represent ±SD. All PCRs were run in triplicate. Abbreviations: EB, embryoid body; ES, human embryonic stem cell line; iPS, human induced pluripotent stem cell lines; Undif. PSCs, undifferentiated pluripotent stem cells.
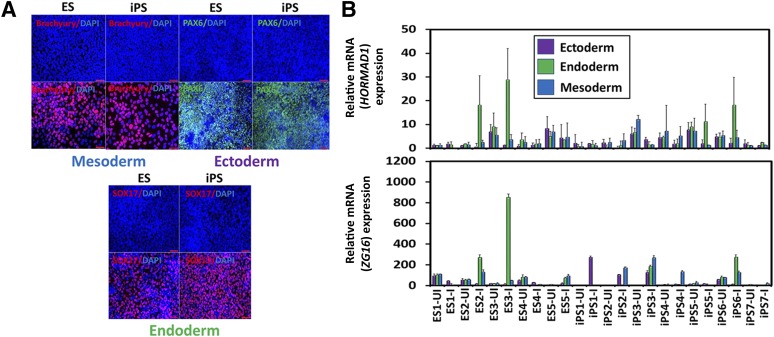
Understanding that different tissues present an array of potentially immunogenic targets, the hPSCs were subjected to directed differentiation into specific early representatives of the three embryonic germ layers characterized by typical gene expression patterns (supplemental online Fig. 2) and protein expression (Fig. 3A; supplemental online Fig. 3) [30, 31]. Again, we found a low expression of HORMAD1, at comparable levels to EBs, that varied across the inter- and intraexperimental hESC and hiPSC lines (Fig. 3B, top). The variable expression of HORMAD1 did not demonstrate any pattern from the uninduced to induced cells (Fig. 3B, top). Additionally, ZG16 expression was higher than in EB expression, and specific cell lines yielded higher levels of expression that differed among the lineages (Fig. 3B, bottom). Certain cell lines in ectoderm, endoderm, and mesoderm differentiation displayed a statistically significant increase in ZG16 expression on induction into each respective lineage, but others showed a significant decrease (Fig. 3B, bottom). This demonstrates that different lines, irrespective of whether they are hESCs or hiPSCs, contain different susceptibilities to differentiation protocols and start to express ZG16 aberrantly. We then tested whether these expression differences on induction would also be observed with full-directed differentiation.
Figure 3.
Semidirected differentiation into early embryonic germ lineages yields heterogeneous HORMAD1 and ZG16 expression. (A): Representative immunofluorescence staining with lineage-specific markers. Top Left: Embryonic stem cell (ESCs) and induced pluripotent stem cells (iPSCs) were differentiated into early mesoderm-like cells and stained for Brachyury (red). Top right: ESCs and iPSCs were differentiated into ectoderm-like cells and stained for PAX6 (green). Bottom: ESCs and iPSCs were differentiated into early endoderm-like cells and stained for SOX17 (red) (supplemental online Figs. 2, 3). Scale bars = 50 µm. (B): Gene expression analysis of HORMAD1 and ZG16 was assessed in each human ESC- and human iPSC-derived trilineage derivative across all lines for UI and I. Quantitative polymerase chain reaction (PCR) for HORMAD1 gene expression was found to be extremely low and variable across all lines, regardless of the induction of lineage (top). ZG16 expression varied in the directionality of differences in that some lines had significant increases on induction into each respective lineage (ectoderm: iPS1 and iPS2; endoderm: ES2, ES3, ES5, iPS3, and iPS6; mesoderm: iPS2, iPS3, and iPS4), but others showed a significant decrease (ectoderm: iPS6; endoderm: ES1; mesoderm: ES1 and ES4), although no clear correlation was found for certain lines or lineages (bottom). Error bars represent ±SD. All PCRs were run in triplicate. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; ES, human embryonic stem cell line; iPS, human induced pluripotent stem cell line; I, induced; UI, uninduced.
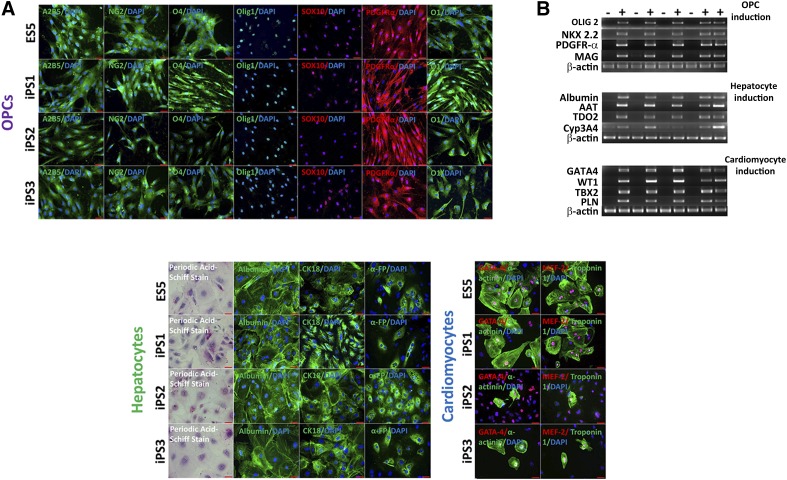
Because multiple hESC and hiPSC lines demonstrated different susceptibilities to the differentiation protocols, we hypothesized that these differences between the cell lines arose during the differentiation by exposure to new molecules that could potentially cause an immune response or interact with ZG16, or both [32]. Therefore, we directed the hESC and hiPSC lines under strict directed differentiation protocols into clinically relevant cell types (specifically, OPCs, hepatocytes, and cardiomyocytes). hESC and hiPSC derivatives were assessed using both immunofluorescence microscopy and gene expression analysis to validate the differentiation protocol (Fig. 4A, 4B).
Figure 4.
Immunocytochemical and reverse transcription-polymerase chain reaction-based characterization of fully differentiated clinically relevant human pluripotent stem cell derivatives. (A): Immunostaining of specific human embryonic stem cell (hESC) and human induced pluripotent stem cell (hiPSC) lines differentiated into three clinically relevant cell types representative of all three germ layers. Top four rows, left to right: OPC (ectoderm) stained with A2B5 (green), neural/glial antigen 2 (green), O4 (green), OLIG1 (green), SOX10 (red), PDGFR-α (red), and O1 (green). Bottom left four rows, left to right: Hepatocytes (endoderm) stained for glycogen synthesis with periodic acid-Schiff stain (pink), albumin (green), cytokeratin 18 (green), and α-fetoprotein (green). Bottom right four rows, left to right: Cardiomyocytes (mesoderm) were stained with GATA4 (red) and α-actinin (green) and myocyte enhancer factor-2 (red) and troponin-1 (green). Scale bars = 50 µm for OPCs, hepatocytes, and cardiomyocytes; scale bars = 100 µm for periodic acid-Schiff stain. (B): Reverse transcription-polymerase chain reaction (PCR) analysis confirmed the expression of lineage-specific genes for differentiated cell types: OPCs (OLIG2, NKX2.2, PDGFR-α, and MAG), hepatocytes (albumin, AAT, TDO2, and Cyp3A4), and cardiomyocytes (GATA4, WT1, TBX2, and PLN). All PCRs were run with β-actin control. Abbreviations: AAT, α1-antitrypsin; Cyp3A4, cytochrome P450; DAPI, 4′,6-diamidino-2-phenylindole; ES, human embryonic stem cell line; GATA-4, GATA binding protein; iPS, induced pluripotent stem; MAG, myelin-associated glycoprotein; NK2.2, NKX homeobox 2; OLIG, oligodendrocyte transcription factor; OPC, oligodendrocyte progenitor cell; PDGFR-α, platelet-derived growth factor receptor-α; TDO2, tryptophan 2,3-dioxygenase.
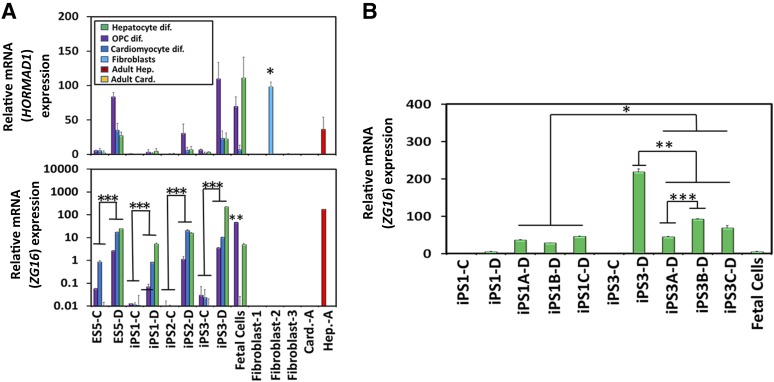
We subsequently examined HORMAD1 and ZG16 gene expression. Although this revealed overall low and heterogeneous expression for HORMAD1, we noted a trend that differentiation increased expression across all cell types for all derivative cell types (Fig. 5A, top). We postulated that because the adult fibroblast-2 line had a significant increase in HORMAD1 expression that was higher than that in the hPSC derivatives (Fig. 5A, top), HORMAD1 would be unlikely to be involved in causing an immune response. Statistical analysis indicated that ZG16 expression was significantly increased with differentiation of every cell line tested into the specific derivatives; the human fetal cells had varied levels of ZG16 compared with the hPSC derivatives, although it is notable that the OPC expression level was significantly higher than all other OPC derivatives (Fig. 5A, bottom). Human fetal cardiomyocytes had no ZG16 expression, and human fetal hepatocytes had intermediate expression (Fig. 5A, bottom). No adult fibroblast lines expressed ZG16, although an adult hepatocyte line displayed high ZG16 expression, with levels equivalent to iPSC3-differentiated hepatocytes (Fig. 5A, bottom). To ascertain whether this significant increase with differentiation is a reproducible phenomenon (and thereby a potential screening tool for lines that have low projected ZG16 expression) or just random intrinsic variation, the hepatocyte differentiation was repeated with the iPS1, and iPS3 lines, at a similar passage number (e.g., within four passages). The previously very-low-expressing iPS1-differentiated line had a significant increase across all replicates compared with the undifferentiated iPS1 cells. The iPS3-differentiated line also had a significant increase across all replicates compared with the undifferentiated iPS3 cells (Fig. 5B). The iPS1 replicates, when averaged together, were again found to be significantly lower than the iPS3-differentiated line when averaged across all three replicates. However, significant variability was also seen in the iPS3-A and iPS3-B replicates, between both themselves and that of the original iPS3 differentiation, thus indicating a high level of variability between the hPSC lines and the intraexperimental replicates (Fig. 5B). Thus, although ZG16 expression is definitively and reproducibly increased with differentiation, our data suggest that the inherent intra- and interline variability and, possibly, clonality issues with hPSC thawing and seeding would preclude this as a useful immunogenicity screening tool (Fig. 5B). These data, thus far, have made it difficult to classify ZG16, because human fetal oligodendrocytes and hepatocytes yielded high levels of ZG16, similar to the levels expressed across the various differentiated hPSC lines, indicating that ZG16 could be a fetally associated antigen. These findings are in contrast to the high level of ZG16 expression from adult hepatocytes, thereby indicating that adult cells are capable of expressing this antigen and eliminating ZG16 as a candidate exclusive fetal antigen.
Figure 5.
ZG16 expression increased significantly on differentiation of human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) into clinically relevant cell derivatives. (A): HORMAD1 expression in hESC and hiPSC derivatives was very low and heterogeneous (top). ZG16 expression analysis resulted in a significant increase across all four lines on differentiation (bottom). ∗, p < .05 for HORMAD1 expression for fibroblast-2 line compared with all other PSC derivatives. ∗∗, p < .05 for human fetal OPCs compared with other OPCs derived from all lines. ∗∗∗, p < .05 for each PSC line undifferentiated compared with differentiated. Error bars represent ±SD. The y-axis is a logarithmic scale, with display units of 100. All polymerase chain reactions were run in triplicate. (B): Gene expression analysis for ZG16 was assessed in hiPSC1- and hiPSC3-derived hepatocytes. The significant increase in ZG16 expression seen during hepatocyte differentiation from control iPSCs is reproducible across both iPS1 and iPS3 lines. A, B, and C listed in the cell line annotations indicate the duplicate differentiation protocol across three replicates. ∗, p < .05 for iPS1-A, -B, and -C differentiated replicates compared with iPS3-A, -B, and -C differentiated replicates; ∗∗, p < .05 for iPS3-D compared with iPS3-A, -B, and -C differentiated replicates; and ∗∗∗, p < .05 for iPS3-A–D compared with iPS3-B–D. ZG16 y-values are in units of hundreds. Error bars represent ±SD. Abbreviations: Card., cardiomyocytes; Card.-A, adult cardiomyocytes; dif, differentiation; ES, human embryonic stem cell line; Hep., hepatocytes; Hep.-A, adult hepatocytes; OPCs, oligodendrocyte progenitor cell; PSC, pluripotent stem cell.
Testing for an In Vitro Acute Immune Response
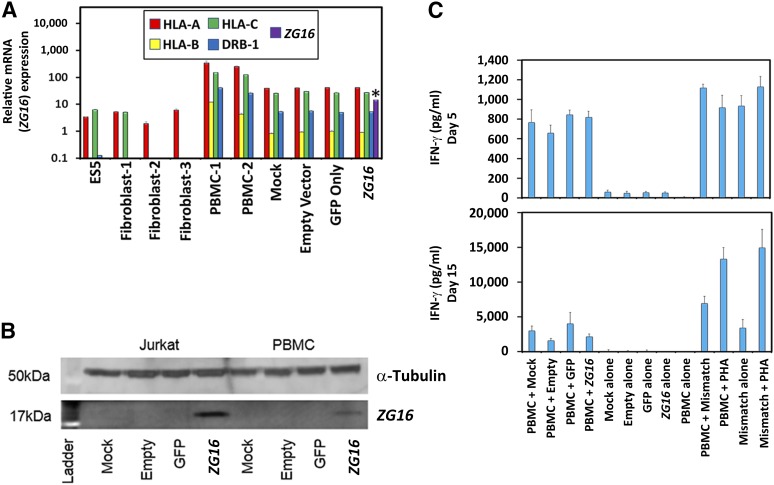
To investigate a functional link between ZG16 expression and an immune response in a human model, ZG16 was transduced into PBMCs and used in a modified PBMC coculture assay with IFN-γ secretion. This assay was then used as a metric of whether ZG16 could be acutely immunogenic [33]. Mismatched PBMC controls and PBMCs that varied only in transduction vector payload composition all displayed high expression levels of major histocompatibility complex (MHC) class I and II (MHC-I and MHC-II), indicating that these cells robustly presented processed peptides of expressed proteins (Fig. 6A). ZG16 was solely and strongly expressed by ZG16-transduced PBMCs (Fig. 6A). The ZG16 expression level was independently confirmed through protein expression analysis (Fig. 6B). PBMCs must be activated to allow transduction, and the subsequent culture with their parental donor PBMCs led to a basal level of IFN-γ release across all coculture conditions in this putative immunogenicity assay (Fig. 6C). No difference was observed between the mock or vector-transduced cells, indicating no increase in reactivity to the presence of any of the vectors within the limits of this assay. Furthermore, these data provide evidence in support of the hypothesis that ZG16 expression does not cause gross changes in MHC presentation sufficient to induce a significant IFN-γ level response similar to that seen in the mismatch control over either 5 or 15 days (Fig. 6C), indicating a negligible acute immune response of ZG16 in vitro.
Figure 6.
Introduction of an in vitro immunogenicity assay to test for a functionality link of human ZG16. (A): PBMCs were analyzed for expression of major histocompatibility complex class I (MHC-I) and class II (MHC-II). Quantitative polymerase chain reaction analysis for HLA-A, -B, -C, and -DRB1 showed that unmatched PBMCs (PBMC-1) (i.e., PBMCs that differed from the control PBMCs used in the modified PBMC coculture assay) and all other PBMCs (PBMC-2) that varied only in what was being transduced all yielded very high expression of MHC-I and MHC-II, providing evidence that these cells would be able to participate in a modified PBMC coculture assay. Only the ZG16 positively transduced line expressed ZG16 to a significant degree. The y-axis is a logarithmic scale with display units of 10,000. Error bars represent ±SD. All polymerase chain reactions were run in triplicate. ∗, p < .05. (B): High levels of ZG16 were only expressed in ZG16 transduced Jurkat cell line and PBMCs as confirmed via Western blot analysis for protein expression. (C): IFN-γ mixed lymphocyte functionality assay was used to test whether viral transduction of PBMCs with ZG16 could cause an acute immune response. Owing to the required activation step of PBMCs to allow successful viral transduction, the PBMCs in coculture with their parental donor PBMCs led to a basal level of self-reactivity that was somewhat decreased from days 5 to 15. No indication was observed that ZG16-positive PBMCs were causing an increased immune response. The y-axis is in units of hundreds. Error bars represent ±SD. Abbreviations: ES, human embryonic stem cell line; GFP, green fluorescent protein; HLA, human leukocyte antigen; IFN-γ, interferon-γ; PHA, phytohemagglutinin; PMBC, peripheral blood mononuclear cell.
Conclusion
In the present study, we performed gene expression analysis across a variety of hESC and hiPSC derivatives with varying levels of differentiation using two putative immunogenicity genes that were previously correlated to an in vivo T-cell-mediated immune response in the mouse [13]. Gene expression differences between the hESCs and hiPSCs, in both their undifferentiated form and a variety of clinically relevant derivatives, indicate that no consistent or specific differences inherent to hESCs or hiPSCs exist. This observation carried through a variety of differentiation representative of the different stages of cell maturation during development. The modified PBMC coculture assay tested for an acute immune response with what we propose to be the only relevant gene capable of potentiating an immune response, ZG16, and was found incapable of producing a significant IFN-γ-based response in PBMCs. Therefore, we conclude that the levels of two putative immunogenicity-related antigens presented in human cells are not capable of producing a significant in vitro acute immunological response.
Supplementary Material
Acknowledgments
We thank Patrick Lee, Cyril Ramathal, Jens Durruthy-Durruthy, and Renee A. Reijo Pera for supplying the mRNA-derived hiPSC line from Stanford; Martin G. Martin for supplying the 2.19, 6.7, and 13.1.0 lines from UCLA; Saravanan Karumbayaram for supplying the multi-integration hiPSC lines from UCLA; and Harvey Herschman, Caius Radu, Kathrin Plath, Patricia Phelps, Heather Christofk, Lily Wu, William Lowry, Lorenz Studer, Konrad Hochedlinger, Marius Wernig, Steven Bensinger, Kenneth Dorshkind, and Lili Yang for their valuable advice.
Author Contributions
J.P.A.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; E.H.G.: conception and design, collection and analyze data, data analysis and interpretation, manuscript writing; A.V.-C.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; J.V.: conception and design, collection and analyze data, manuscript writing; M.H.W. and B.T.: collection and/or assembly of data, data analysis and interpretation; D.B.K. and N.K.: conception and design, data analysis and interpretation, manuscript review; J.A.B.: conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
J.A.B. has multiple financial relationships with Fibrocell Science, Inc. (consultant, sponsored research agreement, royalties), but they do not affect the content of this article. A.V.-C. has compensated research funding from Fibrocell Science, Inc. The other authors indicated no potential conflicts of interest.
References
- 1.Byrne JA. Generation of isogenic pluripotent stem cells. Hum Mol Genet. 2008;17(R1):R37–R41. doi: 10.1093/hmg/ddn053. [DOI] [PubMed] [Google Scholar]
- 2.Chin MH, Pellegrini M, Plath K, et al. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010;7:263–269. doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilic J, Izpisua Belmonte JC. Concise review: Induced pluripotent stem cells versus embryonic stem cells: Close enough or yet too far apart? Stem Cells. 2012;30:33–41. doi: 10.1002/stem.700. [DOI] [PubMed] [Google Scholar]
- 4.Soldner F, Hockemeyer D, Beard C, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T, Wu H, Li Y, et al. Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine-mediated epigenetic modifications during reprogramming to pluripotency. Nat Cell Biol. 2013;15:700–711. doi: 10.1038/ncb2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doi A, Park IH, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim K, Zhao R, Doi A, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister R, Pelizzola M, Kida YS, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchetto MC, Yeo GW, Kainohana O, et al. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stadtfeld M, Apostolou E, Akutsu H, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayshar Y, Ben-David U, Lavon N, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Hu BY, Weick JP, Yu J, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao T, Zhang ZN, Rong Z, et al. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- 14.Araki R, Uda M, Hoki Y, et al. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- 15.Guha P, Morgan JW, Mostoslavsky G, et al. Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell. 2013;12:407–412. doi: 10.1016/j.stem.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Maynard KM, Arvindam U, Cross M, et al. Potentially immunogenic proteins expressed similarly in human embryonic stem cells and induced pluripotent stem cells. Exp Biol Med (Maywood) 2014;239:484–488. doi: 10.1177/1535370214522936. [DOI] [PubMed] [Google Scholar]
- 17.Awe JP, Lee PC, Ramathal C, et al. Generation and characterization of transgene-free human induced pluripotent stem cells and conversion to putative clinical-grade status. Stem Cell Res Ther. 2013;4:87. doi: 10.1186/scrt246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karumbayaram S, Lee P, Azghadi SF, et al. From skin biopsy to neurons through a pluripotent intermediate under Good Manufacturing Practice protocols. Stem Cells Translational Medicine. 2012;1:36–43. doi: 10.5966/sctm.2011-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sommer CA, Stadtfeld M, Murphy GJ, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somers A, Jean JC, Sommer CA, et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan JY, Sriram G, Rufaihah AJ, et al. Efficient derivation of lateral plate and paraxial mesoderm subtypes from human embryonic stem cells through GSKi-mediated differentiation. Stem Cells Dev. 2013;22:1893–1906. doi: 10.1089/scd.2012.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lian X, Hsiao C, Wilson G, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lian X, Zhang J, Azarin SM, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izrael M, Zhang P, Kaufman R, et al. Human oligodendrocytes derived from embryonic stem cells: Effect of noggin on phenotypic differentiation in vitro and on myelination in vivo. Mol Cell Neurosci. 2007;34:310–323. doi: 10.1016/j.mcn.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 25.Bauwens CL, Peerani R, Niebruegge S, et al. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells. 2008;26:2300–2310. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 26.Song Z, Cai J, Liu Y, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009;19:1233–1242. doi: 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- 27.Kurosawa H. Methods for inducing embryoid body formation: In vitro differentiation system of embryonic stem cells. J Biosci Bioeng. 2007;103:389–398. doi: 10.1263/jbb.103.389. [DOI] [PubMed] [Google Scholar]
- 28.Heins N, Englund MC, Sjöblom C, et al. Derivation, characterization, and differentiation of human embryonic stem cells. Stem Cells. 2004;22:367–376. doi: 10.1634/stemcells.22-3-367. [DOI] [PubMed] [Google Scholar]
- 29.Hwang YS, Chung BG, Ortmann D, et al. Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11. Proc Natl Acad Sci USA. 2009;106:16978–16983. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P, Li J, Tan Z, et al. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood. 2008;111:1933–1941. doi: 10.1182/blood-2007-02-074120. [DOI] [PubMed] [Google Scholar]
- 31.Yao S, Chen S, Clark J, et al. Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc Natl Acad Sci USA. 2006;103:6907–6912. doi: 10.1073/pnas.0602280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyd AS, Higashi Y, Wood KJ. Transplanting stem cells: Potential targets for immune attack. Modulating the immune response against embryonic stem cell transplantation. Adv Drug Deliv Rev. 2005;57:1944–1969. doi: 10.1016/j.addr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Kruisbeek AM, Shevach E, Thornton AM. Proliferative assays for T cell function. Curr Protoc Immunol 2004;Chapter 3:Unit 3.12. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.