Abstract
Objectives
To investigate errors identified in SNOMED CT by human reviewers with help from the Abstraction Network methodology and examine why they had escaped detection by the Description Logic (DL) classifier. Case study; Two examples of errors are presented in detail (one missing IS-A relation and one duplicate concept). After correction, SNOMED CT is reclassified to ensure that no new inconsistency was introduced.
Conclusions
DL-based auditing techniques built in terminology development environments ensure the logical consistency of the terminology. However, complementary approaches are needed for identifying and addressing other types of errors.
Keywords: Systematized nomenclature of medicine, Comparative study, Quality assurance, Description logics, Abstraction network
Introduction
SNOMED CT is one of the largest clinical terminologies in the world. The most recent release (July 31, 2009) comprises more than 289,000 active concepts and 1.5 million relations (hierarchical and associative). SNOMED CT concepts are organized into 19 hierarchies, such as “Procedure”, “Clinical finding” and “Body structure.”
Modern terminologies including SNOMED CT and the NCI Thesaurus are created with the support of Description Logics (DL), which ensures the logical consistency of the terminological assertions. However, due to its sheer size and complexity, it is almost unavoidable that SNOMED CT should contain errors, such as inaccurate or incomplete logical definitions (e.g., errors in the nature or in the target of asserted relationships, as well as missing relations).
A number of techniques have been developed for auditing SNOMED CT, based on lexical, structural, and ontological principles. Lexical approaches have been used by [1–2] to suggest missing and erroneous relations based on the compositionality of biomedical terms. Additionally, [2] exploited formal ontological principles. Formal Concept Analysis was employed by [3] to analyze semantic completeness. Based on various structural approaches, [4] detected improper assignment of relationships, redundant concepts, and omission of relationships. Finally, [5] identified redundant and underspecified concepts by detecting equivalent concept definitions. In summary, these approaches applied computational method to the identification of potential errors. This automated process is designed to facilitate the work of human editors (subject matter experts) and it contributes to the quality assurance of biomedical terminologies.
The objective of this paper is to investigate errors identified in SNOMED CT by human reviewers, with help from the Abstraction Network methodology. More specifically, we examine why such errors could not be identified by a Description Logics classifier and propose a strategy for using the Abstraction Network in complement to DL-based techniques for the quality assurance purposes. The contribution of this paper is not to propose novel approaches to identifying errors in SNOMED CT, but rather to tease out differences between existing approaches based on several cases of errors thoroughly investigated.
Background
Description Logic
Description logics (DL) are a family of knowledge representation formalisms often used as ontology languages [6]. Not only does DL provide support for defining concepts, but it also provides methods for reasoning about concepts and their instances. DL reasoning services are carried out by DL classifiers.
The basic inference on concept expression is subsumption, i.e., comparing two classes and checking whether one class is more general than the other. For example, “brain disorder” is more specific than (i.e., subsumes) “disorder,” because “brain disorder” is defined as a “disorder” located to the brain. Another important inference is concept satisfiability. A class is deemed unsatisfiable (i.e., inconsistent) if it cannot possibly have any instances. For example, nothing can be at the same time a procedure and an anatomical structure. If a class “C” were defined as a subclass of both “Procedure” and “Body structure,” while “Procedure” and “Body structure” are defined to be disjoint, a DL classifier would identify “C” as unsatisfiable. The interested reader is referred to [6] for additional details about DL.
There are, however, many different dialects of DL in terms of the set of constructors they offer, resulting in different levels of expressiveness for what can be defined. The expressiveness of the DL also determines the kinds of inference a DL classifier is enabled to perform and the kinds of logical inconsistency it is able to identify. The dialect of DL natively used by SNOMED CT is “EL”, whose expressiveness is relatively limited. For example, EL does not allow disjunction to be stated between classes and the example of unsatisfiability presented earlier could therefore not be identified by the DL classifier used for the creation of SNOMED CT.
From the perspective of error identification in ontologies, two major types of errors can be distinguished. Type I errors are the logical inconsistencies in concept expression that can be detected by DL classifiers (assuming the DL dialect used is expressive enough to state the circumstances under which concepts would be inconsistent, e.g. disjointness). In contrast, Type II errors are those content errors (e.g., wrong relations, missing relations) that would not generate logical conflicts in the DL system. Quality assurance processes in SNOMED CT ensure that all Type I errors have been identified and corrected before the terminology is released to users. All the errors under investigation in this study are therefore Type II errors. (Here, Type I and Type II errors are defined in reference to the level of expressiveness of the EL dialect of DL).
In practice, several views of SNOMED CT are provided to users. The main view is the inferred view, in which all inferences are precomputed and redundant relations removed. The inferred view is automatically derived from the asserted view by a DL classifier. In this work, we analyze the inferred view, but, unlike most users, we also modify the asserted view and use a DL classifier in order to check any suggested changes for consistency.
Abstraction Network
The Abstraction Network (AN) is a structural methodology developed for reducing the complexity of large biomedical terminologies [7]. The AN methodology is based on the associative relationships and their inheritance patterns in the hierarchies of the terminology. It has been applied to auditing SNOMED CT. Here, we give a brief description of its underlying principles and review its application to SNOMED CT. Our examples focus on the “Specimen” hierarchy of SNOMED CT.
AN provides an abstraction of the hierarchical and associative relations of concepts in a SNOMED CT hierarchy. The idea is to partition such concepts into structural uniformity groups (strUGs), and then to refine the partition into semantic uniformity groups (smtUGs). A detailed description can be found in [7–9]1.
A “structural uniformity group (strUG)” is the group of all concepts with exactly the same set of associative relationships. In a graph structure, we use a node to represent a strUG. The label for the strUG node is the set of associative relationships in which its concepts participate.
Five different associative relationships are introduced to the concepts of the “Specimen” hierarchy; they are substance, morphology, procedure, topography, and identity2. For example, the concept “Surgical excision sample” has one relationship procedure pointing to a concept “Excision” (from the “Procedure” hierarchy). Therefore, the concept “Surgical excision sample” is in the strUG{procedure}. Similarly, the concept “Abscess swab” has two relationships procedure and morphology pointing to “Taking of swab” and “Abscess morphology” (from the “Procedure” and “Body structure” hierarchy, respectively). Thus, “Abscess swab” is in the strUG{procedure, morphology}. Note that strUGs do not overlap, because, by construction, one given concept belongs to one and only one strUG corresponding to its relationship pattern. Therefore, the entire set of strUGs forms a partition of the concepts in a given hierarchy of SNOMED CT.
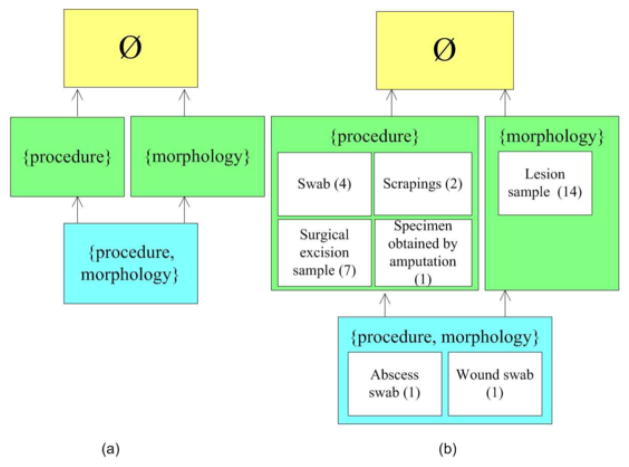
StrUGs can be organized into a graph structure. Hierarchical relations between strUGs are determined by the inclusion of the sets of relationships they represent. For example, the strUG{procedure} subsumes the strUG{procedure, morphology}. Figure 1(a) shows a portion of the graph of strUGs for the “Specimen” hierarchy. Each colored box represents a strUG. The boxes are color-coded to differentiate the levels. Each level corresponds to the number of relations in the strUG. The concepts in the strUG Ø have no associative relationships.
Figure 1.
(a) Portion of the graph of StrUGs for the “Specimen” hierarchy (b) Corresponding portion of the graph of smtUGs
A “semantic uniformity group (smtUG)” is a group of concepts within a structural uniformity group sharing the same lowest common ancestor (LCA). In other words, the smtUG groups concepts with the same associative relationships by hierarchical relations. The label for the smtUG is the LCA from which all other concepts in the smtUG are descendants. A strUG may have more than one LCA, and thus more than one smtUGs. The smtUGs form a semantic subdivision of the strUG, but not necessarily a subpartition of it, since a concept may have more than one LCA.
The graph of strUGs in Figure 1(a) can be refined with the smtUGs contained within each strUG, as shown in Figure 1(b). For example, the strUG{procedure} contains the four smtUGs: smtUG(“Swab”), smtUG(“Scrapings”), smtUG(“Surgical excision sample”) and smtUG(“Specimen obtained by amputation”). The number in the parentheses indicates the number of concepts within a smtUG. For example, in the smtUG(“Surgical excision sample”), there is a total of seven concepts. The six hidden concepts are all subsumed by “Surgical excision sample.”
The strUGs and smtUGs form a graph structure called abstraction network (AN), which hides some of the complexity of the terminology. This abstracted view has proved a useful auditing tool for manual review of biomedical terminologies by subject matter experts.
Auditing Method based on Abstraction Network
Several strategies have been devised to help subject matter experts review parts of SNOMED CT based on the Abstraction Network methodology.
Group-based auditing takes advantage of the grouping of concepts in semantic uniformity groups [7]. All concepts from a given group are reviewed at the same time, making it easier for experts to identify discrepancies among concepts expected to be both structurally and semantically similar. Errors exposed via “group-based auditing” include redundant concepts, erroneous relationships, incorrect IS-A assignments, and other content errors.
Auditing “complex” concepts focuses on those concepts within a structural uniformity group, which belong to several semantic uniformity groups because they have ancestors in several smtUGs [9]. Errors found in such complex concepts include missing child and incorrect parent.
Error concentration based auditing is predicated on the fact that small semantic uniformity groups are more likely to contain errors, because small sets of similar concepts might have received less modeling attention, compared to larger sets (e.g., based on a concept model). The correlation between small smtUG size and error concentration was assessed in [8].
Case study
We selected two of the errors detected in SNOMED CT by subject matter experts with help from the Abstraction Network methodology and reported to the International Health Terminology Standards Development (IHTSDO)3, the organization in charge of SNOMED CT. Our objective in this paper is to investigate these cases and examine how they escaped detection by the DL classifier used to check the logical consistency of SNOMED CT.
DL reasoners are stand-alone tools that point out logical inconsistencies in an ontology. In contrast, the Abstraction Network methodology helps organize the workflow of subject matter experts, in order to focus their attention to parts of the ontology where errors are likely and by grouping the concepts to be audited according to the principles described earlier.
The two errors under investigation were identified in the “Specimen” hierarchy of SNOMED CT. In the first one, “amputation”, it was argued that two sibling concepts actually stand in a subsumption relation. The issue is thus a missing IS-A relation between these two concepts. The second case, “leukocyte”, highlights two concepts that are arguably equivalent, but stand in a IS-A relation.
In addition to discussing the errors, we also want to test the remediation suggested to the IHTSDO. Toward this end, we loaded the asserted version of SNOMED CT in OWL DL into the ontology editor Protégé4 and tested the suggested changes with the DL classifier Fact++5. Our goal is to verify that the proposed changes did not introduce any inconsistencies to SNOMED CT. Classification was performed on a standard desktop machine with the 64-bit Microsoft Windows operating system and 4 GB of RAM. The classification of the OWL version of the SNOMED CT takes about 17 minutes.
Case 1: Amputation
This error was identified by the subject matter expert while examining a group of concepts from the “Specimen” hierarchy corresponding to one particular structural uniformity group, namely the strUG{procedure}. By construction, the concepts naming the smtUGs within a strUG are not expected to stand in any kind of hierarchical relation. The assumption for the subject matter expert reviewing the concepts from a strUG is that they are all expected to be siblings. Therefore, reviewing these concepts as a group makes it easy to identify errors including missing or incorrect parent/child relations, for example.
Figure 2 shows a portion of the inferred view of the SNOMED CT displayed in the CliniClue browser6. The two concepts circled in red, “Specimen obtained by amputation” and “Surgical excision sample”, are siblings. Both of them are in the “Specimen” hierarchy under the root concept “Specimen.” The corresponding target concepts with the relationship procedure are “Amputation” and “Excision,” respectively, in the “Procedure” hierarchy, under the parent concept “Surgical removal” (not shown in the figure). The four concepts “Specimen obtained by amputation”, “Surgical excision sample,” “Amputation” and “Excision” are fully defined.
Figure 2.

“Specimen obtained by amputation” and “Surgical excision sample” displayed in the CliniClue browser
The subject matter expert determined that “Specimen obtained by amputation” is, in fact, a kind of “Surgical excision sample.” The fact that the two concepts were grouped in the strUG{procedure} made it easier for the expert to identify this error. Of note, there was no logical inconsistency in the concept expression and the DL reasoner failed to detect the missing subsumption relation because its absence did not create any kind of conflict in the terminology. One particular reason why no conflict could be identified is because there was a parallel error on the target side. The target concepts “Amputation” and “Excision” are siblings (descendants of “Surgical removal”), while amputation is actually a kind of excision. Because of a missing IS-A relation in parallel on both sides of the procedure relationship, there was no logical error that could be identified by the DL classifier.
From the perspective of the Abstraction Network, both smtUG(“Surgical excision sample”) and smtUG(“Specimen obtained by amputation”) are in the strUG{procedure} (see Figure 1(b)). But the existence – indicated by the expert – of an IS-A relation between these two concepts within the same strUG{procedure} violates the principles under which the strUG was constructed.
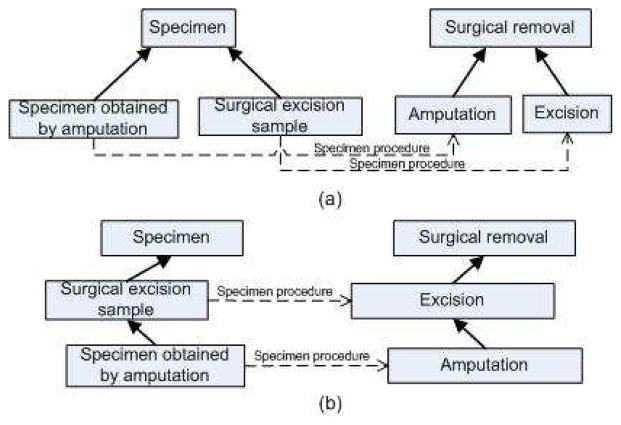
Figure 3 shows the comparison before and after addition of the missing IS-A relations. As a result of this modification, “Specimen obtained by amputation” is now subsumed by “Surgical excision sample”, and the smtUG(“Surgical excision sample”) has gained a new member.
Figure 3.
Parent-child error with “Surgical excision sample” and “Specimen obtained by amputation” (a) Before correction (b) After correction
We modified the target hierarchy (“Procedure”) by making “Surgical Excision” the super class of “Amputation” in our copy of SNOMED CT in Protégé, while leaving the source hierarchy (“Specimen”) unchanged. After reclassification, we saw that the classifier had used the changes we made to the target hierarchy (“Procedure”) to automatically make parallel changes to the source hierarchy (“Specimen”), where “Surgical excision sample” has become the super class of “Specimen obtained by amputation” (Figure 3(b)).
Case 2: Leukocyte
This error was identified by the subject matter expert while examining a group of concepts from the “Specimen” hierarchy corresponding to one particular semantic uniformity group, namely the smtUG(“White blood cell sample”). By construction, concepts within a smtUG are expected to stand in an IS-A relation with the lowest common ancestor after which the smtUG is named. The assumption for the subject matter expert reviewing the concepts from a strUG is that they are all expected to be distinct and descendants of “White blood cell sample”. Therefore, reviewing these concepts as a group makes it easy to identify duplicate concepts, for example.
As shown in Figure 4, “Leukocyte specimen” is one of the children of “White blood cell sample.” The subject matter expert determined that “Leukocyte specimen” and “White blood cell sample” are, in fact, duplicate concepts. The fact that the two concepts were grouped in the smtUG(“White blood cell sample”) made it easier for the expert to identify this error.
Figure 4.
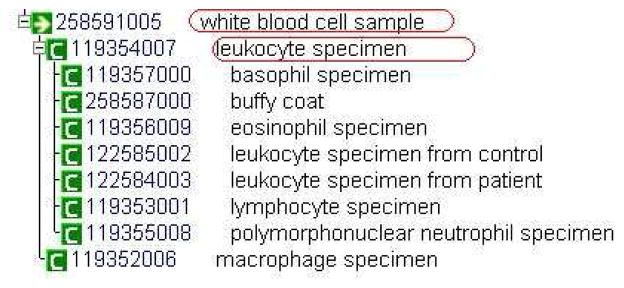
“Leukocyte specimen” and “White blood cell sample” displayed in the CliniClue browser
In DL, concepts exhibiting the same logical definitions are treated as equivalent concepts by the classifier. In this case, the DL classifier did not identify these two concepts as equivalent, because the logical definitions were actually slightly different. “Leukocyte specimen’ is a primitive concept, whereas “White blood cell sample” is fully defined. Because the definition of “Leukocyte specimen” is underspecified (primitive), the DL classifier cannot recognize it as equivalent to the fully defined “White blood cell sample.”
From the perspective of the Abstraction Network, there is no difference between primitive and defined concepts. Only the set of relationships is taken into account during the creation of the groups.
We modified the definition of “Leukocyte specimen” in our copy of SNOMED CT in Protégé, so as to make it fully defined instead of primitive. After reclassification, “White blood cell sample” and “Leukocyte specimen” were indicated as being equivalent concepts.
Discussion
Strengths and limitations of each approach
The main advantage of DL is that it identifies errors completely automatically, while the Abstraction Network (AN) methodology only constrains the workflow of subject matter experts in such a way that it facilitates their work and improves their chances of identifying errors by reducing the complexity of the terminology and by organizing the concepts to be reviewed in small groups, with assumed relations among concepts within and across groups.
Unlike the DL classifier, AN does not rely on defined concepts, but simply takes advantage of the structural properties of concepts, i.e., their sets of relationships. Unlike AN, the DL classifier processes the terminology as a whole and can address remote inconsistencies, whereas experts tend to focus on a small portion of the terminology and may not foresee the consequences of local changes to distant parts of the terminology.
Finally, DL classifiers are limited to the identification of logical inconsistencies. Moreover, they are limited in the type of logical inconsistencies they can identify by the level of expressiveness of the dialect of DL used for creating the ontology [10]. In contrast, subject matter experts guided by the Abstraction Network methodology can address a wider range of issues (i.e., beyond logical inconsistencies) and identify content errors, such as inaccurate and missing relations.
Auditing strategy
The DL classifier is used for detecting logical inconsistencies at the time the terminology is built. The performance of the classifiers has improved tremendously in the past few years and the editors of large terminologies will soon enjoy real-time classification. We recommend the use of the Abstraction Network methodology for targeted auditing, as a possible alternative to dual editing. However, multiple auditing strategies combining lexical, structural and ontological methods are required for quality assurance of large, complex terminologies such as SNOMED CT.
Current developments and future work
One of the limitations of the Abstraction Network methodology is that it relies heavily on the structure of relationships of the concepts and is therefore not applicable to concepts with few or no relationships. In order to address this limitation, we have developed the converse abstraction network [11].
Conclusions
In this study, we examine the differences between two approaches to identifying errors in large biomedical terminologies such as SNOMED CT. On the one hand, Description Logics classifiers can automatically identify logical inconsistencies in the terminology. On the other, the Abstraction Network methodology helps experts perform targeted manual reviews of the terminology by reducing its complexity and grouping the concepts by their structural and semantic properties. We illustrate the differences between the two approaches through two cases of errors identified in SNOMED CT.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM). This work was done while Duo Wei was a visiting fellow at the Lister Hill National Center for Biomedical Communications, NLM, NIH. This work was also partially supported by the NLM under grant R-01-LM008912-01A1.
Footnotes
In our previous work, structural uniformity group is referred to as area, while semantic uniformity group is referred to as partial-area.
The full name of these relationships is specimen substance, specimen source morphology, specimen procedure, specimen source topography, and specimen source identity, respectively.
References
- 1.Campbell KE, Tuttle MS, Spackman KA. A “lexically-suggested logical closure” metric for medical terminology maturity. Proc AMIA Symp. 1998:785–9. [PMC free article] [PubMed] [Google Scholar]
- 2.Ceusters W, Smith B, Kumar A, Dhaen C. Mistakes in medical ontologies: where do they come from and how can they be detected? Stud Health Technol Inform. 2004;102:145–63. [PubMed] [Google Scholar]
- 3.Jiang G, Chute CG. Auditing the semantic completeness of SNOMED CT using formal concept analysis. J Am Med Inform Assoc. 2009;16(1):89–102. doi: 10.1197/jamia.M2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geller J, Perl Y, Halper M, Cornet R. Special issue on auditing of terminologies. J Biomed Inform. 2009;42(3):407–11. doi: 10.1016/j.jbi.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Cornet R, Abu-Hanna A. Auditing description-logic-based medical terminological systems by detecting equivalent concept definitions. Int J Med Inform. 2008;77(5):336–45. doi: 10.1016/j.ijmedinf.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Baader F, Calvanese D, McGuiness D, Nardi D, Patel-Scheneider P, editors. The description logic handbook : theory, implementation, and applications. Cambridge, UK; New York: Cambridge University Press; 2003. [Google Scholar]
- 7.Wang Y, Halper M, Min H, Perl Y, Chen Y, Spackman KA. Structural methodologies for auditing SNOMED. J Biomed Inform. 2007;40(5):561–81. doi: 10.1016/j.jbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Halper M, Wang Y, Min H, Chen Y, Hripcsak G, Perl Y, et al. Analysis of error concentrations in SNOMED. AMIA Annu Symp Proc. 2007:314–8. [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Wei D, Xu J, Elhanan G, Perl Y, Halper M, et al. Auditing complex concepts in overlapping subsets of SNOMED. AMIA Annu Symp Proc. 2008:273–7. [PMC free article] [PubMed] [Google Scholar]
- 10.Rector AL, Brandt S. Why do it the hard way? The case for an expressive description logic for SNOMED. J Am Med Inform Assoc. 2008;15(6):744–51. doi: 10.1197/jamia.M2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei D, Halper M, Elhanan G, Chen Y, Perl Y, Geller J, et al. Auditing SNOMED relationships using a converse Abstraction Network. AMIA Annu Symp Proc. 2009:685–689. [PMC free article] [PubMed] [Google Scholar]