Abstract
Following partial hepatectomy, the liver initiates a regenerative programme involving hepatocyte priming and replication driven by the coordinated actions of cytokine and growth factors. We investigated the mechanisms underlying adiponectin's (Adn) regulation of liver regeneration through modulation of these mediators. Adn–/– mice showed delayed onset of hepatocyte replication, but accelerated cell cycle progression relative to wild-type mice, suggesting Adn has multiple effects fine-tuning the kinetics of liver regeneration. We developed a computational model describing the molecular and physiological kinetics of liver regeneration in Adn–/– mice. We employed this computational model to evaluate the underlying regulatory mechanisms. Our analysis predicted that Adn is required for an efficient early cytokine response to partial hepatectomy, but is inhibitory to later growth factor actions. Consistent with this prediction, Adn knockout reduced hepatocyte responses to interleukin-6 during the priming phase, but enhanced growth factor levels through peak hepatocyte replication. By contrast, supraphysiological concentrations of Adn resulting from rosiglitazone treatment suppressed regeneration by reducing growth factor levels during S phase, consistent with computational predictions. Together, these results revealed that Adn fine-tunes the progression of liver regeneration through dynamically modulating molecular mediator networks and cellular interactions within the liver.
Key points.
Loss of adiponectin delays the initiation of liver regeneration after partial hepatectomy, but later accelerates regeneration.
Loss of adiponectin modulates these regeneration kinetics through decreased hepatocyte response to inflammation and increased growth factor bioavailability.
Increased adiponectin suppresses liver regeneration through decreased growth factor bioavailability.
Our predictive computational model was able to connect these molecular regulatory events to tissue physiology.
Introduction
Liver regeneration is a unique repair mechanism that allows a damaged liver to recover following traumatic or toxic injury or hepatic surgical procedures. This process is clinically important in liver mass recovery in both donor and recipient following live donor liver transplantation. After partial hepatectomy (PHx), normally quiescent hepatocytes are activated to re-enter the cell cycle through a highly synchronized pro-proliferative response, which requires precise timing of cytokine and growth factor (GF) signals. This response is orchestrated through a dynamic pattern of activation and inhibition of a wide range of signalling processes coordinated across multiple cell types in the liver, including hepatocytes, Kupffer cells and hepatic stellate cells (Taub, 2004). Kupffer cells are primary coordinators of the dynamic cytokine microenvironment following tissue damage. Hepatic stellate cells produce growth factors critical to induce hepatocyte replication. Once lost tissue mass is recovered, hepatic stellate cells also produce factors terminating regeneration (Taub, 2004). In addition, signals from extrahepatic tissues, including adipokines, play a role in modulating this coordinated cellular response. Because adipokines originate from outside the liver, treatment of hepatic surgery patients with adipokines is an attractive option to modulate liver regenerative ability following surgical intervention without the complications involved in modifying liver function directly.
One of the factors implicated in modulating both liver cytokine microenvironment and growth factor bioavailability is the serum adipokine adiponectin (Adn) (Yamauchi & Kadowaki, 2013). Adn is a 30 kDa protein produced primarily by adipose tissue that circulates as low molecular weight (trimeric), middle molecular weight (hexameric) and high molecular weight oligomers (Turer & Scherer, 2012). Adn directly sensitizes the body to insulin, and Adn levels are low in patients with Type II diabetes (Kadowaki et al. 2006). It is thought to act in large part through two identified Adn cell surface receptors, Adn receptor (AdipoR) 1 and AdipoR2. Additionally, Adn has been shown to increase acute inflammation (Park et al. 2007; Awazawa et al. 2011) and is known to modulate balances of cytokines and growth factors critical to liver regeneration and repair. There is evidence that Adn also can act through direct binding and inhibition of growth factors. Taken together, these findings suggest Adn may have both pro- and anti-proliferative effects during liver regeneration (Landskroner-Eiger et al. 2009; Kajimura et al. 2013). Consistent with this hypothesis, previous studies suggested both reduced and increased Adn levels can impair regeneration. Adn–/– mice were reported to exhibit a delayed liver regeneration phenotype (Ezaki et al. 2009; Shu et al. 2009), whereas treatment with the anti-diabetic drug rosiglitazone, which is thought to act at least in part by elevating serum Adn (Nawrocki et al. 2006; Yamauchi & Kadowaki, 2013), inhibits liver mass recovery (Turmelle et al. 2006). By contrast, rats with induced diabetes show a delayed initiation of hepatocyte replication after PHx, which is corrected by an increased replication from 36 to 72 h following surgery (Barra & Hall, 1977). We hypothesize that low Adn levels may have contributed to the regeneration dynamics observed in these diabetic animals.
The apparently conflicting actions through which Adn impacts liver regeneration points to non-linear effects that are difficult to parse out with typical over-/under-expression experimental analyses. In this study, we aim to develop a deeper understanding of the multifaceted impact of Adn on liver regeneration using an integrated computational modelling and experimental approach to characterize the molecular mechanisms underlying the Adn-mediated fine-tuning of liver regeneration dynamics.
Molecular mechanisms underlying liver regeneration are both redundant and complex, making prediction of effects of molecular or cellular manipulations difficult. For example, in studies using CI2MDP liposomes to eliminate Kupffer cells from the liver, interleukin 6 (IL-6), tumour necrosis factor alpha (TNFα) and hepatocyte growth factor (HGF) were all significantly decreased; however, mass recovery appeared to be only delayed but not blunted (Meijer et al. 2000). Similarly, mice harbouring a hepatocyte-specific deletion of c-Met were expected to show little or no regeneration following CCl4 injection. These mice, however, showed similar kinetics and magnitude of proliferation following a single CCl4 injection, indicating multiple compensatory mechanisms (Huh et al. 2004). After more extensive injury, the c-Met-deficient mice did show lower recovery, but this was probably due to decreased cell motility rather than decreased proliferation.
Our computational modelling approach was designed to account for the net effect of such molecular redundancies and complex cellular interactions governing liver regeneration. Through model simulations and sensitivity analysis, we investigated how changes to relative balances and timing of multiple regulatory mechanisms contribute to shaping liver regeneration dynamics. We started with a computational model of liver regeneration, recently developed by Furchtgott et al. (2009). This model considers hepatocytes as being distributed across multiple cellular states: quiescent, primed or replicating. The dynamic shift between these states is mediated by factors produced by non-parenchymal cells. Cytokines, such as TNFα and IL-6, produced primarily by Kupffer cells, initiate the JAK-STAT signalling cascade, resulting in activation of the expression of immediate early (IE) genes, which shifts hepatocytes from the quiescent to the primed state. GF produced by hepatic stellate cells stimulate replication of primed hepatocytes. Termination of replication requires renormalization of cytokine and growth factor levels, and buildup of extracellular matrix (ECM). Using this cellular interaction framework, the model acts as a bridge connecting the kinetics of molecular regulation to the regeneration dynamics.
We used Adn–/– and rosiglitazone-treated mice to examine the kinetics of liver regeneration response after PHx through the priming and replicative phase and applied these experimental data to the computational model. We utilized the recently developed pulsatile sensitivity analysis (PSA) to investigate which regulatory balances are critical for the effect of Adn on liver regeneration, and in what temporal intervals the specific changes to regulatory balances have the greatest impact on regeneration.
The experimental data indicated that absence of Adn caused lower STAT3 tyrosine phosphorylation during the first 6 h after PHx, probably associated with decreased hepatocyte priming and causing a 6 h delay in the onset of hepatocyte replication. Lack of Adn, however, also increased GF levels following PHx, accelerating cell cycle progression. When circulating Adn was raised to supraphysiological levels by treatment of wild-type (WT) mice with the anti-diabetic drug rosiglitazone, GF levels were decreased during peak hepatocyte replication and regeneration was suppressed. Our model suggests that Adn modulates hepatocyte priming and GF bioavailability during time windows when regeneration is most sensitive to alterations in these factors.
Methods
Animals
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at Thomas Jefferson University. Jefferson's IACUC is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care and experiments were designed using the Guide for the Care and Use of Laboratory Animals.
Ten- to 12-week-old male Adn–/– mice (B6.129-Adipoqtm1Chan), bred from mice kindly donated by Dr Lawrence Chan, or C57BL/6 J mice (Jackson Laboratories, Bar Harbor, ME, USA) underwent partial hepatectomy based on surgical methods outlined by Mitchell & Willenbring (2008). Briefly, animals were anaesthetized by inhalation of 5% isoflurane in an induction chamber and anaesthetic plane was confirmed by toe pinch. Anaesthesia was maintained during surgery by continual inhalation of 2% isoflurane administered by nose cone. A midline incision was made followed by the sequential ligation and excision of the left lateral and medial lobes of the liver. The abdominal cavity was rinsed with warm lactated Ringer solution, the abdominal muscle layer was sutured and the skin was closed with wound clips. Following surgery, animals were given subcutaneous lactated Ringer solution (1 ml per animal) and placed in a fresh cage under a heat lamp with ad libitum access to hydrogel (Contact ClearH2O, Portland, ME, USA) and food. At specified times after PHx, animals were anaesthetized with isoflurane as described for partial hepatectomy (induction and maintenance). While anaesthetized, animals were weighed and killed. The livers were either immediately (within 10 s) freeze clamped using liquid nitrogen-cooled aluminium clamps as previously described (Crumm et al. 2008), preventing rapid post-mortem changes in cytokine or growth factor levels and protein phosphorylation, or the livers were fixed in 10% neutral buffered formalin (NBF) for assessment of bromodeoxyuridine (BrdU) labelling. For determination of liver to body weight ratios, the liver was dissected out and weighed prior to freeze clamping.
Blood was collected from the tail vein of live animals and from the vena cava under anaesthesia when the animals were killed. Collected blood was incubated at room temperature for 30 min, then centrifuged at 1500 r.p.m. for 5 min. Serum was isolated and flash frozen for further analysis. In some cases, animals were given i.p. injections of BrdU solution (Sigma, St Louis, MO, USA) (150 mg kg−1) in sterile 0.9% saline 2 h prior to being killed. For rosiglitazone treatment, animals were administered rosiglitazone (10 mg kg−1, Cayman Chemical, Ann Arbor, MI, USA) or vehicle (1:1 mixture of 1× PBS and polyethylene glycol; Sigma) by gavage twice daily beginning 2 days before surgery.
Histological analysis
Samples fixed in 10% NBF were paraffin-embedded, sectioned, and stained for haematoxylin and eosin by the Kimmel Cancer Center pathology core facility (Thomas Jefferson University) for analysis of hepatosteatosis. BrdU staining was performed using Impact DAB staining (Vector Laboratories, Burlingame, CA, USA) according to the manufacturer's instructions. For BrdU quantification, five 20× fields were scored per animal.
Biochemical analysis
For Western blotting, tissue lysates were generated by homogenizing frozen tissue in RIPA buffer (Sigma) supplemented with phosphatase and protease inhibitor cocktails (Sigma). Protein was normalized using BCA Protein Assay Reagent (Pierce Biotechnology, Rockford, IL, USA). Then, 20 μg of protein was loaded onto an SDS-PAGE gel and Western blotting for cell cycle markers and pSTAT3/STAT3 was performed as previously described (Crumm et al. 2008). Alternatively, for comparison of growth factor expression time course, 50 μg of protein was resolved by SDS-PAGE, transferred onto nitrocellulose membrane using a Multi-Strip Western blotting approach as described previously (Aksamitiene et al. 2007) and probed with mouse monoclonal antibodies against HGF (SBF5) (Thermo Fisher Scientific, Rockford, IL, USA), FGF-2 (6) (sc-136255; Santa Cruz Biotechnology, Dallas, TX), ANG I (C-1) (sc-74528; Santa Cruz Biotechnology), GAPDH (6C5) (EMD Millipore, Billerica, MA, USA) or rabbit polyclonal antibodies against β-actin (D6A8) (Cell Signaling, Danvers, MA, USA). ELISA kits were used for measurement of Adn (B-Bridge International) and TNFα (eBioscience, San Diego, CA, USA) according to the manufacturer's instructions. Transcription factor binding activity was assessed from nuclear extracts prepared from frozen tissue using a nuclear extraction kit (Origene, Rockville, MD, USA). NF-κB DNA binding activity in 100 μg of nuclear extract was measured using the NF-κB transcription factor assay kit (Cayman Chemical) according to the manufacturer's instructions. Some samples were sent to Raybiotech for analysis of cytokine and chemokine levels using a Quantibody Multiplex ELISA Array (Raybiotech, Norcross, GA).
For RT-PCR analysis, RNA was extracted from frozen tissue using the RNeasy RNA extraction kit (Qiagen, Valencia, CA, USA). Then, 2 μg of RNA was reverse transcribed with EasyScript Plus Reverse Transcriptase (Applied Biological Materials Inc., Richmond, BC, Canada). cDNA was preamplified with TaqMan PreAmp Master Mix (Applied Biosystems, Foster City, CA, USA) and PCR reactions were performed using BioMark Dynamic Arrays (Fluidigm, South San Francisco, CA, USA). Primer sequences are shown in Table1. CT values were calculated using Real-Time PCR Analysis software (Fluidigm) and normalized to the expression of housekeeping genes TBP and β2-microglobulin using the established –ΔΔCT method (Livak & Schmittgen, 2001).
Table 1.
Primer sequences
| Primer | Foreword | Reverse |
|---|---|---|
| SOCS3 | CTACGCATCCAGTGTGAGGG | TGAGTACACAGTCGAAGCGG |
| β2-Microglobulin | GTCGCTTCAGTCGTCAGCAT | TTTCAATGTGAGGCGGGTGG |
| TBP | CCCCTTGTACCCTTCACCAAT | GAAGCTGCGGTACAATTCCAG |
Data were compared using Student's t test on raw data (BrdU incorporation and liver-to-body weight ratios) or log-transformed data (molecular measurements). Paired statistics were used when appropriate. Data are presented as mean ± SEM.
Computational modelling of liver regeneration
Molecular regulation was connected to regeneration phenotype using a previously published model of liver regeneration (Furchtgott et al. 2009). The computational model simulates liver regeneration as a series of regulatory events initiated in non-parenchymal cells and influencing hepatocyte quiescence, priming and replication (shown schematically in Fig. 2A). The initiation of regeneration is governed by a mismatch between ‘metabolic demand’ (M) of the organism and the total number of hepatocytes (N) available to meet this demand. In this scheme, the ‘metabolic load’ per hepatocyte (M/N) increases proportional to the mass of the liver removed, initiating non-parenchymal cell activation following PHx. Once activated, non-parenchymal cells respond to liver damage by early induction of IL-6 (representative of the inflammatory milieu observable following PHx and its effect on hepatocytes), later production of GF (representative of the growth factor environment and effect following PHx) and ECM remodelling. These molecular signals induce hepatocytes to proceed from quiescence (Q) to a primed state (P), from the primed state to a replicating state (R) to recover lost liver tissue, and finally from primed and replicating states back to quiescence. This computational model uses representative molecular components to describe archetypical classes of signalling during liver regeneration. Such an empirical and approximate model allows for investigation into how changes to the magnitude and timing of signalling activation may affect regeneration dynamics. While the metabolic load parameter has no direct molecular correlate, it probably captures the effects of the molecular drivers of liver metabolism and mitochondrial activity such as AMP-activated protein kinase (AMPK) activation or ATP levels or ATP/ADP ratio. Modulation of metabolic load by perturbing this metabolic demand parameter may therefore be considered to reflect a general metabolic challenge.
Figure 2. Identifying the key control factors through sensitivity analysis of a computational model of liver regeneration.
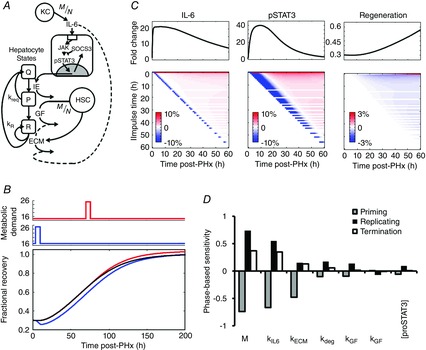
A, schematic representation of the network model of liver regeneration following PHx incorporates contributions of non-parenchymal cells to catalyse hepatocyte replication. B, pulse changes (50% increase for 6 h) to the metabolic demand parameter (M) caused delayed regeneration if administered early after PHx, and enhanced regeneration if administered late after PHx. C, (top) nominal profiles of IL-6, STAT3 and fractional recovery. (Bottom) Heat maps of fractional change from nominal model output. Hour-long, 50% pulsatile decreases in IL-6 production rate (kIL6) caused a temporary decrease in IL-6 levels, independent of when the pulsatile change was introduced (note the consistent blue diagonal). Such a temporary reduction in IL-6 led to a much longer and more pronounced decrease in STAT3 phosphorylation, with earlier IL-6 changes leading to longer STAT3 transients, and ultimately delayed regeneration. The earlier the pulsatile reduction in IL-6 and subsequent transient decrease in STAT3, the longer the regeneration deficit persisted. D, phase-based sensitivity analysis showed that the timing of alterations to the key controlling factors leads to a phase-dependent effect on overall regeneration. In general, changes to these key factors during the priming phase had opposite effects on regeneration as compared to the effect of changes during the replicating and termination phases.
This model includes the JAK-STAT signalling pathway induced by IL-6 and GF produced by non-parenchymal cells as drivers of regeneration and includes ECM as a negative regulator of regeneration. Shifts between hepatocyte states (Q, P, or R) are governed by the following equations:
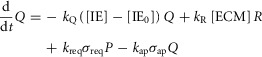 |
(1) |
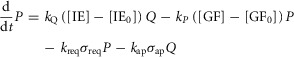 |
(2) |
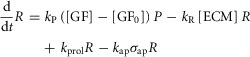 |
(3) |
where IE gene expression catalyses a shift in hepatocytes from the Quiescent state to the Primed state with a rate parameter of kQ. GFs produced in response to metabolic load catalyse a shift in hepatocytes from the Primed state to the Replicating state with a rate parameter of kP. Upon entering the Replicating state, hepatocytes begin to replicate at a rate kprol. However, hepatocytes also return to the Quiescent state due to natural requiescence and ECM buildup (with rate parameters of kreq and kR, respectively).
The parameters σap and σreq in the above equations govern the amount of hepatocytes undergoing apoptosis (or removal by other means from the pool of hepatocytes entering the cell cycle) and requiescence, respectively. They are calculated as sigmoidal functions to account for a switch-like behaviour (eqns 3 and 4):
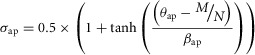 |
(4) |
| (5) |
where θ is the threshold parameter governing at what level a response occurs and β is the response parameter governing how much of a response occurs.
For hepatectomies up to 75–80% σap remains small, but for partial hepatectomies greater than 80% σap becomes large enough to prevent liver regeneration. Similarly, σreq is low when GF levels are high but increases when GF levels return to baseline, capturing the phenomenon that GFs are needed for cells to progress through the cell cycle.
IL-6 produced in response to metabolic load induces IE gene expression through the JAK-STAT signalling cascade, which also includes negative feedback from SOCS-3. IL-6 levels, the JAK-STAT signalling cascade and IE gene levels are calculated using a combination of linear and Michaelis-Menton kinetics (eqns 6–10). GF is modelled as being produced in response to metabolic load and sequestered by ECM (eqn 11). ECM is modelled as constitutively produced (k6) but degraded both consitutively (κECM) and dynamically in response to IL-6-induced matrix metalloproteases (MMPs) (κdeg) (eqn 12). This simulates a continuous steady-state turnover of ECM, which is dynamically regulated during periods of tissue remodelling. All molecules are modelled as being degraded at a rate proportional to the amount present in the liver.
| (6) |
| (7) |
 |
(8) |
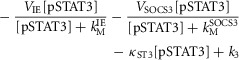 |
(9) |
 |
(10) |
| (11) |
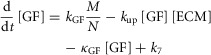 |
(12) |
| (13) |
Parameters for each molecule (XX) include production rate (kXX), degradation rate (κXX) and Michaelis–Menten parameters (VXX and kMXX). Further model parameters include concentration of monomeric STAT3 ([proSTAT3]), rate of GF uptake by ECM (kup) and the parameter governing inhibition of STAT3 phosphorylation by SOCS-3 (kISOCS3). The parameters k1–k7 represent steady-state production or degradation of the molecule which each equation describes.
Model differential equations were solved simultaneously using ode15s in MATLAB (Mathworks, Inc., Natick, MA, USA).
Sensitivity analyses for identifying key factors controlling regeneration phenotype
Parametric sensitivities were estimated based on a dynamic local sensitivity analysis (Zak et al. 2005), phase-based sensitivity (Gunawan & Doyle, 2007) and pulsatile sensitivity methodologies (Perumal & Gunawan, 2011). To calculate the dynamic local sensitivities (DLSs), parameters were changed by ±10% and sensitivity was calculated as the change in overall liver recovery normalized to the number of cells at each time in the nominal regeneration profile divided by the percentage change in the parameter (20%), according to eqn (13):
| (14) |
where N(t) is the nominal fraction of hepatocytes at any given time, and ΔN(t) is the deviation from nominal caused by the parameter change.
Phase-based sensitivities (PBSs) were calculated following the formulations of Gunawan & Doyle (2007) and Perumal & Gunawan (2011). Simulations were run with the value of a single parameter increased by 10% of its nominal value within one of the three phases of regeneration: the priming phase (0–6 h after PHx), the regeneration phase (12–100 h after PHx) and the termination phase (100–200 h after PHx). The simulations were then run again with the same parameter decreased by 10% within the same phase. These two simulations were repeated for every parameter. Sensitivities were estimated as the change in overall liver faction recovered at 300 h after PHx normalized to the nominal regeneration profile divided by the percentage change in parameter value for the phase when the change occurs (20%), as shown in eqn (14):
| (15) |
Pulsatile sensitivities (PSs) were calculated using an approach modified from that of Perumal & Gunawan (2011). In this modified approach, we altered each model parameter by +10% or −10% of the corresponding nominal value at each hour following PHx for 1 h, where τ represents the beginning of the time step where the parameter was changed. We estimated the pulsatile sensitivity at each time point after the pulsatile parametric change (at time τ) according to eqn (15):
| (16) |
While this equation appears similar to that used to calculate DLS, the pulsatile sensitivity values (PSi) take on non-zero values only after the time of the pulse change in the corresponding parameter value.
Parameter estimation to match Adn–/– regeneration phenotype
To estimate parameters characterizing the Adn–/– mice, Sobol sampling was used to search the parameter space of sensitive model parameters (Bratley & Fox, 1988). Each parameter was allowed to vary from its nominal value over approximately one order of magnitude (10×). Simulations were then run with each of 10,000 parameter sets, and the resulting regeneration profiles were compared to the experimental Adn–/– mouse regeneration profile generated in this study. Parameter sets generating similar regeneration profiles were then analysed for common molecular regulation governing tissue behaviour. Search of the parameter space resulted in multiple parameter sets that could simulate regeneration profiles similar to that seen experimentally, but these multiple parameter sets contained similar parameters and caused similar molecular regulation. Therefore, parameter sets generating regeneration profiles similar to that observed in the experiments were further explored using a combination of manual manipulation and local optimization (fminsearch in MATLAB). The parameter set resulting in the lowest mean squared error between simulation and experimental observations of liver regeneration in Adn–/– mice was reported as the parameter set for Adn–/– mice.
Results
Hepatocyte proliferation is delayed after PHx in Adn–/– mice
The dynamics of liver regeneration in Adn–/– and wild-type (WT) mice after PHx were assessed by BrdU pulse labelling and expression of cell cycle marker proteins 24–54 h after PHx. BrdU incorporation increased in WT mice at 30 h after PHx relative to baseline levels. However, Adn–/– mice showed no increase at 30 h (Fig. 1A,B). WT and Adn–/– mice showed similar levels of BrdU incorporation 36 h after PHx (Fig. 1A,B). Adn–/– mice incorporated significantly more BrdU at 42 h after PHx (Fig. 1A,B). By 54 h after PHx, WT and Adn–/– mice again showed no difference in BrdU incorporation. When compared to WT mice, Adn–/– mice also expressed significantly lower levels of G1 phase cell cycle markers proliferating cell nuclear antigen (PCNA) and cyclin D1 at 24 h after PHx (Fig. 1C,D). Both PCNA and cyclin D1 levels were renormalized by 30 h after PHx and PCNA levels remained similar at all subsequent times (Fig. 1C,D). Similarly, Adn–/– mice expressed cyclin A, an important S phase cyclin, at lower levels than WT mice at 30 h after PHx; cyclin A levels were renormalized by 36 h after PHx and remained similar at 42 h (Fig. 1C,D). Liver to body weight ratio was assessed as a measure of liver mass recovery. By 54 h post PHx, both WT and Adn–/– mouse livers had approximately doubled in mass (Fig. 1E). No significant difference was detected between WT and Adn–/– liver mass recovery at this time.
Figure 1. Analysis of hepatocyte replication after PHx in WT and Adn–/– mice.
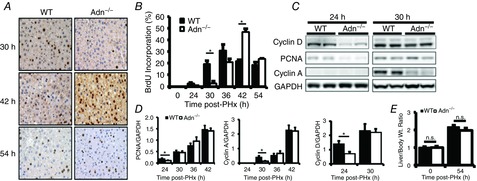
A, representative histological sections from mice at various times after PHx and injected with BrdU 2 h prior to death were stained using BrdU-specific antibodies. B, the percentage of BrdU-positive hepatocytes was calculated by quantifying BrdU-positive nuclei and total nuclei from five representative fields at 20× magnification (n = 4 per group). C, Western blot of representative liver samples probed with antibodies specific for cyclin D1, PCNA, cyclin A and GAPDH. D, quantification of Western blots for cyclin D1, PCNA and cyclin A (n = 3 per group). E, liver to body weight ratio. Data are presented as mean ± SEM. n.s., Not significant, *P < 0.05, ***P < 0.01 Adn–/– significantly different from WT.
Taken together, these results suggest that the onset of hepatocyte proliferation after PHx is delayed in Adn–/– relative to WT mice but that the cell cycle may be accelerated in Adn–/– mice to renormalize regeneration after a delayed cell cycle onset. However, the dynamic changes to molecular balances underlying Adn fine-tuning control of liver regeneration remain unclear.
Pulsatile sensitivity analysis reveals critical time windows of molecular effects on liver regeneration
We used a computational modelling approach to investigate which molecular balances are critical to alter regeneration dynamics and the time windows over which regenerating hepatocytes are responsive to these signals. We initially employed local parametric sensitivity analysis to analyse a recently developed computational model of liver regeneration, shown schematically in Fig. 2A, to identify the regulatory balances that control the dynamics of regeneration (Furchtgott et al. 2009). This dynamic sensitivity metric considers how the time profile of liver regeneration responds to changes in the network parameters. We defined liver regeneration profile as ‘highly sensitive’ to a given parameter if the maximum value of the corresponding normalized sensitivity coefficient had a magnitude greater than 0.15 at any time point. Our analysis identified 12 out of 32 parameters as significantly controlling the dynamics of liver regeneration. These included both molecular parameters – metabolic load (M), IL-6 production rate (kIL6), concentration of monomeric STAT3 ([proSTAT3]), ECM degradation rate by MMPs (κdeg), ECM constitutive degradation rate (κECM), GF production rate (kGF), and GF degradation rate (κGF) – and physiological parameters governing hepatocyte phenotypic state and apoptosis – kP, kR, kprol, θap and βap (Table2).
Table 2.
Normalized sensitivity values of the top-ranked controlling factors
| Parameter | Maximum normalized sensitivity |
|---|---|
| M | 0.34 |
| kIL6 | 0.28 |
| kGF | 0.19 |
| κdeg | 0.16 |
| [proSTAT3] | 0.14 |
| κGF | −0.16 |
| κECM | −0.17 |
| kprol | 0.75 |
| kP | 0.18 |
| βapp | −0.19 |
| θapp | −0.19 |
| kR | −0.65 |
One consideration in the above sensitivity analysis is that the network parameters are altered as a step change throughout the regenerative response time. While the effect on the regeneration is dynamic, it is not possible to deconvolute the changes leading to instantaneous effects versus those altering response at later times. To address this issue, we employed a recently developed impulse sensitivity analysis and modified the scheme to consider finite pulses of parameter changes in defined temporal intervals (Perumal & Gunawan, 2011). The pulsatile sensitivity analysis revealed that magnitude and timing of changes to parameters were both key controlling factors in fine-tuning the regeneration profile. Among the parameters evaluated for their pulsatile sensitivity, the metabolic demand parameter showed a unique sensitivity profile. Changing metabolic demand using a short pulse or longer step increase caused a decreased early regenerative response after PHx, but led to an enhanced regeneration response at later times (Fig. 2B). Such a time interval-dependent effect occurred through multiple processes affected by metabolic demand changes. Within the 0–50 h post-hepatectomy period, additional increases in the metabolic demand led to a transient increase in hepatocyte apoptosis and delayed regeneration, but a renormalization (or moderately enhanced regeneration) caused by increased IL-6 signalling and hepatocyte priming. Additional increase of metabolic demand at later time intervals between 75–150 h after PHx caused increased GF signalling leading to enhanced liver regeneration. In contrast, a 50% impulse decrease in IL-6 production rate caused a transient decrease in IL-6 levels that was quickly renormalized (Fig. 2C). Downstream STAT3 phosphorylation, however, showed a much larger magnitude decrease that persisted for several hours before renormalization. The time window during which these changes consistently resulted in observable changes to regeneration profile were limited to the first 50 h of regeneration (Fig. 2C). In contrast, a 50% impulse increase in GF production rate caused a sustained increase in hepatocyte number when GF production was increased between 0 and 50 h or at certain time points between 50 and 127 h after PHx. These results suggest that fine-tuning signalling dynamics by modulating the timing and temporal balances of non-parenchymal cell activation can have significant functional consequences, with persistent impact on regeneration dynamics and tissue mass recovery.
While all of the molecular parameters apart from metabolic load showed similar pulsatile sensitivity profiles, the key subset identified as significant controlling factors by the parametric sensitivity analysis displayed a phase-dependent sensitivity (Fig. 2D). For example, a transient increase in IL-6 production rate during the priming phase (0–6 h) caused an early increase in IL-6 levels followed by a persistent decrease below nominal levels (Fig. 3A,B). This persistent decrease in IL-6 levels caused a persistently decreasing rate of hepatocyte replication and a blunted overall tissue recovery (Fig. 3C). In contrast, transiently increasing IL-6 production rate during the replication phase (12–100 h) caused IL-6 levels to remain elevated above nominal levels (Fig. 3A,B). This persistent elevation caused a persistently increasing rate of hepatocyte replication and enhanced overall tissue recovery (Fig. 3C). This phase-dependent effect predicts that mistiming of enhanced factor production influences pro- or anti-regenerative effects. Therefore, our computational modelling and sensitivity analysis revealed the dynamic balances of initiation-related and replication-related factors that must be closely regulated to ensure the dynamics and magnitude of the normal liver regeneration profile.
Figure 3. Time-dependent effects of transient increases in IL-6 production rate during different phases of liver regeneration.
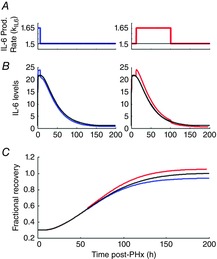
A, transient increases in IL-6 production rate (kIL6) during the priming and replication phase. IL-6 production rate was increased from 0 to 6 h after PHx (left) and 12 to 100 h after PHx (right) to identify phase-dependent effects of IL-6 production rate increases. B, transiently increasing IL-6 production rate during the priming phase (0–6 h, left) caused a transient increase in IL-6 levels followed by a persistent reduction in IL-6 levels. Transiently increasing IL-6 production rate during the replication phase (12–100 h, right) caused a sustained increase in IL-6 levels. C, simulated regeneration based on nominal IL-6 levels (black), increased IL-6 production during priming (blue) and increased IL-6 production during the replication phase (red). Persistent reduction in IL-6 levels from increased production during priming caused a persistent decrease in hepatocyte replication rate and a blunted overall tissue recovery. Sustained increase in IL-6 levels caused a persistent increase in hepatocyte replication rate and an enhanced overall tissue recovery.
Computational modelling of the altered regenerative response in the Adn–/– mice reveals the key controlling molecular regulatory balances
We predicted the factors governing molecular control of the Adn–/– regeneration phenotype by considering simultaneous alterations to multiple molecular parameters including those identified as sensitive in the above analyses (M, kIL6, κIL6, kGF, κGF, κdeg, κECM, kup). Our Monte Carlo approach ensured efficient sampling coverage of the physiologically reasonable parameter space by using a Sobol sampling strategy to modify sensitive parameter values simultaneously (Bratley & Fox, 1988). We analysed the simulation results for similar model parameter values that led to the Adn–/– regeneration phenotype. Our results revealed that the magnitude and timing of IL-6 signalling controlled the priming response of hepatocytes and therefore fine-tuned the timing of initiation of regeneration. Timing and magnitude of the GF peak controlled hepatocyte entry into the replicating phase and therefore fine-tuned the overall tissue regeneration rate and magnitude.
However, modulating these molecular parameters did not adequately account for the observed regeneration profile of Adn–/– mice. In all the simulated scenarios, the hepatocytes entered the cell cycle either too early, renormalized too late, or showed a large increase in cell death in the early phase after PHx coupled with a large overshoot in recovery. These regeneration profiles were inconsistent with the experimental observations in the Adn–/– mice. Because the length of the cell cycle was modelled as lasting approximately 30 h, any molecular changes that allow renormalization of regeneration by 54 h cause regeneration to increase earlier than was seen experimentally in the Adn–/– mice. To account for this difference, we increased the replication rate of hepatocytes in the Adn–/– condition. By increasing hepatocyte replication rate by 15%, the model captured the experimentally observed regenerative profile, including a delay in initiation of regeneration, similar replication by 36 h after PHx, and renormalization by 54 h after PHx (Fig. 4A). Table3 contains the key parameters and their modified values for which the model simulations exhibit an Adn–/– regeneration phenotype that is consistent with the experimental observations.
Figure 4. Modelling the dynamics of the Adn–/– regeneration phenotype.
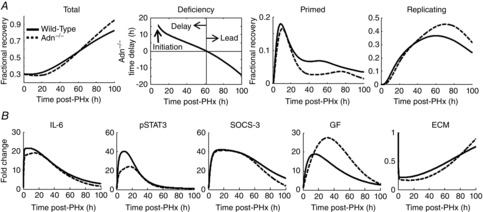
A, simulated profile for Adn–/– mice showed delayed onset of liver regeneration, followed by renormalized liver mass at approximately 60 h after PHx and enhanced recovery thereafter. This profile was mediated by delayed and suppressed priming as well as replication during the first 20 h after PHx and enhanced replication beyond 30 h after PHx. B, the Adn–/– phenotypic changes were governed by decreased IL-6 signalling in the priming phase, causing decreased STAT3 phosphorylation, and increased GF signalling during peak hepatocyte replication.
Table 3.
Modified parameter values
| Parameter | Original value | Adn–/– value |
|---|---|---|
| M | 16.8 | 19.3365 |
| kIL6 | 1.5 | 0.7326 |
| κIL6 | 0.9 | 0.0017 |
| κdeg | 7 | 15.2973 |
| κECM | 33 | 37 |
| kGF | 0.113 | 0.04 |
| κGF | 0.23 | 0.05 |
| kup | 0.06 | 0.0561 |
| kprol | 0.02 | 0.023 |
Simulations with these parameters predicted that one of the key features driving the Adn–/– regeneration phenotype was a slightly decreased IL-6 level, detectable by 3 h after PHx (Fig. 4B). This moderate decrease (∼2% decrease in peak levels) led to a simultaneous decrease in tyrosine phosphorylation of STAT3 by 3 h after PHx (Fig. 4B). Despite this decrease, the levels of phosphorylated STAT3 (pSTAT3) remained at sufficient levels to induce production of suppressor of cytokine signaling-3 (SOCS-3) at levels nearly identical to that in the WT mice (Fig. 4B). The combination of lower IL-6 and normal SOCS-3 synergistically inhibited STAT3 phosphorylation (∼25% decrease in peak levels) and thus its activity, leading to the impaired priming response in simulated Adn–/– mice underlying the delayed regeneration. Note that the IL-6 levels in the model are representative of the production of multiple inflammatory molecules, release, diffusion, receptor binding and cellular response. Hence, the effects of changing the cytokine milieu after PHx may be seen as relatively small changes in many inflammatory signalling levels instead of an isolated change in IL-6 protein levels.
Our simulations pointed to an increase in GF levels, detectable by 12 h after PHx and peaking at approximately 24 h after PHx, as another key feature driving the Adn–/– regeneration profile (Fig. 4B). This increased GF bioavailability stimulated the hepatocytes in the primed state to begin replication. Our analysis predicted that an increased number of replicating hepatocytes in Adn–/– mice, coupled with an increase in proliferation rate, can compensate for the initial delay in regeneration. Similar to the modelled changes in IL-6 levels, increased GF levels in the simulated scenarios do not necessarily represent a single growth factor but rather reflect a strengthening of the growth factor milieu and their effects on hepatocyte replication. Based on the model predictions, we postulate that an increased bioavailability of growth factors associated with cell cycle progression may also contribute to the enhanced cell cycle rate seen in Adn–/– mice.
Biochemical analysis revealed an altered balance of cytokine and growth factor profiles in Adn–/– mice, consistent with computational model predictions
We next analysed biological correlates of the predicted control factors from the computational analysis. We focused on cytokine production and response during priming (0–6 h after PHx) and growth factor levels leading up to and during peak hepatocyte replication (6–42 h after PHx). We evaluated the changes in the inflammatory cytokines TNFα and IL-6 as well as several growth factors implicated in liver repair: hepatocyte growth factor (HGF), angiogenin-1 (Ang-1) and fibroblast growth factor 2 (FGF-2 or bFGF). TNFα and IL-6 are the main inflammatory-type molecules identified as priming hepatocytes to enter the cell cycle. HGF is a potent mitogen and strongly contributes to hepatocyte entry into the cell cycle (Taub, 2004; Michalopoulos, 2007). Ang-1 contributes to regulation of angiogenesis in a variety of pathological conditions and has been shown to be involved in several processes involved in liver recovery from hepatectomy, including wound healing and negative regulation of inflammation (Pan et al. 2012; Lee et al. 2014). In contrast to these two growth factors, FGF-2 is not typically associated with liver repair, and genetic deletion does not impair regeneration after PHx (Sturm et al. 2004). When FGF-2 is deleted, however, VEGF increases after PHx above that in WT mice, indicating that FGF-2 may act synergistically with VEGF to maintain liver architecture, activate non-parenchymal cells and induce hepatocyte replication.
Levels of TNFα protein, a driver of priming following PHx, were measured in liver tissue lysates. TNFα levels declined 1 h after PHx in Adn–/– mice (Fig. 5A). Both WT and Adn–/– mice showed reduced TNFα levels by 3 h after PHx that remained reduced relative to baseline levels 6 h after PHx (Fig. 5A). No difference in liver TNFα levels was noted between WT and Adn–/– mice at 3 or 6 h after PHx (Fig. 5A). Our data are consistent with membrane-bound TNFα present at baseline being cleaved and degraded following receptor activation. Serum IL-6 levels were significantly elevated relative to baseline levels at 1, 3 and 6 h after PHx in Adn–/– mice and at 3 h after PHx in WT mice, with the peak observed levels occurring 3 h after PHx (Fig. 5B). No significant differences in IL-6 levels were noted between WT and Adn–/– mice (Fig. 5B).
Figure 5. Altered TNFα and IL-6 signalling and enhanced growth factor kinetics following PHx in the Adn–/– mice.
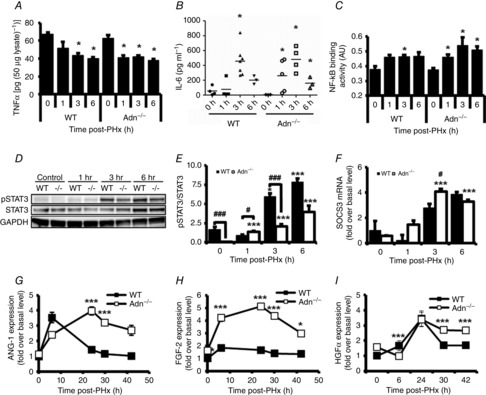
A, TNFα protein levels after PHx were measured in liver lysates by enzyme-linked immunosorbent assay (ELISA) and normalized to total sample protein levels. B, serum IL-6 levels were analysed by Quantibody Multiplex ELISA Array. Each point represents an individual animal and horizontal bars represent mean IL-6 levels at each time point. C, NF-κB DNA binding activity in liver nuclear extracts. D, Western blot of representative liver samples probed with antibodies specific for phosphor-STAT3 (Tyr 705), STAT3 protein and GAPDH. E, quantification of pSTAT3 normalized to total STAT3 protein (n = 3 per group). F, SOCS-3 transcripts were assessed by RT-PCR and normalized to WT control transcript levels. G, quantification of Western blots of liver lysates for ANG-1. H, quantification of Western blots of liver lysates for FGF-2. I, quantification of Western blots of liver lysates for HGFα. Data are presented as mean ± SEM. *P < 0.05, ***P < 0.01 compared with control of respective genotype; #P < 0.05, ###P < 0.01 Adn–/– different from WT at same time point (n = 3 per group).
We assessed intracellular response to these cytokines by measuring NF-κB DNA binding and tyrosine 705 phosphorylated STAT3 (pSTAT3), important mediators of TNFα and IL-6 action, respectively (Fausto et al. 2006). A significant increase in NF-κB DNA binding activity was observed 1 h after PHx in Adn–/– mice (Fig. 5C), simultaneous with the observed reduction in tissue levels of TNFα protein (Fig. 5A). NF-κB activity remained elevated at 3 and 6 h after PHx in Adn–/– mice. Although NF-κB was significantly elevated only at 3 h after PHx in WT mice, no significant differences in NF-κB DNA binding activity were detected between WT and Adn–/– mice (Fig. 5C). Although pSTAT3 levels at baseline were low in both genotypes, Adn–/– mice had significantly lower baseline pSTAT3 levels than WT mice (Fig. 5D,E). pSTAT3 levels increased relative to baseline levels in Adn–/– but not WT mice 1 h after PHx (Fig. 5D,E). Liver pSTAT3 was significantly elevated in WT mice relative to baseline at 3 and 6 h after PHx (Fig. 5D,E), coinciding with elevated serum IL-6 levels (Fig. 5B). Despite similar IL-6 levels in WT and Adn–/– mice at 3 and 6 h after PHx (Fig. 5B), pSTAT3 was significantly lower in Adn–/– mice than WT mice at 3 h after PHx (Fig. 5D,E), possibly leading to reduced hepatocyte priming.
To assess factors that could contribute to these reduced pSTAT3 levels, we analysed transcript levels of SOCS-3, which codes for an inhibitor of STAT3 phosphorylation (Fausto et al. 2006). At 3 h after PHx, SOCS-3 transcripts in Adn–/– mice were elevated fourfold over baseline levels and were significantly higher than in WT mice (Fig. 5F), coincident with reduced pSTAT3 in Adn–/– mice. To test the model prediction of higher growth factor signalling in Adn–/– mice, we measured levels of multiple growth factors known to influence liver regeneration: Ang-1, FGF-2 and HGF. We observed that Adn–/– mice showed significantly elevated and persistent Ang-1 levels at 24 and 30 h after PHx (Fig. 5G). Adn–/– mice also showed FGF-2 levels higher than WT mice at all times examined after PHx (Fig. 5H). Expression of HGF proceeded similarly in WT and Adn–/– mice up to 24 h after PHx. The increase in HGF levels persisted in Adn–/– mice, however, at both 30 and 42 h after PHx (Fig. 5I), suggesting a sustained HGF signal in Adn–/– mice. Thus, our results show decreased STAT3 phosphorylation during the priming phase coupled with sustained elevation in several growth factors, consistent with our model predictions of their putative contributions to the altered Adn–/– regeneration phenotype. Increased levels of SOCS-3 may contribute to decreased STAT3 phosphorylation, which may induce lower hepatocyte priming. Additionally, elevated growth factors in Adn–/– mice coincident with accelerated cell cycle progression provide a potential mechanism underlying the observed acceleration of cell cycle progression in Adn–/– animals.
Adn can directly inhibit growth factor-mediated proliferation in part through direct binding of growth factors and inhibiting their association with their cognate receptors. This has been shown in vitro for platelet-derived growth factor BB (PDGF BB), heparin-binding epidermal growth factor-like growth factor (HB-EGF) and FGF-2 (Arita et al. 2002; Wang et al. 2005; Fayad et al. 2007). We therefore investigated how Adn levels changed in WT mice following PHx. We found that serum Adn decreased significantly during the onset of S phase in WT mice, 30 h after PHx, and remained low through 48 h after PHx (Fig. 6).
Figure 6. Adn serum levels following PHx in WT mice.
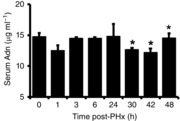
Adn decreased during onset of S phase (30 h after PHx) and remained decreased at 42 and 48 h after PHx.
Computational analysis predicts that overexpression of Adn disrupts regeneration by dysregulating the cytokine and growth factor profiles
We speculated that Adn-mediated fine-tuning of liver regeneration may be non-linear, with increasing Adn levels leading to profoundly different effects than that of lowering Adn levels by the same degree. We explored this possibility using the computational model by simulating the putative effects of increased Adn levels during liver regeneration. We evaluated the effect of deviations to parameters in opposite direction to those required for matching the Adn–/– regeneration. This scenario approximated an increase of Adn to twice the normal physiological levels of serum Adn prior to PHx. The resulting regeneration profile showed that increased Adn led to an initially accelerated regeneration (6–12 h after PHx) followed by a suppression of tissue mass recovery (Fig. 7A). The underlying molecular changes, however, were not merely the opposite of that of the Adn–/– scenario. Our simulation results revealed that increasing Adn levels will lead to a relatively minor decrease in IL-6 signalling and STAT3 phosphorylation during the first 12 h after PHx, without significant effect on the hepatocyte priming response during this time (Fig. 7B). Increasing Adn levels also lengthened the priming response by sustaining IL-6 signalling and STAT3 phosphorylation after PHx, detectable by 48 h after PHx. In spite of this increased priming response, our simulations predicted that GF levels will be decreased at all times after PHx, counteracting any potentially beneficial effects of increased priming and leading to deficient regeneration in mice with increased Adn levels.
Figure 7. Modelling the regeneration dynamics corresponding to the rosiglitazone-induced supraphysiological Adn phenotype.
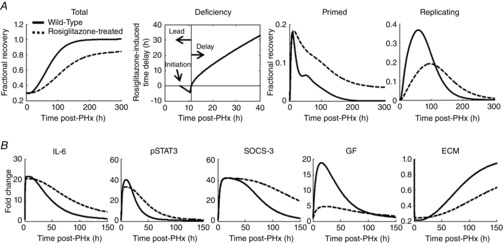
A, the key controlling factors corresponding to the Adn–/– phenotype were altered in the opposite manner to simulate the effect of an increase in Adn levels. The simulated profile for rosiglitazone-treated mice showed suppressed liver regeneration. Although rosiglitazone treatment initially caused a slight lead in liver recovery due to increased priming, ultimately reduced replication suppressed liver recovery from PHx. B, these phenotypic changes were governed by sustained IL-6 signalling but decreased GF signalling throughout regeneration.
Rosiglitazone-induced supraphysiological levels of Adn inhibited hepatocyte replication in WT but not Adn–/– mice
We tested the model predictions on the effects of elevation of Adn on hepatocyte replication after PHx by pharmacologically increasing serum Adn levels in WT and Adn–/– mice. We utilized rosiglitazone, an anti-diabetic drug known to elevate serum Adn levels (Nawrocki et al. 2006; Tao et al. 2010). Animals were administered rosiglitazone (10 mg kg−1) or vehicle by gavage twice a day during the 2 days preceding PHx. Blood samples were taken before treatment and at harvest, and livers were assessed for BrdU incorporation and cyclin A expression at the peak of S phase, 36 h after PHx. Rosiglitazone treatment was associated with a 60% elevation in serum Adn in WT mice relative to controls both before surgery and at harvest (Fig. 8A). No serum Adn was detected in Adn–/– mice at any time, which is consistent with previous studies of rosiglitazone effects in Adn–/– mice (Tao et al. 2010). Rosiglitazone treatment was associated with significant reductions in both BrdU incorporation (Fig. 8B) and cyclin A protein levels (Fig. 8C) 36 h after PHx in WT mice compared to vehicle-treated controls. Adn–/– mice showed no differences relative to vehicle-treated controls (Fig. 8C,D). Cyclin D1 protein levels were also reduced in rosiglitazone-treated WT mice relative to Adn–/– mice (at a 90% confidence level) (Fig. 8D).
Figure 8. Effects of rosiglitazone treatment on hepatocyte replication after PHx in WT and Adn –/– mice.
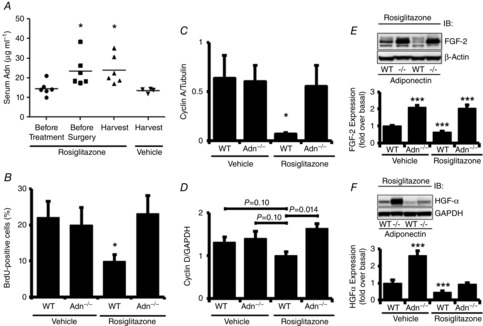
A, serum samples taken from tail blood of WT mice at various times were assessed for Adn by ELISA. B, the percentage of BrdU-positive hepatocytes was calculated from liver sections stained for BrdU and quantifying BrdU-positive nuclei and total nuclei from five representative fields at 20× magnification (n = 4 per group). C, quantification of Western blots for cyclin A (n = 3 per group). D, quantification of Western blots for cyclin D1 (n = 3 per group). E, Western blot of representative liver samples probed with antibodies specific for FGF-2 and β-actin. (Lower) Quantification of Western blots for FGF-2 (n = 3 per group). F, Western blot of representative liver samples probed with antibodies specific for HGF-α and β-actin. (Lower) Quantification of Western blots for HGF-α (n = 3 per group). Data are presented as mean ± SEM. ***P < 0.01, *P < 0.05 significantly different from vehicle-treated WT mice. ###P < 0.01 compared to vehicle-treated Adn–/– mice.
We additionally investigated the effects of elevated Adn on growth factors at 36 h after PHx. Consistent with the model-based predictions, rosiglitazone treatment decreased both FGF-2 and HGF levels in WT animals at 36 h after PHx (Fig. 8E,F). Adn–/– animals, however, showed a more complex response with no change to FGF-2 levels but a marked decrease in HGF levels (Fig. 8E,F). This indicates that in addition to stimulating Adn to decrease HGF levels, rosiglitazone may both indirectly (through Adn) and directly inhibit HGF production. This is in direct conflict with a recent study reporting that rosiglitazone induces HGF production in vitro in isolated lung fibroblasts (Bogatkevich et al. 2012). Other studies, however, have found that rosiglitazone treatment in vitro inhibits HGF production by patient-derived primary effusion lymphoma cells (Bhatt et al. 2010). Together with our data, these studies indicate potential cell or tissue type-specific effects of rosiglitazone treatment on HGF production.
Our data show that rosiglitazone treatment elevates serum Adn levels in WT mice and, consistent with model-based predictions, inhibits PHx-induced GF bioavailability, cyclin A expression and BrdU incorporation in WT mice. Rosiglitazone-treated Adn–/– mice, on the other hand, show no inhibition of cyclin A expression or BrdU incorportation but a differential GF response to PHx, with normal levels of FGF-2 and low levels of HGF (compared to untreated Adn–/– mice) at 36 h after PHx. These results suggest that elevated Adn inhibits hepatocyte proliferation through decreasing growth factor response to PHx but that rosiglitazone has one or more additional inhibitory effecta on some growth factor levels that are independent of Adn.
Discussion
The PHx studies reported here demonstrate that Adn–/– mice have a delayed onset of hepatocyte proliferation compared to WT mice after PHx but show no difference in BrdU incorporation or liver mass recovery 54 h after PHx. This suggests that the loss of Adn suppresses the processes associated with priming of the liver, at least in part through a deregulation of STAT3 signalling. Cell cycle kinetics, however, eventually accelerate to normalize the regenerative response. Our model analysis suggests that this acceleration is probably due to sustained increases in critical pro-proliferative growth factors.
During hepatocyte priming, Adn–/– mice have similar serum IL-6 levels to WT mice; however, they have reduced pSTAT3, coupled with increased expression of the STAT3 inhibitor SOCS-3. The computational model suggests that small deficiencies in IL-6 signalling transduction during the priming phase (modelled as a ∼2% decrease in IL-6 levels) in Adn–/– mice may be responsible for larger decreases in downstream STAT3 phosphorylation (up to ∼25% decrease, based on the model). Two clear implications arise from these results. The first is that subtle, unobservable changes in upstream signalling may cause large, significant changes downstream. Therefore, it is important to consider systems-level interactions when unravelling complex disease phenotypes. The second implication is that interventions with relatively small effect on targeted upstream regulators may be effective at renormalizing the altered regeneration phenotypes.
Our data showing delayed onset of hepatocyte proliferation after PHx are consistent with previous reports involving Adn–/– mice (Ezaki et al. 2009; Shu et al. 2009). In addition, the latter authors also observed reduced STAT3 activation coordinated with SOCS3 upregulation at 24 and 48 h after PHx in Adn–/– mice relative to WT. However, STAT3 phosphorylation at these times is much lower than STAT3 phosphorylation at 3 and 6 h after PHx (Aoyama et al. 2009), and the functional importance of the later phosphorylation remains unclear.
Additionally, other signalling processes may contribute to the observed profile. Adn may also regulate changes in liver ceramide levels after hepatectomy (Correnti et al. 2014). TNFα is a potent activator of sphingomyelinase, which hydrolyses sphingomyelin to ceramide. Therefore, inflammatory signals early after PHx probably promote increases in cellular ceramide, which have been observed after PHx (Alessenko et al. 1999). We expect this effect to be enhanced in Adn–/– mice because of the absence of Adn receptor-dependent ceramidase activity (Holland et al. 2011). Increased ceramide levels have also recently been linked to elevated levels of the tyrosine phosphatase SHP-1 (Gopalan et al. 2013). Increased SHP-1, which can dephosphorylate STAT3, provides an additional potential mechanism for the abrogated STAT3 signalling observed in Adn–/– mice.
While hepatocytes are in the replicating stage of regeneration, Adn–/– mice have sustained higher levels of growth factors that are known drivers of regeneration as well as growth factors not typically associated with regeneration. HGF is one of only a few potent mitogens which can induce hepatocyte proliferation without the benefit of cofactors (Court et al. 2002). It is produced predominantly in hepatic stellate cells, can be bound to the ECM and is released from ECM MMPs produced by non-parenchymal cells. HGF signals through the c-Met receptor in hepatoctyes to stimulate regeneration (Taub, 2004; Michalopoulos, 2007). The sustained increase in HGF suggests hepatocytes from Adn–/– mice receive a more sustained growth signal, which may both promote cell cycle entry and contribute to the accelerated cell cycle progression in Adn–/– mice. Ang-1 is one of the highest expressed genes in activated hepatic stellate cells in vitro and contributes to vascularization in tissues (Jiang et al. 2006; Pan et al. 2012). Higher Ang-1 levels probably correspond to increased tissue remodelling in Adn–/– mice to maintain liver architecture during later periods of enhanced regeneration. In contrast to these two growth factors, FGF-2 has been shown to have little effect during liver regeneration in WT animals. It is therefore not surprising that FGF-2 levels were not altered after PHx in WT mice. In contrast, Adn–/– mice expressed elevated FGF-2 following PHx, suggesting that Adn negatively regulates the FGF-2 response. Also, an earlier report of an increase in VEGF after PHx in mice carrying a genetic deletion of FGF-2 suggests that FGF-2 can act similarly to VEGF to regulate liver structure and non-parenchymal cell activity. By measuring these three growth factors, we were able to characterize classical growth factor signalling to hepatocytes, remodelling growth factor signalling influencing non-parenchymal cell activity, and compensatory or additional growth factor signalling (Sturm et al. 2004). The sustained bioavailability of these growth factors in Adn–/– mice suggests that cells producing these growth factors (predominantly hepatic stellate cells) may be constitutively activated or activated to an alternative phenotype following PHx in the absence of Adn (Jiang et al. 2006; Friedman, 2008).
A recent study suggests that Adn may inhibit hepatic stellate cell activation and induce apoptosis by binding to AdipoR1 and AdipoR2, inducing activation of PPAR-α (Ding et al. 2005). It is possible that the absence of Adn removes this inhibition on stellate cell activation, thus enabling stellate cell-produced factors to persist longer in the liver, including the growth factors FGF-2, Ang-1 and HGF. The altered dynamics of growth factor signalling during the first 20 h after PHx, however, indicates that the modulatory effect of Adn on stellate cells is more complex than a simple activation/deactivation relationship.
Adn has also been shown to directly inhibit growth factor-mediated proliferation in part through direct binding of growth factors and inhibiting their association with their cognate receptors (Fayad et al. 2007; Wang et al. 2005). We observed significant decreases in serum Adn during the onset of S phase in WT mice, 30 h after PHx. While these decreases were modest, because serum Adn levels are tightly regulated, small decreases in Adn may have larger effects on sequestering GFs (Nawrocki et al. 2006). Although GFs were higher in Adn–/– mice, we noted no differences in cyclin D1 expression between genotypes at this time, suggesting the effect of elevated Adn is to block hepatocyte cell cycle after G1, potentially at the G1/S transition. We have investigated intracellular pathways classically activated by growth factors (ERK, JNK, Akt) but have found no differential regulation between WT and Adn–/– mice.
Rosiglitazone also modulates liver regeneration, probably in part through raising serum Adn. Rosiglita-zone-treated mice show higher levels of serum Adn and opposite changes in the progression of regenera-tion to that observed in Adn–/– mice. Rosiglitazone-treated mice have lower growth factor levels and lower regeneration markers than WT mice. Our data show that deficient regeneration is associated with a significant elevation of serum Adn in WT mice and is abrogated in the Adn–/– mice, suggesting that Adn is required for this effect. This is also consistent with a growing body of literature demonstrating that Adn is required for the full beneficial effects of rosiglitazone treatment in diabetic patients (Combs et al. 2002; Nawrocki et al. 2006; Hoo et al. 2007; Tao et al. 2010). Rosiglitazone, however, may have an additional inhibitory effect on HGF production that is independent of Adn.
These results have further implications for the systemic effects of rosiglitazone (and possibly other drugs of the glitazone class that act through increasing Adn levels). Rosiglitazone, which probably has tissue-specific effects, does not specifically target the liver. Previous studies have shown that rosiglitazone treatment increases the risk of myocardial infarction and subsequent death from cardiovascular causes in humans (Nissen & Wolski, 2007), decreases the extent of lung injury in animals (Honiden & Gong, 2009) and may be protective in cancer (Monami et al. 2008), in addition to blunting liver repair as we have shown in the present study and has been shown previously (Turmelle et al. 2006). Additionally, our study suggests that Adn is required for the suppressive effect of rosiglitazone on liver regeneration. It is possible that the sustained inflammatory response to injury and reduced GF response that we observed may parallel the effect of rosiglitazone on other tissues as well, which may also be mediated by Adn.
Our integrated experimental and computational modelling demonstrates that Adn regulates liver regeneration through modulating multiple opposing hepatocyte signalling inputs from non-parenchymal cells governing the rate of progression through the cell cycle, cytokine signalling and growth factor bioavailability. Adn probably fine-tunes the dynamics of regeneration by enhancing the onset of hepatocyte proliferation during the priming phase by increasing STAT3 phosphorylation but suppressing overall liver regeneration through sequestration of GFs and decreasing GF persistence in the liver.
Glossary
- Adn
adiponectin
- Adn–/–
adiponectin knockout
- Ang-1
angiogenin-1
- β
response parameter
- CCl4
carbon tetrachloride
- DLS
dynamic local sensitivity
- ECM
extracellular matrix
- FGF-2 or bFGF
fibroblast growth factor 2
- GF
growth factor
- HGF
hepatocyte growth factor
- IE
immediate early
- IL-6
interleukin 6
- JAK
Janus kinase
- kiSOCS3
SOCS3 inhibition parameter of STAT3 production
- kMXX
Michaelis–Menten constant
- kprol
replication rate
- kP
P to R rate parameter
- kQ
Q to P rate parameter
- kR
R to Q rate parameter
- kup
GF uptake rate by ECM
- kXX
XX production rate
- k1–k7
steady-state molecular production rates
- κXX
XX degradation rate
- M
metabolic demand
- M/N
metabolic load
- N
number of hepatocytes
- P
primed
- PBS
phase-based sensitivity
- PHx
partial hepatectomy
- PS
pulsatile sensitivity
- PSA
pulsatile sensitivity analysis
- pSTAT3
phosphorylated STAT3
- proSTAT3
monomeric STAT3
- θ
threshold parameter
- Q
quiescent
- R
replicating
- SHP-1
Src homology region 2 domain-containing phosphatase-1
- SOCS-3
suppressor of cytokine signalling 3
- STAT3
signal transducer and activator of transcription 3
- TNFα
tumour necrosis factor alpha
- VEGF
vascular endothelial growth factor
- VXX
Michaelis–Menten maximum reaction rate
- WT
wild-type
Additional information
Competing interests
The authors declare that no conflicts of interest exist.
Author contributions
R.V. and J.B.H. conceived and designed the research. J.M.C., J.B.H., D.C. and R.V. designed the experimental procedures. J.M.C., E.A., A.S. and D.C. performed the experiments. R.V., B.O. and D.C. designed the computational analysis. D.C. performed the computational analysis. D.C., J.M.C., B.O., J.B.H. and R.V. contributed to writing the paper.
Funding
Funding for this work was provided by the National Institutes of Health, National Institute on Alcohol Abuse and Alcoholism grants R01 AA018873, R21 AA022417 and T32 AA007463.
Acknowledgements
We thank Dr Lawrence Chan for kindly donating the Adn–/– mice used in this study and Dr Periwal for providing his MATLAB code.
Author's present address
J. M. Correnti: Division of Gastroenterology, University of Pennsylvania, Philadelphia, PA 19107, USA. Edita Aksamitiene: Department of Otolaryngology – Head & Neck Surgery, Thomas Jefferson University, Philadelphia, PA 19107, USA.
References
- Aksamitiene E, Hoek JB, Kholodenko B. Kiyatkin A. Multistrip Western blotting to increase quantitative data output. Electrophoresis. 2007;28:3163–3173. doi: 10.1002/elps.200700002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessenko AV, Platonova LV, Sakevarashvili GR, Khrenov AV, Shingarova LN, Shono NI. Galperin EI. Role of endogenous TNF-α and sphingosine in induced DNA synthesis in regenerating rat liver after partial hepatectomy. Biochemistry (Mosc) 1999;64:890–895. [PubMed] [Google Scholar]
- Aoyama T, Ikejima K, Kon K, Okumura K, Arai K. Watanabe S. Pioglitazone promotes survival and prevents hepatic regeneration failure after partial hepatectomy in obese and diabetic KK-A(y) mice. Hepatology. 2009;49:1636–1644. doi: 10.1002/hep.22828. [DOI] [PubMed] [Google Scholar]
- Arita Y, Kihara S, Ouchi N, Maeda K, Kuriyama H, Okamoto Y, Kumada M, Hotta K, Nishida M, Takahashi M, Nakamura T, Shimomura I, Muraguchi M, Ohmoto Y, Funahashi T. Matsuzawa Y. Adipocyte-derived plasma protein adiponectin acts as a platelet-derived growth factor-BB-binding protein and regulates growth factor-induced common postreceptor signal in vascular smooth muscle cell. Circulation. 2002;105:2893–2898. doi: 10.1161/01.cir.0000018622.84402.ff. [DOI] [PubMed] [Google Scholar]
- Awazawa M, Ueki K, Inabe K, Yamauchi T, Kubota N, Kaneko K, Kobayashi M, Iwane A, Sasako T, Okazaki Y, Ohsugi M, Takamoto I, Yamashita S, Asahara H, Akira S, Kasuga M. Kadowaki T. Adiponectin enhances insulin sensitivity by increasing hepatic IRS-2 expression via a macrophage-derived IL-6-dependent pathway. Cell Metab. 2011;13:401–412. doi: 10.1016/j.cmet.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Barra R. Hall JC. Liver regeneration in normal and alloxan-induced diabetic rats. J Exp Zool. 1977;201:93–99. doi: 10.1002/jez.1402010111. [DOI] [PubMed] [Google Scholar]
- Bhatt AP, Bhende PM, Sin SH, Roy D, Dittmer DP. Damania B. Dual inhibition of PI3K and mTOR inhibits autocrine and paracrine proliferative loops in PI3K/Akt/mTOR-addicted lymphomas. Blood. 2010;115:4455–4463. doi: 10.1182/blood-2009-10-251082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogatkevich GS, Highland KB, Akter T. Silver RM. The PPARγ agonist rosiglitazone is antifibrotic for scleroderma lung fibroblasts: mechanisms of action and differential racial effects. Pulm Med. 2012;2012:545172. doi: 10.1155/2012/545172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratley P. Fox B. Algorithm 659 implementing Sobol's quasirandom sequence generator. ACM Trans Math Software. 1988;14:88–100. [Google Scholar]
- Combs TP, Wagner JA, Berger J, Doebber T, Wang WJ, Zhang BB, Tanen M, Berg AH, O'Rahilly S, Savage DB, Chatterjee K, Weiss S, Larson PJ, Gottesdiener KM, Gertz BJ, Charron MJ, Scherer PE. Moller DE. Induction of adipocyte complement-related protein of 30 kilodaltons by PPARγ agonists: a potential mechanism of insulin sensitization. Endocrinology. 2002;143:998–1007. doi: 10.1210/endo.143.3.8662. [DOI] [PubMed] [Google Scholar]
- Correnti JM, Juskeviciute E, Swarup A. Hoek JB. Pharmacological ceramide reduction alleviates alcohol-induced steatosis and hepatomegaly in adiponectin knockout mice. Am J Physiol Gastrointest Liver Physiol. 2014;306:G959–973. doi: 10.1152/ajpgi.00395.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court FG, Wemyss-Holden SA, Dennison AR. Maddern GJ. The mystery of liver regeneration. Br J Surg. 2002;89:1089–1095. doi: 10.1046/j.1365-2168.2002.02166.x. [DOI] [PubMed] [Google Scholar]
- Crumm S, Cofan M, Juskeviciute E. Hoek JB. Adenine nucleotide changes in the remnant liver: an early signal for regeneration after partial hepatectomy. Hepatology. 2008;48:898–908. doi: 10.1002/hep.22421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Saxena NK, Lin S, Xu A, Srinivasan S. Anania FA. The roles of leptin and adiponectin: a novel paradigm in adipocytokine regulation of liver fibrosis and stellate cell biology. Am J Pathol. 2005;166:1655–1669. doi: 10.1016/S0002-9440(10)62476-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki H, Yoshida Y, Saji Y, Takemura T, Fukushima J, Matsumoto H, Kamada Y, Wada A, Igura T, Kihara S, Funahashi T, Shimomura I, Tamura S, Kiso S. Hayashi N. Delayed liver regeneration after partial hepatectomy in adiponectin knockout mice. Biochem Biophys Res Commun. 2009;378:68–72. doi: 10.1016/j.bbrc.2008.10.176. [DOI] [PubMed] [Google Scholar]
- Fausto N, Campbell JS. Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- Fayad R, Pini M, Sennello JA, Cabay RJ, Chan L, Xu A. Fantuzzi G. Adiponectin deficiency protects mice from chemically induced colonic inflammation. Gastroenterology. 2007;132:601–614. doi: 10.1053/j.gastro.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furchtgott LA, Chow CC. Periwal V. A model of liver regeneration. Biophys J. 2009;96:3926–3935. doi: 10.1016/j.bpj.2009.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan A, Yu W, Sanders BG. Kline K. Eliminating drug resistant breast cancer stem-like cells with combination of simvastatin and γ-tocotrienol. Cancer Lett. 2013;328:285–296. doi: 10.1016/j.canlet.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Gunawan R. Doyle FJ., 3rd Phase sensitivity analysis of circadian rhythm entrainment. J Biol Rhythms. 2007;22:180–194. doi: 10.1177/0748730407299194. [DOI] [PubMed] [Google Scholar]
- Holland WL, Miller RA, Wang ZV, Sun K, Barth BM, Bui HH, Davis KE, Bikman BT, Halberg N, Rutkowski JM, Wade MR, Tenorio VM, Kuo MS, Brozinick JT, Zhang BB, Birnbaum MJ, Summers SA. Scherer PE. Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat Med. 2011;17:55–63. doi: 10.1038/nm.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honiden S. Gong MN. Diabetes, insulin, and development of acute lung injury. Crit Care Med. 2009;37:2455–2464. doi: 10.1097/CCM.0b013e3181a0fea5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoo RL, Chow WS, Yau MH, Xu A, Tso AW, Tse HF, Fong CH, Tam S, Chan L. Lam KS. Adiponectin mediates the suppressive effect of rosiglitazone on plasminogen activator inhibitor-1 production. Arterioscler Thromb Vasc Biol. 2007;27:2777–2782. doi: 10.1161/ATVBAHA.107.152462. [DOI] [PubMed] [Google Scholar]
- Huh CG, Factor VM, Sanchez A, Uchida K, Conner EA. Thorgeirsson SS. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc Natl Acad Sci U S A. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Parsons CJ. Stefanovic B. Gene expression profile of quiescent and activated rat hepatic stellate cells implicates Wnt signaling pathway in activation. J Hepatol. 2006;45:401–409. doi: 10.1016/j.jhep.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K. Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura D, Lee HW, Riley KJ, Arteaga-Solis E, Ferron M, Zhou B, Clarke CJ, Hannun YA, Depinho RA, Guo EX, Mann JJ. Karsenty G. Adiponectin regulates bone mass via opposite central and peripheral mechanisms through FoxO1. Cell Metab. 2013;17:901–915. doi: 10.1016/j.cmet.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landskroner-Eiger S, Qian B, Muise ES, Nawrocki AR, Berger JP, Fine EJ, Koba W, Deng Y, Pollard JW. Scherer PE. Proangiogenic contribution of adiponectin toward mammary tumor growth in vivo. Clin Cancer Res. 2009;15:3265–3276. doi: 10.1158/1078-0432.CCR-08-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kim KW, Min KM, Kim KW, Chang SI. Kim JC. Angiogenin reduces immune inflammation via inhibition of TANK-binding kinase 1 expression in human corneal fibroblast cells. Mediators Inflamm. 2014;2014:861435. doi: 10.1155/2014/861435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Meijer C, Wiezer MJ, Diehl AM, Schouten HJ, Schouten HJ, Meijer S, van Rooijen N, van Lambalgen AA, Dijkstra CD. van Leeuwen PA. Kupffer cell depletion by CI2MDP-liposomes alters hepatic cytokine expression and delays liver regeneration after partial hepatectomy. Liver. 2000;20:66–77. doi: 10.1034/j.1600-0676.2000.020001066.x. [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C. Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- Monami M, Lamanna C, Marchionni N. Mannucci E. Rosiglitazone and risk of cancer: a meta-analysis of randomized clinical trials. Diabetes Care. 2008;31:1455–1460. doi: 10.2337/dc07-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki AR, Rajala MW, Tomas E, Pajvani UB, Saha AK, Trumbauer ME, Pang Z, Chen AS, Ruderman NB, Chen H, Rossetti L. Scherer PE. Mice lacking adiponectin show decreased hepatic insulin sensitivity and reduced responsiveness to peroxisome proliferator-activated receptor gamma agonists. J Biol Chem. 2006;281:2654–2660. doi: 10.1074/jbc.M505311200. [DOI] [PubMed] [Google Scholar]
- Nissen SE. Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. N Engl J Med. 2007;356:2457–2471. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- Pan SC, Wu LW, Chen CL, Shieh SJ. Chiu HY. Angiogenin expression in burn blister fluid: implications for its role in burn wound neovascularization. Wound Repair Regen. 2012;20:731–739. doi: 10.1111/j.1524-475X.2012.00819.x. [DOI] [PubMed] [Google Scholar]
- Park PH, McMullen MR, Huang H, Thakur V. Nagy LE. Short-term treatment of RAW264.7 macrophages with adiponectin increases tumor necrosis factor-α (TNF-α) expression via ERK1/2 activation and Egr-1 expression: role of TNF-α in adiponectin-stimulated interleukin-10 production. J Biol Chem. 2007;282:21695–21703. doi: 10.1074/jbc.M701419200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal TM. Gunawan R. Impulse parametric sensitivity analysis. Proc 18th IFAC World Congr. 2011;18:9686–9690. [Google Scholar]
- Shu RZ, Zhang F, Wang F, Feng DC, Li XH, Ren WH, Wu XL, Yang X, Liao XD, Huang L. Wang ZG. Adiponectin deficiency impairs liver regeneration through attenuating STAT3 phosphorylation in mice. Lab Invest. 2009;89:1043–1052. doi: 10.1038/labinvest.2009.63. [DOI] [PubMed] [Google Scholar]
- Sturm J, Keese M, Zhang H, Bonninghoff R, Magdeburg R, Vajkoczy P, Dono R, Zeller R. Gretz N. Liver regeneration in FGF-2-deficient mice: VEGF acts as potential functional substitute for FGF-2. Liver Int. 2004;24:161–168. doi: 10.1111/j.1478-3231.2004.0896.x. [DOI] [PubMed] [Google Scholar]
- Tao L, Wang Y, Gao E, Zhang H, Yuan Y, Lau WB, Chan L, Koch WJ. Ma XL. Adiponectin: an indispensable molecule in rosiglitazone cardioprotection following myocardial infarction. Circ Res. 2010;106:409–417. doi: 10.1161/CIRCRESAHA.109.211797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- Turer AT. Scherer PE. Adiponectin: mechanistic insights and clinical implications. Diabetologia. 2012;55:2319–2326. doi: 10.1007/s00125-012-2598-x. [DOI] [PubMed] [Google Scholar]
- Turmelle YP, Shikapwashya O, Tu S, Hruz PW, Yan Q. Rudnick DA. Rosiglitazone inhibits mouse liver regeneration. FASEB J. 2006;20:2609–2611. doi: 10.1096/fj.06-6511fje. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lam KS, Xu JY, Lu G, Xu LY, Cooper GJ. Xu A. Adiponectin inhibits cell proliferation by interacting with several growth factors in an oligomerization-dependent manner. J Biol Chem. 2005;280:18341–18347. doi: 10.1074/jbc.M501149200. [DOI] [PubMed] [Google Scholar]
- Yamauchi T. Kadowaki T. Adiponectin receptor as a key player in healthy longevity and obesity-related diseases. Cell Metab. 2013;17:185–196. doi: 10.1016/j.cmet.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Zak DE, Stellingb J. Doyle FJ., III Sensitivity analysis of oscillatory (bio)chemical systems. Comput Chem Eng. 2005;29:663–673. [Google Scholar]


