Abstract
Common scab of potatoes is a disease, which is difficult to manage due to complex interactions of the pathogenic bacteria (Streptomyces spp.) with soil, microbial community and potato plants. In Bohemian-Moravian Highlands in the Czech Republic two sites (Vyklantice and Zdirec) were selected for a study of common scab disease suppressivity. At both sites, a field with low disease severity occurs next to one with high severity and the situation was regularly observed over four decades although all four fields undergo a crop rotation. In the four fields, quantities of bacteria, actinobacteria and the gene txtB from the biosynthetic gene cluster of thaxtomin, the main pathogenicity factor of common scab, were analyzed by real-time PCR. Microbial community structure was compared by terminal fragment length polymorphism analysis. Soil and potato periderm were characterized by contents of carbon, nitrogen, phosporus, sulphur, calcium, magnesium, and iron. Quality of organic matter was assessed by high performance liquid chromatography of soil extracts. The study demonstrated that the suppressive character of the fields is locally specific. At Zdirec, the suppressivity was associated with low txtB gene copies in bulk soil, while at Vyklantice site it was associated with low txtB gene copies in the tuberosphere. The differences were discussed with respect to the effect of abiotic conditions at Zdirec and interaction between potato plant and soil microbial community at Vyklantice. Soil pH, Ca soil content or cation concentrations, although different were not in the range to predict the disease severity. Low severity of common scab was associated with low content of soil C, N, C/N, Ca and Fe suggesting that oligotrophic conditions may be favorable to common scab suppression.
Introduction
The common scab of potatoes is an important soil-born disease with worldwide occurrence. It has been rated among the top five diseases of potatoes by seed producers in the USA [1]. The disease affects tuber quality due to superficial and pitted lesions that form around the site of infection [2]. The infection is caused by actinobacteria from the genus Streptomyces that possess a large (325–660 kb) pathogenicity island (PAI) in their genomes. The most important pathogenicity determinant is a phytotoxin thaxtomin synthetized from two amino-acid precursors via a condensation step catalyzed by nonribosomal peptide synthetase coded by txtAB genes. These genes are used for determination and quantification of pathogens responsible for the disease [3, 4, 5].
The distribution, severity and incidence of common scab have been widely studied in relationship to physico-chemical and microbial soil characteristics but the disease is still difficult to control. Traditional control strategies of common scab such as irrigation and reduced soil pH are not sufficient and often fail because the involved relationships are very complex. Several studies indicated that control measures may be soil specific [5], though it is unclear whether this is due to physical soil properties, or soil microbial flora [1].
Naturally occurring disease-suppressive soils provide a valuable resource for implementation of a sustainable alternative to current control strategies [6]. Yet more information about those soils functioning needs to be collected because different, often locally specific, mechanisms lead to soil suppressiveness. Those include situations: (i) the pathogen does not establish or persist at the site, (ii) the pathogen establishes but causes little or no damage, or (iii) the pathogen establishes and causes disease for a while but thereafter the disease ceases, although the pathogen may persist in the soil [7].
Fields suppressive to common potato scab were identified at various locations, for example in central Washington, Grand Rapids and Becker, MN, East Lansing, MI [8]. In all of those sites the suppressivity was attributed to biological interactions between antagonistic microflora and pathogens mediated via antibiotic production or enzymatic activity [9]. That type of suppressivity can establish after many years of potato (or other crop) monoculture because new protective biotic interactions are stimulated over time by continuous co-occurrence of pathogens and competing microorganisms. However, the mechanisms of suppression vary and particularly in soils undergoing regular crop rotation the suppressive character may be related to geological origin and soil physico-chemical characteristics [9].
The Bohemian-Moravian Highlands represent an important potato growing area in central Europe, where particularly seed potatoes, representing production of about 70 thousand of metric tons are produced and distributed in Europe and near East. The certification for seed potatoes requires less than 5% of surface irregularities. Soil conditions in the area do not belong to those favorable to common scab because they are usually characterized by pH between 5–6 and relatively high spring and summer precipitation with annual average about 800 mm. However, common potato scab is widely spread producing significant damage to potatoes and consequently causing economic losses.
Long history of potato growing in the area enabled to identify fields with low incidence of common potato scab. In this study, two sites Vyklantice and Zdirec where a field with low potato scab severity lies next to a field with high disease severity were selected. Those two sites undergo a regular four crop rotation, while the differences in severity of the disease between the respective fields have been constant over four decades.
The goal of the study was to determine and prove the suppressive or conducive character of the two nearby fields at each site. The approach was based on a field experiment, in which three potato cultivars susceptible to common scab were grown at the two sites (four fields) in a Latin square design with four replicates. The three cultivars were selected to account for the variability of relationships between potatoes and pathogens. The soil suppressivity or conducivity of the studied fields were determined by relationships between common scab severity, quantity of thaxtomine biosynthetic gene txtB, soil and periderm chemical characteristics, quality of organic matter and microbial community. Differences between those relationships determined in fields with low and high severity of common scab were used to predict mechanisms involved in disease suppressivity and to identify soil descriptors, either abiotic or biotic, associated with soil health and suitability for potato growing.
Methods
Ethics Statement
The field study did not involve vertebrates, endangered or protected species; it was not carried out in a protected area. The experiments were carried out on private arable land with the permission of the owners, the companies Vysocina Vyklantice a.s. (fields VL, VH) and Vesa Ceska Bela a.s. (fields ZL, ZH). No other permissions were required.
Sites
Vyklantice and Zdirec are sites where two fields (VL, VH, and ZL, ZH, resp.) about 100m distant differ in common scab severity (marked by L and H for low and high scab severity at two sites Vyklantice, Zdirec (V, Z) by observation over 30 years. The two sites differ in environmental factors, while the two fields within each site are similar in geological context, soil type, climate and management (Table 1). All studied fields were regularly planted under four-field crop rotation system including rape, clover, potatoes, and grains (wheat, oats) in the past two decades.
Table 1. Site characteristics.
|
Vyklantice
|
Zdirec
|
|||
|---|---|---|---|---|
| VL | VH | ZL | ZH | |
| coordinates | 49°33'48"N, 15°03'31"E | 49°33'43"N, 15°03'18"E | 49°37'37"N, 15°37'56"E | 49°37'48"N, 15°37'58"E |
| altitude [MAMSL] | 600 | 590 | 510 | 510 |
| clay [%] | 3 | 0 | 12 | 6 |
| silt [%] | 16 | 30 | 56 | 56 |
| sand [%] | 72 | 62 | 29 | 35 |
| gravel [%] | 9 | 8 | 3 | 3 |
| CEC [cmol/kg] | 13.3 | 16 | 15.4 | 20.1 |
Field experiment
The field experiment was conducted in 2010. Potatoes were planted on May 27 and the samples of bulk soil, tuberosphere soil and potatoes were collected after 80 days. Three cultivars susceptible to common scab Agria, David and Valfi were used. Potatoes were all certified seed tubers (common scab below 5% surface). Four replicates of each cultivar at each field were used and arranged in a Latin square design (3 cultivars × 4 plots plus 4 bulk soils at each field and site, 16 samples per field × 4 fields). Each plot was planted with 3 rows of 12 potato plants (36 plants) separated by 50 cm of bare soil. The effect of the site and field (selectively also the effect of a cultivar) on common scab severity was assessed. At all fields, soil chemical characteristics were determined by total soil C, N, S, P, Ca, Mg, Fe content, potato chemical characteristics were determined by N, P, Ca, Mg, Fe periderm content. Microbial characteristics were described by actinobacterial diversity, quantity of bacteria, actinobacteria and thaxtomin biosynthetic gene txtB in both soil and potato periderm.
Sampling
One potato plant growing in the center of each plot was sampled. Tuberosphere soil samples were collected no further than 3 mm from a potato tuber. A tuber was located by careful uncovering the top soil surrounding the plant, slightly pressed to the remaining soil and taken out. The socket remaining in soil after potato extraction was carefully scratched by a sharp spoon to collect a thin layer of soil. Soil was also collected from the tuber itself if any soil remained attached on the tuber in a thin layer. Bulk soil was collected at a distance of cca 30 cm from the closest plant within each plot using a small sterile spade. To demonstrate the effect of potato plants on microbial community, results were calculated either for all soil samples (identified “soil”) or separately for bulk or tuberosphere soil. Potatoes from the central plant were collected and washed in distilled water. All potatoes were carefully pealed by a potato peeler taking approximately 1 mm thick periderm samples, the peels were homogenized and mixed and subsamples were taken for further analyses. Common scab severity was evaluated on 20 potatoes per plot using a 9 degree scale [10]. Potatoes used for evaluation were those of the collected plant and several more plants from each plot to achieve at least 20 measurements per plot.
Soil and potato periderm analyses
To determine total soil C, N, and S content 2-gram aliquots of homogenized soil samples from both bulk and tuberosphere were dried, milled, and analyzed using Vario MAX CNS analyzer (Elementar Analysensysteme, Hanau, Germany). To determine all other soil elements soil subsamples were leached with boiling nitro-hydrochloric acid (aqua regia) and assessed by optical emission spectroscopy with inductively coupled plasma (ICP-OES) in Aquatest company (Prague, Czech Republic). The analyses of potato periderm were performed by the service laboratory of the Institute of Botany (Trebon, Czech Republic). For total nitrogen analysis, 1–3 mg dried periderm was mineralized by modified Kjeldahl method in H2SO4 with catalyzer at 360°C. For total phosphorus analysis, 20 mg dried periderm was sequentially decomposed by HNO3 and HClO4 [11]. In the mineralized samples, both N and P were determined by flow injection analysis with spectrophotometric detection using FIA Lachat QC 8500 analyzer (Lachat Instruments, Hach Company, Loveland, CO). Cation contents in periderm were determined by atomic absorption spectrometry using AAS spectrometer ContrAA 700 (Analytik Jena, Jena, Germany) after mineralization with nitro-hydrochloric acid.
DNA extraction
Soil samples from tuberosphere and bulk soil were homogenized and subsamples of 0.5 g were used for DNA extraction by method SK described by Sagova-Mareckova et al. [12]. Briefly, the method is based on bead-beating and phenol/chloroform extraction followed by purification with CaCl2 and a GeneClean Turbo kit (MP Biomedicals, Santa Ana, CA). For DNA extraction from potato periderm, 3 g of periderm samples were fine cut in sterile Petri dish, homogenized, and 0.3 g subsample was processed in the same way to obtain total periderm DNA.
HPLC analysis of extractable low-molecular-weight soil metabolites
Soil samples for HPLC analyses were collected from the tuberosphere of Agria cultivar and bulk soil from the same plots (8 samples per field, 32 in total). Approximately 50 ml of soil were extracted with approximately 50 ml of methanol-water-acetic acid (80:19:1, vol/vol/vol) at room temperature for 1 to 2 days. The extracts were filtered and vacuum concentrated to remove organic solvent. The resulting aqueous suspension was chromatographed over Amberlite XAD 1180 (5g of aqueous suspension; diameter of glass column, 2 cm). Fifty ml of water (MilliQ quality) was used as effluent for the first (hydrophilic) fraction; 50 ml of absolute ethanol was used for the second (lipophilic) fraction. The ethanolic fraction was evaporated and diluted in methanol at a concentration of 5 mg ml-1. The analysis was performed using Acquity UPLC system with the 2996 PDA detector (Waters, Milford, MA). The column was a Phenomenex Luna 5 µm C18(2) 100 Å, 100 × 4.6 mm, equipped with SecurityGuard cartridge C18, 4 × 3.0 mm ID (Phenomenex, Torrance, CA). The separation was run in air-conditioned room at 20°C, and the flow rate was 1.0 ml min-1. Solvent A was ultrapure water; solvent B was acetonitrile. The gradient started with 10% of solvent B for 1 min and then increased linearly to 100% of B within 8 min. The final concentration was held for a further 3.5 min. Twenty µl of each sample were injected. The spectra were recorded from 194 to 800 nm in 1 nm intervals and data points recorded once per second were exported from MassLynx software (Waters) in text format for further processing.
Terminal restriction fragment length polymorphism analysis (T-RFLP)
Soil and periderm DNA samples were analyzed by amplification of genes for 16S rRNA using universal forward primer 16Seu27f (5´-AGAGTTTGATCMTGGCKCAG-3´) [13] 5-end labelled with 6-FAM (6-carboxyfluorescein) and Actinobacteria-specific reverse primer 16Sact1114r (5´-GAGTTGACCCCGGCRGT-3´) [14]. Reaction mixture contained in a total volume of 50 µl: 1× polymerase buffer, 1.5 mM MgCl2, 400 nM of each primer, 0.2 mM of each dNTP, 0.6 mg/ml BSA, 1 U Taq polymerase (TopBio, Prague, Czech Republic). PCR amplification was performed in C1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA) and the PCR program consisted of an initial denaturation at 94°C for 2 min, followed by 35 cycles of 94ºC for 45 s, annealing at 57ºC for 45 s, extension at 72ºC for 1 min 30 s, and a final extension step at 72ºC for 5 min. Labelled PCR products were purified using QIAquick PCR purification kit (Qiagen, Hilden, Germany). Labelled PCR amplicons of the 16S rRNA gene region were cleaved by AluI restriction endonuclease for 4 h at 37ºC with subsequent additions of the enzyme in two equal aliquots at the beginning and in the middle. After inactivation of the restriction enzymes and purification with Sigma Spin Post-Reaction Clean-Up Columns (Sigma-Aldrich, St. Louis, MO), the samples were subjected to fragment analysis on a 96-capillary sequencer (Applied Biosystems, Foster City, CA) in Genomac company (Prague, Czech Republic). T-RFLP analyses were performed in one run for all samples. Crude profiles were filtered to the largest T-RF peaks representing 95% of the total of peak heights (areas) to eliminate the noise intermittently exceeding the cut-off limit.
Quantitative PCR
Quantifications were performed with primers eub338f (5’-ACTCCTACGGGAGGCAGCAG-3) [15] and eub518r (5’-ATTACCGCGGCTGCTGG-3’) [16] amplifying a 197 bp fragment of the 16S rRNA gene from Bacteria, act235f (5’-CGCGGCCTATCAGCTTGTTG-3’) [17] and eu518r yielding 280 bp product for Actinobacteria, and StrepF (5’-GCAGGACGCTCACCAGGTAGT-3’) and StrepR (5’-ACTTCGACACCGTTGTCCTCAA-3’) yielding 72 bp amplicon of the thaxtomin biosynthetic gene txtB [18], respectively. The analyses were done on a StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA) using 96-well plates with GoTaq qPCR Master Mix (Promega, Madison, WI) containing SYBR Green as a double-stranded DNA binding dye. The reaction mixture contained in a total volume of 15 µl: 1× GoTaq qPCR Master Mix, 0.2 µM primers, and 0.2–2 ng diluted DNA sample. For all of the mentioned targets the PCR cycling protocol consisted of initial denaturation at 95°C for 10 min, followed by 45 cycles of 95°C for 15s, 60°C for 30s and 72°C for 30s. Melting curves were recorded to ensure qPCR specificity. Baseline and threshold calculations were performed with the StepOne v. 2.2.2 software. The inhibition was tested by serial DNA dilution from each site. All qPCR measurements were done in duplicate.
The qPCR standards were prepared by cloning the fragments of the target genes in pGEM-T Easy vector system (Promega). After PCR verification and isolation of the cloned constructs by Pure Yield Plasmid Miniprep System (Promega), a linear standard was prepared by cleaving with SalI enzyme (New England Biolabs, UK) in a 200 µl reaction mixture containing 1× reaction buffer, 2 µg circular plasmid, and 20 U restriction endonuclease for 2h in 37°C. The linearized plasmid DNA was purified by phenol-chloroform extraction. The aliquots of linearized and purified standard diluted to 20 ng/µl were stored in-70°C.
Data analysis
The differences between suppressive and conducive fields were tested by ANOVA and Welch’s two sample t-test (allowing differences between variability of variables), which aims at detection of differences in mean values. All variables were log-transformed to make their distribution more similar to a normal distribution. The HPLC absorption signals recorded at the wavelength 210 nm were baseline corrected and normalized by division through the median of the absorption values. For the T-RFLP and HPLC profiles, the Manhattan metric (sum of absolute differences) was used to calculate the distance matrices. The distance matrices were plotted by Sammon’s Multidimensional Scaling [19]. Two tests based on distance matrices was used. While PERMANOVA [20] tests that the within group distances are on average shorter than the between group distances (aiming at detection of different mean profiles), dispersion test (denoted later as ‘disp’) introduced in Gijbels and Omelka [21] tests that the average within group distances differ among the groups (aiming at detecting of different dispersions of the samples). The correlation coefficients at different fields were compared through the permutation test introduced in Omelka and Pauly [22]. All statistical calculations were done in the R-computing environment [23].
Results
Common scab
At both sites, Vyklantice (V) and Zdirec (Z), the two fields (VL, VH, and ZL, ZH) differed significantly in common scab severity (VL x VH p < 0.001, ZL x ZH p < 0.001) (Table 2). The fields VH and ZH had higher common scab severity than the respective fields VL and ZL at the same site (Table 2).
Table 2. Biological characteristics.
| VL | VH | ZL | ZH | p-values | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | mean | S.E. | mean | S.E. | mean | S.E. | mean | S.E. | overall ANOVA | VL vs. VH | ZL. vs. ZH | |
| Soil | ||||||||||||
| bacteria | 16 | 10.30 | 0.06 | 10.10 | 0.04 | 10.10 | 0.06 | 10.20 | 0.04 | 0.073 | 0.059 | 0.058 |
| actinobacteria | 16 | 9.57 | 0.05 | 9.53 | 0.03 | 9.57 | 0.03 | 9.75 | 0.04 | 0.001 | 0.534 | 0.001 |
| txtB.total (bulk and tuberosphere) | 16 | 4.38 | 0.07 | 4.49 | 0.12 | 4.93 | 0.19 | 5.57 | 0.14 | < 0.001 | 0.436 | 0.013 |
| txtB.bulk | 4 | 4.53 | 0.04 | 3.97 | 0.07 | 4.00 | 0.22 | 4.93 | 0.06 | < 0.001 | 0.002 | 0.02 |
| txtB.tuberosphere | 12 | 4.33 | 0.09 | 4.67 | 0.13 | 5.80 | 0.14 | 5.27 | 0.15 | < 0.001 | 0.044 | 0.015 |
| Periderm | ||||||||||||
| actinobacteria | 12 | 7.82 | 0.12 | 8.74 | 0.17 | 9.40 | 0.13 | 9.27 | 0.11 | < 0.001 | < 0.001 | 0.45 |
| txtB.periderm | 12 | 6.10 | 0.05 | 7.18 | 0.16 | 7.31 | 0.15 | 7.51 | 0.12 | < 0.001 | < 0.001 | 0.312 |
| Common scab severity | 12 | 2.34 | 0.14 | 4.06 | 0.14 | 4.23 | 0.15 | 6.65 | 0.15 | < 0.001 | < 0.001 | < 0.001 |
Quantity of bacteria, actinobacteria (16S rRNA gene) and txtB gene in soil and periderm. Means and respective standard errors (gene copies per gram soil, log10 transformed). Significantly higher values are in bold.
Soil and periderm biological characteristics
In bulk soil, the field VL had significantly higher txtB gene copies than VH (p = 0.002), while ZL had significantly lower txtB gene copies than ZH (p = 0.020) (Table 2). In tuberosphere soil, the field VL had significantly lower txtB gene copies than the field VH(p = 0.003), while ZL had significantly higher txtB gene copies than ZH (p = 0.015). In periderm, the field VL had significantly lower txtB gene copies than the field VH (p < 0.001), while ZL did not differ from ZH. Also, the field VH had significantly higher actinobacteria in soil than the field VL, while the field ZH had significantly higher actinobacteria in periderm than the field ZL (Table 2).
Soil and periderm chemical characteristics
The field VH had significantly higher pH and contents of soil (bulk and tuberosphere) C, N, P, Ca, Fe and periderm P, Ca, Fe, while it had significantly lower content of soil Mg than the field VL. The field ZH had significantly higher pH and contents of soil (bulk and tuberosphere) C, N, S, Mg, Ca, Fe and periderm Ca, while it had significantly lower soil P than the field ZL (Table 3).
Table 3. Chemical characteristics in soil and periderm.
| VL | VH | ZL | ZH | p-values | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | mean | S.E. | mean | S.E. | mean | S.E. | mean | S.E. | overall ANOVA | VL vs. VH | ZL. vs. ZH | |
| Soil | ||||||||||||
| pH | 16 | 5.61 | 0.03 | 6.43 | 0.03 | 6.87 | 0.03 | 7.01 | 0.03 | < 0.001 | < 0.001 | < 0.001 |
| C (%) | 16 | 1.28 | 0.03 | 2.04 | 0.03 | 1.67 | 0.04 | 2.33 | 0.05 | < 0.001 | < 0.001 | < 0.001 |
| N (%) | 16 | 0.146 | 0.003 | 0.214 | 0.003 | 0.178 | 0.003 | 0.221 | 0.003 | < 0.001 | < 0.001 | < 0.001 |
| S (mg.kg-1) | 16 | 744.0 | 23.1 | 702.0 | 16.8 | 244.0 | 6.5 | 305.0 | 3.3 | < 0.001 | 0.151 | < 0.001 |
| P (mg.kg-1) | 16 | 921.0 | 13.8 | 1180.0 | 12.8 | 858.0 | 20.3 | 722.0 | 7.4 | < 0.001 | < 0.001 | < 0.001 |
| Mg (g.kg-1) | 16 | 12.40 | 0.22 | 11.80 | 0.18 | 3.78 | 0.18 | 4.56 | 0.06 | < 0.001 | 0.043 | 0.001 |
| Ca (g.kg-1) | 16 | 1.89 | 0.10 | 3.20 | 0.13 | 2.34 | 0.11 | 4.15 | 0.10 | < 0.001 | < 0.001 | < 0.001 |
| Fe (g.kg-1) | 16 | 43.00 | 0.63 | 46.80 | 0.87 | 22.50 | 0.89 | 27.10 | 0.38 | < 0.001 | 0.001 | < 0.001 |
| Periderm | ||||||||||||
| N (g.kg-1) | 12 | 23.40 | 1.01 | 25.80 | 0.73 | 23.30 | 0.83 | 23.50 | 0.80 | 0.105 | 0.068 | 0.835 |
| P (g.kg-1) | 12 | 1.95 | 0.10 | 2.19 | 0.12 | 2.71 | 0.10 | 3.18 | 0.20 | < 0.001 | 0.136 | 0.054 |
| Ca (mg.kg-1) | 12 | 0.57 | 0.06 | 1.04 | 0.10 | 0.68 | 0.07 | 1.00 | 0.09 | < 0.001 | 0.001 | 0.008 |
| Mg (mg.kg-1) | 12 | 1.09 | 0.03 | 1.04 | 0.02 | 1.12 | 0.03 | 1.07 | 0.04 | 0.264 | 0.146 | 0.433 |
| Fe (mg.kg-1) | 12 | 0.37 | 0.03 | 0.51 | 0.05 | 0.70 | 0.09 | 0.71 | 0.15 | 0.007 | 0.035 | 0.966 |
Means and respective standard errors. Significantly higher values are in bold.
Actinobacterial communities
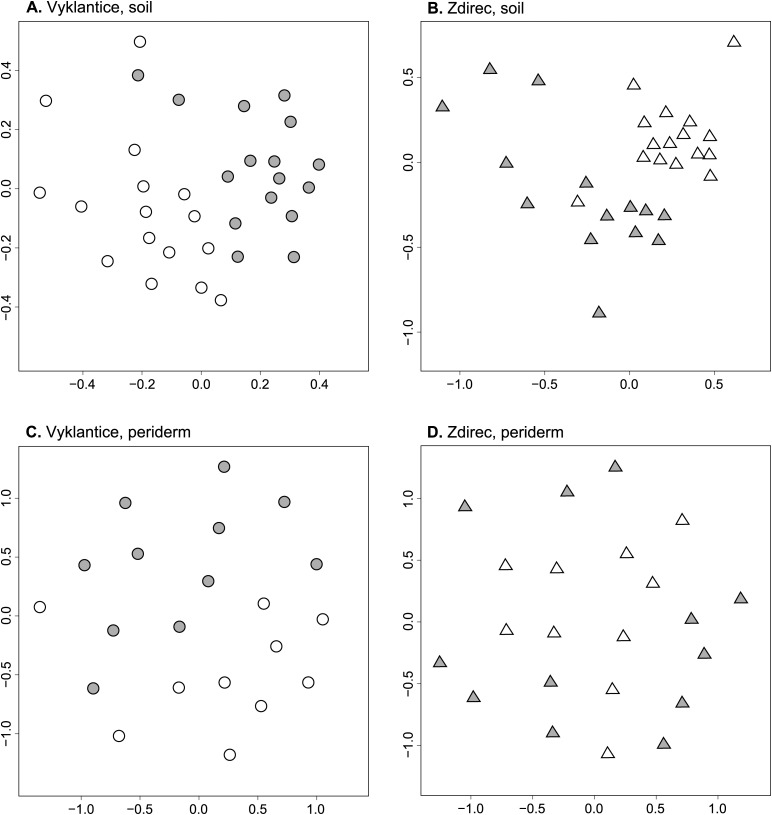
T-RFLP profiles of actinobacteria in soil samples differed significantly between sites Vyklantice and Zdirec (p-permanova < 0.001, p-disp = 0.003, not presented in a figure) and between the fields within each site (Fig. 1; p-permanova < 0.001 both sites, p-disp = 0.042 Zdirec). T-RFLP profiles of actinobacteria in periderm samples did not differ between sites Vyklantice and Zdirec but differed significantly between fields (Vyklantice: p-permanova = 0.009, Zdirec: p-permanova = 0.019 and p-disp = 0.017). T-RFLP profiles of periderm samples were significantly different between cultivars at both sites (Vyklantice: p-disp = 0.005; Zdirec: p-permanova = 0.022; not presented in a figure).
Figure 1. T-RFLP profiles of the communities of actinobacteria sampled at Vyklantice and Zdirec experimental sites from tuberosphere soil (A,B) and potato periderm (C,D).
Sammon’s multidimensional scaling was based on Manhattan distance matrices calculated for the baseline filtered and normalized T-RFLP profiles. The symbols represent the profiles of actinobacteria at the fields with low (open symbols) and high (closed symbols) common scab severity.
Low-molecular-weight compounds
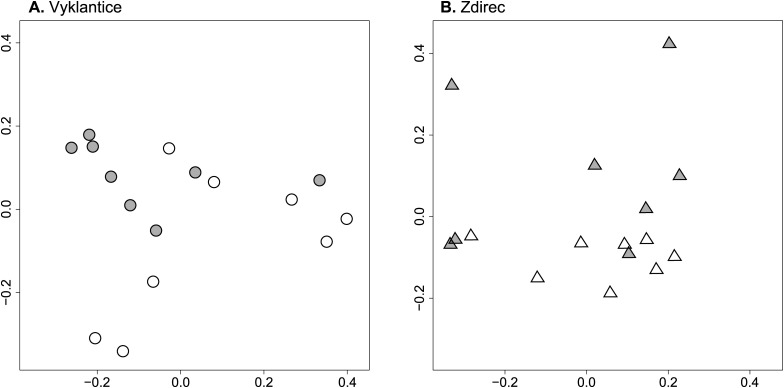
The HPLC profiles of extractable low-molecular-weight compounds of soil samples were significantly different between sites (p-permanova = 0.009) and fields in Vyklantice (p-permanova = 0.033) and Zdirec (p-disp = 0.049) (Fig. 2).
Figure 2. HPLC-UV profiles of extractable low-molecular-weight compounds at Vyklantice (A) and Zdirec (B) sites.
Sammon’s multidimensional scaling was based on Manhattan distance matrices calculated from normalized signal at 210 nm (for details, see Materials and Methods). The symbols represent the profiles of compounds extracted from the fields with low (open symbols) and high (closed symbols) common scab severity.
Discussion
Common scab severity differed between the two fields at each site, while the pathogen identified by the thaxtomin biosynthetic gene txtB was always present. Therefore, we considered the fields with low common scab severity suppressive to the disease. Both proposed suppressive fields represent the long-term/natural type of disease suppressivity functioning over long time in spite of regular crop rotation [24].
This type of suppression is likely a function of the broad physical and chemical characteristics of the soil not only of soil microbial communities [25]. In our study, the fields with higher severity of common scab had in common significantly higher content of soil C, N, C/N, Ca, Fe and higher pH. High soil pH and Ca were associated with high disease severity in other studies [5, 26, 27] but it was demonstrated that even this relationship was site specific. The observed threshold of pH for low disease soils was established by Lacey et al. [28] as a limit to common scab severity below 5.0–5.2 but the soil pH of all studied fields was above this value. Also, common scab was not observed on potatoes grown in soil with combined exchangeable Ca, Mg and K at 12 cmol/kg or less [28] but in our fields only the conducive soil of the ZH field had cation concentration above this value. So, the lower soil pH or cation concentration were not likely the cause of suppressivity in our fields with low disease severity.
The two suppressive fields at both sites had lower Ca in periderm, the field in Vyklantice also periderm Fe. For accumulation of Ca in plant cells induced by thaxtomin it was established that increase in infected cells is rather an effect than a cause of infection because this toxin induced a rapid Ca2+ influx and cell death in Arabidopsis thaliana cell suspensions [29]. Fe ions represent one of redox transformations brokers [30]. Increased iron may be a result of cross talk between iron and manganese, iron and zinc, manganese and phosphate, and copper and zinc, among the many documented interactions, but may be also related to reactive oxygen species (for iron and manganese) as a result of pathogen stress [30]. Since Ca is closely related to soil pH and Fe may be rather a result of infection, just low soil C, N, and C/N remain potentially associated with suppressivity to common scab at our sites. That result was complemented by specifically high soil Mg and P contents in the suppressive fields at Vyklantice and Zdirec, respectively.
Soil organic matter (SOM) can enhance disease-suppressive activities of soil microbial communities [31–33]. In our study, soil organic matter content was high in conducive soils but the HPLC profiles of low molecular weight compounds differed between the fields with proposed suppressivity. Therefore it showed that quality rather than quantity of SOM was related to suppressivity in our fields.
Soil actinobacterial communities were significantly different between the two fields with low and high common scab severity at both sites. That is consistent with previous findings at similar disease suppressive sites, where differences between microbial community structures were assigned the primary cause of disease suppressivity [6, 8]. The T-RFLP method which was used here can demonstrate general differences between actinobacterial communities so we propose that the determined differences must be due to many taxa at various taxonomic levels. The suppressive character of soils was often associated with multiple species [6, 8, 34]. Yet, since soil microbial communities are site specific [35, 36] it may be difficult to show the taxa particularly responsible for soil suppressivity [8, 34].
Periderm actinobacterial communities differed significantly between the fields and potato cultivars at both sites but not between the two sites. That showed the importance of the effect of cultivars in spite of the differences in the soil microbial communities, which was found previously [37]. It was also proposed that the cultivar distinctive microbial community may be related to cultivars’ specific traits including resistance to pathogens [38, 39].
In our study it seemed that the character of suppressivity differed between the two sites. Firstly, the bulk soil at the suppressive field in Zdirec (ZL) had lower number of txtB gene copies than the conducive field (ZH), while in Vyklantice the suppressive field (VL) had higher number of txtB gene copies than the conducive field (VH). Secondly, the tuberosphere was the opposite because the suppressive field in Zdirec (ZL) had significantly higher txtB gene copies than the conducive field (ZH), while the suppressive field in Vyklantice had significantly lower txtB gene copies than the conducive field (VH). Therefore it seemed that at Zdirec the soil conditions directly suppressed the pathogen development because in the suppressive field it was low in bulk soil but increased in the tuberosphere. This may be possibly related to increased phosphorus concentration in the suppressive soil there because P content was previously often associated with low disease severity [40, 41]. In the contrary at Vyklantice the interaction between the potato plant and microbial community may be responsible for the suppression because the pathogen populations decrease in the tuberosphere. Different microbial taxa were found to participate in suppression of diseases either directly or indirectly by enhancing plant nutrition by acquiring limiting nutrients or producing beneficial enzymes (e.g. [24, 42]) and consequently supporting the plant self-protection capabilities. Also actinobacteria, namely nonpathogenic streptomycetes, were suggested to participate in common scab suppression [43].
In conclusion, we demonstrated that indeed the suppressive character of fields is locally specific. In our study it seemed that soil pH, calcium content or cation concentrations were not predicting the disease severity. Microbial communities had a site specific character corresponding to the particular field in soil and corresponding to potato cultivar in periderm. Regarding the chemical soil characteristics it seemed that oligotrophic conditions possibly complemented by specific quality of organic matter are favorable to disease suppression because C, N, Ca and Fe were less in soils with low common scab severity. The ratio between the number of txtB gene copies in soil and common scab severity was a good indicator of suppressivity. In the future studies, the effect of soil P and Mg on suppression of the disease in the respective situations may be applied to evaluate the use in management of the disease severity.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The supporting information files including results of all analyses in replicates, T-RFLP and HPLC profiles are available under Dryad Digital Repository entry doi:10.5061/dryad.332dj.
Funding Statement
The study was supported by the Ministry of Agriculture of the Czech Republic, grant No. QJ1210359 and project MZe RO0414 (http://www.mze.cz), and Czech Science Foundation, grant No. P201/11/P290 (http://www.gacr.cz). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dees MW, Wanner LA (2012) In Search of Better Management of Potato Common Scab. Potato Res 55: 249–268. 10.1007/s11540-012-9206-9 [DOI] [Google Scholar]
- 2. Archuleta JG, Easton GD (1981) The cause of deep-pitted scab of potatoes. Am Potato J 58: 385–392. 10.1007/BF02852950 [DOI] [Google Scholar]
- 3. Kers JA, Cameron KD, Joshi MV, Bukhalid RA, Morello JE, et al. (2005) A large, mobile pathogenicity island confers plant pathogenicity on Streptomyces species. Mol Microbiol 55: 1025–1033. 10.1111/j.1365-2958.2004.04461.x [DOI] [PubMed] [Google Scholar]
- 4. Hiltunen LH, Laakso I, Chobot V, Hakala K, Weckman A, Valkonen JPT (2006) The influence of thaxtomins in different combinations and concentrations on growth of micropropagated potato shoot cultures. J Agric Food Chem 54: 3372–3379. 10.1021/jf060270m [DOI] [PubMed] [Google Scholar]
- 5. Lazarovits G, Hill J, Patterson G, Kenneth L, Conn KL, Crump NS (2007) Edaphic soil levels of mineral nutrients, pH, organic matter, and cationic exchange capacity in the geocaulosphere associated with potato common scab. Phytopathology 97: 1071–1082. 10.1094/PHYTO-97-9-1071 [DOI] [PubMed] [Google Scholar]
- 6. Rosenzweig N, Tiedje JM, Quensen JF III, Meng Q, Hao JJ (2012) Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses. Plant Dis 96: 718–725. 10.1094/PDIS-07-11-0571 [DOI] [PubMed] [Google Scholar]
- 7. Baker KF, Cook RJ (1974) Biological control of plant pathogens. San Francisco: WH Freeman and Company; 10.2307/3758248 [DOI] [Google Scholar]
- 8. Meng QX, Yin JF, Rosenzweig N, Douches D, Hao JJ (2012) Culture-based assessment of microbial communities in soil suppressive to potato common scab. Plant Dis 96: 712–717. 10.1094/PDIS-05-11-0441 [DOI] [PubMed] [Google Scholar]
- 9. Almario J, Kyselková M, Kopecký J, Ságová-Marečková M, Muller D, et al. (2013) Assessment of the relationship between geologic origin of soil, rhizobacterial community composition and soil receptivity to tobacco black root rot in Savoie region (France). Plant Soil 371: 397–408. 10.1007/s11104-013-1677-1 [DOI] [Google Scholar]
- 10. Wenzl H, Demel J (1967) Bildskalen für die Beurteilung von Kartoffelschorf und Rhizoctonia-Pocken. Der Pflanzenarzt 20: 77–78. [Google Scholar]
- 11. Kopáček J, Hejzlar J (1995) Semi-micro determination of total phosphorus in soils, sediments, and organic materials: A simplified perchloric acid digestion procedure. Commun Soil Sci Plant Anal 26: 1935–1946. 10.1080/00103629509369419 [DOI] [Google Scholar]
- 12. Ságová-Marečková M, Čermák L, Novotná J, Plháčková K, Forstová J, et al. (2008) Innovative methods for soil DNA purification tested in soils of widely differing characteristics. Appl Environ Microbiol 74: 2902–2907. 10.1128/AEM.02161-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cermak L, Kopecky J, Novotna J, Omelka M, Parkhomenko N, et al. (2008) Bacterial communities of two contrasting soils reacted differently to lincomycin treatment. Appl Soil Ecol 40: 348–358. 10.1016/j.apsoil.2008.06.001 [DOI] [Google Scholar]
- 14. Kyselková M, Kopecký J, Felföldi T, Čermák L, Omelka M, et al. (2008) Development of a 16S rRNA gene-based prototype microarray for the detection of selected actinomycetes genera. Antonie Van Leeuwenhoek 94: 439–453. 10.1007/s10482-008-9261-z [DOI] [PubMed] [Google Scholar]
- 15. Lane D (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. West Sussex: John Wiley & Sons; pp. 115–175. [Google Scholar]
- 16. Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stach JEM, Maldonado LA, Ward AC, Goodfellow M, Bull AT (2003) New primers for the class Actinobacteria: application to marine and terrestrial environments. Environ Microbiol 5: 828–841. 10.1046/j.1462-2920.2003.00483.x [DOI] [PubMed] [Google Scholar]
- 18. Qu X, Wanner LA, Christ BJ (2008) Using the TxtAB Operon to Quantify Pathogenic Streptomyces in Potato Tubers and Soil. Phytopathology 98: 405–412. 10.1094/PHYTO-98-4-0405 [DOI] [PubMed] [Google Scholar]
- 19. Venables WN, Ripley BD (2002) Modern applied statistics with S. New York: Springer; 10.1007/978-0-387-21706-2 [DOI] [Google Scholar]
- 20. McArdle BH, Anderson MJ (2001) Fitting multivariate models to community data: A comment on distance based redundancy analysis. Ecology 82: 290–297. 10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2 [DOI] [Google Scholar]
- 21. Gijbels I, Omelka M (2013) Testing for Homogeneity of Multivariate Dispersions Using Dissimilarity Measures. Biometrics 69: 137–145. 10.1111/j.1541-0420.2012.01797.x [DOI] [PubMed] [Google Scholar]
- 22. Omelka M, Pauly M (2012) Testing equality of correlation coefficients in two populations via permutation methods. J Stat Plan Infer 142: 1396–1406. 10.1016/j.jspi.2011.12.018 [DOI] [Google Scholar]
- 23. R Core Team (2014) R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- 24. Kyselkova M, Moenne-Loccoz Y (2012) Pseudomonas and other Microbes in Disease-Suppressive Soils. In: Lichtfouse E, editor. Organic Fertilisation, Soil Quality and Human Health 93. Sustainable Agriculture Reviews 9. Springer Science+Business Media B.V. pp. 93–140. 10.1007/978-94-007-4113-3_5 [DOI] [Google Scholar]
- 25. Kinkel LL, Bakker MG, Schlatter DC (2011) A Coevolutionary Framework for Managing Disease-Suppressive Soils. Annu Rev Phytopathol 49: 47–67. 10.1146/annurev-phyto-072910-095232 [DOI] [PubMed] [Google Scholar]
- 26. Lambert DH, Powelson ML, Stevenson WR (2005) Nutritional interactions influencing diseases of potato. Am J Potato Res 82: 309–319. 10.1007/BF02871961 [DOI] [Google Scholar]
- 27. Davis JR, McDole RE, Callihan RH (1976) Fertilizer effects on common scab of potato and the relation of calcium and phosphatephosphorus. Phytopathology 66: 1236–1241. 10.1094/Phyto-66-1236 [DOI] [Google Scholar]
- 28. Lacey MJ, Wilson CR (2001) Relationship of common scab incidence of potatoes grown in Tasmanian ferrosol soils with pH, exchangeable cations and other chemical properties of those soils. J Phytopathol 149: 679–683. 10.1046/j.1439-0434.2001.00693.x [DOI] [Google Scholar]
- 29. Errakhi R, Dauphin A, Meimoun P, Lehner A, Reboutier D, et al. (2008) An early Ca2+ influx is a prerequisite to thaxtomin A-induced cell death in Arabidopsis thaliana cells. J Exp Bot 59: 4259–4270. 10.1093/jxb/ern267 [DOI] [PubMed] [Google Scholar]
- 30. Merchant SS (2010) The Elements of Plant Micronutrients. Plant Physiology 154: 512–515. 10.1104/pp.110.161810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bailey K, Lazarovits G (2003) Suppressing soil-borne diseases with residue management and organic amendments. Soil Tillage Res 72: 169–180. 10.1016/S0167-1987(03)00086-2 [DOI] [Google Scholar]
- 32. Bonanomi G, Antignani V, Pane C, Scala F (2007) Suppression of soilborne fungal diseases with organic amendments. J Plant Pathol 89: 311–324. [Google Scholar]
- 33. Noble R, Coventry E (2005) Suppression of soil-borne plant diseases with composts: a review. Biocontrol Sci Tech 15: 3–20. 10.1080/09583150400015904 [DOI] [Google Scholar]
- 34. Kyselková M, Almario J, Kopecký J, Ságová-Marečková M, Haurat J, et al. (2014) Evaluation of rhizobacterial indicators of tobacco black root rot suppressiveness in farmers’ fields. Environ Microbiol Rep 6: 346–353. 10.1111/1758-2229.12131 [DOI] [PubMed] [Google Scholar]
- 35. Yergeau E, Bezemer TM, Hedlund K, Mortimer SR, Kowalchuk GA, et al. (2010) Influences of space, soil, nematodes and plants on microbial community composition of chalk grassland soils. Environ Microbiol 12: 2096–2106. 10.1111/j.1462-2920.2009.02053.x [DOI] [PubMed] [Google Scholar]
- 36. Schlatter DC, DavelosBaines AL, Xiao K, Kinkel LL (2013) Resource use of soilborne Streptomyces varies with location, phylogeny, and nitrogen amendment. Microb Ecol 66: 961–971. 10.1007/s00248-013-0280-6 [DOI] [PubMed] [Google Scholar]
- 37. Weinert N, Meincke R, Gottwald C, Heuer H, Schloter M, et al. (2010) Bacterial diversity on the surface of potato tubers in soil and the in£uence of the plant genotype. FEMS Microbiol Ecol 74: 114–123. 10.1111/j.1574-6941.2010.00936.x [DOI] [PubMed] [Google Scholar]
- 38. Mazzola M (2002) Mechanisms of natural soil suppressiveness to soilborne diseases. Anton Leeuw 81:557–564. 10.1023/A:1020557523557 [DOI] [PubMed] [Google Scholar]
- 39. Meera MS, Shivanna MB, Kageyama K, Hyakumachi M (1995) Responses of cucumber cultivars to induction of systemic resistance against anthracnose by plant growth promoting fungi. Eur J Plant Pathol 101:421–430. 10.1007/BF01874856 [DOI] [Google Scholar]
- 40. Davis JR, McDole RE, Callihan RH (1976) Fertilizer Effects on common scab of potato and the relation of calcium and phosphate-phosphorus. Phytopathology 66: 1404–1410. 10.1094/Phyto-66-1236 [DOI] [Google Scholar]
- 41. Kristufek V, Divis J, Dostalkova I, Kalcik J (2000) Accumulation of mineral elements in tuber periderm of potato cultivars differing in susceptibility to common scab. Potato Res 43: 107–114. 10.1007/BF02357951 [DOI] [Google Scholar]
- 42. Tokala RK, Strap JL, Jung CM, Crawford DL, Salove MH, et al. (2002) Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum). Appl Environ Microbiol 68: 2161–2171. 10.1128/AEM.68.5.2161-2171.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lorang JM, Anderson NA, Lauer FI, Wildung DK (1989) Disease decline in a Minnesota potato scab plot. Am Potato J 66: 531. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The supporting information files including results of all analyses in replicates, T-RFLP and HPLC profiles are available under Dryad Digital Repository entry doi:10.5061/dryad.332dj.