Abstract
Loss of primary cilia is a key feature of von Hippel-Lindau tumor suppressor (VHL)-associated pathology. Although VHL-deficiency predisposes cells to precipitous cilia disassembly in response to growth factor cues, it does not affect ciliogenesis. Here, using a siRNA-based screen to find genes that are essential for ciliogenesis only in the presence of the VHL tumor suppressor gene product pVHL, we identify ubiquitin-specific protease (USP)8. The pVHL-dependency of USP8 for ciliogenesis is directly linked to its function as a HIF1α deubiquitinating enzyme. By counteracting pVHL-mediated ubiquitination of HIF1α, USP8 maintains a basal expression of HIF1α and HIF transcriptional output in normoxia, including the repression of Rabaptin5, which is essential for endosome trafficking-mediated ciliogenesis.
Keywords: ciliogenesis, endosome trafficking, HIF1α, pVHL, USP8
Introduction
Primary cilia are microtubule-based organelles that serve key functions in the perception of environmental cues and in cell signaling 1. Mounting evidence links ciliary dysfunction or absence of primary cilia to a wide variety of diseases 2. The signaling pathways dynamically regulating the assembly and disassembly of this organelle in response to environmental cues remain poorly understood.
The von Hippel-Lindau (VHL) syndrome is caused by functional inactivation of the VHL tumor suppressor gene product pVHL and is associated with the development of cystic kidney lesions and clear cell renal cell carcinoma (ccRCC) 3. The development of cysts in VHL disease is linked, at least in part, to defects in primary cilium maintenance as a result of VHL loss-associated precipitous resorption of primary cilia in response to growth factors 4 5. Also in vivo, loss of primary cilia and cyst formation requires combined inactivation of Vhl and Pten 5.
While a role for pVHL in the normal control of cilia resorption in response to growth factor stimulation is emerging, pVHL appears to be dispensable for ciliogenesis triggered by growth factor withdrawal at least in normal cells 4. This notwithstanding, VHL-deficiency could still alter certain dependencies of cells on processes normally essential for ciliogenesis by alleviating the requirement for certain essential ciliogenesis genes. Considering that a prime function of pVHL is to act as an E3 ubiquitin ligase 3, we embarked on screening a short-interfering RNA (siRNA) library targeting members of the Ubiquitin Specific Protease (USP) family to identify genes essential for ciliogenesis only in VHL-proficient but not VHL-deficient mammalian cells and identified USP8.
Results and Discussion
USP8 function is required for ciliogenesis depending on VHL status
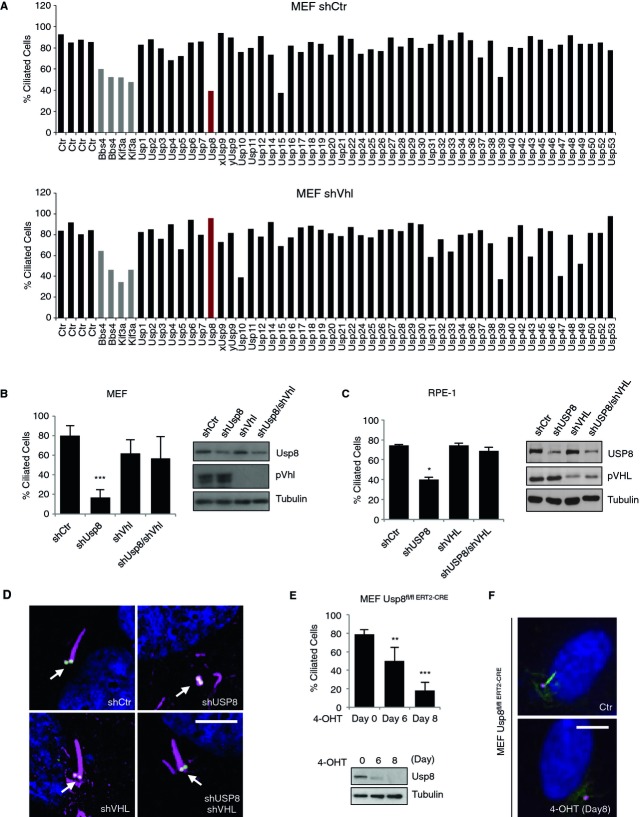
We engineered SV40 largeT-Antigen-immortalized mouse embryonic fibroblasts (MEFs) to stably express either a non-silencing short-hairpin RNA (shRNA) (MEF shCtr) or an shRNA targeting Vhl (MEF shVhl). Stable cell pools were transfected with siRNA pools (four non-redundant siRNAs for each gene) targeting 48 members of the Usp family. The percentage of ciliated cells was assessed after serum starvation in the transfected cells by staining for acetylated tubulin (Fig1A and supplementary Table S1). In the primary screen performed in MEF shCtr, downregulation of three genes, encoding Usp8, Usp15 and Usp39, caused a significant reduction in cilia formation. Validation experiments in the same cells confirmed this observation only for Usp8 and Usp39 (supplementary Fig S1A and B). Rescreening of the Usp siRNA library in MEF shVhl revealed that the phenotype observed for Usp8 was rescued in pVhl-depleted cells, suggesting a pVhl-dependency of Usp8 for cilia formation (Fig1A). Several other Usps including Usp10, Usp31, Usp43, Usp47 and Usp49 displayed a converse behavior or were like Usp39 essential irrespective of Vhl status (Fig1A) and were thus not further pursued.
Figure 1. pVHL-dependency of USP8 function for primary cilia formation.
- Graphs depicting the percentage of ciliated cells in wild-type (shCtr) and pVhl-deficient (shVhl) immortalized MEFs after siRNA knockdown of Usp genes. Control (Ctr) bars on the left represent cells transfected with non-silencing siRNA. Bbs4 and Kif3a are positive controls.
- Frequency of ciliated wild-type MEFs infected with shRNAs against Vhl (shVhl), Usp8 (shUsp8) or both (shUsp8/shVhl). A non-silencing hairpin (shCtr) serves as a control. Knockdown efficiency was verified by Western blotting.
- Frequency of ciliated wild-type hTERT-immortalized RPE-1 cells infected as in (B). Knockdown efficiency was verified by Western blotting.
- Representative immunofluorescence staining of acetylated tubulin, γ-tubulin (basal body, indicated with a white arrow) and DAPI of RPE-1 cells described in (C).
- Percentage of ciliated cells in Usp8fl/fl ERT2-CRE MEFs treated with 4-hydroxytamoxifen (4-OHT) for the indicated times to induce excision of Usp8. Deletion of Usp8 was verified by Western blot.
- Representative immunofluorescence staining of acetylated tubulin, γ-tubulin and DAPI of MEF cells described in (E).
Data information: Bar graphs, mean ± s.d. of (N = 2 (A, MEF shCtr), N = 1 (A, MEF shVhl), N = 3 (B, C, E)). Student's t-test: *, P < 0.05; **, P < 0.01; ***, P < 0.001. N = number of independent experiments performed in triplicates. Scale bar, 10 μm.Source data are available online for this figure.
Stable downregulation of Usp8 and pVhl either individually or in combination in immortalized MEFs (Fig1B) and in human retinal epithelial RPE-1 cells (Fig1C and D) further corroborated the pVhl-dependency of Usp8 for cilia formation. Immunoblotting confirmed the downregulation of USP8 and pVHL where expected (Fig1B and C). Staining for γ-tubulin, a marker for the basal body, remained unaffected by USP8-deficiency in RPE-1 cells (Fig1D). A second shRNA targeting Usp8/USP8 in MEFs and RPE-1 cells confirmed a requirement of this enzyme for ciliogenesis (supplementary Fig S1C and D). Finally, tamoxifen-induced genetic elimination of Usp8 in MEFs derived from Usp8fl/fl mice that express the tamoxifen-inducible Cre recombinase fused to a mutated ligand-binding domain of the human estrogen receptor ERT2 (MEF Usp8fl/fl ERT2-CRE) 6 resulted, likewise, in a failure of such cells to form primary cilia in response to growth factor deprivation (Fig1E and F). When tested, tamoxifen did neither negatively affect ciliogenesis nor Usp8 expression (supplementary Fig S1E). This data provide genetic evidence for a role of Usp8 in cilia formation and further suggest that depletion of pVhl phenotypically suppresses the requirement of Usp8 function for ciliogenesis.
USP8 binds to and deubiquitinates HIF1α
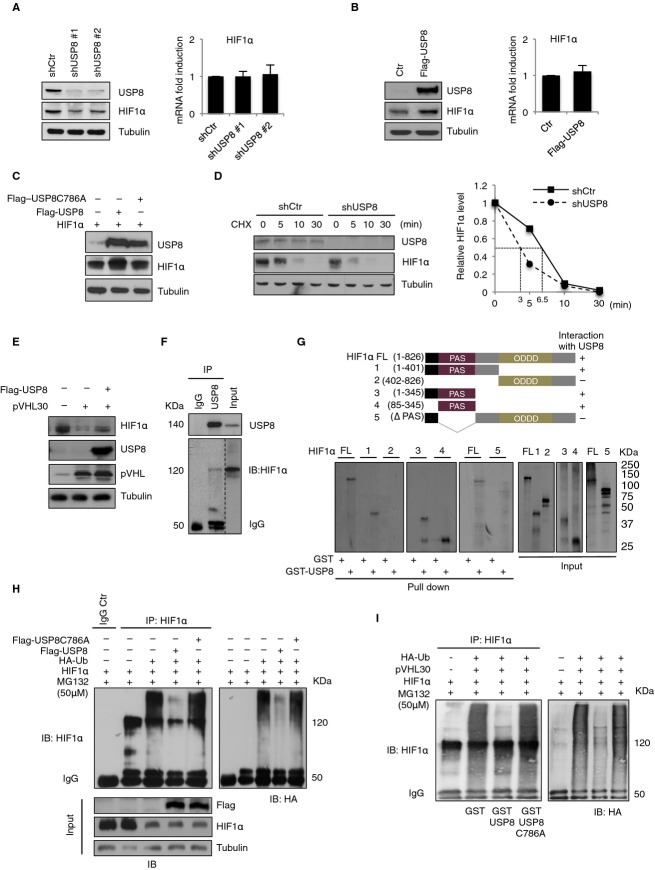
One possible explanation of the observed phenotypic suppression effect is that pVHL and USP8 regulate the ubiquitination state and hence the stability of one or more common targets that are directly required for primary cilia formation. As pVHL E3 ligase targets hypoxia-inducible factor (HIF)α for ubiquitin-mediated degradation 3, we asked whether USP8 functions as a deubiquitination enzyme for HIF1α. Downregulation of USP8 in VHL-positive HEK293T cells with two distinct shRNAs decreased HIF1α protein levels with no effect on HIF1α mRNA levels (Fig2A). Consistent with this observation, the activity of a HIF promoter-luciferase reporter construct 7 was also reduced upon downregulation of USP8 or HIF1α in RPE-1 cells cultured in normoxia or hypoxia (supplementary Fig S2A and B). In the same cells the expression of HIF target genes was also reduced (supplementary Fig S2C–E). Overexpression of a Flag-tagged species of USP8 increased endogenous HIF1α protein but not mRNA levels (Fig2B). Also exogenously produced HIF1α levels rose in the presence of USP8 wild-type but not of a catalytically-inactive mutant (Flag-USP8C786A) (Fig2C), suggesting that the USP8-induced increase in HIF1α abundance depends on USP8 enzymatic activity. USP8 depletion reduced the half-life of endogenous HIF1α in a cycloheximide-chase experiment (Fig2D). In addition, both endogenously and exogenously produced HIF1α were downregulated upon overexpression of pVHL, an effect relieved by coexpression of USP8 (Fig2E and supplementary Fig S2F). Consistent with the above, endogenous HIF1α coimmunoprecipitated with USP8 from HEK293T cell lysates (Fig2F). These results indicate that USP8 binds to and stabilizes HIF1α.
Figure 2. USP8 stabilizes HIF1α by deubiquitination.
- A, B Whole-cell lysates from HEK293T either infected with (A) shRNAs against USP8 (#1 or #2) or (B) transiently transfected with Flag-USP8 vector were processed for Western blotting with indicated antibodies and for qRT-PCR.
- C Cell lysates of HEK293T cells transiently transfected with Flag-USP8 and Flag-USP8C786A in combination with HIF1α were processed for Western blotting with indicated antibodies.
- D RPE-1 cells infected with shRNAs were treated with cycloheximide (CHX, 0.05 mg/ml) and harvested at indicated times. Left panel: immunoblot of USP8 and HIF1α. Right panel: quantification of HIF1α relative to tubulin protein levels.
- E Western blot analysis of endogenous HIF1α in HEK293T transiently transfected with pVHL either alone or in combination with Flag-USP8.
- F Immunoprecipitation with either rabbit control IgG or anti-USP8 antibody from HEK293T cells treated by hypoxia. The immunoprecipitates (IP) and the lysate input were blotted with anti-HIF1α antibody (right, short exposure separated by a dashed line) and then with anti-USP8 antibody.
- G Schematic representation of different HIF1α fragments used in USP8 binding assay. [35S]methionine-labeled in vitro-translated HIF1α fragments (Input: FL, 1, 2, 3, 4, 5) were incubated with bacterially expressed GST or GST-USP8 bound to glutathione sepharose 4B beads. HIF1α fragments retained on beads were detected by autoradiography (Pulldown: FL, 1, 2, 3, 4, 5).
- H In vivo deubiquitination assay using HEK293T cells transfected with HIF1α, HA-Ubiquitin, Flag-USP8 and Flag-USP8C786A plasmids and treated with proteasome inhibitor MG132. In all 5 lanes, identical amounts of HIF1α expression plasmid were transfected. In lanes 3-5 additional plasmids were added as indicated, enhancing the total amount of plasmids in these transfection mixes. In lanes 1 and 2 no additional empty plasmid was added resulting in a reduced amount of total plasmid in the transfection mixes compared to lanes 3-5. HIF1α was immunoprecipitated with either mouse control IgG or anti-HIF1α antibody. The immunoprecipitated HIF1α and the input were immunoblotted with the indicated antibodies.
- I In vitro deubiquitination assay of USP8 using HEK293T cells transfected with HIF1α, HA-Ubiquitin and pVHL vectors, and treated as in (H). Ubiquitinated HIF1α was immunoprecipitated, incubated with bacterially purified GST, GST-USP8 or the catalytically inactive GST-USP8C768A protein, and then immunoblotted with anti-HIF1α antibody.
Data information: Bar graphs, mean ± s.d. (N = 3 (A, B). N = number of independent experiments.).Source data are available online for this figure.
We mapped the domain of HIF1α required for USP8 binding by assessing the ability of various in vitro-translated HIF1α fragments to bind to bacterially-expressed glutathione S-transferase-USP8 (GST-USP8) fusion proteins. As shown in Fig2G and supplementary Fig S3A, the PERN-ARNT-SIM (PAS) domain 8 of HIF1α is both necessary and sufficient for complex formation with USP8 in vitro. Thus, USP8 and pVHL bind to distinct segments of HIF1α involving HIF1α's PAS and oxygen-dependent degradation (ODD) domains, respectively. HIF2α, which also harbors a PAS domain, bound likewise to GST-USP8 in vitro (supplementary Fig S3B). Moreover, a stable proline-mutant derivative of HIF2α, HIF2α(P405A/P531A), coimmunoprecipitates with USP8 (supplementary Fig S3C) and HIF2α levels, like HIF1α levels, were reduced upon depletion of USP8 in HEK293T cells (supplementary Fig S3D). Hence, USP8 functions as a DUB for HIF1α and likely also for HIF2α.
Next we examined whether USP8 serves as a deubiquitinase (DUB) for HIF1α in cells. Immunoprecipitates of ectopically-produced HIF1α from HEK293T cells expressing hemagglutinin (HA)-tagged ubiquitin revealed high levels of HIF1α ubiquitination in the presence of the proteasome inhibitor MG132 (Fig2H). Importantly, HIF1α ubiquitination levels were significantly diminished by coexpression of wild-type Flag-USP8 but not a catalytically-inactive Flag-USP8C786A mutant (Fig2H). Immunoprecipitates of HIF1α were specific as no such bands were detectable in the IgG control (Fig2H). Similarly, GST-USP8 wild-type fusion protein purified from bacteria (supplementary Fig S3E) deubiquitinated in vivo-ubiquitinated HIF1α that was derived from HEK293T cells coexpressing HIF1α, pVHL and HA-ubiquitin and treated with MG132 (Fig2I). The catalytically inactive GST-USP8C786A fusion protein was inactive in this assay (Fig2I and supplementary Fig S3F). HIF1α was previously reported to be targeted by the USP family member USP20 (VDU2) 9. Although overexpression of USP20 was shown to increase HIF target gene expression, the effects on endogenous HIF1α protein levels were not assessed. As shown in supplementary Fig S4, neither overexpression of USP20 nor its efficient downregulation in HEK293T cells resulted in measurable effects on endogenous HIF1α levels. In contrast, downregulation of USP8, done in parallel, caused a decrease in HIF1α. These results demonstrate that USP8 functions as a DUB for HIF1α both in vivo and in vitro under normoxic condition.
Functional relationship between USP8, HIF1α and Rabaptin5 in ciliogenesis
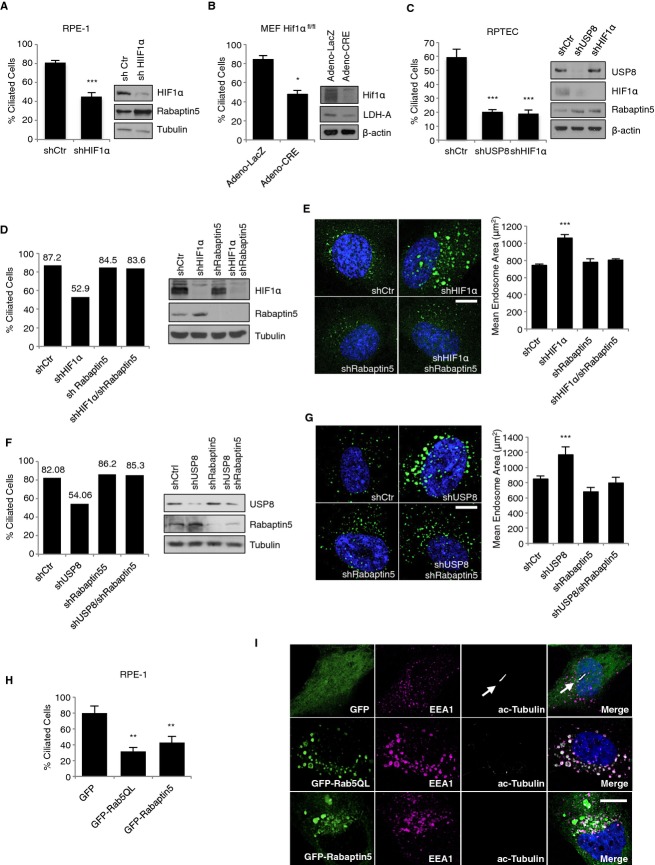
The identification of USP8 as a pVHL-dependent ciliogenesis factor, raised the possibility that the reduction of primary cilia observed in cells silenced by USP8 and rescued by the additional knockdown of pVHL is a result of altered HIF1α levels and the ensuing changes in HIF-dependent transcription of genes whose products are critical for cilia formation. To test this, the primary cilia forming capacity of RPE-1 cells silenced for HIF1α was determined. The number of cells able to form cilia was significantly decreased in HIF1α-depleted cells (Fig3A). Moreover, MEFs genetically-deleted for Hif1α also failed to form cilia (Fig3B). Immunoblotting revealed that HIF1α was absent where expected. Furthermore, the expression of Rabaptin5, a gene product known to be suppressed by HIF1α 10, increased under these conditions, while expression of LDH-A that is activated by HIF1α, decreased (Fig3A and B). Finally, depletion of USP8 or HIF1α negatively affected ciliogenesis in renal proximal tubular epithelial cells (RPTECs) (Fig3C). This data suggest a critical role for HIF1α in the process of ciliogenesis. Depletion of HIF2α neither affected cilia formation in RPE-1 cells to a major extent nor did it enhance the ciliogenesis defect of HIF1α-depleted cells (supplementary Fig S5A). This may be explained by the low levels of HIF2α compared to HIF1α mRNA expression in these cells (supplementary Fig S5B) and the known context-dependent functions of these factors 8.
Figure 3. HIF1α-mediated Rabaptin5 repression is critical for ciliogenesis and early endosome fusion.
- Frequency of ciliated RPE-1 cells infected with lentivirus encoding a control non-silencing hairpin (shCtr) and an shRNA against HIF1α. Whole-cell lysates were immunoblotted with antibodies targeting HIF1α and Rabaptin5.
- Frequency of ciliated isogenic immortalized HIF1αfl/fl MEFs infected with virus encoding Adeno-Cre recombinase or Adeno-LacZ. Hif1α excision was verified by Western blotting with antibodies against HIF1α and LDH-A.
- Percentage of ciliated RPTEC cells infected with lentivirus encoding shRNAs against USP8 or HIF1α and corresponding Western blots.
- Frequency of ciliated RPE-1 cells infected with lentivirus encoding the indicated shRNAs and corresponding Western blots.
- Fluorescence images of RPE-1 cells described in C and stained with early endosome antigen 1 (EEA1) antibody and DAPI. Image analysis and quantification of the mean endosome area of early endosomes was assessed using custom algorithms written in MatLab (see Supplementary Materials and Methods).
- Frequency of ciliated RPE-1 cells infected with lentivirus encoding the indicated shRNAs and corresponding Western blots.
- Fluorescence images of RPE-1 cells described in (E), and stained and analyzed as in (D).
- Frequency of ciliated RPE-1 cells transiently transfected with GFP, GFP-Rab5QL and GFP-Rabaptin5 vectors.
- Representative immunofluorescence staining of RPE-1 cells described in G and stained with acetylated tubulin, EEA1 antibodies and DAPI (white arrows indicate primary cilium).
Data information: Bar graphs, mean ± s.d. (N = 3 (A, B, C), N = 2 (D, G)). N = number of independent experiments performed in triplicates. Student's t-test: *, P < 0.05; **, P < 0.01; ***, P < 0.001. In (E), (F) and (I): Scale bar, 10 μm.Source data are available online for this figure.
Rabaptin5 is a critical Rab5 effector that promotes Rab5-mediated early endosome fusion, which in turn, accelerates endocytosis 11. HIF has been shown to suppress Rabaptin5 transcription thereby negatively affecting endocytic trafficking 10. Moreover, endocytic trafficking is critical for cilia formation 12 13 and a survey of the expression of certain components of the early endosome fusion complex revealed that only Rabaptin5 levels were affected upon depletion of USP8 or HIF1α (supplementary Fig S5C and D). Given this and the fact that USP8 stabilizes HIF1α, it is conceivable that changes in either HIF1α or USP8 levels would directly translate into changes of HIF-dependent Rabaptin5 transcriptional output and consequent endocytic trafficking in the context of ciliogenesis. We first knocked down HIF1α and Rabaptin5 individually or in combination in RPE-1 cells and measured the ability of cells to form primary cilia. Strikingly, the reduction of cilia formation caused by downregulation of HIF1α in RPE-1 cells was rescued by co-depletion of Rabaptin5 (Fig3D). Similar results were obtained in MEFs (supplementary Fig S6). Downregulation of HIF1α was in each system accompanied by increased expression of Rabaptin5 (Fig3D and, supplementary Fig S6). As for USP8 depletion 6, depletion of HIF1α caused a large early endosome phenotype that was rescued by codepletion of Rabaptin5 as evidenced by quantifying early endosome mean area (Fig3E and supplementary Fig S7A). Thus, it appears that Rabaptin5 is a key downstream effector of HIF1α's ciliogenesis-promoting function. Furthermore, these results provide a potential explanation why USP8 function in ciliogenesis is pVHL-dependent.
Also the primary cilia formation defect and early endosome phenotype seen in USP8-depleted cells was rescued by co-depletion of Rabaptin5 (Fig3F and G). The early endosome phenotype caused by USP8 depletion was also rescued by co-depletion of pVHL (supplementary Fig S7B–D). Together, these results suggest a critical role for an USP8-HIF1α-Rabaptin5 axis in primary cilia formation.
One would predict that activation of HIF1α in USP8-depleted cells would reverse the associated ciliogenesis defect. Indeed, stabilization of HIF1α in RPE-1 cells through different means including hypoxia (supplementary Fig S8A), treatment of cells with prolyl hydroxylase inhibitor (supplementary Fig S8B) or overexpression of a stable mutant form of HIF1α that lacks the ODD domain (HIF1α-ΔODD) (supplementary Fig S8C), rescued in each case the ciliogenesis defect induced by knockdown of USP8. Hypoxia also rescued the ciliogenesis defect associated with depletion of HIF1α (supplementary Fig S8D). Furthermore, depletion of USP8 failed to affect ciliogenesis in VHL-deficient RCC4 renal carcinoma cells (RCC4-Ctr) but affected this process in RCC4 cells engineered to re-express wild-type pVHL (RCC4-VHL) (supplementary Fig S8E). This data further support the notion that a critical target of USP8 in the process of ciliogenesis is HIF1α.
Finally, to assess whether changes in Rabaptin5 levels and Rab5 activity are indeed critical for ciliogenesis, we overexpressed a constitutively active form of Rab5 (Rab5Q79L) or Rabaptin5 as green-fluorescent protein (GFP) fusion proteins alongside with GFP alone as a control and scored for the formation of primary cilia. Importantly, these fusion proteins, when overexpressed, reduced cilia formation (Fig3H). GFP expression alone did not significantly impact ciliogenesis irrespective of expression levels (supplementary Fig S9A and 14). Expression of these proteins caused also, as expected, aberrantly enlarged early endosomes (Fig3I) 11 15. The results were also confirmed in MEFs (supplementary Fig S9B and C). These results link the early endosome fusion process to the assembly of primary cilia.
Disruption of USP8 or HIF1α function impairs endocytic recycling
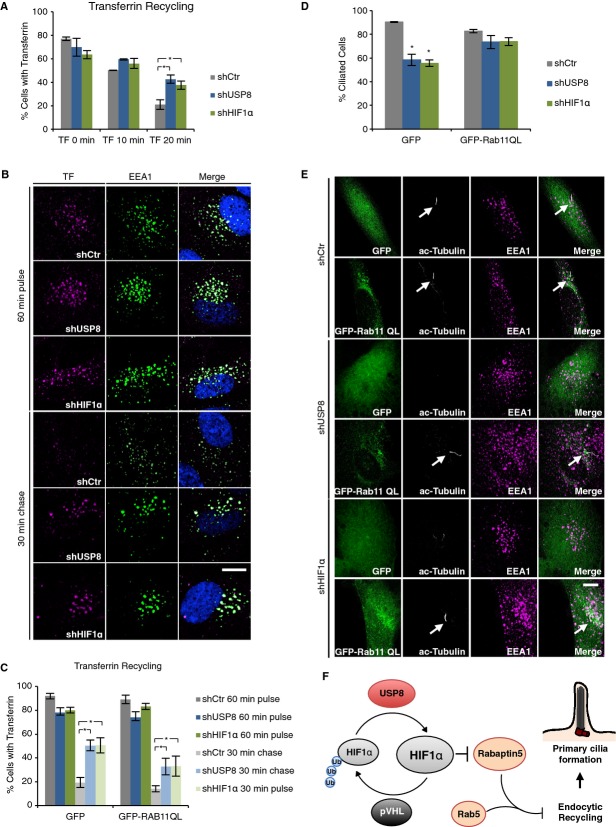
Enlarged early endosomes caused by expression of Rab5Q79L have been shown to negatively influence endosome recycling as scored by recycling of transferrin from early endosomes back to the cell surface 15. As endosome recycling was previously shown to be required for ciliogenesis 13, we monitored the endocytic recycling rate of transferrin in RPE-1 cells lacking USP8 or HIF1α. Interestingly, the depletion of USP8 or HIF1α resulted in evident slowing of transferrin recycling, as compared to transferrin retention in control cells (Fig4A), and concomitant accumulation of transferrin in the enlarged early endosomes (Fig4B). Endocytic recycling mediated by Rab8 and Rab11 have been shown to regulate ciliogenesis insofar as overexpression of dominant-negative forms of these two small GTPases inhibit ciliogenesis 12 13. Moreover, Rab11 inactivation was previously shown to negatively affect transferrin recycling to the plasma membrane 16. Therefore, we tested whether the ciliogenesis defect observed in cells depleted for USP8 or HIF1α can be rescued, at least in part, by overexpression of a constitutively-active form of Rab11 fused to GFP (GFP-Rab11Q70L) in RPE-1 cells. Strikingly, overexpression of GFP-Rab11Q70L restored, partially, both transferrin recycling kinetics and ciliogenesis (Fig4C–E). These findings demonstrate that the balance of HIF1α ubiquitination mediated by USP8 and pVHL is critical to control the dynamics of recycling endosomes and ensuing primary cilia formation.
Figure 4. Loss of USP8 or HIF1α impairs endocytic recycling required for ciliogenesis.
- Kinetic analysis of transferrin recycling measured by flow cytometry. RPE-1 cells infected with virus expressing the indicated shRNAs were incubated for 60 min with fluorescently labeled transferrin and then chased with excess of unlabeled transferrin for indicated times.
- RPE-1 cells described in (A), were fixed and stained with EEA1 antibody and DAPI.
- Kinetic analysis of transferrin recycling of RPE-1 infected as in (A) for shRNA expression and transfected with GFP or GFP-Rab11QL.
- Percentage of ciliated RPE-1 cells described in (C).
- Representative immunofluorescence staining of RPE-1 cells described in (C) (white arrows indicate primary cilium).
Data information: Bar graphs, mean ± s.d. (N = 3 (A, C, D)). N = number of independent experiments performed in triplicates. Student's t-test: *, P < 0.05. In (B) and (E): Scale bar, 10 μm.
Our functional screen for genes essential for ciliogenesis only in VHL-proficient but not VHL-deficient cells identified USP8 as a DUB for HIF1α. By reversing pVHL-mediated ubiquitination, USP8 balances against overly fast degradation of HIF1α, providing a mechanism of homeostastic control over HIF1α in normoxia. That HIF1α is a deubqiuitination target of USP8 and an essential ciliogenesis factor might explain the pVHL-dependency of USP8 function for ciliogenesis. By deubiquitinating HIF1α, USP8 sustains basal HIF1α expression under normoxic conditions thereby preventing derepression of Rabaptin5. As an activator of Rab5, increased Rabaptin5 promotes early endosome fusion, impairs endocytic recycling and impedes ciliogenesis (Fig4F). In hypoxic or VHL-negative ccRCC cells, Rabaptin5 protein levels are low owing to high levels of HIFα 10, thus allowing cilia formation. Previous studies conducted in various cell contexts revealed different effects of oxygen-pathway components on ciliary dynamics insofar that in primary cells, depletion of pVHL failed to affect ciliogenesis 4, while in VHL-deficient ccRCC cells the extent of cilia formation upon reexpression of pVHL and/or HIFα subunits varied 17 18, possibly due to different spectra of cancer mutations in these cells. The identification of a USP8-pVHL-HIF1α ubiquitination circuit offers now a new avenue to further elucidate ciliary dynamics in normal and cancer cells.
Materials and Methods
Screening procedures
An arrayed library containing pooled siRNAs (4 oligos per gene) targeting 48 USP genes (Qiagen, Hilden, Germany) (see Supplementary Table S1) was screened in wild-type and pVHL deprived MEFs mediated by shRNA. Cells were transfected by electroporation and plated into 96-well optical bottom plates (Nunc) after transfection. The number of ciliated cells was determined 72 h post siRNA treatment of which 48 h were under serum starvation (0% FCS) to induce ciliogenesis. Cilia and nuclei were stained by immunofluorescence with anti-acetylated tubulin antibody and DAPI, respectively. Two randomly chosen images were acquired per well with a BD Pathway 855 screening microscope (BD Biosciences, San Jose, CA, USA) and manually analyzed by counting cilia and nuclei.
In vivo deubiquitination assays
HEK293T cells were transfected with pCMV-Flag-USP8 or pCMV-Flag-USP8C786A, pcDNA3-HA-Ubiquitin and pCMV-HIF1α, and treated for 4 h with 50 μM MG132 (Sigma, St. Louis, MO, USA) before harvesting. Cell extracts were incubated with 5 μg of mouse anti-HIF1α antibody or the corresponding IgG control for 1 h in TNN buffer and then over night with proteinG-bound Sepharose beads (GE Healthcare, Little Chalfont, UK). The immunocomplexes were washed in TNN buffer (250 mM NaCl, without DTT) and separated by SDS-PAGE.
Transferrin recycling assay
RPE-1 cells were seeded in triplicates and 24 h later serum starved using serum-free DMEM/F12 medium for 48 h. Cells were incubated for 1 h at 37°C with serum-free medium containing 5 μg/ml human transferrin-Alexa Fluor 647. After cells were washed with ice-cold PBS, chased in serum-free medium containing 10 μg/ml of unlabeled holo-transferrin at 37°C for the indicated times, washed again with ice-cold PBS, acid-stripped for 3 min and trypsinized with 10X trypsin. Cells were then fixed with PFA on ice, pelleted and resuspended in PBS.
For each condition 6,000 cells were analysed on a FACSCalibur flow cytometer. (See also supplementary Materials and Methods).
Acknowledgments
We thank all members of our laboratory for scientific discussion and technical support. The authors are grateful to the Light Microscopy Centre ETH Zurich, in particular to Peter Horvath, who performed the analysis and quantification of early endosomes, and Joachim Hehl for help with microscopy. We thank Ari Helenius' group for technical help with transferrin recycling experiments and in particular Thomas Heger for the support in setting up the primary screen. Almut Dufner, Juan Bonifacino and Urs Greber for providing reagents, and Claudio Thoma and Michael Hell for reading and editing the manuscript. This work was supported by a grant from the SNF to W. K.
Author contributions
AT, IA and WK designed the project. AT, IA, performed most of the experiments and SM carried out biochemical analysis of HIF1α. DJ contributed to the in vitro binding assay. PK provided Usp8fl/fl mice, MEF Usp8fl/fl ERT2-CRE cells and Flag-CMV-5b-USP8 plasmid. AT and WK analyzed all data and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://embor.embopress.org
Supplementary Informations
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9
Supplementary Table S1
Review Process File
Source Data Figure 1A
Source Data Figure 1B 1E
Source Data Figure 2
Source Data Figure 3
Source Data Figure S2A S3E S4A S5A
References
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG. Von Hippel-Lindau disease. Ann Rev Pathol. 2007;2:145–173. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- Thoma CR, Frew IJ, Hoerner CR, Montani M, Moch H, Krek W. pVHL and GSK3beta are components of a primary cilium-maintenance signalling network. Nat Cell Biol. 2007;9:588–595. doi: 10.1038/ncb1579. [DOI] [PubMed] [Google Scholar]
- Frew IJ, Thoma CR, Georgiev S, Minola A, Hitz M, Montani M, Moch H, Krek W. pVHL and PTEN tumour suppressor proteins cooperatively suppress kidney cyst formation. EMBO J. 2008;27:1747–1757. doi: 10.1038/emboj.2008.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niendorf S, Oksche A, Kisser A, Löhler J, Prinz M, Schorle H, Feller S, Lewitzky M, Horak I, Knobeloch K-P. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol Cell Biol. 2007;27:5029–5039. doi: 10.1128/MCB.01566-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 2005;6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Roche O, Yan MS, Finak G, Evans AJ, Metcalf JL, Hast BE, Hanna SC, Wondergem B, Furge KA, Irwin MS, Kim WY, Teh BT, Grinstein S, Park M, Marsden PA, Ohh M. Regulation of endocytosis via the oxygen-sensing pathway. Nat Med. 2009;15:319–324. doi: 10.1038/nm.1922. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Vitale G, Ullrich O, Zerial M. Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell. 1995;83:423–432. doi: 10.1016/0092-8674(95)90120-5. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Knodler A, Feng S, Zhang J, Zhang X, Das A, Peranen J, Guo W. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci USA. 2010;107:6346–6351. doi: 10.1073/pnas.1002401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhns S, Schmidt KN, Reymann J, Gilbert DF, Neuner A, Hub B, Carvalho R, Wiedemann P, Zentgraf H, Erfle H, Klingmüller U, Boutros M, Pereira G. The microtubule affinity regulating kinase MARK4 promotes axoneme extension during early ciliogenesis. J Cell Biol. 2013;200:505–522. doi: 10.1083/jcb.201206013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz MS, Burk RD. Primary cilium formation requires von hippel-lindau gene function in renal-derived cells. Cancer Res. 2006;66:6903–6907. doi: 10.1158/0008-5472.CAN-06-0501. [DOI] [PubMed] [Google Scholar]
- Esteban MA, Harten SK, Tran MG, Maxwell PH. Formation of primary cilia in the renal epithelium is regulated by the von Hippel-Lindau tumor suppressor protein. J Am Soc Nephrol. 2006;17:1801–1806. doi: 10.1681/ASN.2006020181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Informations
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9
Supplementary Table S1
Review Process File
Source Data Figure 1A
Source Data Figure 1B 1E
Source Data Figure 2
Source Data Figure 3
Source Data Figure S2A S3E S4A S5A