Abstract
Background
Thoracic tumor, especially lung cancer, ranks as the top cancer mortality in most parts of the world. Lung adenocarcinoma is the predominant subtype and there is increasing knowledge on therapeutic molecular targets, namely EGFR, ALK, KRAS, and ROS1, among lung cancers. Lung cancer cell lines established with known clinical characteristics and molecular profiling of oncogenic targets like ALK or KRAS could be useful tools for understanding the biology of known molecular targets as well as for drug testing and screening.
Materials and methods
Five new cancer cell lines were established from pleural fluid or biopsy tissues obtained from Chinese patients with primary lung adenocarcinomas or malignant pleural mesothelioma. They were characterized by immunohistochemistry, growth kinetics, tests for tumorigenicity, EGFR and KRAS gene mutations, ALK gene rearrangement and OncoSeq mutation profiling.
Results
These newly established lung adenocarcinoma and mesothelioma cell lines were maintained for over 100 passages and demonstrated morphological and immunohistochemical features as well as growth kinetics of tumor cell lines. One of these new cell lines bears EML4-ALK rearrangement variant 2, two lung cancer cell lines bear different KRAS mutations at codon 12, and known single nucleotide polymorphism variants were identified in these cell lines.
Discussion
Four new lung adenocarcinoma and one mesothelioma cell lines were established from patients with different clinical characteristics and oncogenic mutation profiles. These characterized cell lines and their mutation profiles will provide resources for exploration of lung cancer and mesothelioma biology with regard to the presence of known oncogenic mutations.
Keywords: lung adenocarcinomas, oncogenic mutations, EGFR, ALK, KRAS
Introduction
Despite knowledge on known therapeutic targets like epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) gene rearrangement, and continuous efforts in clinical trials, lung cancer remains the top cause of cancer morbidity and mortality in most parts of the world.1,2 Lung adenocarcinomas arising from female nonsmokers have emerged as a prominent subgroup among lung cancer patients, especially in Asia, compared to lung cancer arising from smokers and squamous cell carcinoma as in the past.3,4 Oncogenic driver mutations have been identified and they are present in mutually exclusive ways in individual lung tumors. These oncogenic mutations include EGFR, KRas (KRAS), ALK, MET, etc.5 Some of these oncogenic mutations possess therapeutic implications for being the targets of actions for small molecule tyrosine kinase inhibitors (TKI).6 Even with the presence of these molecular targets, however, there are mutant tumors that do not respond to the corresponding targeted agents, or mutant tumors that do respond initially followed subsequently by development of resistance to the corresponding targeted agents. A better understanding of the mechanisms of actions of these therapeutic oncogenic driver mutations in lung cancer may allow for identification of new molecular targets for better diagnostics and targeted therapy of lung cancer.
We have previously reported the establishment of lung cancer cell lines with known clinical properties like sex, smoking habits, and ethnicity.7 In this study, we have further established five new cell lines with four being adenocarcinomas and one being a malignant pleural mesothelioma (MPM) cell line. They carry different molecular properties and together with the four published before, represent the heterogeneity of thoracic tumors.7 The aim of this study was to establish and to characterize new cancer cell lines from Chinese patients with known clinical characteristics, tumor staging, prior therapy, and known oncogenic mutation profiles so that these new lung cancers and mesotheliomas could provide defined materials for research in lung cancer and mesothelioma. The morphological and growth kinetics properties as well as oncogenic driver mutation status of the cell lines were described. Mutation profiling was performed to document the single nucleotide polymorphism (SNP) variants of these newly established lung cancer and MPM cell lines.
Materials and methods
Method of culture, medium, and supplement
All clinical specimens were collected after informed written consents had been given by the respective subjects. The study was approved by the University of Hong Kong/Hong Kong Hospital Authority Hong Kong West Cluster Institutional Review Board/Ethics Committee (UW 11 – 326).
Pleural effusions were all obtained with aseptic techniques and centrifuged to collect malignant cells. For the patient with mesothelioma, tumor deposits on pleura were biopsied under direct vision through flexirigid pleuroscopy. The pieces of fresh pleural biopsy were minced with pairs of fine scissors under aseptic conditions. Collected tissue or cancer cells were seeded into two different culture media, namely, ACL4 or RPMI supplemented with 10% fetal bovine serum, in order to maximize the chance of establishment. ACL4 medium was prepared according to the protocol published by Oie et al.8 Penicillin and Streptomycin (105 U) were added as antibiotic prophylaxis for the first passage and both were omitted from the culture medium recipe from the second passage onwards. The cells were then incubated in 37°C and 5% carbon dioxide. Attached cells reaching confluent density were subcultured and growth was maintained in either ACL4 or RPMI medium.
Growth kinetics study
In studying the growth kinetics of these newly established cell lines, 105 cells of each cell line were seeded into 25-cm2 culture flasks with 5 mL of growth medium. Cells were counted on days 1, 2, 4, 6, 8, 10, and 12 on triplicate flasks.
Test for tumorigenicity in nude mice
Tumor cells were harvested and counted with supravital staining. At least 2×105 cells were resuspended in 0.5 mL of growth medium. The whole cell suspension was then injected beneath the dorsal skin of two nude mice. The mice were maintained and observed for up to 6 months for any development of xenograft tumor. Xenografts were harvested before they reached an external diameter of 1 cm. The harvested xenografts were fixed in 4% formalin and were then processed into paraffin blocks with tissue sections cut and examined to confirm the presence of tumor xenograft as well as analyzed for their morphological characteristics. If no obvious tumor development was observed at the end of 6 months, the corresponding mice were sacrificed and dissected to look for tumor involvement in internal organs by gross and microscopic examination. For those lines with no xenograft development, a gradation of inoculums containing 2×106, 2×107, and 2×108 tumor cells were performed further on two separate mice for each inoculum dosage.
Morphological characteristics and immunohistochemical studies
For each newly established cell line, cultured tumor cells were centrifuged, fixed, and embedded into paraffin cell blocks according to routine cytology procedures. The cultured cells and xenografts were characterized for their morphological characteristics by routine hematoxylin and eosin stain, mucin content, and immunohistochemistry with a panel of monoclonal antibodies against cytokeratins (CK7 [M7018; DAKO {Agilent Technologies, Santa Clara, CA, USA}], CK20 [M7019; DAKO]) and the thyroid transcription factor TTF-1 (M3575; DAKO). All cell lines were subjected to DNA fingerprinting with markers STR D11S922, D11S4099, and D11S4152.
Epidermal growth factor receptor (EGFR) mutation at exons 18–21 and KRAS mutation detection
Genomic DNA was extracted from freshly harvested cells, and extracted DNA was analyzed for mutations in exons 18–21 of the EGFR gene and codons 12, 13, and 61 of the KRAS gene by direct sequencing according to previous protocol.9 The methods and primers used for polymerase chain reaction (PCR)-based sequencing detection were described elsewhere.9
ALK rearrangement and variant detection
The Vysis ALK FISH dual color break apart rearrangement probe (Abbott Vysis 05J89-001; Abbott Laboratories, Abbott Park, IL, USA) was applied according to manufacturer’s instructions. The FISH detection was performed on unstained 5-μm-thick paraffin-embedded formalin-fixed cytospin sections. Ten microliters of ALK probe was administered to each air-dried slide, and the slides were then denatured at 94°C for 4 minutes, then hybridized overnight at 37°C in Vysis HYBrite (Abbott Laboratories). The sections were then mounted with 10 μL of VECTASHIELD mounting medium (Vector Laboratories, Inc., Burlingame, CA, USA) before being ready for examination and scoring of results. Scoring of FISH study results were performed with Leica DMR fluorescence microscope (Leica Microsystems, Wetzlar, Germany). The FISH signals were evaluated by TSKW who was blinded to the nature of the cell lines. At least 100 nonoverlapping and intact tumor nuclei were evaluated. The tumor cells were considered to be positive for FISH study when the red and green probe signals were wider apart than the distance of two nuclei or when the corresponding paired green signal was lost for any indexed red signal.
Immunohistochemistry was performed on unstained 4-μm paraffin-embedded cell block sections. The primary antibody for ALK (Thermo Fisher Scientific, Waltham, MA, USA) was used with a dilution of 1:500. Reverse transcription-PCR combined with direct sequencing of the PCR products were performed for any cell line with ALK rearrangement with FISH to identify EML4-ALK variants.10
Karyotyping
Metaphase preparation
To accumulate metaphases, cells were treated with colcemid (Sigma-Aldrich Co, St Louis, MO, USA; 0.03 mg/mL) for 3 hours. Chromosome spreads were prepared as previously described.11 Slides with cells in metaphase were left for aging at room temperature for up to 7 days prior to spectral karyotyping (SKY) procedures.
SKY
The slides were treated with DNase-free RNase solution (0.1 mg/mL) at 37°C for 1 hour. After washing in 2X SSC for 10 minutes, the slides were dehydrated successively in 70%, 85%, and 95% ethanol at room temperature for 2 minutes each, and were then air-dried with nitrogen gas. The slides were treated with proteinase K solution (0.02 μg/mL) at 37°C for 15 minutes and washed in 2X SSC for 10 minutes. The slides were then fixed in 2% paraformaldehyde and were washed in 2X SSC for 10 minutes each, followed by successive dehydration again in 70%, 85%, and 95% ethanol for 2 minutes. The slides were placed in 70% formamide/2X SSC at 80°C for 4 minutes and again were successively dehydrated in 70%, 85%, and 95% ethanol for 2 minutes each. The SKY probes (SkyPaintTM mixture; Applied Spectral Imaging, Migdal HaEmek, Israel), after prior denaturation at 80°C for 7 minutes and being placed at 37°C for 1 hour in a water bath, were then applied to denatured slides, which were then covered by coverslips, followed by sealing rubber cement and then were further incubated at 37°C for more than 36 hours in a humidified chamber. The recommendations of the SKY probe manufacturer were followed for SKY signal detection. SKY image capturing and karyotyping were performed using the SkyVision Imaging System equipped with a Zeiss Axioplan 2 fluorescence microscope (Zeiss Germany, Jena, Germany). Twenty-five to thirty metaphases from each cell line were analyzed using SKY. Karyotypes were analyzed using international nomenclature system.12
Oncogenic molecular profiling with RainDance OncoSeq analysis
Three micrograms of genomic DNA was extracted from each cell line and optical density was measured with 260/280 ratio to be 1.8 or more before proceeding with further tests. The RainDance OncoSeq predesigned panel (RainDance Technologies, Inc., Lexington, KY, USA) used, which covered 142 oncogenes and included downstream Solexa next-generation sequencing with a coverage depth of 200×. The FastQC package v0.11.2 was used to assess the read quality. Filtered read sequences were mapped to the human reference genome (hg19) with Burrows–Wheeler Alignment v0.7.9a,13 and PCR as well as optical duplicates were marked using Picard v1.105. Variants were called using FreeBayes v0.9.1414 with a quality score cutoff of ≥30 and a minimum coverage of ten per sample. Variant annotation was performed with ANNOVAR15 using GALAXY16 and functional predictions were conducted using SIFT17,18 and PROVEAN.19
Results
Clinicopathological features of the patients from which cell lines were established (Table 1)
Table 1.
A table summarizing the clinical characteristics of patients from which these new cell lines were derived and established
| Cell lines | FA34 | FA44 | FA49 | FA98 | PME1 |
|---|---|---|---|---|---|
| Sex/age (years) | Female/34 | Male/58 | Male/53 | Male/80 | Male/73 |
| Smoking habits | Lifetime nonsmoker | Chronic smoker | Chronic smoker | Chronic smoker | Ex-smoker |
| Tissue origin | Pleural fluid | Pleural fluid | Pleural fluid | Pleural fluid | Pleural fluid |
| Tumor cell type | Adenocarcinoma | Adenocarcinoma | Adenocarcinoma | Adenocarcinoma | Malignant pleural mesothelioma |
| Staging | IV | IV | IV | IV | IV |
| Prior treatment | Paclitaxel, pemetrexed, cisplatinum, erlotinib | Paclitaxel, carboplatin | Paclitaxel, carboplatin | Empirical erlotinib | No treatment prior to collection of biopsy specimen |
| Tumorigenicity in nude mice | Yes | Yes | Yes | Yes | No |
| Potential oncogenic mutations | EML4-ALK translocation variant 2 | KRAS mutation at codon 12 (G215T) | KRAS mutation at codon 12 (G219A) | EGFR and KRAS wild type |
FA34 was grown from the malignant pleural effusion of a 34-year-old nonsmoking lady with primary adenocarcinoma of the right lower lobe (T3N2M1b). She presented with pleural effusion and had tried multiple lines of systemic chemotherapy, including paclitaxel, pemetrexed, and TKI treatment (despite the fact that she was known to have EGFR wild type lung tumor). Computerized tomography (CT) of thorax and abdomen showed no other tumor site. The patient passed away in 2008 with progressive disease despite different lines of drug therapy, but the cultured cells continue to proliferate to beyond the 100th passage, and it was only after the patient passed away that EML4-ALK translocation became known to be present in up to 7% of non-small cell lung cancer.20 The established line was tested to harbor EML4-ALK translocation with variant type 2 of exon 20: exon 20 rearrangement (Table 1).
FA44 came from the pleural fluid of a 58-year-old male chronic smoker who presented with right pleural effusion with confirmed malignant pleural effusion with metastatic adenocarcinoma. CT scan revealed a contrast-enhancing tumor in the right lower lobe and metastatic nodules in the right middle and left lower lobes of the lungs. There was no other mass lesion in the liver, spleen, or kidneys. The patient had received paclitaxel/carboplatin for six cycles with subsequent stable disease maintained for 5 months. He subsequently went into progressive disease with increasing recurrent right pleural effusion and enlargement of the primary tumor as shown on CT scan. Pleural fluid was obtained on this disease progression. His general condition deteriorated quickly and did not allow time for consideration of salvage chemotherapy.
FA49 was also taken from the pleural fluid of a 53-year-old chronic smoker who was diagnosed to have stage IV adenocarcinoma of lung with primary tumor in the right lower lobe. He also received paclitaxel and carboplatin for six cycles with partial response maintained for 3 months after chemotherapy, followed by progressive disease with enlargement of primary tumor, increasing right pleural effusion, and development of new cervical lymphadenopathy. The right pleural fluid was obtained with therapeutic pleurocentesis at that juncture.
FA98 was cultured from an 80-year-old male chronic smoker presented with right pleural effusion and CT finding of a right hilar mass with right upper lobe collapse. The patient opted for, because of his age, upfront EGFR-TKI and not chemotherapy despite the fact that EGFR mutation status was not known for him at the time of diagnosis. The patient was started on erlotinib and he decided to maintain it despite the fact that EGFR mutation tests came back to show no mutation subsequently. His general condition continued to deteriorate with rapid accumulation of right pleural effusion requiring multiple pleurocentesis, where aspirated pleural fluid was saved for culture. He finally succumbed after 4 weeks of empirical erlotinib.
PME1 was derived from a 73-year-old man who had retired from incinerator service with exposure to asbestos; he presented with right pleural effusion and nodular thickening of right pleura and ascites. CT scan revealed bilateral nodular pleural thickening, right pleural effusion, and ascites. Repeated diagnostic pleural tapping only showed atypical cells not diagnostic of malignant effusion. Flexirigid pleuroscopy was performed with pleural biopsy taken, which confirmed on both light and electronic microscopy to be MPM. The patient received six cycles of pemetrexed and cisplatin with stable disease maintained for the subsequent 7 months, after which the disease progressed with increasing bilateral pleural effusion and ascites and he finally succumbed.
FA34, FA44, FA49, and FA98 were derived from pleural fluid adenocarcinoma cells taken from patients who had received prior treatment with either platinum-based chemotherapy (for FA34, FA44, and FA49) or empirical EGFR-TKI treatment (for FA98) before EGFR mutation status was known. PME1 was taken from patient alongside diagnostic pleuroscopy and thus mesothelioma cells were treatment-naïve.
Cell culture characteristics and authentication
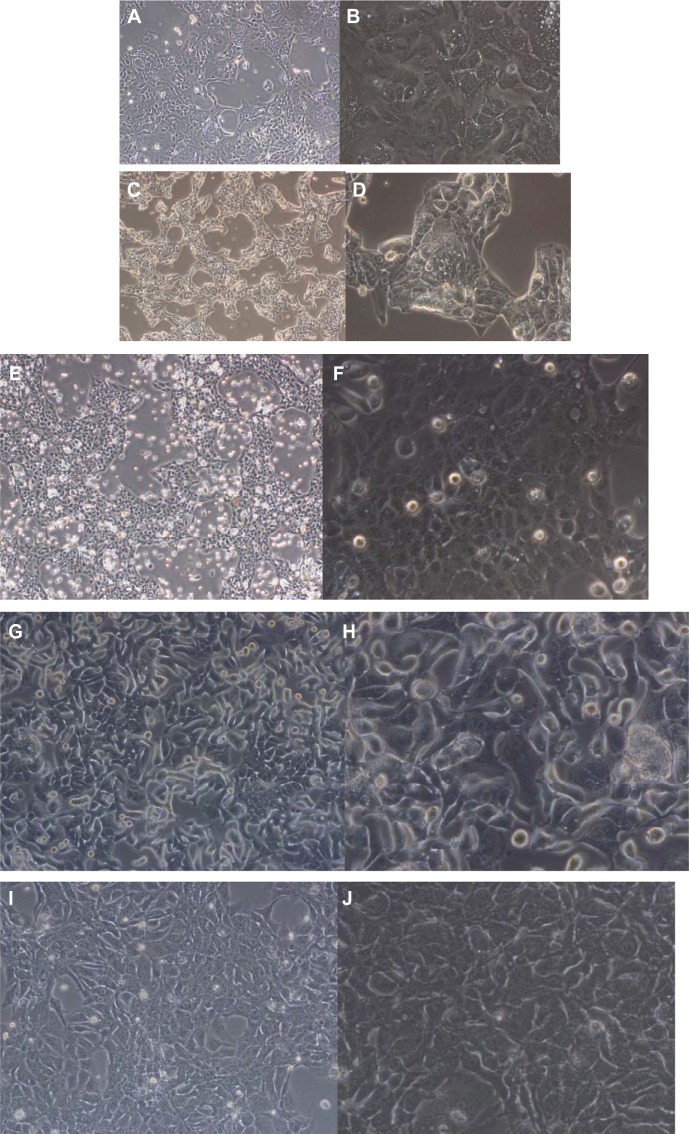
All cell lines were maintained for over 100 passages. The five cell lines demonstrated different morphological properties. All the cell lines demonstrated tightly packed attached cells that tended to pile up before reaching confluency (Figure 1). This feature might be related to the formation of cell clusters in these cell lines before spreading to form confluent tumor cell layers. FA44, FA49, FA98, and PME1 showed fairly uniform populations of cells while FA34 showed slightly more variation in tumor cell size.
Figure 1.
Photomicrographs showing morphological and immunohistochemical properties of different cell lines.
Notes: Side-by-side photomicrographs at 40× and 200× magnification, respectively, for each of FA34 (A and B), FA44 (C and D), FA49 (E and F), FA98 (G and H), and PME1 (I and J) cell lines.
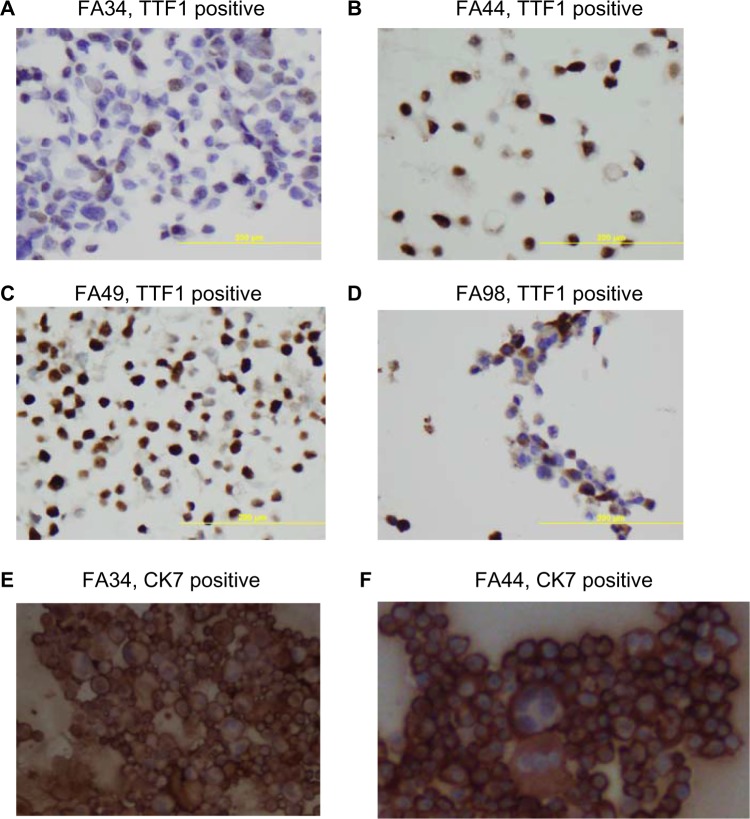
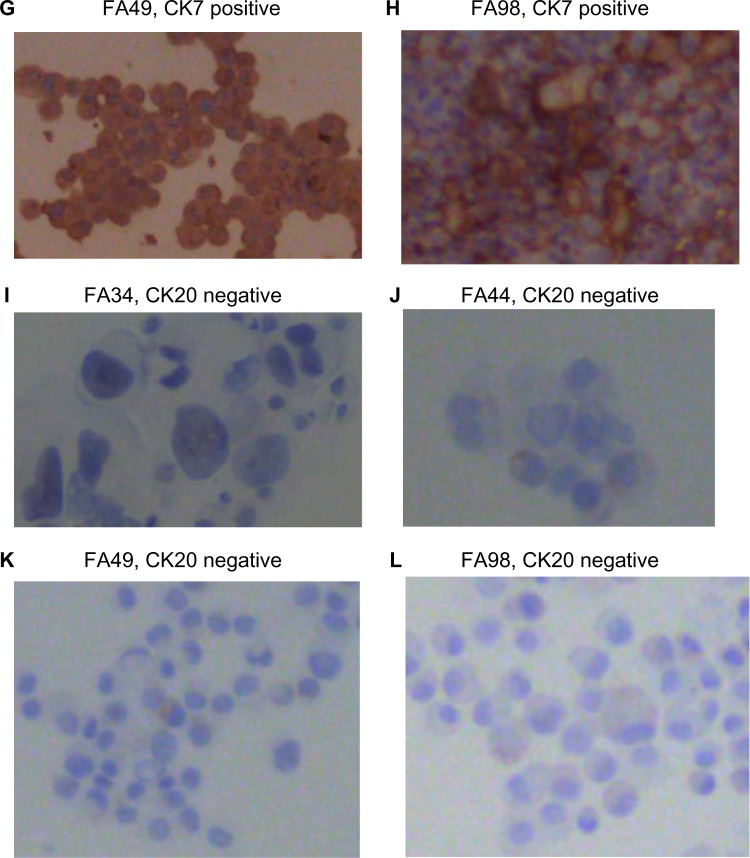
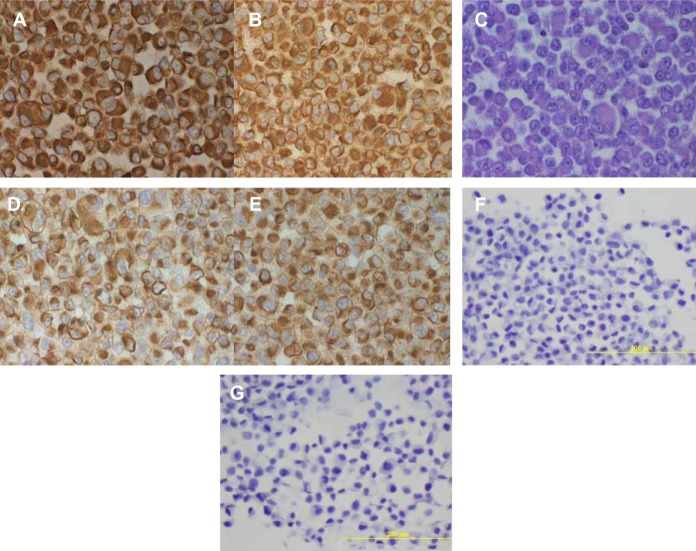
All the lung adenocarcinoma cell lines were stained positive for CK7 but stained negative for CK20, which is the typical immunohistochemical profile for carcinoma of pulmonary origin. The lung adenocarcinoma cell lines were also all positive for CK7 and TTF1, and negative for CK20 (Figure 2). Napsin A was stained positive for FA44 but negative for other lung adenocarcinoma cell lines in this report (Figure 3). PME1 was also stained positive for anti-vimentin A and B but negative for mucin, WT1, and calretinin (Figure 4), which again suggested mesothelial origin. All the cell lines showed different fragments on DNA fingerprinting with markers STR D11S922, D11S4099, and D11S4152.
Figure 2.
Immunohistochemical staining with TTF1, CK7, and CK20 on paraffin cell blocks for all four lung adenocarcinoma cell lines.
Notes: (A–D) TTF1 positive staining for each cell line, (E–H) CK7 positive staining for each line, and (I–L) CK20 negative staining for each line.
Figure 3.
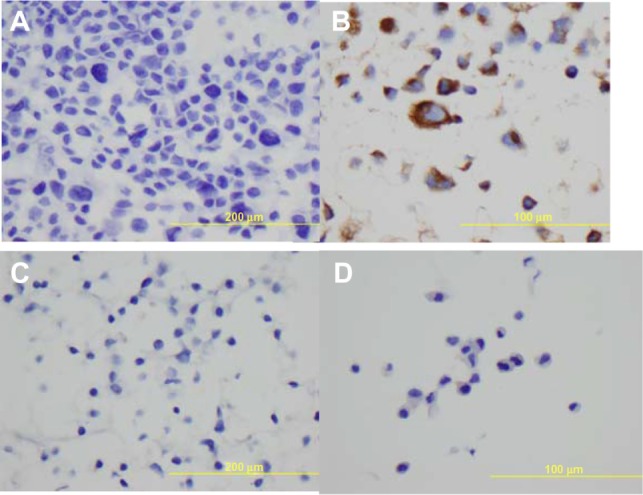
Immunohistochemical staining with Napsin A on paraffin cell blocks for all the four lung adenocarcinoma cell lines.
Notes: (A) FA34, (B) FA44, (C) FA49, and (D) FA98.
Figure 4.
Immunohistochemical staining on paraffin cell blocks from PME1.
Notes: Staining with (A) anti-vimentin a, (B) anti-vimentin b, (C) mucin negative, (D) anti-CK a, (E) anti-CK b, (F) calretinin negative, and (G) WT1 negative.
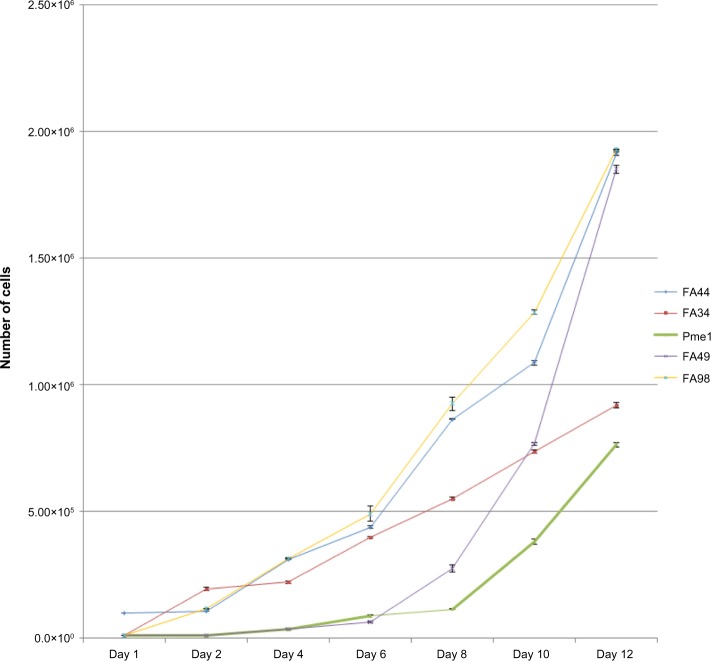
Growth kinetic study
FA44, FA49, FA98, and PME1 showed a short lag phase of up to 2 days. FA98 grew faster than others (Figure 5). FA34 showed an initial lag phase of around 5 days followed by a more accelerated phase (Figure 5). FA44, FA49, FA98, and PME1 showed similar doubling times of about 22 hours; FA34 showed a doubling time of around 34 hours.
Figure 5.
Growth curves of all the cell lines showing the initial lag phase followed by accelerated growth phase in most cell lines with FA44, FA49, and FA98 showing the fastest growth rate compared to FA34 and PME1.
Test for tumorigenicity in nude mice
All adenocarcinoma cell lines, FA34, FA44, FA49, and FA98, formed xenografts of 1 cm in about 6–8 weeks after subcutaneous inoculation of 106 cells onto the back of a nude mouse for each cell line. The xenografts obtained from these cell inoculations could result in tumors again after retransplantation (passaging) into separate nude mice. The time for xenograft detection was 3–4 weeks after injection (when a subcutaneous bulge became visible), and by 6–8 weeks after inoculation the xenograft would reach the external dimension of 1 cm for xenograft harvesting. PME1 failed to produce any xenograft at 12 weeks after tumor cell injection with at least three separate attempts for each dose of tumor cell injection. Xenografts obtained from FA34, FA44, FA49, and FA98 were sectioned and reviewed, and they all showed xenografts predominantly containing the inoculated tumor cells.
EGFR and KRAS mutation status
All of the cell lines were tested wild type for EGFR mutation at exons 18–21. FA44 and FA49 were both KRAS mutations at codon 12 with G215T and G219A, respectively. No KRAS mutation was found at codons 12, 13, or 61 in FA34, FA98, and PME1.
ALK rearrangement and variant detection
FISH for ALK rearrangement were tested for all these new cell lines, with FA34 showing ALK rearrangement (Figure 6) according to standard scoring criteria while all the others showed no ALK rearrangement. ALK immunohistochemical staining for sections from FA34 cytospin cell blocks also showed strong positive staining when compared to other cell lines and the kit control. Reverse transcription PCR showed transcripts compatible with E20: A20 translocation, ie, EML4-ALK variant 2.
Figure 6.
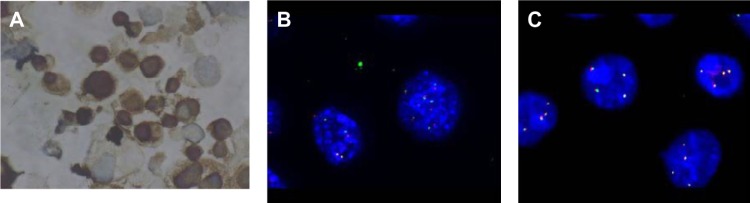
ALK immunohistochemistry and FISH.
Notes: (A) Paraffin cell block section of FA34 showed positive ALK immunohistochemical staining; (B) ALK rearrangement with FISHfor FA34; and (C) FA44 as a control showing no ALK rearrangement with FISH.
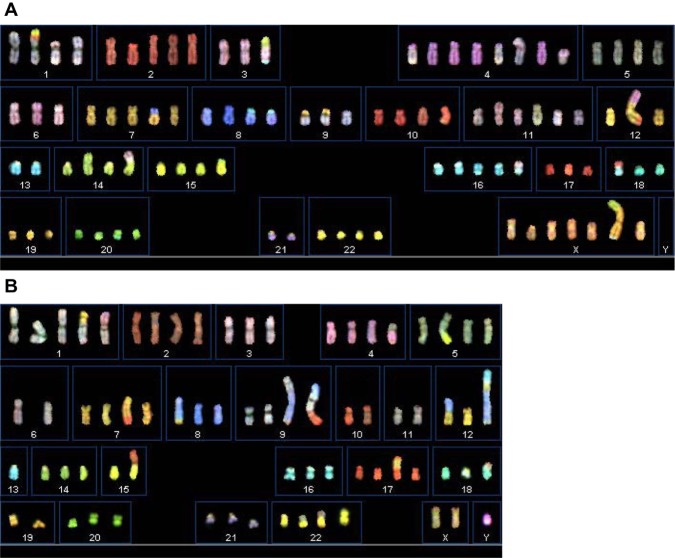
Karyotypes of the cell lines analyzed using SKY
We analyzed the karyotypes of the cancer cell lines at late passages (greater than passage 50) using SKY (Figure 7). All of these newly established lung cancer cell lines showed specific clonal chromosomal aberrations (Table 2). Numerically, FA34b is near-tetraploid; FA44a, FA49, FA98, and PME1 are near-triploid.
Figure 7.
Spectral karyotyping images of all the cell lines.
Notes: (A) FA34; (B) FA44; (C) FA49; (D) FA98; and (E) PME1.
Table 2.
Karyotype descriptions in newly established lung cancer cells lines analyzed using spectral karyotyping
| Cell lines | Karyotype |
|---|---|
| FA34 | 90∼96<4n>, XXXX, +del(X)(p1?), +del(X)(q2?), +der(X)t(X;14)(p22.3;q11)t(X;18)(q28;?), der(1)t(1;17)(p2?;?)t(14;17)(?;?), del(1)(q2?), del(1)(q3?), +del(2)(q14), −3, der(3)t(3;14)(p11;q?)t(3;13)(q26;q?), +der(4)t(1;4)(p11;q11)×2, +der(4)t(1;4)(?;p12)t(1;4)(q11;q13), +der(4;5) (p10;p10), −6, +der(7)t(7;8)(p11;p11), der(8;18)(q10;p10)×2, −9, del(9)(q3?), der(9)t(9;19)(p11;p11)del(9)(q3?)×2, del(10)(p11)dup(10)(q?), der(10)t(1;10)(?;q26), +i(11)(q10), +der(11)t(1;11)(p11;q11), −12, der(12)t(4;12)(q?;p13)t(12;17)(q24.3;?), −13, −13, i(14)(q10), der(14) t(11;14)(q12;p13), der(15)t(15;20)(p11;?), +der(16)t(11;16)(p11;p11), −17, +der(18)t(17;18)(q12;p11.3), −19, del(19)(p11), −21, −21[cp30] |
| FA44a | 68∼75<3n>, XXY, +dic(1;19)(p11;q13.4), +der(1)t(1;4)(p11;q11), +2, +4, der(4;5)(p10;p10), +5, +der(5)t(5;15)(q2?;q1?), −6, +der(7) t(7;17)(q3?;q?), der(8)t(7;8)(?;q2?), +9, der(9)t(8;9)(q?;q34)dup(8)(q?), der(9)t(8;9)(?;p24)t(9;10)(q3?;q11), −10, 11, der(12)t(8;12) (q11;p11), der(12)t(12;13)(p1?;q2?)t(8;12)(q?;q2?)dup(8)(q?), −13, −13, −15, der(10;15)(q10;q10), +der(17)t(12;17)(q?;p1?)t(12;17)(q?;?), del(18)(p11), der(18)t(11;18)(?;p11.3), −19, del(19)(p11), +22, der(22)t(19;22)(?;p11), der(22)t(5;22)(?;p11), der(22)t(11;22)(?;p11), i(22) (q10)[cp27] |
| FA49 | 60∼65<3n>, der(X)t(X;15)(p11;q1?)t(X;9)(q2?;p11), der(X)t(X;9)(q2?;p11), Y, +Y, der(2)t(2;14)(p1?;q2?)t(2;7)(q2?;?), +3, der(3)t(3;15) (q11;q?)t(13;15)(q1?;q?), der(3;17)(p10;q10), der(4)t(4;20)(q11;?), i(4)(q10), +del(5)(q12)×2, der(5;15)(q10;q10), der(5)t(5;22)(q11;q?) t(4;22)(q?;q?), der(6)t(6;20)(q27;?), der(6;11)(p10;p10), del(6)(p12), +8, der(8;11)(q10;p10), dup(8)(q?), der(8)t(8;13)(q11;q?)t(13;17) (q?;q11), −9, der(9)t(9;12)(q11;?), +10, der(10)t(7;10)(p11;p11)×2, +12, del(12)(p11), der(12)t(12;22)(p13;q12)t(12;21)(q13;q21), −13, −13, der(13)t(10;13)(q12;p11)t(8;13)(q21;q13), −14, −15, der(15;18)(q10;q10), −17, del(18)(q12), −19, −20, der(20)t(7;20)(q36;q21), −21, −22, der(21;22)(q10;q10), der(22)t(13;22)(q21;q13)[cp25] |
| FA98 | 58∼−62<3n>, XXY, i(1)(q10), −2, −3, −4, +5, −6, −7, +8, der(9;22)(q10;q10), −10, −11, −13, +15, der(15;19)(q10;p10)×2, −16, −18, −20, der(22)t(9;22)(q11;q13)[cp30] |
| PME1 | 68∼−74<3n>, XX, −Y, del(1)(p31), der(2;19)(p10;?q10), −3, +4, der(4)t(4;9)(q2?;?)×2, der(5)t(5;18)(p15;q12), +i(6)(p10), +9, del(9) (q12)×2, −0, der(12)t(9;12)(q11,p13), del(13)(q?), +14, der(14)t(2;14)(q22;q32)×2, +der(17)t(Y;17)(q12;q24), del(18)(q11)×2, del(19) (p11)del(19)(q11), der(19)t(2;19)(p12;p13), −22[cp30] |
Oncogenic molecular profiling
Out of 142 cancer genes profiled with RainDance OncoSeq, 42 gene variants representing variants in 27 genes were identified in different cell lines (Table 3). With the SIFT17,18 and PROVEAN19 softwares, detected variants were predicted to have damaging/deleterious or tolerated/neutral effects on the resulting protein structure or functions. We also examined the list for mutations that had been reported in the COSMIC database (v69).
Table 3.
Summary of mutations of the five cell lines analyzed with targeted sequencing (RainDance OncoSeq)
| Chr | Position (hg19) | Ref/Alt alleles | dbSNP ID | Gene | Amino acid change | FA34 | FA44 | FA49 | FA98 | PME1 | PROVEAN Prediction | SIFT Prediction | COSMIC v69 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3,328,358 | T/C | rs870124 | PRDM16 | NM_022114: exon9: c.T1597C: p.S533P | CC | CC | CC | CC | CC | Neutral | Tolerated | N |
| 2 | 29,416,572 | T/C | rs1670283 | ALK | NM_004304: exon29: c.A4381G: p.I1461V | CC | CC | CC | CC | CC | Neutral | Tolerated | N |
| 2 | 220,462,640 | G/T | rs681747 | STK11IP | NM_052902: exon1: c.G2T: p.R1M | TT | TT | TT | TT | TT | Neutral | Damaging | N |
| 2 | 220,473,355 | G/A | rs673951 | STK11IP | NM_052902: exon15: c.G1687A: p.V563I | GA | GG | AA | AA | AA | Neutral | Tolerated | N |
| 2 | 220,476,443 | C/T | rs627530 | STK11IP | NM_052902: exon18: c.C2255T: p.S752F | TT | TT | TT | TT | TT | Neutral | Tolerated | N |
| 3 | 14,187,449 | G/T | rs2228001 | XPC | NM_001145769: exon16: c.C2704A: p.Q902K | TT | TT | TT | TT | TT | Neutral | Tolerated | N |
| 3 | 14,199,887 | G/A | rs2228000 | XPC | NM_001145769: exon9: c.C1385T: p.A462V | AA | GA | AA | GA | AA | Neutral | Tolerated | N |
| 5 | 112,176,756 | T/A | rs459552 | APC | NM_001127511: exon14: c.T5411A: p.V1804D | AA | AA | TA | AA | TA | Neutral | Tolerated | N |
| 7 | 2,968,245 | C/T | rs75403455 | CARD11 | NM_032415: exon13: c.G1741A: p.A581T | CT | CC | CC | CC | CC | Neutral | Tolerated | Y |
| 7 | 55,229,255 | G/A | rs2227983 | EGFR | NM_005228: exon13: c.G1562A: p.R521K | GA | GG | GA | AA | AA | Neutral | Tolerated | N |
| 8 | 30,999,280 | G/T | rs1801195 | WRN | NM_000553: exon26: c.G3222T: p.L1074F | TT | TT | TT | TT | GG | Neutral | Tolerated | N |
| 8 | 145,737,816 | C/T | rs4251691 | RECQL4 | NM_004260: exon18: c.G3014A: p.R1005Q | CT | CC | CC | CT | CC | N/A | N/A | Y |
| 8 | 145,741,443 | C/T | N/A | RECQL4 | NM_004260: exon5: c.G1060A: p.V354I | CT | CC | CC | CC | CC | N/A | N/A | N |
| 8 | 145,741,702 | C/G | rs4244612 | RECQL4 | NM_004260: exon5: c.G801C: p.E267D | CG | CC | CG | CG | CC | N/A | N/A | N |
| 8 | 145,742,514 | A/G | rs2721190 | RECQL4 | NM_004260: exon4: c.T274C: p.S92P | GG | AG | GG | GG | AG | N/A | N/A | N |
| 9 | 139,410,076 | T/G | N/A | NOTCH1 | NM_017617: exon11: c.A1762C: p.T588P | TG | TG | TG | TT | TG | Deleterious | Damaging | N |
| 9 | 139,410,120 | T/G | N/A | NOTCH1 | NM_017617: exon11: c.A1718C: p.D573A | TG | TG | TT | TT | TG | Deleterious | Tolerated | N |
| 9 | 139,413,211 | T/G | N/A | NOTCH1 | NM_017617: exon6: c.A931C: p.T311P | TG | TG | TG | TT | TT | Deleterious | Damaging | N |
| 11 | 108,114,749 | G/A | rs79075295 | ATM | NM_000051: exon6: c.G566A: p.R189K | GA | GG | GA | GA | GA | Neutral | Damaging | Y |
| 11 | 108,183,167 | A/G | rs659243 | ATM | NM_000051: exon40: c.A5948G: p.N1983S | GG | GG | GG | GG | GG | Neutral | Tolerated | N |
| 13 | 28,537,317 | G/A | rs1805107 | CDX2 | NM_001265: exon3: c.C877T: p.P293S | AA | AA | AA | GG | GG | Neutral | Tolerated | N |
| 13 | 28,624,294 | G/A | rs1933437 | FLT3 | NM_004119: exon6: c.C680T: p.T227M | AA | GA | AA | AA | AA | Neutral | Tolerated | N |
| 13 | 32,906,729 | A/C | rs144848 | BRCA2 | NM_000059: exon10: c.A1114C: p.N372H | CC | CC | AA | AA | AA | Neutral | Tolerated | Y |
| 14 | 75,513,883 | T/C | rs175081 | MLH3 | NM_001040108: exon2: c.A2476G: p.N826D | CC | CC | CC | CC | CC | Neutral | Tolerated | N |
| 15 | 40,477,831 | G/A | rs1801376 | BUB1B | NM_001211: exon8: c.G1046A: p.R349Q | GA | AA | GA | GG | AA | Neutral | Tolerated | N |
| 15 | 74,328,116 | A/G | rs743580 | PML | NM_033250: exon7: c.A2170G: p.S724G | AG | AA | AA | GG | AG | Neutral | Tolerated | N |
| 15 | 74,328,141 | G/T | rs743581 | PML | NM_033250: exon7: c.G2195T: p.G732V | GT | GG | GG | GG | GG | Neutral | Tolerated | N |
| 15 | 91,312,426 | C/T | rs55880859 | BLM | NM_000057: exon11: c.C2371T: p.R791C | CT | CC | CC | CC | CC | Deleterious | Damaging | N |
| 16 | 3,860,746 | T/G | N/A | CREBBP | NM_001079846: exon3: c.A833C: p.Q278P | TG | TT | TT | TT | TT | Neutral | Tolerated | N |
| 16 | 15,841,509 | G/T | N/A | MYH11 | NM_002474: exon19: c.C2329A: p.H777N | GT | GG | GG | GG | GG | Deleterious | Tolerated | N |
| 16 | 23,646,942 | T/C | rs3809683 | PALB2 | NM_024675: exon4: c.A925G: p.I309V | TC | TT | TT | TT | TT | Neutral | Tolerated | N |
| 16 | 27,448,894 | A/C | N/A | IL21R | NM_021798: exon4: c.A238C: p.T80P | AC | AA | AC | AC | AA | Deleterious | Damaging | N |
| 16 | 89,838,136 | T/C | rs56369086 | FANCA | NM_000135: exon23: c.A2101G: p.K701E | TC | TT | TT | TT | TT | Neutral | Tolerated | N |
| 16 | 89,849,480 | C/T | rs2239359 | FANCA | NM_000135: exon16: c.G1501A: p.G501S | TT | CT | TT | CT | TT | Neutral | Tolerated | N |
| 16 | 89,866,043 | T/C | rs7190823 | FANCA | NM_000135: exon9: c.A796G: p.T266A | CC | CC | CC | CC | CC | Neutral | Tolerated | N |
| 17 | 37,884,037 | C/G | rs1058808 | ERBB2 | NM_004448: exon27: c.C3508G: p.P1170A | GG | CG | CC | CC | CC | Neutral | Damaging | N |
| 17 | 41,223,094 | T/C | rs1799966 | BRCA1 | NM_007297: exon14: c.A4696G: p.S1566G | CC | TC | TC | TC | CC | Neutral | Tolerated | N |
| 17 | 41,244,000 | T/C | rs16942 | BRCA1 | NM_007297: exon9: c.A3407G: p.K1136R | CC | TC | TC | TC | CC | Neutral | Tolerated | Y |
| 17 | 41,244,435 | T/C | rs16941 | BRCA1 | NM_007297: exon9: c.A2972G: p.E991G | CC | TC | TC | TC | CC | Deleterious | Damaging | N |
| 17 | 41,244,936 | G/A | rs799917 | BRCA1 | NM_007297: exon9: c.C2471T: p.P824L | AA | GA | GA | GA | AA | Neutral | Tolerated | Y |
| 17 | 59,763,347 | A/G | rs4986764 | BRIP1 | NM_032043: exon19: c.T2755C: p.S919P | GG | GG | GG | GG | GG | Neutral | Tolerated | N |
| 17 | 63,533,087 | C/G | rs145353986 | AXIN2 | NM_004655: exon7: c.G1807C: p.A603P | GG | GG | CC | CC | CC | Neutral | Tolerated | N |
Note: Only mutations predicted to affect the protein coding sequences are listed.
Abbreviations: Chr, chromosome; dbSNP ID, database of SNP identification; N, no; N/A, not applicable; Ref/Alt, reference/alternate; Y, yes.
Two STK11/P variants (serine/threonine kinase 11 interacting protein, NM_052902: exon15: c.G1687A: p.V563I) with predicted deleterious effects were found in FA34 and FA44. BLM (Bloom syndrome, RecQ helicase-like, NM_000057: exon11: c.C2371T: p.R791C), RECQL4 (NM_004260: exon5: c.G1060A: p.V354I), and ERBB2 (NM_004448: exon27: c.C3508G: p.P1170A) variants were found in FA34 alone. An ATM variant (ATM serine/threonine kinase, NM_000051:exon6:c.G566A:p.R189K) with potential deleterious effects was found in FA44, together with a tolerated variant of EGFR at exon 13 (NM_005228: exon13: c.G1562A: p.R521K), and FLT3 (fms-related tyrosine kinase 3, NM_004119: exon6: c.C680T: p.T227M), and another variant of ERBB2 was found at the same genetic locus as in FA34 above. A Notch1 variant (NM_017617: exon11: c.A1762C: p.T588P) was found in FA98. A WRN variant (Werner syndrome, RecQ helicase-like, NM_000553: exon26: c.G3222T: p.L1074F) was found in PME1.
Discussion
The five lung adenocarcinoma and mesothelioma cell lines were newly established from lung cancer and mesothelioma patients with advanced-stage disease, and each of them carries their respective oncogenic driver mutation, including two new lung adenocarcinoma cell lines with two different KRAS mutations at codon 12, and one new lung adenocarcinoma cell line with EML4-ALK translocation. All the new adenocarcinoma cell lines were CK7 and TTF1 positive on immunohistochemical staining and at the same time CK20 negative, which is the typical immunohistochemical profile for lung adenocarcinoma. The combination of TTF1- and napsin A-positive staining has been shown to give a sensitivity of 79.2% in demonstrating cells of primary lung adenocarcinoma origin.21 The mesothelioma cell line was in addition stained positive for anti-vimentin and calretinin, again the typical immunohistochemical profile for mesothelioma cells.
FA44 and FA98 showed a shorter initial lag phase and they formed confluent cell layers, while FA34 and PME1 displayed tumor cell aggregation. The tendency for tumor cells to form clusters is apparently not associated with tumorigenicity in nude mice, as all four lung adenocarcinomas but not the mesothelioma cell line resulted in formation of tumor xenografts in nude mice (Table 1).
ALK rearrangement variant 2 was found in FA34 which originated from a young lady, who was a nonsmoker, with lung adenocarcinomas with metastatic pleural effusion that demonstrated no response to multiple lines of systemic chemotherapy and EGFR-TKI. At the time of presentation, crizotinib was not yet available, so had not been considered. Lung adenocarcinoma cell lines bearing EML4-ALK rearrangement were seldom found in the literature and the more-commonly used ones were NCI-H3122 and DFCI032.22 EML4-ALK rearrangement was reported to be more-commonly found in young patients, especially nonsmokers, with lung adenocarcinoma.20 The FA34 ALK rearranged lung adenocarcinoma cell line reported in this study will be useful available material for further study of EML4-ALK translocated lung adenocarcinomas of Chinese origin for further study in the biology and drug response for EML4-ALK lung cancer.
Two different KRAS mutations at codon 12 were found in two lung adenocarcinoma cell lines, FA44 and FA49. These two cell lines were derived from male smokers, and were the typical subtype of lung adenocarcinomas for smokers who were not responsive to platinum-based chemotherapy. KRAS mutant lung cancer cell lines were found in the NCI-H series, namely H23, H358, H441, and H1792, which all bear codon 12 mutations.23 The two newly established lines in this report, FA44 and FA49, would add further available lung adenocarcinoma cell lines of Chinese origin for further in vitro study into the understanding of KRAS mutant lung cancer.
Each of the cell lines showed complex structural chromosomal aberrations, yet shared no common structural exchanges (derivative chromosomes), indicating the complex nature of chromosome aberrations in cancer development. However, losses or partial deletions of chromosome 13 were observed in all five cancer cell lines. Chromosome 13 is known to harbor RB1, a tumor suppressor gene, the deletion of which may contribute to tumor growth in lung cancer.24 The composite karyotypes (indicated by “cp” at the ends of descriptions) of the cell lines suggest the high levels of genomic instability of the cell lines.
Among the 42 genes with mutation or SNP identified from the RainDance OncoSeq panel, there were genes commonly found to be mutated/variant in lung cancer while some other mutant/variants could be the characteristics of individual cancer cell lines. We used the SIFT17,18 and PROVEAN19 softwares to predict whether these mutations could give rise to damaging or deleterious effects to the resultant expression.
Notch1 was found to be a variant in FA49, FA98, and PME1. Notch1 is believed to control cell proliferation and apoptosis. Notch1 upregulation and expression were found to have inhibitory effects on tumor cell growth in adenocarcinoma cells.25 The growth of mesothelioma cells has been shown to be affected by aberrant Notch1 signaling.26 The two adenocarcinoma cell lines, FA49 and FA98, as well as PME1 in the mesothelioma cell line, possess variants in Notch1 and could be used as an in vitro model to further study the role of Notch1 in lung adenocarcinoma and mesothelioma cell growth.
STK11/P (or LKB1) was found to be mutated in all five cancer cell lines. LKB1 is known to be a tumor suppressor gene that encodes for a serine/threonine kinase, which is involved in the adenosine monophosphate-dependent protein kinase (AMPK) pathways for the regulation of intracellular energy level.27 Mutation or variants of LKB1 have been reported in genomic studies on lung cancers, and the presence of LKB1 mutation may cause dysregulation of these AMPK pathways and thus could result in disinhibition of energy-consuming anabolic processes and eventually promote tumor growth.28,29 AXIN2 is another known tumor suppressor gene on the Wnt/β-catenin pathway and a variant (NM_004655: exon7: c.G1807C: p.A603P) was found in both FA34 and FA98. AXIN2 polymorphism was reported to be associated with risk of developing lung cancer, at least in Japanese30 and Turkish groups.31
Two different ERBB2 variants were found in FA34 and FA44, whereas an EGFR variant was also present in FA44. Both EGFR and ERBB2 (also known as HER2/neu) belong to the same family of erbB receptor tyrosine kinases. Both ERBB2 gene (which encodes for HER2) and EGFR mutation at exons 18–21 are major proliferative drivers that activate downstream growth signaling pathways.32 However, the ERBB2 variants were found at exon 27, which differs from the mutations at exon 20 that have been found to correlate with clinical characteristics and therapeutic response;33 the same for EGFR mutation at exon 13 in this study, which was different from the usual region of mutations within exons 18–21.32
ATM protein plays a role in the DNA damage response signaling pathway.34 It acts as a link between DNA damage, cell cycle progression, and cell death.34 FA44 was found to carry ATM mutation and thus could be a cell line model for in vitro drug testing and targeting.35
BLM and RECQL4 were found to be variants in FA34. Both BLM and RECQL4 encode for RecQ helicase which is important for the proper maintenance of telomere length.36 IL21R (interleukin 21 receptor, NM_021798: exon4: c.A238C: p.T80P) was another variant found among these newly established cell lines. Aberrant IL21R may interfere with the action of IL21, which itself could be tumor suppressive.37,38
A BRCA1 variant (breast cancer 1, early onset, NM_007297: exon9: c.A2972G: p.E991G) with potential deleterious effects was found in FA34 and PME1. BRCA1 is known to be involved not only in breast cancer but also in lung cancer, where it has been found to be associated with smoking status, histological features, as well as sensitivity towards platinum-based chemotherapy.39
Mutations in MPM are thought to be scant, are seldom identified to be of significant functional implication, and are mostly believed to be epigenetic oncogenic changes.40,41 In PME1, however, we did identify WRN _ENREF_33 mutations or SNP, which may represent the scant oncogenic genomic changes in mesothelioma.42
Although the panel of RainDance Oncoseq was limited to the detection of genetic variants/mutations in a panel of 140 known oncogenic mutations, it served as a screening tool for potential oncogenic targets within those cancer cell lines. The molecular profiles identified in this study shared similarity as well as difference with recent molecular landscapes identified by next-generation sequencing. EGFR, KRAS, and STK11/P have been identified to be commonly variant in cytological specimens of lung adenocarcinomas43 and in lung adenocarcinoma tissue (STK11/P and ATM).44 Mutations of all these potential therapeutic oncology targets – namely ALK rearrangement, KRAS mutations, ERBB2, Notch1, BRCA1, BLM, and related RecQL4, IL-21R, STK11/P, and ATM – were reported in various frequencies in lung cancer before.45,46 This demonstrated the representativeness of these new lung cancer and mesothelioma cell lines and their potential usefulness for therapeutic drug testing.
In summary, we have established four new primary lung adenocarcinoma cell lines and one new malignant mesothelioma cell line from Chinese patients with different clinical characteristics, cell culture properties, and oncogenic mutation profiles. These new cancer cell lines could add to the present pools of publicly available lung cancer and mesothelioma cell lines, the majority of which were derived from patients of non-Asian origin, serving as potentially useful tools for functional studies and drug sensitivity testing in lung cancer and malignant mesothelioma.
Acknowledgments
The work described in this paper was supported in part by the Lee Wing Tat Cardio-thoracic Medicine Education and Research Fund 2008 – 2010, the Hong Kong SK Yee Medical Foundation 2010 (Project no. 210210, for EGFR and KRAS mutation tests) and 2012 (Project no. 212207, for ALK rearrangement test), the Shun Tak District Min Yuen Tong of Hong Kong 2012, and the Hong Kong Pneumoconiosis Compensation Fund Board Research Grant 2013.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 2.Larsen JE, Minna JD. Molecular biology of lung cancer: clinical implications. Clin Chest Med. 2011;32(4):703–740. doi: 10.1016/j.ccm.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brownson RC, Chang JC, Davis JR. Gender and histologic type variations in smoking-related risk of lung cancer. Epidemiology. 1992;3(1):61–64. doi: 10.1097/00001648-199201000-00012. [DOI] [PubMed] [Google Scholar]
- 4.Zang EA, Wynder EL. Differences in lung cancer risk between men and women: examination of the evidence. J Natl Cancer Inst. 1996;88(3–4):183–192. doi: 10.1093/jnci/88.3-4.183. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 6.Nebhan C, Pao W. Further advances in genetically informed lung cancer medicine. J Thorac Oncol. 2013;8(5):521–522. doi: 10.1097/JTO.0b013e31828f583c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam DC, Girard L, Suen WS, et al. Establishment and expression profiling of new lung cancer cell lines from Chinese smokers and lifetime never-smokers. J Thorac Oncol. 2006;1(9):932–942. [PubMed] [Google Scholar]
- 8.Oie HK, Russell EK, Carney DN, Gazdar AF. Cell culture methods for the establishment of the NCI series of lung cancer cell lines. J Cell Biochem Suppl. 1996;24:24–31. doi: 10.1002/jcb.240630504. [DOI] [PubMed] [Google Scholar]
- 9.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12(5):1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 10.de Figueiredo-Pontes LL, Wong DW, Tin VP, et al. Identification and characterization of ALK kinase splicing isoforms in non-small-cell lung cancer. J Thorac Oncol. 2014;9(2):248–253. doi: 10.1097/JTO.0000000000000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng W, Tsao SW, Lucas JN, Leung CS, Cheung AL. A new method for improving metaphase chromosome spreading. Cytometry A. 2003;51(1):46–51. doi: 10.1002/cyto.a.10004. [DOI] [PubMed] [Google Scholar]
- 12.Shaffer LG, Slovak ML, Campbell LJ, editors. An international system for human cytogenetic nomenclature (2009) Basel; Unionville, CT: Karger; 2009. [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [q-bio.GN] 2012 [Google Scholar]
- 15.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goecks J, Nekrutenko A, Taylor J, Galaxy Team Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 18.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue):W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PloS One. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 21.Ye J, Findeis-Hosey JJ, Yang Q, et al. Combination of napsin A and TTF-1 immunohistochemistry helps in differentiating primary lung adenocarcinoma from metastatic carcinoma in the lung. Appl Immunohistochem Mol Morphol. 2011;19(4):313–317. doi: 10.1097/PAI.0b013e318205b059. [DOI] [PubMed] [Google Scholar]
- 22.Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14(13):4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunaga N, Shames DS, Girard L, et al. Knockdown of oncogenic KRAS in non-small cell lung cancers suppresses tumor growth and sensitizes tumor cells to targeted therapy. Mol Cancer Ther. 2011;10(2):336–346. doi: 10.1158/1535-7163.MCT-10-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelley MJ, Nakagawa K, Steinberg SM, Mulshine JL, Kamb A, Johnson BE. Differential inactivation of CDKN2 and Rb protein in non-small-cell and small-cell lung cancer cell lines. J Natl Cancer Inst. 1995;87(10):756–761. doi: 10.1093/jnci/87.10.756. [DOI] [PubMed] [Google Scholar]
- 25.Wael H, Yoshida R, Kudoh S, Hasegawa K, Niimori-Kita K, Ito T. Notch1 signaling controls cell proliferation, apoptosis and differentiation in lung carcinoma. Lung Cancer. 2014;85(2):131–140. doi: 10.1016/j.lungcan.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Graziani I, Eliasz S, De Marco MA, et al. Opposite effects of Notch-1 and Notch-2 on mesothelioma cell survival under hypoxia are exerted through the Akt pathway. Cancer Res. 2008;68(23):9678–9685. doi: 10.1158/0008-5472.CAN-08-0969. [DOI] [PubMed] [Google Scholar]
- 27.Conde E, Suarez-Gauthier A, García-García E, et al. Specific pattern of LKB1 and phospho-acetyl-CoA carboxylase protein immunostaining in human normal tissues and lung carcinomas. Hum Pathol. 2007;38(9):1351–1360. doi: 10.1016/j.humpath.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Marks K, Cowley GS, et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov. 2013;3(8):870–879. doi: 10.1158/2159-8290.CD-13-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shackelford DB, Abt E, Gerken L, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23(2):143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanzaki H, Ouchida M, Hanafusa H, et al. Single nucleotide polymorphism of the AXIN2 gene is preferentially associated with human lung cancer risk in a Japanese population. Int J Mol Med. 2006;18(2):279–284. [PubMed] [Google Scholar]
- 31.Gunes EG, Pinarbasi E, Pinarbasi H, Silig Y. Strong association between lung cancer and the AXIN2 polymorphism. Mol Med Rep. 2009;2(6):1029–1035. doi: 10.3892/mmr_00000210. [DOI] [PubMed] [Google Scholar]
- 32.Siegelin MD, Borczuk AC. Epidermal growth factor receptor mutations in lung adenocarcinoma. Lab Invest. 2014;94(2):129–137. doi: 10.1038/labinvest.2013.147. [DOI] [PubMed] [Google Scholar]
- 33.Mazieres J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol. 2013;31(16):1997–2003. doi: 10.1200/JCO.2012.45.6095. [DOI] [PubMed] [Google Scholar]
- 34.Khalil HS, Tummala H, Hupp TR, Zhelev N. Pharmacological inhibition of ATM by KU55933 stimulates ATM transcription. Exp Biol Med (Maywood) 2012;237(6):622–634. doi: 10.1258/ebm.2012.011378. [DOI] [PubMed] [Google Scholar]
- 35.Storozhuk Y, Hopmans SN, Sanli T, et al. Metformin inhibits growth and enhances radiation response of non-small cell lung cancer (NSCLC) through ATM and AMPK. Br J Cancer. 2013;108(10):2021–2032. doi: 10.1038/bjc.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rezazadeh S. On BLM helicase in recombination-mediated telomere maintenance. Mol Biol Rep. 2013;40(4):3049–3064. doi: 10.1007/s11033-012-2379-0. [DOI] [PubMed] [Google Scholar]
- 37.Spolski R, Leonard WJ. Interleukin-21: basic biology and implications for cancer and autoimmunity. Annu Rev Immunol. 2008;26:57–79. doi: 10.1146/annurev.immunol.26.021607.090316. [DOI] [PubMed] [Google Scholar]
- 38.Sondergaard H, Coquet JM, Uldrich AP, et al. Endogenous IL-21 restricts CD8+ T cell expansion and is not required for tumor immunity. J Immunol. 2009;183(11):7326–7336. doi: 10.4049/jimmunol.0902697. [DOI] [PubMed] [Google Scholar]
- 39.Bonanno L, Favaretto A, Rosell R. Platinum drugs and DNA repair mechanisms in lung cancer. Anticancer Res. 2014;34(1):493–501. [PubMed] [Google Scholar]
- 40.Tallet A, Nault JC, Renier A, et al. Overexpression and promoter mutation of the TERT gene in malignant pleural mesothelioma. Oncogene. 2014;33(28):3748–3752. doi: 10.1038/onc.2013.351. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi M, Takeuchi T, Ohtsuki Y. Establishment of three novel human malignant pleural mesothelioma cell lines: morphological and cytogenetical studies and EGFR mutation status. Anticancer Res. 2008;28(1A):197–208. [PubMed] [Google Scholar]
- 42.Korhonen MK, Vuorenmaa E, Nyström M. The first functional study of MLH3 mutations found in cancer patients. Genes Chromosomes Cancer. 2008;47(9):803–809. doi: 10.1002/gcc.20581. [DOI] [PubMed] [Google Scholar]
- 43.Scarpa A, Sikora K, Fassan M, et al. Molecular typing of lung adenocarcinoma on cytological samples using a multigene next generation sequencing panel. PloS One. 2013;8(11):e80478. doi: 10.1371/journal.pone.0080478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greulich H. The genomics of lung adenocarcinoma: opportunities for targeted therapies. Genes Cancer. 2010;1(12):1200–1210. doi: 10.1177/1947601911407324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]