Abstract
Hepatocellular carcinoma (HCC) is the most commonly diagnosed form of liver cancer with high morbidity and mortality. Copy number variation analysis (CNV) of human HCC revealed that leukocyte specific protein-1 (LSP1) had the highest number of cases with CNV. LSP1, a F-actin binding protein, is expressed in hematopoietic cells and interacts with Kinase Suppressor of Ras (KSR), a scaffold for the ERK/MAPK pathway. The expression of LSP1 in liver and its role in normal hepatocellular function and carcinogenesis remains unknown. Therefore, LSP1 mRNA and protein levels were analyzed in normal hepatocytes in culture, rat liver following partial hepatectomy (PHx), and hepatoma cell lines. In culture and after PHx, LSP1 increased after the termination of hepatocyte proliferation. To investigate LSP1 function in HCC, shRNA was utilized to stably knock down LSP1 expression in the JM1 rat hepatoma cell line. Loss of LSP1 in JM1 cells resulted in dramatic upregulation of cyclin D1 and pERK2, increased cell proliferation and migration. Co-immunoprecipitation and immunofluoresence analysis displayed an interaction and co-localization between LSP1, KSR and F-actin in the JM1 cells and liver during regeneration. Conversely, expression of LSP1 in JM2 rat hepatoma cell line led to decreased proliferation. Enhanced expression of LSP1 in mouse hepatocytes during liver regeneration following injection of an LSP1 expression plasmid also led to decreased hepatocyte proliferation.
Conclusion
LSP1 is expressed in normal hepatocytes and liver following PHx after the termination of proliferation. In rat hepatoma cell lines and mouse liver in vivo, LSP1 functions as a negative regulator of proliferation and migration. Given the high frequency of LSP1 CNV in human HCC, LSP1 may be a novel target for diagnosis and treatment of HCC.
Keywords: Hepatocellular carcinoma, partial hepatectomy, liver regeneration, cell proliferation and migration
Introduction
Hepatocellular carcinoma (HCC), the most common form of liver cancer, is characterized with high rates of mortality. The 5-year survival rate of individuals with HCC is relatively low at approximately 15%, which stems in part from the lack of effective treatment modalities. HCC is often resistant to current anticancer therapies and underlying diseases of the liver, such as cirrhosis, can limit the use of chemotherapeutic agents leading to increased lethality of HCC. An effective therapeutic alternative is surgical tumor resection; however this can only be implemented in patients with localized disease and liver transplant can only be performed in patients that meet strict criteria (1). Since there is only a basic understanding of the molecular, cellular and environmental processes that lead to this disease and limited treatment options, additional studies must be conducted to gain a better understanding of the development and progression of HCC in order to develop novel therapeutic modalities to combat this lethal disease.
In a previous publication from our laboratory, copy number variation (CNV) analysis was performed on 98 human HCC samples. The results revealed a portion of the gene for leukocyte-specific protein (LSP1), an intracellular F-actin binding protein expressed in neutrophils, macrophages and endothelial cells, is deleted or amplified (in its carboxy-terminal F-actin binding site) in a majority of HCCs evaluated (51 of 98 cases) (2, 3). All of the deletions and amplifications of LSP1 affected the C-terminal F-actin binding region, indicating alteration of this portion of the LSP1 gene may play an important role in the development or progression of HCC (2). There are no previous reports defining a role for LSP1 in the liver or establishing the expression of LSP1 in normal liver cells. Therefore, given the very high frequency of LSP1 CNV in human HCC, the expression and function of LSP1 in normal hepatocytes should be fully elucidated.
Previous studies have demonstrated that mice deficient in LSP1 display accelerated skin wound healing and that LSP1 functions to negatively regulate migration of neutrophils (4). LSP1 acts as a scaffold through KSR for the extracellular signal-regulated kinase/mitogen activated protein kinase pathway (ERK/MAPK) and targets proteins of this pathway to the actin cytoskeleton (5). Signaling through the ERK/MAPK pathways leads to a variety of cellular processes including migration, proliferation, differentiation, survival and is a key signaling pathway in the progression of proliferating hepatocytes through G1 phase of the cell cycle during liver regeneration (6). LSP1 specifically binds to Kinase Suppressor of Ras (KSR), a key regulator of cellular growth due to its function as a scaffold for MAPK and Raf kinase (7, 8). Therefore, loss of LSP1 function may remove suppressing effects on KSR and ERK/MAPK signaling, leading to aberrant hepatocyte proliferation and facilitation of HCC development. In the present study, we demonstrate that LSP1 is expressed in cultured hepatocytes and after the termination of proliferation. Further, the rat hepatoma cell line JM1 expresses LSP1 protein, which, through co-immunoprecipitation analysis, interacts with KSR and F-actin in these cells. Loss of LSP1 expression in the hepatoma cell line leads to increased migration and proliferation, suggesting that LSP1 acts as a negative regulator for these processes. Expression of LSP1 by suitable expression vectors both in vitro (JM2 rat hepatoma cell line) and in vivo (rat liver regeneration) leads to decreased proliferation. Therefore, loss of LSP1 expression and function could promote HCC development and metastasis.
Materials and Methods
Results
LSP1 is expressed in primary hepatocytes in culture
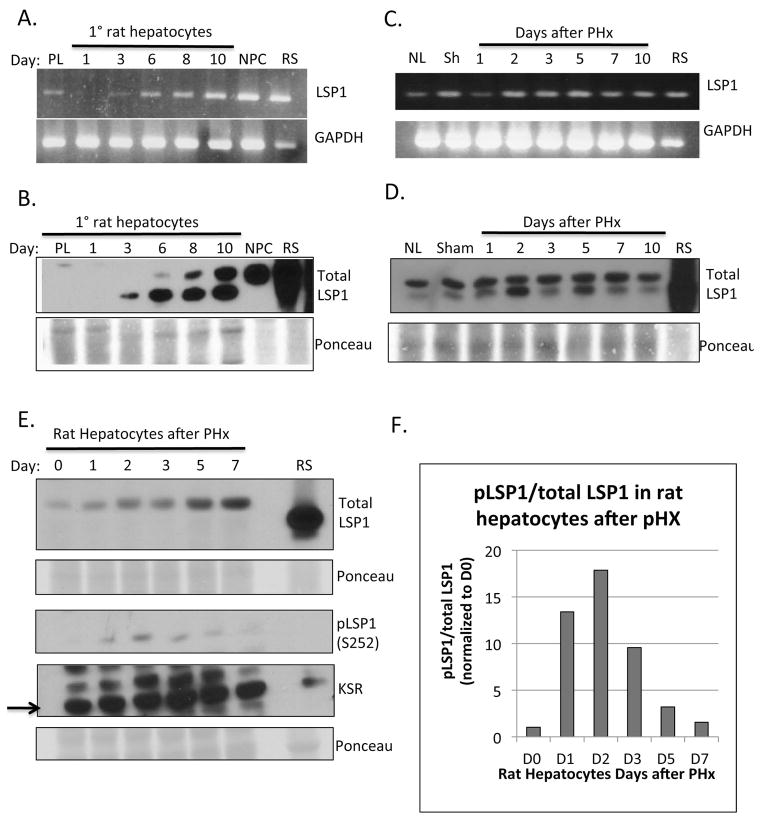
We demonstrate that LSP1 is expressed in primary rat hepatocytes in culture. (Figure 1A and B). Expression of LSP1 protein is not seen in the hepatocyte pellet after isolation by collagenase perfusion or on day 1 in culture but begins to be expressed starting on day 3 in culture and steadily increases to day 10 in culture (Figure 1A and B). The peak of hepatocyte proliferation occurs between days 2 and 4 in culture with growth factors HGF and EGF and after day 4 in culture proliferation starts to decrease (Supplemental Figure 1) (9). The correlation between LSP1 expression and decreased proliferation rates suggests that LSP1 is functioning at the termination stage of hepatocyte proliferation in culture.
Figure 1. LSP1 expression in primary rat hepatocytes in culture, rat liver and hepatocytes after PHx.
Primary rat hepatocytes were cultured in the presence of HGF and EGF and harvested at various time points for A. mRNA expression using semi-quantitative RT-PCR (upper panel, LSP1 and bottom panel GAPDH as loading control), and B. protein expression by western blot (upper panel, total LSP1, bottom panel, Ponceau S stain, loading control). PL: Hepatocyte Pellet; NPC: Non-parenchymal cell fraction; RS: Rat spleen, used as positive control. Whole rat liver was harvested at various times after PHx and LSP1 expression was analyzed using C. RT-PCR (upper panel, LSP1, bottom panel GAPDH) and D. western blot of LSP1 (upper panel) and Ponceau S (bottom panel, loading control). NL: Normal whole liver lysate; RS: Rat spleen. Rat hepatocytes were isolated from liver after PHx by collagenase perfusion and analyzed by E. western blot for total LSP1 (upper panel), phophoLSP1 (S252) (3rd panel down) and KSR (4th panel down). F. Quantification of phosphoLSP1 to total LSP1 in the rat hepatocytes after PHx. For details of interpretation, see Results.
LSP1 expression in liver regeneration
Next, we analyzed the expression of LSP1 during liver regeneration using the commonly utilized 2/3 partial hepatectomy (PHx) model (10, 11). LSP1 mRNA and protein expression was observed at low levels in whole lysates from normal resting liver and 1 day after PHx (Figure 1C and D), which corresponds to the peak of hepatocyte proliferation after PHx in the rat (10, 11). Increased LSP1 expression is observed by western blot and immunofluorescence on days 2 and 5 after PHx. (Figure 1D and 2). Day 2 after PHx, hepatocyte proliferation in the rat is decreased (10, 11) and this corresponds with the increased LSP1 expression, which suggests that LSP1 may play a role in the termination of proliferation in vivo during regeneration. Since LSP1 expression was measured in whole liver after PHx, we are unable to determine which specific cell types of the liver are contributing to the signal. This is because LSP1 is heavily expressed in the non-parenchymal hepatic cell populations (see NPC in Fig. 1B). Therefore, we isolated rat hepatocytes from PHx livers by collagenase perfusion at various time points after PHx and measured LSP1 expression. Western blot analysis for total LSP1 revealed that LSP1 is expressed at low levels at time 0 but steadily increases until day 7. (Figure 1E) Analysis of phopho-LSP1 levels indicates a peak of expression at day 2 after PHx in the hepatocytes. (Figure 1E and F). Upon phosphorylation of LSP1 at serine 252, LSP1 localizes to the F-actin filaments (3) which corresponds to the co-localization of F-actin and LSP1 observed in rat liver after PHx (Figure 2). Western blot analysis of total LSP1 in isolated NPCs after PHx showed a marked increase in expression on day 3 after PHx. (Supplemental Figure 2)
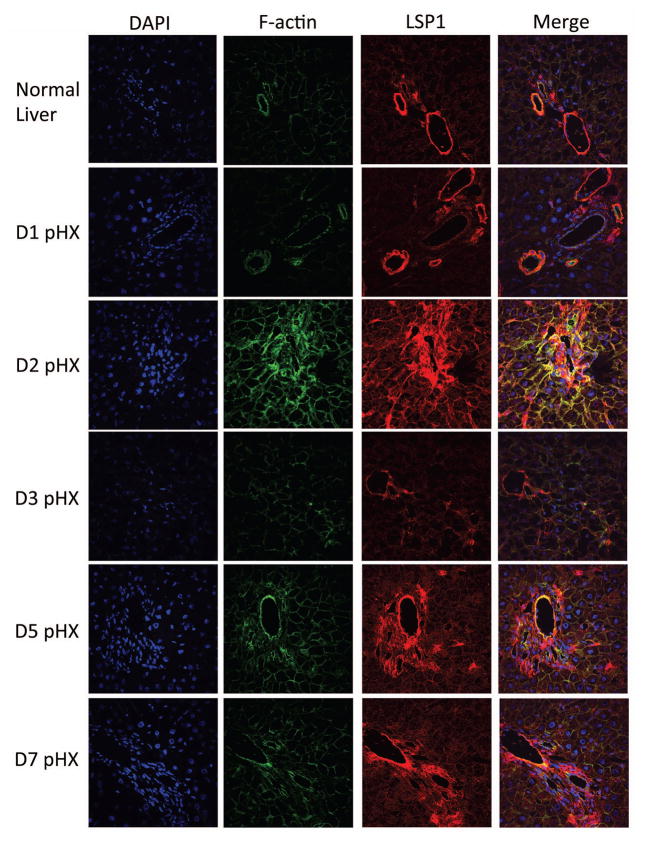
Figure 2. Immunofluoresence of LSP1 and F-actin in rat liver after PHx.
Rat liver was harvested at various time points after PHx and was probed for LSP1 and F-actin. First panel is DAPI to stain the nuclei, second panel shows the F-actin signal, third panel shows the signal for total LSP1 and the last panel displays a merge of the DAPI, F-actin, and total LSP1 signals. 600x magnification. For details of interpretation, see Results.
Since LSP1 is an F-actin binding protein in hematopoietic cells, we utilized immunofluorescence to determine if LSP1 and F-actin co-localize in hepatocytes during liver regeneration. LSP1 and F-actin do co-localize at day 2 after PHx as indicated by the yellow signal observed in the merged images and this co-localization appears to be hepatocellular, since the size, shape and morphological characteristics of the LSP1 positive cells is consistent with hepatocytes (Figure 2). Strong expression of LSP1 in the vascular endothelium and portal mesenchyme (Figure 2) is expected since previous literature has demonstrated LSP1 expression in endothelial and mesenchymal cells (12, 13).
LSP1 has been shown to interact with Kinase Suppressor of Ras (KSR), a ERK/MAPK pathway scaffold, and target these proteins to the actin cytoskeleton (5). Therefore, we also measured the expression of KSR in the isolated rat hepatocytes after PHx. KSR expression is observed in normal resting hepatocytes and it slightly increases towards the end of regeneration.
LSP1 expression in hepatoma cell lines and interactions with ERK/MAPK scaffold Kinase Suppressor of Ras (KSR)
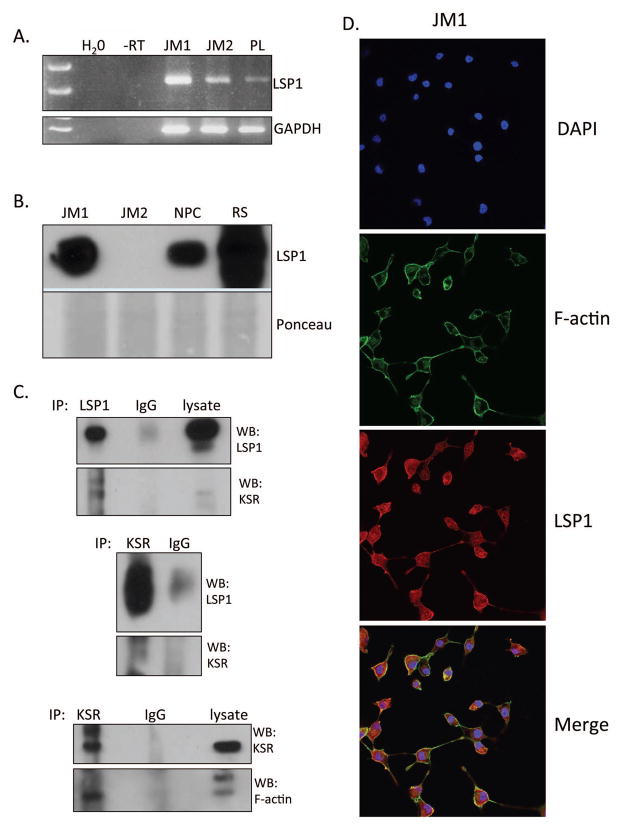
Expression of LSP1 was measured in two rat hepatoma cell lines, JM1 and JM2 (14). JM1 and JM2 cells exhibit different morphological and functional characteristics. Both JM1 and JM2 cells expressed LSP1 mRNA but only JM1 cell line expressed LSP1 protein (Figure 3A and B). Immunofluorescence images demonstrate that LSP1 co-localizes with F-actin in the JM1 cell line as well (Figure 3D). Next, we utilized co-immunoprecipitation to determine if LSP1 interacts with KSR and F-actin in the JM1 cell line. The results indicate that LSP1 does interact with KSR and KSR interacts with F-actin in these cells (Figure 3C). These findings demonstrate that LSP1 is expressed in a rat hepatoma cell line and targets the ERK/MAPK scaffold KSR to F-actin filaments.
Figure 3. LSP1 expression in rat hepatoma cell lines and analysis of the interaction between LSP1, KSR and F-actin.
JM1 and JM2 cell line were analyzed for expression of LSP1 A. mRNA by RT-PCR and B. protein by western blotting. C. Co-immunoprecipitation analysis was performed using the JM1 cell line for interactions between LSP1, KSR and F-actin. Upper panel is an IP of LSP1 and western blot of LSP1 (top) and KSR (bottom). Middle panel is an IP of KSR and western blot of LSP1 (top) and KSR (bottom). Bottom panel is an IP of KSR and western blot of KSR (top) and F-actin (bottom). D. Immunofluorescence images of JM1 cells for DAPI (top panel), F-actin (2nd panel from top), LSP1 (3rd panel from top), and merge (bottom panel). Images were taken at 400x magnification.
Loss of LSP1 expression results in increased proliferation and migration
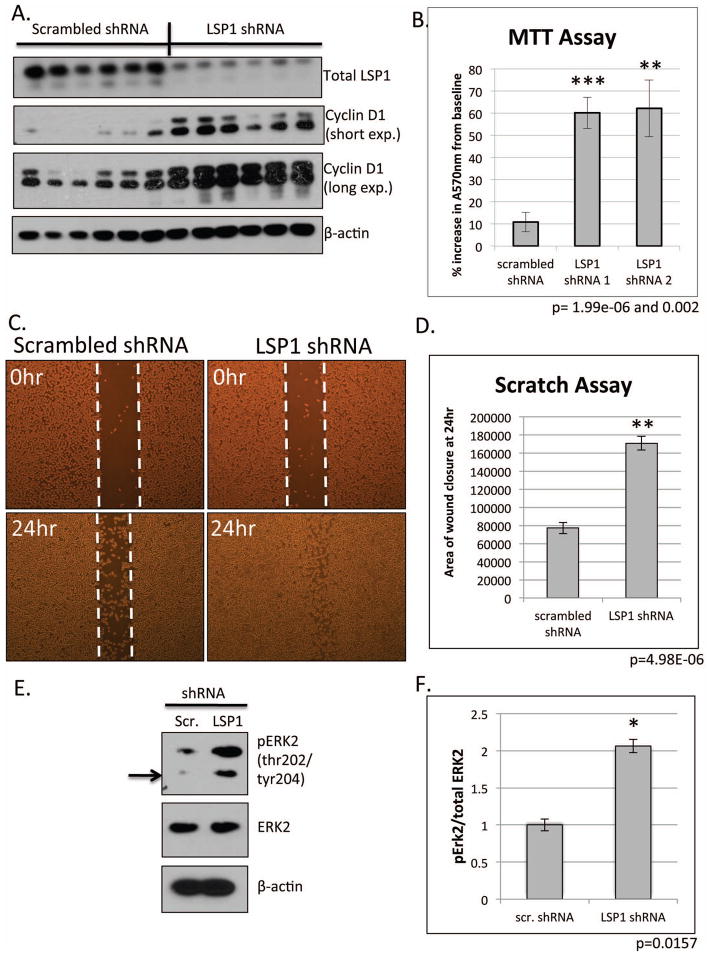
Since LSP1 protein is expressed in JM1 cell line, we utilized these cells as a model to measure the functional significance of the loss of LSP1 in HCC. We transfected the JM1 cell line with short hairpin RNA (shRNA) in order to knock down expression of LSP1 and then measured the effect of loss of LSP1 on proliferation and migration. Since the shRNA vector contained a puromycin resistance gene, we created a stable cell line and first measured the expression of LSP1. Western blot analysis demonstrates a marked decrease in LSP1 protein expression in the LSP1 shRNA transfected cell in comparison to the scrambled shRNA control cells (Figure 4). Next, using immunoblotting, we measured the expression of the commonly utilized proliferation marker, cyclin D1. LSP1 shRNA JM1 cells exhibited increased levels of cyclinD1 in comparison to scrambled control indicating an increased proliferation in the LSP1 shRNA cells (Figure 4A). These findings were further confirmed by MTT assay in which two LSP1 shRNA cell lines demonstrated a 3 and 4-fold increase in absorbance in comparison to scrambled controls indicated an increase in cell numbers (Figure 4B). Since LSP1 is known to negatively regulate migration in leukocytes and endothelial cells (3, 12), we measured migration of LSP1 shRNA cells using a “monolayer scratch” assay and a transwell migration assay. Loss of LSP1 expression led to increased migration into the scratch with an approximately 50% increase in wound closure in the LSP1 shRNA JM1 cells compared to scrambled shRNA cells (Figure 4C and D, Supplemental Figure 3) as well as increased migration across the transwell membrane (Supplemental Figure 4). To determine the mechanism by which LSP1 knockdown resulted in increased proliferation and migration, we analyzed expression of downstream signaling pathways. Since LSP1 is known to interact with KSR and the MEK/ERK pathway signaling proteins, we analyzed expression of phosphorylated ERK2. Loss of LSP1 expression resulted in increased phophorylated ERK2 indicating increased ERK2 activation and suggesting that the increased proliferation and migration occurs through an ERK activated mechanism (Figure 4E and F).
Figure 4. Functional analysis of loss of LSP1 expression in JM1 hepatoma cell line.
JM1 cells were transfected with LSP1 shRNA and a stable cell line was created. A. Western blot analysis of the stable LSP1 shRNA JM1 cell line for total LSP1 (top panel), cyclin D1 (2nd and 3rd panel), and β-actin (bottom panel, loading control). B. MTT assay of scrambled shRNA control cells and two LSP1 shRNA stable clones (LSP1 shRNA 1 and LSP1 shRNA 2). n=18 (experiment repeated at least two independent times), p=1.99e-06 and p=0.002, respectively. C. Representative bright field images of migration “scratch” assay at time 0 (upper panels) and 24 hours post scratch (bottom panel). D. Quantification of area of wound closure from scratch assay. n=6, p=4.98e-06. E. Representative western blot of phophoERK2 (top panel) and total ERK2 (middle panel) in JM1 LSP1 shRNA stable cell lines. F. Quantification of pERK2 and total ERK2. n=7, p=0.0157.
Enhanced expression of LSP1 leads to decreased proliferation in vitro and in vivo
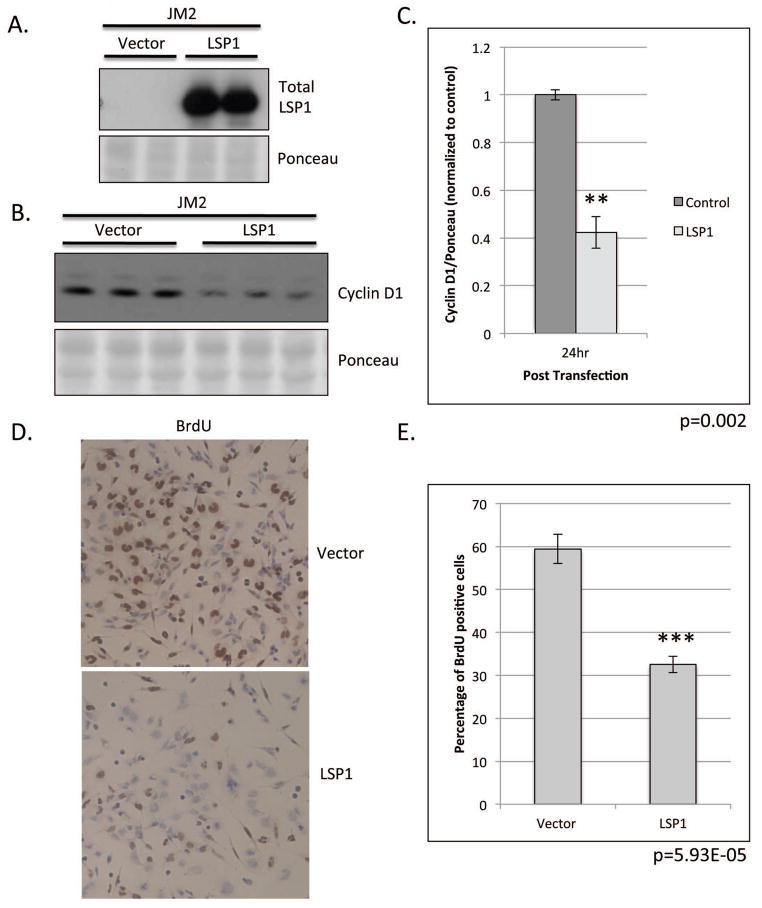
JM2 rat hepatoma cell line does not express LSP1 protein (Figure 3B). Therefore we utilized these cells to study the role of LSP1 expression on proliferation. After transient transfection of JM2 cells with LSP1 cDNA, we observed a significant decrease in the expression of the proliferation marker, cyclin D1 (Figure 5B and C). Furthermore, LSP1 expression in these normally deficient cells led to an approximate 50% decrease in BrdU labeling indicating a decrease in the rate of proliferation. (Figure 5D and E).
Figure 5. Functional analysis of LSP1 expression in JM2 rat hepatoma cell line.
JM2 cells were transiently transfected with LSP1 cDNA and pExpress-1 plasmid (control). A. Western blot analysis of total LSP1 expression (top panel) at 24 hours post transfection in transiently transfected JM2 cell line. Ponceau S (bottom panel) loading control. B. Western blot analysis of cyclin D1 (top panel) expression at 24 hours post transfection. (Ponceau S (bottom panel), loading control). C. Quantification of cyclin D1 protein expression from B. n=3, p=0.002. D. Representative images of BrDU staining in transiently transfected JM2, 100x magnification. E. Quantification of BrDU labeling in D. n=5 per condition, p=5.93E-05.
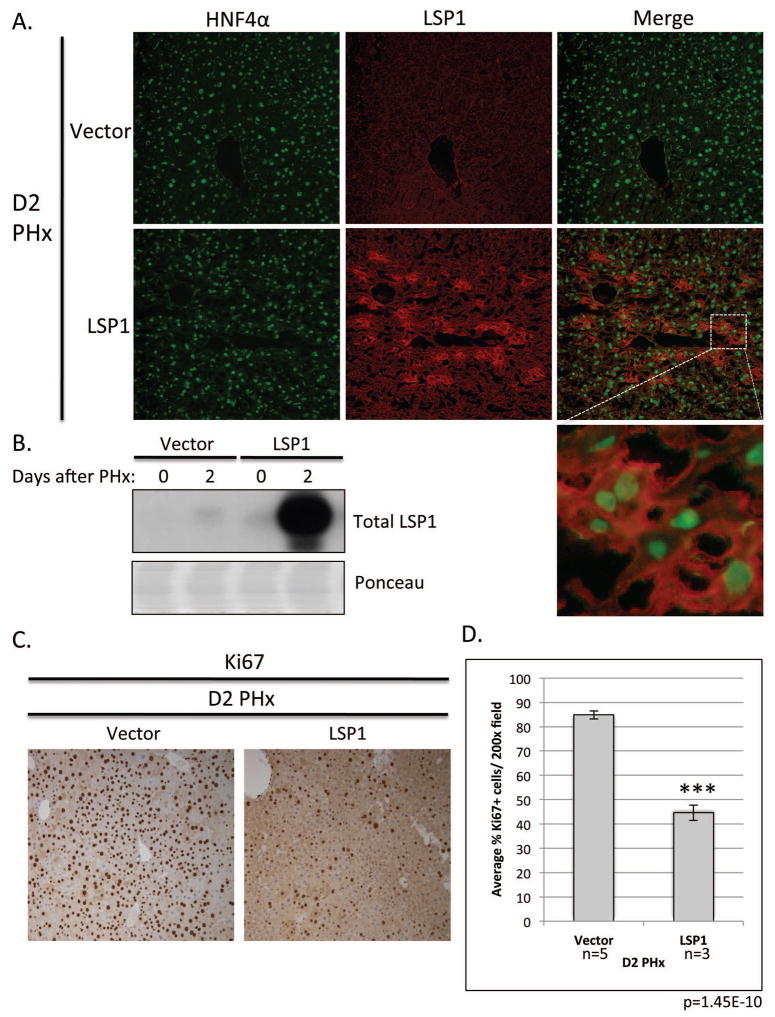
To demonstrate a role for LSP1 in the regulation of proliferation in vivo, we performed a hydrodynamic tail vein injection of LSP1 cDNA plasmid into male FVB mice and immediately performed a 2/3 partial hepatectomy. To show that the livers, specifically the hepatocytes, were successfully expressing LSP1 following injection, we performed western blot analysis for LSP1 expression and immunofluorescence staining of LSP1 and Hepatocyte Nuclear Factor 4α (HNF4α), a marker of hepatocytes. The LSP1 injected animals expressed LSP1 and the LSP1 expression occurred in the cells also expressing HNF4α demonstrating that a majority of the cells expressing LSP1 are hepatocytes. (Figure 6A and B). Though there were variations of the percentage of hepatocytes with high expression of LSP1 from one lobule to another, on average, about 30% of hepatocytes were found to have high LSP1 expression following the hydrodynamic injection. The peak of hepatocyte proliferation after PHx in mice is on day 2 (15). Therefore we harvested the livers on day 2 and performed immunohistochemical staining for the proliferation marker Ki67. The livers of the LSP1 injected mice displayed approximately 50% less Ki67 labeled hepatocytes in comparison to the vector control injected mice suggesting that LSP1 plays a role in regulating the proliferation of hepatocytes in vivo (Figure 6C and D). The results show that enhanced expression of LSP1 at the early (proliferative) stages of liver regeneration has inhibitory effects on hepatocyte proliferation.
Figure 6. In vivo expression of LSP1 in a mouse PHx model.
Hydrodynamic tail vein injection was utilized to express LSP1 in vivo during PHx. A. Immunofluoresecence images of mouse liver at D2 after PHx (vector control (top panel), and LSP1, (bottom panel)) for HNF4α (left panel), LSP1 (middle panel), and merge (right panel). Images were taken at 200x magnification. Inset is shown to demonstrate LSP1 and HNF4α expression is present in the hepatocytes. B. Western blot analysis of total LSP1 expression (top panel) and ponceau (loading control) in total liver lysate from injected mice at Day 0 and 2 after PHx. C. Representative images of Ki67 staining of the injected mouse liver tissue on day 2 after PHx. Right image, vector control and left image, LSP1 injected animal. All images were taken at 200x magnification D. Quantification of the percentage of Ki67 positive hepatocytes in mouse liver tissue on D2 PHx. At least three random fields per slide were quantified using Image J software. Vector control mice n=5, LSP1 mice n=3. p=1.45E-10.
Discussion
A previous study from our laboratory demonstrated that in human HCC, the LSP1 gene had the highest number of cases with CNV (2); however the expression and functional role of LSP1 in both normal liver and HCC is unknown. The finding arrived as a surprise since there is no literature on LSP1 in hepatic biology. In order to place the role of LSP1 in the perspective of normal liver biology, and thus get a framework for understanding its role in hepatic neoplasia, we embarked upon the studies described above, to provide a baseline of LSP1 functions in normal liver and obtain evidence from experimental models as to its functional significance. As a first step in this study, we aimed to determine if LSP1 is expressed in liver, specifically in the hepatocytes, and the role LSP1 plays in hepatocyte growth regulation. LSP1 is intensely expressed in endothelial cells and cells of hematopoietic origin. Thus, studies of LSP1 in whole liver tissue are not likely to be informative since hepatocyte expression of LSP1 is likely to be overwhelmed by the intense expression of LSP1 in hepatic macrophages and endothelial cells. Because of this, we concentrated our studies in hepatocytes in primary culture or in hepatocytes isolated by collagenase perfusion at different stages of regeneration. Our results indicate that LSP1 expression is not observed in normal hepatocytes until day 3 in culture and expression increases over time. Since hepatocytes in culture experience the peak of proliferation between days 2–4 and the highest level of LSP1 expression is observed on day 10, it is reasonable to conclude that LSP1 may be associated with a termination signal for hepatocyte proliferation (9). Analysis of LSP1 expression during liver regeneration demonstrates that the peak of expression occurs on day 7 (Fig. 1E) providing further support for the role of LSP1 as associated with inhibition of hepatocyte growth (10, 11). Measurement of phophoLSP1 in regenerating hepatocytes also reveals increased expression on day 2 after PHx. Phosphorylation of LSP1 at serine 252 corresponds with increased binding to the F-actin filaments.
Our findings revealed changes in LSP1 expression and association with its binding targets (F-actin filaments) during rat liver regeneration. Immunofluorescence analysis shows that LSP1 co-localizes with hepatocyte F-actin on day 2 following PH, which suggests that the ability of LSP1 to bind to the actin filaments may be vital for its role in controlling proliferation. In the CNV study of HCC, all of the amplifications and deletions of LSP1 affected the C-terminal region of the gene, which encodes for the F-actin binding elements of LSP1 (2). We have assumed that amplification of the C-terminal portion of LSP1 may create a situation of a dominant negative protein containing only the C-terminal, which may interfere with the binding of the complete protein. This further supports the role of LSP1 in terminating proliferation and that loss of its ability to bind to F-actin by deletion of c-terminal region could lead to aberrant hepatocyte cell division, leading to or enhancing carcinogenesis.
Since LSP1 CNV was discovered in human HCC and our experimental studies were carried out in the rat,, we measured the expression of LSP1 in two distinct rat hepatoma cell lines, JM1 and JM2 (14). JM1 cells expressed LSP1 RNA and protein as demonstrated by RT-PCR, western blot and immunofluoresence. To demonstrate how LSP1 functions in these cells, co-immunoprecipitation and immunofluoresence displayed that LSP1 interacts and co-localizes with F-actin and the ERK/MAPK pathway scaffold KSR, respectively. KSR functions by targeting MEK1 and ERK2 to the actin cytoskeleton. In unstimulated cells, this complex is mainly localized to the cytoplasm. Upon growth factor stimulation, KSR translocates to the plasma membrane thereby targeting ERK2 and its direct activator MEK1 to the plasma membrane (5, 8). KSR functions to both potentiate and attenuate ERK cascade activation thereby regulating the intensity and duration of ERK pathway signaling from the plasma membrane during growth factor stimulation. The ERK/MAPkinase pathway transmits signals from the plasma membrane to a plethora of cytoplasmic and nuclear targets that affect downstream functions such as proliferation, differentiation and survival. Scaffolding proteins, such as LSP1 and KSR, target the ERK cascade to the appropriate intracellular location, which ensures the correct response of the ERK pathway to various extracellular signals (8). Therefore, in HCC, if LSP1 functions as a growth suppressor and there is a loss of LSP1 expression, this could affect the ERK/MAPK pathway leading to aberrant proliferation or migration. Our findings in Figure 4 demonstrate that loss of LSP1 is indeed associated with enhanced migration, enhanced proliferation rate, evidenced by both increased in cell numbers and dramatic increase in Cyclin D1 in the rat JM1 HCC cells. We demonstrated that the loss of LSP1 expression in hepatoma cells leads to increased ERK2 activation indicating that the observed increase in proliferation and migration is through an ERK2 dependent pathway. Previous literature has demonstrated that increased LSP1 negatively regulates migration of leukocytes and melanoma cell lines (3, 16, 17) and our data supports this notion in hepatocytes as well. Expression of LSP1 in the LSP1 deficient JM2 cell line and in mouse livers in vivo after PHx led to decreased proliferation as evidenced by decreased cyclin D1 levels and BrdU incorporation as well as decreased Ki67 staining. These results corroborate a role for LSP1 in the regulation of hepatocellular proliferation both in vitro and in vivo.
The literature documents the presence of several isoforms for LSP1 in different cell types as a result of alternative splicing (18). As shown in Fig. 1D, there are two protein isoforms in whole liver homogenates. We also noticed that Fig. 1E demonstrates that hepatocytes express only the higher molecular weight isoform. However, only the lower molecular weight mRNA is expressed in hepatocytes in supplemental Fig. S5. The significance of this is not clear. There are multiple isoforms described for LSP1 in the literature (13, 18) and the importance of this should be further investigated.
There have been several recent studies identifying point mutations and large deletions in HCC (19). Driver and “passenger” mutations have been identified and correlated with HCC outcomes. Large size copy number variations (CNV: deletions or amplifications) have also been identified (19). We had concentrated our study (2) in identifying small size CNV, that would otherwise be missed in investigating approaches that would detect CNV of only large size. The latter would be likely to contain many genes in each CNV, thus making it difficult to identify the specific genes whose smaller size CNV would affect neoplastic behavior. LSP1 had the largest number of CNV (51 of 98 cases). In view of this, we believe that elucidation of its function in normal hepatocyte biology is important. We do not believe that genomic alterations in LSP1 alone would be necessarily sufficient to drive a hepatocyte into neoplasia. The high frequency of the LSP1 CNV, however, suggests LSP1 loss of function is important, and that, along with other genomic changes, LSP1 certainly adds to the neoplastic behavior and may be sufficient to convert a low level neoplastic clone into one of a higher malignant potential.
In summary, LSP1 is expressed at the time of cessation of growth in hepatocytes in culture and increases gradually toward the end of liver regeneration. LSP1 functions as a regulator of hepatocyte proliferation and migration most likely by interacting and negatively regulating the function of the ERK/MAPK scaffold KSR. Future studies are aimed at elucidating if loss and overexpression of LSP1 will affect liver carcinogenesis in vivo. Previous studies with a LSP1 global knockout mouse model have shown increased skin wound healing indicating an increase in regenerative capacity of these mice (4). Understanding the function of LSP1 in liver regeneration and cancer may lead to the development of novel target therapies for HCC.
Supplementary Material
Supplemental Figure 1: Quantification of BrdU labeling in primary rat hepatocytes in culture. Primary rat hepatocytes were pulsed with 2μl of BrdU solution in 1ml of complete growth media supplemented with HGF and EGF. Every two days, cells were fixed in 10% formalin and stained with BrdU antibody. Percentage of BrdU positive hepatocytes were quantified by counting cells in at least three random fields per well.
Supplemental Figure 2: Total and phospho LSP1 expression in rat non-parenchymal cells (NPC) after PHx. Western blot analysis of total LSP1 expression (top panel) and phosphoLSP1 expression (3rd panel) in rat NPC after PHx. Ponceau S stain (loading control).
Supplemental Figure 3: Live cell imaging of migration scratch assay of JM1 LSP1 shRNA stable cell line. Left panel: scrambled shRNA stable cell line. Right panel: LSP1 shRNA stable cell line. Images were taken every 2 hours for 18 hours. 100x magnification.
Supplemental Figure 4: Transwell Migration Assay of JM1 LSP1 shRNA stable cell line. A. Representative images of 0.1% Coomassie blue/10% methanol/10% acetic acid stained cells that migrated to the bottom of the transwell membrane. Images were taken using Olympus Provis inverted microscope at a 100x magnification. Left panel: scrambled shRNA stable cells. Right panel: LSP1 shRNA JM1 stable cell line. B. Quantification of the number of migrating cells. At least 3 random fields per membrane were counted using Image J software (NIH).
Supplemental Figure 5: RT-PCR of rat hepatocytes in culture and rat hepatoma cell lines for LSP1 isoforms. Primers were designed for the two most common isoforms of LSP1. Top panel: Isoform 1. Bottom panel: Isoform 2.
Acknowledgments
This work was supported by Grant NIH CA103958 (GKM), by funds from the Cleveland Foundation (GKM), and a grant from NHLBI T32HL094295 (KK).
We would like to thank Dr. Liang-I Kang, Anne Orr, Meagan Haynes, Michael Ding and Jun Yan Tao for technical assistance and Dr. Jan Jongstra for generously providing the LSP1 antibody and his invaluable advice.
Abbreviations
- HCC
Hepatocellular Carcinoma
- CNV
Copy number variation
- LSP1
Leukocyte Specific Protein-1
- KSR
Kinase Suppressor of Ras
- ERK/MAPK
Extracellular signal related kinase/mitogen-activated protein kinase
- PHx
Partial Hepatectomy
- NPC
non-parenchymal cells
- RS
rat spleen
- PL
Hepatocyte pellet
- RT-PCR
reverse transcriptase-polymerase chain reaction
References
- 1.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6(9):674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 2.Nalesnik MA, Tseng G, Ding Y, Xiang GS, Zheng ZL, Yu Y, et al. Gene deletions and amplifications in human hepatocellular carcinomas: correlation with hepatocyte growth regulation. Am J Pathol. 2012;180(4):1495–1508. doi: 10.1016/j.ajpath.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jongstra-Bilen J, Jongstra J. Leukocyte-specific protein 1 (LSP1): a regulator of leukocyte emigration in inflammation. Immunol Res. 2006;35(1–2):65–74. doi: 10.1385/IR:35:1:65. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Jiao H, Stewart TL, Lyons MV, Shankowsky HA, Scott PG, et al. Accelerated wound healing in leukocyte-specific, protein 1-deficient mouse is associated with increased infiltration of leukocytes and fibrocytes. J Leukoc Biol. 2007;82(6):1554–1563. doi: 10.1189/0507306. [DOI] [PubMed] [Google Scholar]
- 5.Harrison RE, Sikorski BA, Jongstra J. Leukocyte-specific protein 1 targets the ERK/MAP kinase scaffold protein KSR and MEK1 and ERK2 to the actin cytoskeleton. J Cell Sci. 2004;117(Pt 10):2151–2157. doi: 10.1242/jcs.00955. [DOI] [PubMed] [Google Scholar]
- 6.Talarmin H, Rescan C, Cariou S, Glaise D, Zanninelli G, Bilodeau M, et al. The mitogen-activated protein kinase kinase/extracellular signal-regulated kinase cascade activation is a key signalling pathway involved in the regulation of G(1) phase progression in proliferating hepatocytes. Mol Cell Biol. 1999;19(9):6003–6011. doi: 10.1128/mcb.19.9.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wortzel I, Seger R. The ERK Cascade: Distinct Functions within Various Subcellular Organelles. Genes Cancer. 2011;2(3):195–209. doi: 10.1177/1947601911407328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKay MM, Ritt DA, Morrison DK. Signaling dynamics of the KSR1 scaffold complex. Proc Natl Acad Sci U S A. 2009;106(27):11022–11027. doi: 10.1073/pnas.0901590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Block GD, Locker J, Bowen WC, Petersen BE, Katyal S, Strom SC, et al. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J Cell Biol. 1996;132(6):1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276(5309):60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 11.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176(1):2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Cara DC, Kaur J, Raharjo E, Mullaly SC, Jongstra-Bilen J, et al. LSP1 is an endothelial gatekeeper of leukocyte transendothelial migration. J Exp Med. 2005;201(3):409–418. doi: 10.1084/jem.20040830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misener VL, Hui C, Malapitan IA, Ittel ME, Joyner AL, Jongstra J. Expression of mouse LSP1/S37 isoforms. S37 is expressed in embryonic mesenchymal cells. J Cell Sci. 1994;107(Pt 12):3591–3600. doi: 10.1242/jcs.107.12.3591. [DOI] [PubMed] [Google Scholar]
- 14.Novicki DL, Jirtle RL, Michalopoulos G. Establishment of two rat hepatoma cell strains produced by a carcinogen initiation, phenobarbital promotion protocol. In Vitro. 1983;19(3 Pt 1):191–202. doi: 10.1007/BF02618059. [DOI] [PubMed] [Google Scholar]
- 15.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213(2):286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, Hayashi H, Harrison R, Chiu B, Chan JR, Ostergaard HL, et al. Modulation of Mac-1 (CD11b/CD18)-mediated adhesion by the leukocyte-specific protein 1 is key to its role in neutrophil polarization and chemotaxis. J Immunol. 2002;169(1):415–423. doi: 10.4049/jimmunol.169.1.415. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Zhang Q, Aaron R, Hilliard L, Howard TH. LSP1 modulates the locomotion of monocyte-differentiated U937 cells. Blood. 2000;96(3):1100–1105. [PubMed] [Google Scholar]
- 18.Thompson AA, Omori SA, Gilly MJ, May W, Gordon MS, Wodd WJ, Jr, et al. Alternatively spliced exons encode the tissue-specific 5′ termini of leukocyte pp52 and stromal cell S37 mRNA isoforms. Genomics. 1996;32(3):352–357. doi: 10.1006/geno.1996.0129. [DOI] [PubMed] [Google Scholar]
- 19.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Quantification of BrdU labeling in primary rat hepatocytes in culture. Primary rat hepatocytes were pulsed with 2μl of BrdU solution in 1ml of complete growth media supplemented with HGF and EGF. Every two days, cells were fixed in 10% formalin and stained with BrdU antibody. Percentage of BrdU positive hepatocytes were quantified by counting cells in at least three random fields per well.
Supplemental Figure 2: Total and phospho LSP1 expression in rat non-parenchymal cells (NPC) after PHx. Western blot analysis of total LSP1 expression (top panel) and phosphoLSP1 expression (3rd panel) in rat NPC after PHx. Ponceau S stain (loading control).
Supplemental Figure 3: Live cell imaging of migration scratch assay of JM1 LSP1 shRNA stable cell line. Left panel: scrambled shRNA stable cell line. Right panel: LSP1 shRNA stable cell line. Images were taken every 2 hours for 18 hours. 100x magnification.
Supplemental Figure 4: Transwell Migration Assay of JM1 LSP1 shRNA stable cell line. A. Representative images of 0.1% Coomassie blue/10% methanol/10% acetic acid stained cells that migrated to the bottom of the transwell membrane. Images were taken using Olympus Provis inverted microscope at a 100x magnification. Left panel: scrambled shRNA stable cells. Right panel: LSP1 shRNA JM1 stable cell line. B. Quantification of the number of migrating cells. At least 3 random fields per membrane were counted using Image J software (NIH).
Supplemental Figure 5: RT-PCR of rat hepatocytes in culture and rat hepatoma cell lines for LSP1 isoforms. Primers were designed for the two most common isoforms of LSP1. Top panel: Isoform 1. Bottom panel: Isoform 2.