Summary
Transforming growth factor-beta (TGF-β) suppresses T cell function to maintain self-tolerance and to promote tumor immune evasion. Yet how Smad4, a transcription factor component of TGF-β signaling, regulates T cell function remains unclear. Here we have demonstrated an essential role for Smad4 in promoting T cell function during autoimmunity and anti-tumor immunity. Smad4 deletion rescued the lethal autoimmunity resulting from transforming growth factor-beta receptor (TGF-βR) deletion and compromised T-cell-mediated tumor rejection. While Smad4 was dispensable for T cell generation, homeostasis and effector function, it was essential for T cell proliferation following activation in vitro and in vivo. The transcription factor Myc was identified to mediate Smad4-controlled T cell proliferation. This study thus reveals a requirement of Smad4 for T-cell-mediated autoimmunity and tumor rejection, which is beyond the current paradigm. It highlights a TGF-βR-independent role for Smad4 in promoting T cell function, autoimmunity and anti-tumor immunity.
Introduction
Transforming growth factor-beta (TGF-β) is important to maintain immune homeostasis and self-tolerance (Li et al., 2006b) and to promote tumorigenesis by inhibiting tumor immune surveillance (Wahl et al., 2006; Yang et al., 2010). Deletion of TGF-β1 leads to multifocal autoimmune disease (Shull et al., 1992), which underscores its essential role in suppressing immune cell function. As a pleiotropic cytokine, TGF-β regulates various immune cell types including CD4+ and CD8+ T cells, natural killer cells, CD8αα+ T cells, B cells, and myeloid cells to control immunity (Cazac and Roes, 2000; Fridlender et al., 2009; Konkel et al., 2011; Laouar et al., 2005; Li et al., 2006a; Liu et al., 2008; Marie et al., 2005; Yang et al., 2008a). T cells are the critical target of TGF-β as disruption of TGF-β signaling specifically in T cells results in early lethal autoimmunity similar to what observed in germ-line TGF-β1-deficienct mice (Li et al., 2006b; Liu et al., 2008; Marie et al., 2006). In addition, defective TGF-β signaling in T cells leads to enhanced tumor rejection (Gorelik and Flavell, 2001). TGF-β controls T cell response through several mechanisms. It suppresses T cell proliferation and effector T cell function (Li et al., 2006b). In addition, TGF-β promotes the generation, maintenance and function of Foxp3-expressing regulatory T (Treg) cells that potently suppress immune response (Rubtsov and Rudensky, 2007). In the absence of TGF-β receptor (TGF-βR), T cells proliferate aberrantly in response to self-antigen (Zhang and Bevan, 2012). Such uncontrolled proliferation leads to uninhibited T helper-1 (Th1) cell-associated responses. The overwhelming Th1 cell cytokine production impairs Treg cell function to further enhance inflammatory response, culminating into a systemic inflammation syndrome and lethal autoimmunity (Ishigame et al., 2013). Impaired TGF-β signaling contributes to inflammatory diseases and autoimmunity. Aberrantly enhanced TGF-β signaling in the tumor microenvironment thwarts immune surveillance and promotes cancer development. Therefore, in order to understand the etiology of autoimmunity and cancer and to devise effective therapies against these diseases, it is important to reveal how TGF-β functions in immune cells, especially in T cells.
TGF-β-activated pathways have been well-characterized in non-T cells (Shi and Massague, 2003). TGF-β signaling is predominantly mediated by a Smad transcription factor-dependent pathway. Upon ligand binding, TGF-βR phosphorylates and activates receptor-associated Smad2 and Smad3, which subsequently associate with a co-Smad, Smad4, to control target gene expression (Attisano and Wrana, 2002; Massague, 1998). Mitogen activated protein kinases (MAPKs), including TGF-β-activated kinase 1 (TAK1), also mediate TGF-β signaling independent of Smad proteins (Derynck and Zhang, 2003). While the importance of TGF-β in controlling T cell function has been recognized for two decades, it is not until recent years that studies begin to delineate the contributions of Smad-dependent and –independent pathways to T cell function. Conforming to the paradigm established in non-T cells, Smad2 and Smad3 are redundantly required for TGF-β suppressed T cell proliferation and Th cell differentiation, and for TGF-β promoted Treg cell generation (Gu et al., 2012; Takimoto et al., 2010). However TAK1 contributes to the homeostasis of thymic-derived Treg cells (Gu et al., 2012). Nevertheless, how Smad4 controls T cell function remains unclear.
Based on the current understanding of TGF-β signaling, one would predict that T cell specific deletion of Smad4, a co-Smad protein central for TGF-β signaling in non-T cells, would ablate TGF-β signaling in T cells and phenotypically resemble T cell specific TGF-βR-deficient mice (Li et al., 2006b; Liu et al., 2008; Marie et al., 2006). Yet, mice with T cell specific deletion of Smad4 are grossly normal without apparent T cell activation (Yang et al., 2008b). In addition, when these mice are on a specific genetic background, they spontaneously develop cancer but not autoimmunity (Hahn et al., 2011; Kim et al., 2006). Similarly, people with germ-line mutations of Smad4 predispose to familial juvenile polyposis and gastrointestinal cancers (Howe et al., 1998; Miyaki and Kuroki, 2003) but not autoimmune disease. These findings question the relationship between Smad4 and TGF-β signaling in T cells according to established notions. At least two possible explanations may account for these observations. One is that other co-regulator(s) for Smad2 and Smad3, such as TIF1γ (He et al., 2006), may compensate for the loss of Smad4 in T cells and that Smad4 is dispensable for T cell function. The other is, while being involved in TGF-β-mediated suppression of T cell function, Smad4 also plays a critical role in promoting T cell function and the loss of Smad4 impairs T cell function to offset the effects of TGF-βR deficiency. Using genetic models, we provide evidence to support the latter explanation and reveal an essential role for Smad4 in T cell function during autoimmunity and tumor immunity.
Results
Smad4 deletion rescues lethal autoimmunity resulted from TGF-βR deficiency
To address whether Smad4 is dispensable for TGF-β signaling or has previously unappreciated function to counter-balance TGF-β-dependent function in T cells, we generated T cell specific Tgfbr2−/− Smad4−/− mice in which T cells were deficient in both TGF-βRII and Smad4 by crossing Cd4-cre mice(Lee et al., 2001) with Tgfbr2fl/fl and Smad4fl/fl mice (Chu et al., 2004; Chytil et al., 2002). If Smad4 is dispensable for T cell function, one would predict that T cell specific Tgfbr2−/− Smad4−/− mice develop early lethal autoimmunity as in Cd4-cre-Tgfbr2fl/fl mice (Li et al., 2006a; Marie et al., 2006). However, if Smad4 plays a role in counter-balancing TGF-β-dependent function, one will predict that Smad4 deletion would ameliorate the lethal autoimmunity in Cd4-cre-Tgfbr2fl/fl mice to display a phenotype similar to Cd4-cre-Smad4fl/fl mice. We found that, unlike Cd4-cre-Tgfbr2fl/fl mice that succumbed to a systemic, lympho-proliferative, autoimmune syndrome by four weeks after birth, T cell specific Tgfbr2−/− Smad4−/− mice survived well beyond four months of age (Fig. 1a). While Cd4-cre-Tgfbr2fl/fl mice were runted, T cell specific Tgfbr2−/− Smad4−/− mice were similar in size as wild-type littermates (data not shown). Smad4 deletion in T cells therefore rescues early lethality of Cd4-cre-Tgfbr2fl/fl mice.
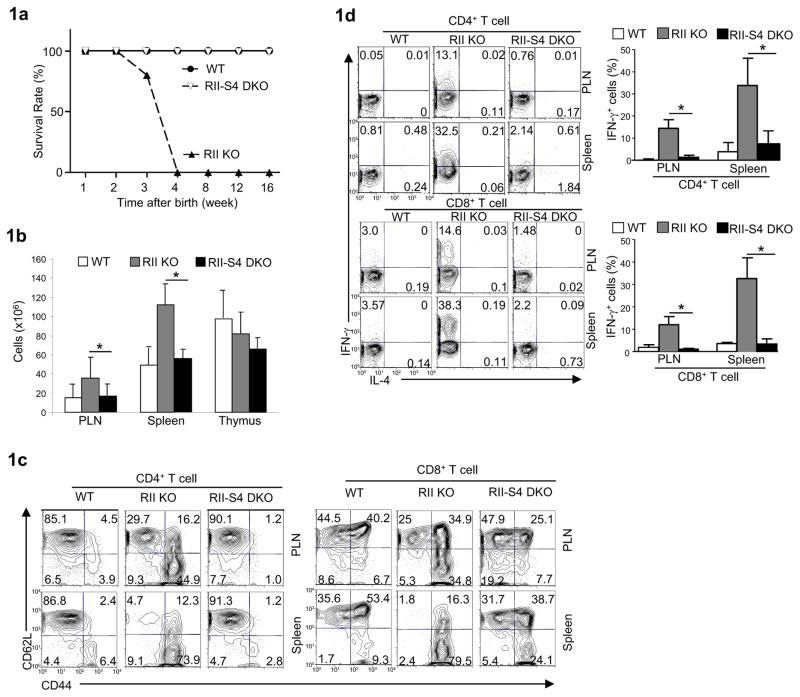
Fig. 1. Analysis of wild-type (WT), Cd4-cre-Tgfbr2fl/fl (RII KO) and Cd4-cre- Tgfbr2fl/fl-Smad4fl/fl (RII-S4 DKO) mice.
(a) The survival rate of different strains at different time after birth.
(b) Total lymphocytes recovered from the peripheral lymph-nodes (PLN), spleens and thymus of different strains. Means ± SD of five sets of mice are shown. (*P<0.05)
(c) CD62L and CD44 expression on peripheral T cells in different strains assessed by flow-cytometry. Representative results of at least three experiments are shown.
(d) IFN-γ and IL-4 production in peripheral T cells in different strains assessed by flow-cytometry. Representative results of at least three experiments (left) and means ± SD of five sets of mice (right) are shown. (*P<0.05)
See also Supplemental Figure 1.
Abrogation of TGF-βR in T cells results in perturbed development of thymocytes (Li et al., 2006a) and spontaneous activation of peripheral T cells (Li et al., 2006a; Marie et al., 2006). We assessed if deletion of Smad4 corrected these abnormalities. We first confirmed that Smad4 was efficiently deleted in Tgfbr2−/− Smad4−/− T cells and that the phosphorylation of Smad2, a hallmark for the activation of TGF-β signaling, was abrogated in these T cells (Supplemental Fig. 1a). The distribution of DN, DP, CD4+ SP, and CD8+ SP thymocyte populations appeared normal in T cell specific Tgfbr2−/− Smad4−/− mice (Supplemental Fig. 1b). The expression of CD5, CD24 and CD69 was similar between Tgfbr2−/− Smad4−/− and wild-type thymocytes (Supplemental Fig. 1c). These findings suggest a normal thymocyte development in T cell specific Tgfbr2−/−Smad4−/− mice. In contrast to the increased lymphocyte numbers found in the periphery of Cd4-cre-Tgfbr2fl/fl mice, normal numbers of lymphocytes were recovered in T cell specific Tgfbr2−/−Smad4−/− mice (Fig. 1b). Importantly, while CD4+ and CD8+ T cells from Cd4-cre-Tgfbr2fl/fl mice were spontaneously activated (Fig. 1c) and produced large amounts of interferon-γ (IFN-γ) (Fig. 1d), T cells in T cell specific Tgfbr2−/−Smad4−/− mice maintained a naïve state (Fig. 1c) with minimal production of effector cytokine (Fig. 1d), similar to wild-type controls. Smad4 is thus required for the spontaneous T cell activation due to defective TGF-β signaling in vivo.
Cell-intrinsic properties of Tgfbr2−/−Smad4−/− T cells are normal under steady state
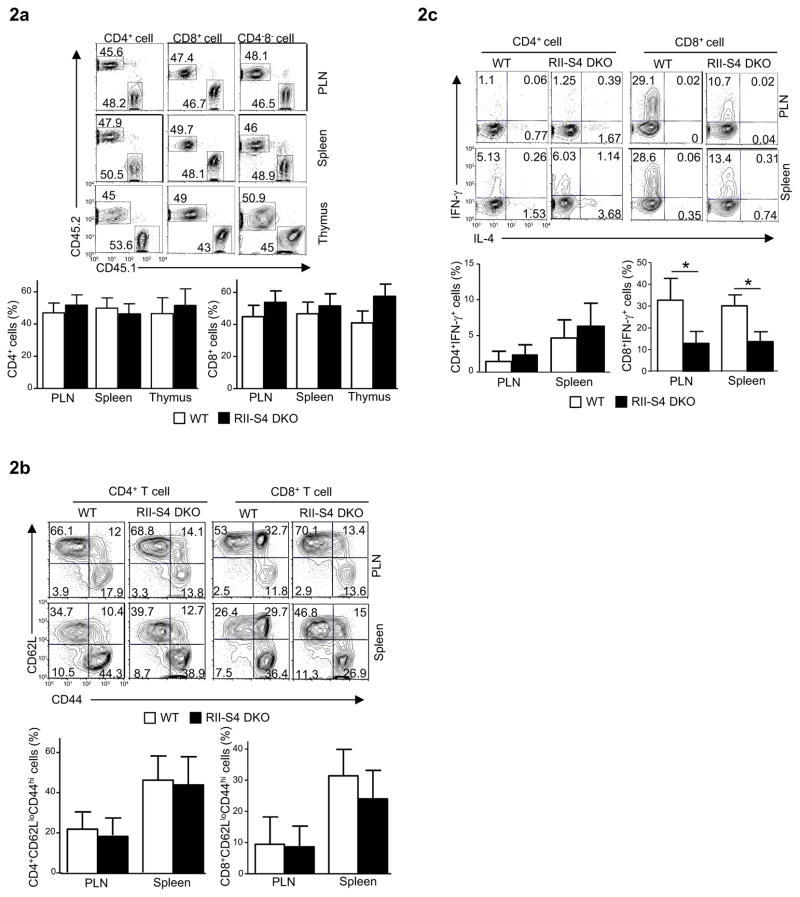
Because T cell-extrinsic factors, such as the cytokine milieu and natural killer T (NKT) cell activation, may influence T cell phenotypes when TGF-β signal is disrupted (Li et al., 2006a; Liu et al., 2008; Marie et al., 2006), we investigated if Tgfbr2−/−Smad4−/− T cells had cell-intrinsic defects not revealed in T cell specific Tgfbr2−/−Smad4−/− mice. We mixed equal numbers of bone marrow cells isolated from wild-type (CD45.1+) and T cell specific Tgfbr2−/−Smad4−/− (CD45.2+) mice and then transferred them into irradiated recipient mice deficient in recombination activated gene-1 (Rag1−/−). In the mixed bone marrow chimera, Tgfbr2−/−Smad4−/− thymocytes were generated to a similar extent as their wild-type counterparts (Fig. 2a) with normal expression of maturation markers (Supplemental Fig. 2). In addition, the percentages of CD62LloCD44hi effector T cells were comparable between wild-type and Tgfbr2−/−Smad4−/− T cells in the same hosts (Fig. 2b), suggesting that Tgfbr2−/−Smad4−/− T cells were not spontaneously activated in the chimeric mice. In agreement with this observation, the fractions of IFN-γ producing CD4+ Tgfbr2−/−Smad4−/− T cells were similar to that of wild-type counterparts. The fractions of IFN-γ producing CD8+ Tgfbr2−/−Smad4−/− T cells however were lower than that of wild-type cells (Fig. 2c). Therefore, while TGF-βR-deficient T cells are activated in vivo in a cell intrinsic fashion ((Li et al., 2006a) and data not shown), Smad4 deletion corrects such a phenotype of these T cells.
Fig. 2. Cell-intrinsic property of Tgfbr2−/−Smad4−/− T cells.
(a) The percentages of T cells in the peripheral lymph-nodes (PLN), spleens and thymuses of mixed-bone-marrow-chimeras reconstituted with bone marrow cells from wild-type (CD45.1+) and Cd4-cre-Tgfbr2fl/fl-Smad4fl/fl (RII-S4 DKO, CD45.2+) mice, assessed by flow-cytometry.
(b) CD62L and CD44 expression on CD4+ (left) and CD8+ (right) T cells of WT and RII-S4 DKO origins in the chimeras described in a, assessed by flow-cytometry.
(c) IFN-γ and IL-4 expression in T cells of WT and RII-S4 DKO origins in the chimeras described in a, assessed by flow-cytometry.
For a, b and c, representative flow-cytometry results of at least three experiments (upper panels) and means ± SD of five sets of mice (lower panels) are shown. (*P<0.05)
See also Supplemental Figure 2.
Homeostasis of Tgfβr2−/−Smad4−/− Treg cells is defective
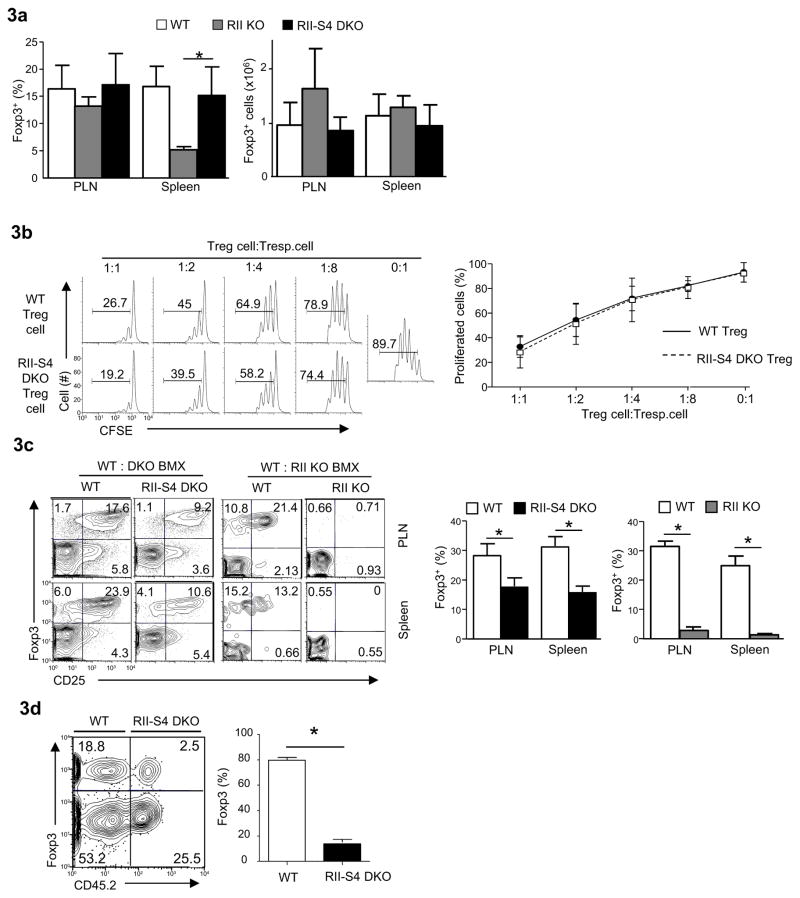
Impaired generation and function of Treg cells is one of the important mechanisms contributing to the lethal autoimmunity and spontaneous T cell activation in Cd4-cre-Tgfbr2fl/fl mice (Li et al., 2006a; Marie et al., 2006). We investigated how Treg cells were affected in T cell specific Tgfbr2−/−Smad4−/− mice. Under steady state, similar frequencies and numbers of Foxp3+ Treg cells were found in T cell specific Tgfbr2−/−Smad4−/− and wild-type mice (Fig. 3a). In addition, the suppressive function of Tgfbr2−/−Smad4−/− Treg cells was comparable to wild-type Treg cells (Fig. 3b). Nonetheless, upon closer examination, we found that the cell intrinsic properties of Tgfbr2−/−Smad4−/− Treg cells were impaired. By creating and analyzing mixed bone marrow chimeric mice, we found that, although Tgfbr2−/−Smad4−/− Treg cells were much more efficiently generated than Tgfbr2−/− Treg cells in the chimeras (Fig. 3c), the percentage of Tgfbr2−/−Smad4−/− Treg cells was lower than that of wild-type counterparts in the same hosts (Fig. 3c). In addition, we investigated how Tgfbr2−/−Smad4−/− Treg cells were maintained under inflammatory conditions, because IFN-γ is important to disrupt the homeostasis of Treg cells in the absence of TGF-β signaling (Ishigame et al., 2013). The non-inflammatory environment in T cell specific Tgfbr2−/−Smad4−/− mice precluded us from addressing this question; we therefore created inflammatory conditions in vivo by using a T-cell-induced acute-graft versus-host disease (aGvHD) model. The same numbers of wild-type (CD45.1+) and Tgfbr2−/−Smad4−/− (CD45.2+) T cells of C57BL/6 background were mixed at a ratio of 1 to 1 and then transferred into irradiated IL2rg−/− mice of Balb/c background to induce strong inflammatory aGvHD response. Large amounts of IFN-γ were produced by T cells in the recipient mice (data now shown). Under such condition, the homeostasis of Tgfbr2−/−Smad4−/− Treg cells was greatly impaired when compared to co-existing wild-type Treg cells (Fig. 3d). Collectively, these observations suggest that deletion of Smad4 is not able to restore normal Treg cell homeostasis in the absence of TGF-βR.
Fig. 3. Defective homeostasis of Tgfbr2−/−Smad4−/− Treg cells.
(a) The percentages (left) and absolute numbers (right) of Foxp3+ Treg cells detected in CD4+ T cells in different strains using flow-cytometry and live cell counting. Means ± SD of five sets of mice are shown. (*P<0.05)
(b) Immune-suppressive activity of purified WT and Cd4-cre-Tgfbr2fl/fl-Smad4fl/fl (RII-S4 DKO) Treg cells (CD4+CD25+) assessed by in vitro suppression assay. Representative results (left) and means ± SD (right) of three experiments are shown.
(c) The percentages of Foxp3+ Treg cells in CD4+ T cells in the periphery of mixed-bone-marrow-chimeras containing both wild-type (CD45.1+) and DKO (CD45.2+) T cells as in Fig. 2 or of chimeras reconstituted with bone marrow cells from wild-type (CD45.1+) and Cd4-cre-Tgfbr2fl/fl (RII KO, CD45.2+) mice, assessed by flow-cytometry. All flow cytometry plots are representative of at least three experiments. All bar graphs are means ± SD of five mice of one experiment of two. (*P<0.05).
(d). The compositions of Treg cell populations in the Rag2−/−Il2rg−/− mice (Balb/c) that have received equal numbers of CD4+ T cells from wild-type mice (CD45.1+, C57BL/6) and DKO mice (CD45.2+, C57BL/6). Representative results of at least three experiments (left) and means ± SD of five recipient mice (right) in one experiment of three are shown. (*P<0.05)
The proliferation of Tgfbr2−/−Smad4−/− T cells is impaired
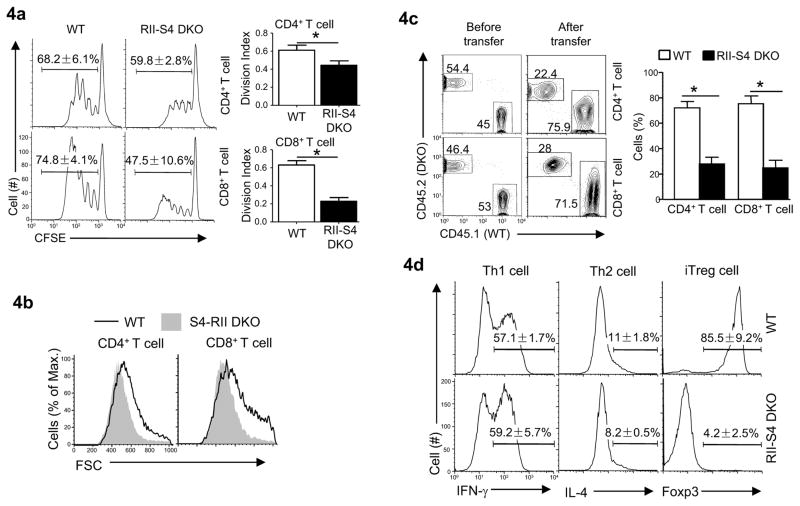
The findings described above prompted us to investigate whether the function of Tgfbr2−/−Smad4−/− non-Treg cells was impaired. In the absence of TGF-βR, aberrant T cell proliferation in response to self-antigen and overly exuberant Th1 cell differentiation lead to lethal autoimmunity in mice (Ishigame et al., 2013; Li et al., 2006b; Marie et al., 2006; Zhang and Bevan, 2012). Therefore, aberrant T cell activation, proliferation and Th cell differentiation are the key elements for uncontrolled T cell function in the absence of TGF-β signal. Tgfbr2−/−Smad4−/− T cells may be defective in these processes. Upon T cell receptor (TCR) ligation, Tgfbr2−/−Smad4−/− T cells up-regulated the activation markers similarly to wild-type cells (Supplemental Fig. 3a). Nonetheless, fewer numbers of Tgfbr2−/−Smad4−/− T cells were recovered compared to that of wild-type T cells three days after TCR ligation (Supplemental Fig. 3b). Such a reduction was likely due to impaired proliferation of Tgfbr2−/−Smad4−/− T cells (Fig. 4a), as these T cells survived similarly as wild-type T cells (Supplemental Fig. 3c) with comparable expression of the genes controlling cell survival (Supplemental 3d). In addition, the cell sizes of Tgfbr2−/−Smad4−/− T cells measured by forward-scatter by flow-cytometry were smaller than that of wild-type T cells after being activated in vitro (Fig. 4b), suggesting a defect in the growth of such T cells. The defective expansion of Tgfbr2−/−Smad4−/− T cell population could also be observed during a GvHD response in vivo (Fig. 4c), an allo-immune response that shares many features with autoimmune response (Shlomchik, 2007; Welniak et al., 2007). While Tgfbr2−/−Smad4−/− T cells could be differentiated into Th1 and Th2 cells similarly to wild-type T cells (Fig. 4d and Supplemental Fig. 3e), they failed to differentiate into Foxp3+ Treg cells in the presence of TGF-β (Chen et al., 2003) (Fig. 4d). Lack of TGF-β-induced Treg cell differentiation agreed with and contributed to the observation that fewer Tgfbr2−/−Smad4−/− than wild-type Treg cells were recovered in mixed bone marrow chimeras and during GvHD response (Fig. 3c and 3d). Therefore, Tgfbr2−/−Smad4−/− T cells are defective in activation-induced proliferation.
Fig. 4. Defective function of Tgfbr2−/−Smad4−/− T cells.
(a) Anti-CD3 and anti-CD28 activated proliferation of T cells isolated from wild-type (WT) and Cd4-cre-Tgfbr2fl/fl-Smad4fl/fl (RII-S4 DKO) mice assessed by flow-cytometry of CFSE dilution (left). Division index was calculated using FlowJo software (right). Means ± SD of three experiments are shown. (*P<0.05)
(b) T cell sizes assessed by flow-cytometry 48 hours after anti-CD3 and anti-CD28 stimulation.
(c) The percentages of T cells in Rag2−/−Il2rg−/− recipient mice (Balb/c) transferred with splenocytes from wild-type (CD45.1+) and Cd4-cre-Tgfbr2fl/fl-Smad4fl/fl (RII-S4 DKO, CD45.2+) mice (C57BL/6) assessed by flow-cytometry. Representative results (left) and means ± SD of five mice in one experiment of two (right) are shown.
(d) IFN-γ, IL-4 and Foxp3 expression in CD4+ T cells assessed by flow-cytometry after being differentiated under Th1, Th2 and iTreg cell polarizing conditions. Means ± SD of three experiments are shown.
See also Supplemental Figure 3.
Smad4 is required for T cell function in inflammation and cancer
Aforementioned observations suggest that Smad4 plays an essential role in promoting T cell function. To test this possibility, we investigated whether the activation, proliferation and/or differentiation of T cells were affected in the absence of Smad4. Naïve T cells isolated from Cd4-cre-Smad4fl/fl mice were activated normally with efficient up-regulation of activation markers (Supplemental Fig. 4a). Fewer Smad4-deficient than -sufficient T cells were recovered three days post activation (Supplemental Fig. 4b). While Smad4-deficient T cells survived similarly as wild-type T cells (Supplemental Fig. 4c) with comparable expression of genes that control T cell survival (Supplemental Fig. 4d), the proliferation of Smad4-deficient T cells was impaired following TCR stimulation in vitro (Fig. 5a). In addition, the sizes of activated Smad4-deficient T cells were smaller than that of Smad4-sufficient T cells in vitro (Fig. 5b) indicating that activation induced growth of Smad4-deficient T cells was impaired. While Th1 and Th2 cell differentiation and IL2 production of Smad4-deficient CD4+ T cells were largely normal (Supplemental Fig. 4e, 4f, and 4g), TGF-β-induced Treg cell differentiation of these cells was defective (Supplemental Fig. 4e), which is in agreement with a previous report (Yang et al., 2008b) and suggests that Smad4 indeed mediates TGF-β signaling in T cells.
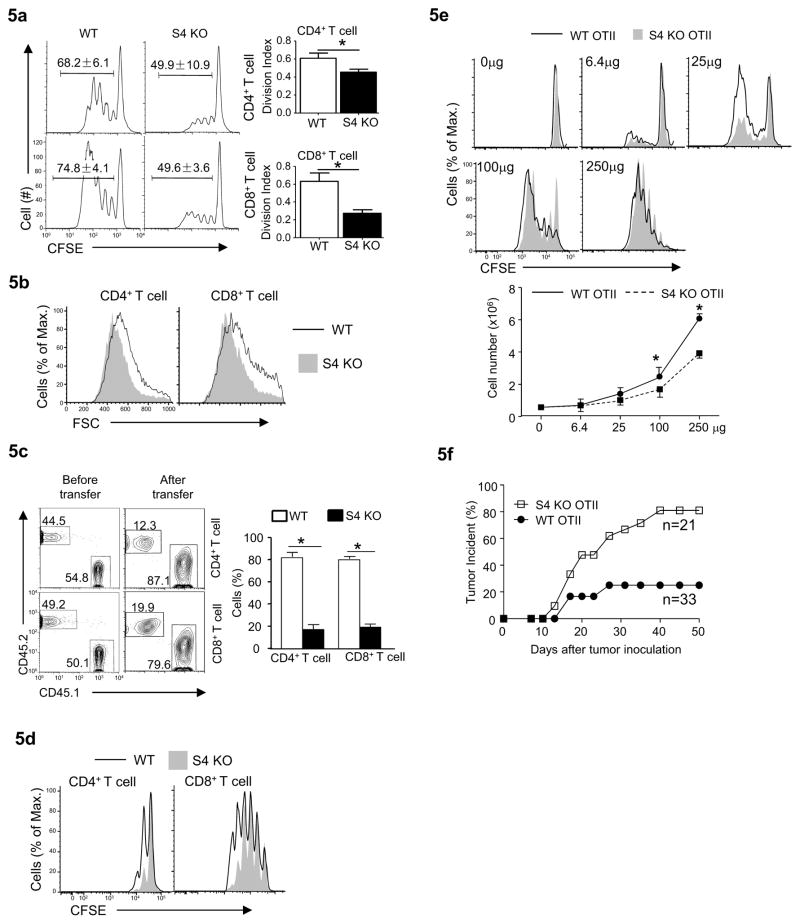
Fig. 5. Smad4 is required for T cell function.
(a) The proliferation of T cells isolated from wild-type and Cd4-cre-Smad4fl/fl (S4 KO) mice assessed as in Fig. 4a. Means ± SD of three experiments are shown (*P<0.05).
(b) T cell sizes assessed by flow-cytometry 48 hours after anti-CD3 and anti-CD28 stimulation. Results are representative of at least three experiments.
(c) The fractions of T cells in Rag2−/−Il2rg−/− recipient mice (Balb/c) that have received splenocytes from wild-type (CD45.1+) and Cd4-cre-Smad4fl/fl (S4 KO, CD45.2+) mice (C57BL/6) assessed by flow-cytometry. Representative flow-cytometry results (left) and means ± SD of five mice in one experiment representative of two (right) are shown.
(d) CD4+ and CD8+ T cells were isolated from wild-type (CD45.1+CD45.2+) and Cd4-cre-Smad4fl/fl mice (S4 KO, CD45.2+) mice. Cells of different origins were mixed and labeled with CFSE, and transferred into sub-lethally irradiated wild-type (CD45.1+) mice. The proliferation of transferred cells in the recipients was assessed 5 days post transfer. Results are representative of at least three experiments.
(e) CD4+ T cells were isolated from Cd4-cre-Smad4fl/+-OTII (WT OTII, CD45.1+CD45.2+) and Cd4-cre-Smad4fl/fl-OTII mice (S4 KO OTII, CD45.2+) mice, mixed at the ratio of 1 to 1, labeled with CFSE, and then transferred into syngeneic wild-type mice (CD45.1+). The proliferation and the numbers of transferred cells of different origins in the spleens were assessed 60 hours after the recipient mice were injected with different doses of OVA proteins (as indicated). Representative flow-cytometry results of at least three experiments are shown. Means ± SD of the cell numbers are shown. (* P<0.05)
(f) The incidence of tumor development (≥3mm in diameter) in WT OTII and Cd4-cre-Smad4fl/fl-OTII (S4 KO OTII) mice inoculated s.c. with OVA-expressing melanoma cells (20,000 cells/injection). Combined results of three independent experiments are plotted.
See also Supplemental Figure 4.
Similar to Tgfbr2−/−Smad4−/− T cells, Smad4−/− T cells expanded poorly when compared to wild-type cells during a GvHD response in vivo (Fig. 5c). This observation could be due to impaired proliferation driven by lymphopenic condition and/or by cognate antigen stimulation. Smad4-deficient T cells proliferated less than wild-type T cells when transferred into sub-lethally irradiated syngeneic recipients, (Fig. 5d), suggesting Smad4 is required for lymphopenia-driven T cell proliferation. To test how Smad4 deletion may affect T cell proliferation in response to cognate antigen, we crossed Cd4-cre-Smad4fl/fl mice with OTII TCR transgenic mice to obtain Cd4-cre-Smad4fl/fl-OTII T cells that respond specifically to peptide 323–339 of chicken ovalbumin protein (OVA). Compared to co-transferred wild-type OTII cells, Smad4-deficient OTII cells proliferated less in response to OVA protein stimulation in the immune competent syngeneic hosts (Fig. 5e and Supplemental Fig. 4h).
Smad4 mutation has been associated with cancers in human and mice (Hahn et al., 2011; Howe et al., 1998; Kim et al., 2006; Miyaki and Kuroki, 2003). The immune etiology is not entirely clear, although enhanced Th cell responses may contribute to cancer development (Hahn et al., 2011; Kim et al., 2006). The finding that Smad4-deficient T cells proliferated poorly to cognate antigen stimulation suggests Smad4 may be required for T cell mediated tumor rejection. Indeed, while the growth of engrafted OVA-expressing B16 melanoma was controlled effectively in wild-type OTII TCR transgenic (Tg) recipient mice, a large percentage of Cd4-cre-Smad4fl/fl-OTII mice developed melanoma after tumor cell engraftment (Fig. 5f).
The above mentioned observations suggest that, although Smad4 is largely dispensable for T cell activation and Th cell differentiation, it is required for T cell proliferation in response to cognate-antigen to promote T cell mediated inflammatory disease and tumor rejection.
Myc transcription factor is important for Smad4-controlled T cell proliferation
We further investigated the molecular mechanisms underlying the defective growth and proliferation of activated Smad4-deficient T cells. Because Myc is a central regulator for T cell growth and proliferation (Nie et al., 2012; Wang et al., 2011) and Smad4 has been suggested to promote Myc expression in tumor cells (Lim and Hoffmann, 2006), we hypothesized that the expression of Myc was dysregulated in Smad4-deficient T cells. Indeed, following activation, Myc expression in Tgfbr2−/−Smad4−/− and Smad4−/− T cells was lower than in wild-type T cells (Fig. 6a and 6b), suggesting that Myc functions downstream of Smad4 to control T cell proliferation. To test whether Myc downregulation is functionally important for defective proliferation of Smad4-defiicent T cells, we expressed Myc in activated Smad4-deficient T cells using retrovirus-mediated gene delivery. Ectopic Myc expression largely restored the proliferation of Smad4-deficient T cells after activation in vitro (Fig. 6c) as well as in vivo during a GvHD response (Fig. 6d and Supplemental Fig. 5), indicating that Myc is an important Smad4 downstream target to control T cell proliferation, whereas Myc-independent mechanisms might also be involved. Smad4 may promote Myc expression by binding to a TGF-β independent site in the Myc locus in T cells (Lim and Hoffmann, 2006). Using chromatin-immuno-precipitation (ChIP) assay, we found that Smad4 binding was enriched at this site in T cells (Fig. 6e). Therefore, one of the important mechanisms by which Smad4 promotes T cell proliferation is through regulating Myc expression.
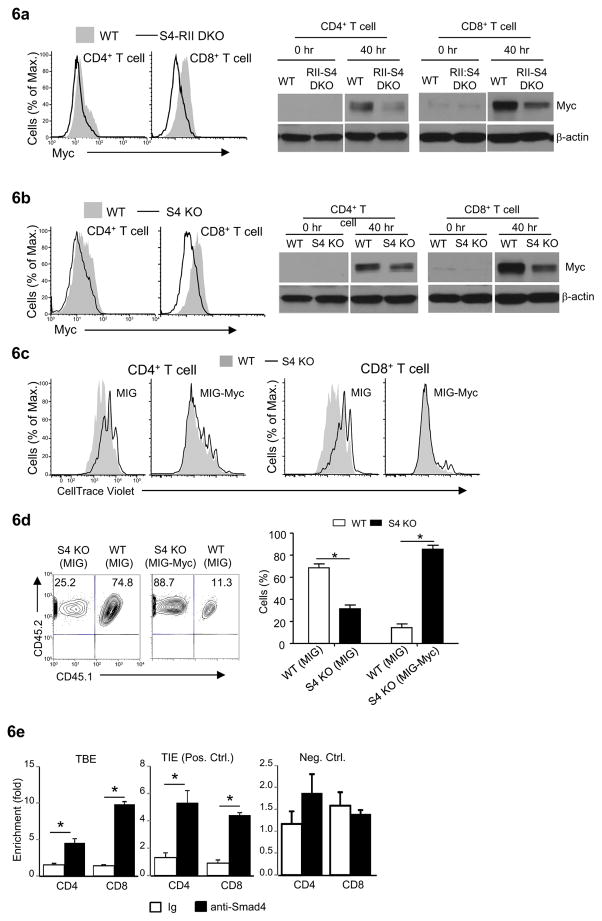
Fig. 6. Smad4 controls T cell proliferation through Myc.
(a and b) Myc expression in T cells assessed by flow-cytometry (left) and immuno-blotting (right). Representative results of at least three experiments are shown.
(c) The proliferation of wild-type and Cd4-cre-Smad4fl/fl (S4 KO) CD4+ and CD8+ T cells that were transduced (GFP+) with MSCV-IRES-EGFP (MIG) or MSCV-Myc-IRES-EGFP (MIG-Myc), assessed by flow-cytometry of CellTrace violet dilution. Results are representative of at least three experiments.
(d) The fractions of T cells in Rag2−/−Il2rg−/− recipient mice (Balb/c) received MIG and MIG-Myc transduced (GFP+) CD4+ T cells from wild-type (CD45.1+) and Cd4-cre-Smad4fl/fl (S4 KO, CD45.2+) mice (C57BL/6) assessed by flow-cytometry. Representative results (left) and means ± SD of five mice in one experiment of three are shown.
(e) ChIP analysis of Smad4 binding to TGF-β-independent elements (TBE) in Myc locus in freshly isolated T cells. TGF-β-inhibitory elements (TIE) and an irrelevant site in Myc locus was used as positive (Pos. Ctrl.) and negative (Neg. Ctrl.) controls respectively. Means ± SD of triplicates in one experiment of at least three are shown (*P<0.05).
See also Supplemental Figure 5.
Discussion
TGF-β suppresses autoimmunity and promotes tumorigenesis by regulating T cell function. Nonetheless, how Smad4 (a central component for TGF-β signaling) is involved in T cell function during autoimmunity and malignancies is unclear. T cell specific deletion of Smad4 is associated with cancer but not with autoimmunity (Hahn et al., 2011; Kim et al., 2006). Here, we found that Smad4 was essential for the proliferation of T cells and Myc expression. Importantly, deletion of Smad4 in T cells rescued early lethal autoimmunity in mice whose T cells lack TGF-βR and led to impaired tumor rejection. These findings therefore reveal a previously unappreciated requirement of Smad4 to promote T cell function for autoimmunity and tumor immune surveillance. The information gained from this study sheds light on TGF-β signaling and Smad function, and provides insights into the control of T cell function, autoimmunity and cancer.
Aberrant T cell proliferation, uncontrolled T cell activation and effector function contribute to various types of inflammatory diseases. Therefore, inhibiting the proliferation and pro-inflammatory function of T cells is effective to treat such illness. Current strategies aiming to interfere with pro-inflammatory T cell function often lead to the ablation of lymphocyte population and abrogation of effector T cell function, which could be detrimental to the patients. It is therefore important to identify pathways that are essential for autoimmunity development but largely dispensable for T cell homeostasis. By studying Cd4-cre-Tgfbr2fl/fl mice whose T cells lack TGF-β signaling, a pathway frequently impaired in various autoimmune and inflammatory diseases (Li et al., 2006b), we found that Smad4 deletion corrected the lethal autoimmunity in these mice. While Smad4-deficient T cells were generated, maintained, activated and differentiated normally, their proliferation was impaired. Therefore, one of the key steps leading to autoimmunity in mice lacking TGF-β signaling is the aberrant T cell proliferation, which is a prerequisite for the uncontrolled T cell activation and differentiation in these mice as suggested by a previous study (Zhang and Bevan, 2012). More importantly, this observation suggests that modestly inhibiting T cell proliferation could be sufficient to prevent autoimmunity and that Smad4 may be a promising target for treating autoimmunity without the drastic measures of ablating T cell population or function.
Treg cells are critical to maintain self-tolerance and immune homeostasis. Impaired Treg cell function has been implicated in nearly every autoimmune and inflammatory disease. It is therefore thought that defective Treg cell function will invariantly lead to inflammatory disorders. We found that, although Smad4 deletion rescued the early lethal autoimmune syndrome and restored naive T cell phenotype in mice whose T cells lacking TGF-βR, the homeostasis of Tgfbr2−/−Smad4−/− Treg cells was defective especially under inflammatory conditions. These findings indicate that impaired Treg cell function may not always lead to inflammation and autoimmunity when the function of conventional T cells is defective albeit to a modest extent.
This study sheds light on the etiology of certain cancers. People with loss function mutation of Smad4 are predisposed to the development of familial juvenile polyposis and gastric-intestinal cancers at older age (Howe et al., 1998; Miyaki and Kuroki, 2003). The immune etiology of such cancers is not entirely clear. While increased Th cell differentiation of Smad4-deficient T cells is associated with cancer development (Hahn et al., 2011; Kim et al., 2006), other mechanisms may also contribute. The current study provides another plausible immune mechanism for the cancer development in people with Smad4 mutation. We found that Smad4-deficient cells proliferated poorly in response to cognate and tumor antigen and showed reduced ability to reject engraft tumor cells. Therefore, Smad4 mutation impairs T cell mediated tumor rejection to allow tumor growth. These new insights may facilitate the development of effective anti-cancer drug. Blocking TGF-β signaling has been explored as an immunotherapy against cancers (Bierie and Moses, 2006; Yingling et al., 2004). The observation that Smad4-deficient T cells are functionally defective even when TGF-β signaling is abrogated suggests that anti-TGF-β signaling immunotherapy may be tumor type specific and may not be efficacious when Smad4 function is perturbed.
We found that Smad4-deficient T cells proliferated poorly compared to wild-type T cells even in the absence of TGF-β signaling. It suggests that the proliferation of T cells is controlled by Smad4-dependent, TGF-β-independent pathways. Smad4 is a hub integrating multiple signaling pathways. Smad4 mediates the signaling of TGF-β superfamily members including TGF-βs, bone morphogenetic proteins (BMPs), activins and growth differentiation factors (GDFs) (Massague et al., 2005). In addition, Smad4 is involved in Wnt and Notch signaling (Massague, 2012). Moreover, Smad4 interacts with critical cell cycle regulators, such as E2Fs, that do not mediate specific upstream signaling (Taylor and Wrana, 2008). Therefore, Smad4 may control T cell proliferation through many diverse pathways independent of TGF-β. BMP signaling may be involved in this process as it has been suggested that BMP signaling is important for T cell proliferation (Yoshioka et al., 2012). We found that Smad4 is required for optimal Myc expression in T cells. Previous studies show that, through a TGF-β-independent mechanism, Smad4 promotes Myc expression in tumor cells by cooperating with TCF1 (Lim and Hoffmann, 2006), a factor important to mediate Notch and Wnt signal in T cells (Staal et al., 2008; Weber et al., 2011). Because Notch and Wnt signal promotes cell proliferation (Artavanis-Tsakonas et al., 1999; Staal et al., 2008), Smad4 may control T cell proliferation also through Notch and Wnt-TCF1 signal axis. Therefore, Smad4 may promote T cell function by integrating multiple pathways independent of TGF-β. The question of what the Smad4-dependent pathways are and how precisely they contribute to T cell function through Smad4 in vitro and in vivo is of interest and warrants further investigation in order to understand the etiology of and devise therapies against diseases including autoimmunity and cancer.
Experimental Procedures
Mice
Smad4fl/fl, Tgfbr2fl/fl, Cd4-cre, Rag1−/−, OTII and CD45.1 congenic wild-type mice were on the C57BL/6 background. Rag2−/−Il2rg−/− mice were on the BALB/c background. All mice were housed and bred in specific pathogen–free conditions in the animal facility at the University of North Carolina at Chapel Hill. All mouse experiments were approved by Institution Animal Care and Use Committee of the University of North Carolina.
Flow-cytometry and cell sorting
Lymphocytes were isolated from the various organs of age- and sex-matched mice 3–16 weeks of age. Fluorescence-conjugated anti-CD4 (GK1.5), anti-CD8 (53–6.7), anti-CD5 (53–7.3), anti-CD24 (30-F1), anti-CD25 (PC61.5), anti-CD44 (IM7), anti-CD69 (H1.2F3), anti-CD62L (MEL-14), anti-CD45.1 (A20), anti-CD45.2 (104), anti-IFN-γ (XMG1.2), anti-IL-4 (11B11), anti-IL-2 (JES6-5H4), and anti-Foxp3 (FJK-16s) (eBioscience), Myc (9E10, Santa Cruz) as well as Annexin V and 7-amino-actinomycin D (BD Biosciences) were used. For intracellular cytokine staining, lymphocytes were stimulated for 4 hours with 50 ng/ml of PMA (phorbol 12-myristate 13-acetate) and 1 μM ionomycin in the presence of brefeldin A. Stained cells were analyzed on an LSRII (BD Biosciences) or were sorted on a MoFlo (DakoCytomation; Beckman Coulter).
T cell activation, proliferation, differentiation, and Treg suppression assay
Naïve (CD62LhighCD44low) T cells were sorted from the peripheral lymph-nodes and/or spleens of mice. Cells were then activation by stimulation via the TCR by anti-CD3 (145-2C11; BioXCell) and anti-CD28 (37.51; BioXCell). For proliferation assays, cells were labeled with CFSE (carboxyfluorescein diacetate succinimidyl ester) or CellTrace Violet (BD Biosciences) and then cultured under the appropriate conditions. Proliferation was assessed by the dilution of live dye with flow-cytometry 72–96 hours after T cell activation. Th1 cells were differentiated in the presence of 20ng/ml rIL-12 (R&D systems) and 20μg/ml anti-IL-4 (11B11, BioXcell). Th2 cells were differentiated in the presence of 20ng/ml rIL-4 (R&D systems) and 20μg/ml anti-IFN-γ (XMG1.2, BioXcell). Treg cells were differentiated in the presence of 2ng/ml rTGF-β1 (R&D systems). To assess the efficacy of Treg-mediated immune suppression in vitro, 2x104 sorted CD4+CD25−CD45RBhi responder T cells were labeled with CFSE and mixed with varying amounts (as indicated) of CD4+CD25+ Treg suppressor cells. Cell mixtures were stimulated with soluble anti-CD3 antibody (1μg/ml) in the presence of 1x105 irradiated (3000 cGy) T-cell depleted splenocytes as APC. The proliferation of responder cells was assessed by CFSE dilution detected by flow-cytometry 72 hours post stimulation.
Mixed–bone marrow chimeras
Bone marrow cells were isolated from the femur bones of sex- and age-matched Cd4-cre-Tgfbr2fl/fl (CD45.2+) or Cd4-cre-Tgfbr2fl/fl-Smad4fl/fl (CD45.2+) mice and wild-type (CD45.1+) mice. Bone marrow cells (1 × 106) from each donor were mixed and transferred into sub-lethally irradiated (400 cGy) Rag1−/− recipient mice. T cell populations of each donor origin were determined in the recipients 8–12 weeks after transfer.
Mouse model of the graft-versus-host response
Total splenocytes from sex- and age-matched Cd4-cre-Tgfbr2fl/fl-Smad4fl/fl (CD45.2+) or Cd4-cre–Smad4fl/fl (CD45.2+) mice and wild-type (CD45.1+) mice of C57BL/6 background were mixed at a ratio of 1:1 and then transferred into sub-lethally irradiated Rag2−/−Il2rg−/− mice on the BALB/c background by retro-orbital injection. Recipient mice were monitored and euthanized at the appropriate time. T cells from the spleens of recipient mice were collected and subjected to immunological analysis.
Immunoblot analysis
Protein extracts were resolved by 4–12% SDS-PAGE (Invitrogen), then were transferred to a polyvinylidene fluoride membrane (Millipore) and analyzed by immuno-blotting with the following antibodies: anti-Smad4 (#9515, Cell Signaling), anti-phospho-Smad2 (D43B4, Cell Signaling), anti-c-Myc (D84C12; Cell Signaling), and anti-β-actin (I-19; Santa Cruz).
ChIP assay
The ChIP assay was done according to the protocol of Upstate Biotechnology. Cells were crosslinked by 1% formaldehyde and were lysed in lysis buffer. Lysates were sonicated with a Bioruptor sonicator to shear genomic DNA into fragments. Chromatin prepared from 2 × 106 cells were subjected to immunoprecipitation overnight at 4 °C with goat anti-Smad4 (sc-1909; Santa Cruz) or normal goat immunoglobulin G (sc-2028; Santa Cruz). Quantitative real-time PCR was done to determine the relative abundance of target DNA. Specific primers for analysis of the binding of Smad4 to target loci are:
TBE: TGGCATATTCTCGCGTCTAGC and AGGAGTCTCTGCCGGTCTACA;
TIE: CTTTATATTCCGGGGGTCTGC and GCAATGGGCAAAGTTTCCCA;
Negative control: GGGTACATGGCGTATTGTGT and TCGGCTGAACTGTGTTCTTG.
Statistical analysis
Data from at least three sets of samples were used for statistical analysis. Statistical significance was calculated by Student’s t-test. A P value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank E. Robertson and E. Bikoff (University of Oxford, UK) for Smad4fl/fl mice and H. Moses (Vanderbilt University, USA) for Tgfbr2fl/fl mice, N. Fisher and J. Kalnitsky (University of North Carolina Flow-cytometry facility supported in part by an NCI Center Core Support Grant (P30CA016086) to the UNC Lineberger Comprehensive Cancer Center) for cell sorting; M. Su (University of North Carolina) for discussions. This study is supported by the NIH (R01AI097392), National Multiple Sclerosis Society (RG4654), and the University Cancer Research Fund (Y.Y.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. Signal transduction by the TGF--beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- Bierie B, Moses HL. Tumour microenvironment: TGF-beta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- Cazac BB, Roes J. TGF--beta receptor controls B cell responsiveness and induction of IgA in vivo. Immunity. 2000;13:443–451. doi: 10.1016/s1074-7613(00)00044-3. [DOI] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF--beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. Differential requirements for Smad4 in TGF-beta-dependent patterning of the early mouse embryo. Development. 2004;131:3501–3512. doi: 10.1242/dev.01248. [DOI] [PubMed] [Google Scholar]
- Chytil A, Magnuson MA, Wright CV, Moses HL. Conditional inactivation of the TGF--beta type II receptor using cre:Lox. Genesis. 2002;32:73–75. doi: 10.1002/gene.10046. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF--beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, Worthen GS, Albelda SM. Polarization of tumor-associated neutrophil phenotype by TGF--beta: “N1” versus “N2” TAN. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik L, Flavell RA. Immune-mediated eradication of tumors through the blockade of transforming growth factor-beta signaling in T cells. Nat Med. 2001;7:1118–1122. doi: 10.1038/nm1001-1118. [DOI] [PubMed] [Google Scholar]
- Gu AD, Wang Y, Lin L, Zhang SS, Wan YY. Requirements of transcription factor Smad-dependent and -independent TGF--beta signaling to control discrete T-cell functions. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1108352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JN, Falck VG, Jirik FR. Smad4 deficiency in T cells leads to the Th17-associated development of premalignant gastroduodenal lesions in mice. J Clin Invest. 2011;121:4030–4042. doi: 10.1172/JCI45114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGF-beta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Howe JR, Roth S, Ringold JC, Summers RW, Jarvinen HJ, Sistonen P, Tomlinson IP, Houlston RS, Bevan S, Mitros FA, et al. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science. 1998;280:1086–1088. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- Ishigame H, Zenewicz LA, Sanjabi S, Licona-Limon P, Nakayama M, Leonard WJ, Flavell RA. Excessive Th1 responses due to the absence of TGF--beta signaling cause autoimmune diabetes and dysregulated Treg cell homeostasis. Proc Natl Acad Sci U S A. 2013;110:6961–6966. doi: 10.1073/pnas.1304498110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BG, Li C, Qiao W, Mamura M, Kasprzak B, Anver M, Wolfraim L, Hong S, Mushinski E, Potter M, et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature. 2006;441:1015–1019. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- Konkel JE, Maruyama T, Carpenter AC, Xiong Y, Zamarron BF, Hall BE, Kulkarni AB, Zhang P, Bosselut R, Chen W. Control of the development of CD8alphaalpha+ intestinal intraepithelial lymphocytes by TGF--beta. Nat Immunol. 2011;12:312–319. doi: 10.1038/ni.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laouar Y, Sutterwala FS, Gorelik L, Flavell RA. Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat Immunol. 2005;6:600–607. doi: 10.1038/ni1197. [DOI] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S. A Critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006a;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006b;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- Lim SK, Hoffmann FM. Smad4 cooperates with lymphoid enhancer-binding factor 1/T cell-specific factor to increase c-myc expression in the absence of TGF--beta signaling. Proc Natl Acad Sci U S A. 2006;103:18580–18585. doi: 10.1073/pnas.0604773103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang P, Li J, Kulkarni AB, Perruche S, Chen W. A critical function for TGF--beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat Immunol. 2008;9:632–640. doi: 10.1038/ni.1607. [DOI] [PubMed] [Google Scholar]
- Marie JC, Letterio JJ, Gavin M, Rudensky AY. TGF--beta1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J Exp Med. 2005;201:1061–1067. doi: 10.1084/jem.20042276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie JC, Liggitt D, Rudensky AY. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-beta receptor. Immunity. 2006;25:441–454. doi: 10.1016/j.immuni.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Massague J. TGF--beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Massague J. TGF-beta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Miyaki M, Kuroki T. Role of Smad4 (DPC4) inactivation in human cancer. Biochem Biophys Res Commun. 2003;306:799–804. doi: 10.1016/s0006-291x(03)01066-0. [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc Is a Universal Amplifier of Expressed Genes in Lymphocytes and Embryonic Stem Cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov YP, Rudensky AY. TGF-beta signalling in control of T-cell-mediated self-reactivity. Nat Rev Immunol. 2007;7:443–453. doi: 10.1038/nri2095. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF--beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Shlomchik WD. Graft-versus-host disease. Nat Rev Immunol. 2007;7:340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol. 2008;8:581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- Takimoto T, Wakabayashi Y, Sekiya T, Inoue N, Morita R, Ichiyama K, Takahashi R, Asakawa M, Muto G, Mori T, et al. Smad2 and Smad3 are redundantly essential for the TGF--beta-mediated regulation of regulatory T plasticity and Th1 development. J Immunol. 2010;185:842–855. doi: 10.4049/jimmunol.0904100. [DOI] [PubMed] [Google Scholar]
- Taylor IW, Wrana JL. SnapShot: The TGF-beta pathway interactome. Cell. 2008;133:378, e371. doi: 10.1016/j.cell.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Wahl SM, Wen J, Moutsopoulos N. TGF--beta: a mobile purveyor of immune privilege. Immunol Rev. 2006;213:213–227. doi: 10.1111/j.1600-065X.2006.00437.x. [DOI] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welniak LA, Blazar BR, Murphy WJ. Immunobiology of allogeneic hematopoietic stem cell transplantation. Annu Rev Immunol. 2007;25:139–170. doi: 10.1146/annurev.immunol.25.022106.141606. [DOI] [PubMed] [Google Scholar]
- Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, Carbone DP, Matrisian LM, Richmond A, Lin PC, Moses HL. Abrogation of TGF- beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008a;13:23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Pang Y, Moses HL. TGF--beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010;31:220–227. doi: 10.1016/j.it.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008b;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling JM, Blanchard KL, Sawyer JS. Development of TGF--beta signalling inhibitors for cancer therapy. Nature reviews Drug discovery. 2004;3:1011–1022. doi: 10.1038/nrd1580. [DOI] [PubMed] [Google Scholar]
- Yoshioka Y, Ono M, Osaki M, Konishi I, Sakaguchi S. Differential effects of inhibition of bone morphogenic protein (BMP) signalling on T-cell activation and differentiation. Eur J Immunol. 2012;42:749–759. doi: 10.1002/eji.201141702. [DOI] [PubMed] [Google Scholar]
- Zhang N, Bevan MJ. TGF--beta signaling to T cells inhibits autoimmunity during lymphopenia-driven proliferation. Nat Immunol. 2012;13:667–673. doi: 10.1038/ni.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.