Abstract
Paper-based devices serve to address many analytical questions both inside and outside of the laboratory setting. For the first time, yeast were used to construct a whole-cell, paper-based biosensor device. This Biologically-based Paper Analytical Device (BioPAD) is sensitive to antibiotics in the tetracycline family, and it could potentially address questions of pharmaceutical quality as well as antibiotic contamination in liquids. Our BioPAD can qualitatively discriminate the presence/absence of doxycycline over a range of 30 – 10,000 μg/mL. In an analysis of a doxycycline dosage form (tablet) commonly used for malaria prophylaxis, BioPADs identified the presence of antibiotic with 92% and 95% sensitivity, evaluated by eye and computer-assisted image analysis respectively, with no false positives by either method. BioPADs were found to remain viable for at least 415 days when stored at 4ºC. This research demonstrates the utility of whole yeast cells in paper-based pharmaceutical testing, and it highlights the potential for the development of yeast-based BioPADs to address a range of qualitative analytical questions, especially in low resource settings.
Keywords: Biosensor, paper-based, pharmaceutical, yeast, antibiotic
INTRODUCTION
Paper-based tests, which have a long history of use, can provide answers to many analytical problems outside of the laboratory, and they can be especially useful in developing countries or settings where resources are limited[1]. Recent examples include paper-based, colorimetric tests for glucose, proteins[2], liver function[3] and beta-lactam antibiotics[4]. These tests are generally rapid and user-friendly, but they rely on the recognition of chemical motifs and are therefore not molecule specific. Paper-based tests incorporating antibodies have become a staple of medical diagnostic testing, as is exemplified by the multitude of pregnancy tests now available. The use of biological components like antibodies or enzymes allows tests to be highly specific for analyte, but fabrication requires isolation steps, and these purified biological components can be unstable during long term storage. The use of whole microorganisms eliminates the need for isolated components, simplifying biosensor device fabrication and potentially increasing test longevity.
The primary aim of this research is to harness the robust biological recognition and response intrinsic to living cells by creating whole cell biosensor that incorporate genetically engineered yeast into paper analytical devices (PADs), thus making “BioPADs.” Living organisms like yeast possess the innate capacity to respond to many pharmaceuticals including antibiotics[5, 6], and they can be made responsive to others through genetic engineering[7]. Redirecting this responsive genetic machinery to produce a reporter molecule transforms yeast into a sensor for specific medications. Incorporating these biosensors into a paper-based test could produce an inexpensive and specific test, increasing the repertoire of current paper-based tests.
While several whole-cell biosensors have been developed[8, 9], most are bacteria based, and those converted to paper-based tests are limited to bacterial systems that detect arsenic[10] and quorum sensing molecules[11]. These examples showed that reporters could be produced by bacterial biosensors on a paper substrate. However, yeast offer some advantages for use in a biosensor including a) tolerance to pH and temperature fluctuations, b) established procedures for long term storage, c) ability to survive over long periods of time in a dried state, d) an extensive genetic toolkit, e) a non-threatening public perception, and f) eukaryotic nature, such that response to many pharmaceutical agents and/or toxic substances is similar to higher eukaryotes[7]. Previously developed yeast biosensors have relied on electrode measurements of solution pH or oxygen levels that reflect the increased metabolism of a substrate in the presence of the analyte. For example, as yeast metabolize glucose, the drop in pH of the surrounding solution is measured to reflect glucose concentration. The concentrations of small molecules, such as glucose, galactose, and copper, have also been measured by these means[12–14]. This manuscript demonstrates the feasibility of using yeast as the whole cell biosensor embedded in paper and improves upon previous microbial biosensors[10, 11] on paper by a) defining test zones to combine multiple tests on one device, b) trapping cells onto paper with a hydrogel matrix, c) evaluating visual interpretation of tests used with a pharmaceutical dosage form and d) conducting a long-term study of storage time after which the test remains viable.
EXPERIMENTAL
Materials and Methods can be found in the Electronic Supplementary Material (EMS).
RESULTS
We envisioned that an analytical device composed of simple components, i.e., yeast, paper and hydrogel, would be a new and useful tool to address analytical questions in low resource settings. In developing this paper-based biosensor we wanted a test design that was easy to make and easy to use. We also recognized at the outset that a yeast strain to be used should respond to analyte with robust expression of a reporter molecule, while tightly repressing reporter expression in the absence of analyte. Since the ultimate goal was to produce a device that could be used in a resource limited setting, we chose to use yeast able to produce a visible reporter signal, eliminating the need for specialized equipment for reporter detection. Additionally, the analyte recognized needed to be relevant to an analytical problem in a low resource setting. We wanted to embed these into paper using a “yeast-friendly” environment that would fix yeast cells to a location on paper while allowing the analyte to pass through the matrix. Finally, we hoped to determine a straightforward method of preservation that would allow tests to remain viable during storage and transport.
BioPAD yeast component
The yeast incorporated into the BioPAD produce reporter in response to antibiotics in the tetracycline family. In the absence of tetracycline antibiotics, repressors bind a series of tetracycline operators placed in tandem upstream of the promoter region of the reporter gene and block transcription. Tetracycline antibiotics, when present, interact with both activators and repressors, causing binding of the activator to the Tet operators to become favored and transcription of the reporter is up-regulated in this tightly regulated system[15, 16].
BioPAD test design considerations
Two test designs, exposing yeast to analyte through either a lateral flow design [4] (Figure 1a) or a stationary system (Figure 1d, ESM Fig. S1), were employed in this study. Images of lateral flow test regions following exposure to either 0 or 1mg/mL doxycycline are shown in Figures 1b and 1c, respectively. Yeast in the stationary format, exposed to a range of doxycycline concentrations, are shown in Figure 1e. While the stationary BioPADs provide a well-defined testing area, the lateral flow BioPADs are more user-friendly (wicking replaces the need for sample deposition) and therefore are the more desirable format for a qualitative analytical tool for use outside of the laboratory setting. In this study, stationary tests were used to define a responsive range of BioPADs and lateral flow testing was employed to determine presence/absence of active ingredient in doxycycline pills.
Figure 1.
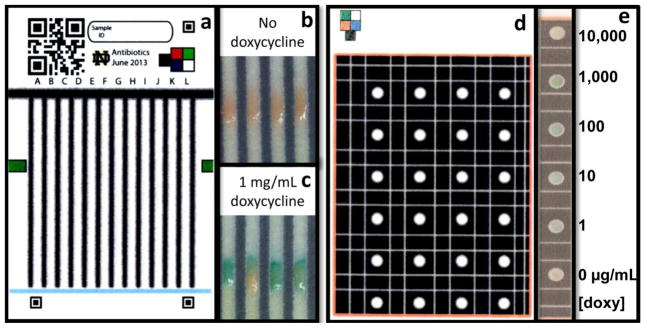
The BioPAD lateral flow test platform (6 x 11 cm) (a) is described in Weaver et al.[4] QR codes are inserted for test identification, corner fiducials and color box are for computer image analysis, currently in development. Following color development, test regions containing yeast in the lateral flow BioPAD lack blue pigment following exposure to media only (b) while tests exposed to media with doxycycline appear blue (c). The stationary BioPAD test platform (8.5 x 11cm) isolates testing regions using wax barriers (d). Hydrophilic lines or “moats” are included to redirect liquid that may potentially contaminate a neighboring test region. Stationary BioPAD test spots exposed to doxycycline concentrations ranging from 0–10,000μg/mL and following color development are shown in image e
Growth phase considerations
Yeast liquid cultures of logarithmic (log), late log, and approaching stationary growth phase were exposed to doxycycline in concentrations of 0.1, 1, 10, 100 and 1,000 μg/mL overnight at 30ºC. Log phase yeast having an OD600nm of 1.2 were found to be most responsive to doxycycline, especially at 100 μg/mL concentrations, while BioPADs fabricated with yeast at an OD600nm of 4.2 were generally least responsive (ESM Fig. S2a). Therefore, subsequent BioPAD fabrication was carried out using log phase yeast. In preliminary studies we found that two OD600nm units of yeast at log phase, concentrated and embedded into paper in a hydrogel matrix were sufficient to visualize reporter production in response to 1-10,000 μg/mL analyte (not shown); this amount of yeast was used for fabrication of all further BioPADs unless otherwise indicated.
BioPAD response
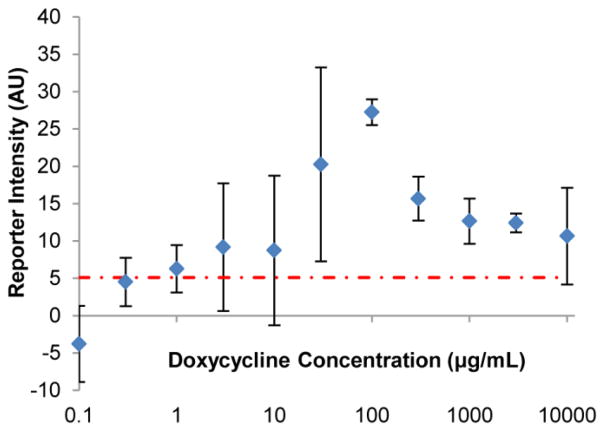
To test the responsive range of stationary BioPADs, these tests were exposed to 0 – 10,000 μg/mL doxycycline. At concentrations between 1 and 10 μg/mL, BioPAD response was too variable to clearly indicate the presence of doxycycline, while following exposures from 30 – 10,000 μg/mL, reporter intensities could be distinguished from negative controls with a >95% confidence level, with the strongest color response at 100 μg/mL (Figure 2). We noted the moderate loss of signal at higher doxycycline concentrations, and hypothesized that it might be due to yeast death induced by doxycycline.
Figure 2.
BioPAD reporter intensity in response to doxycycline exposure. The increase in reporter intensity of stationary BioPAD in response to doxycycline/media mixture, as compared to media only samples, is reported over a range of concentrations (note log scale on X axis). These data show that doxycycline concentrations between 30–10,000 μg/mL can be distinguished from negative controls (Student T-test p < 0.05). The standard deviation of media only samples is indicated by the dashed red line
To test the effects of high levels of doxycycline on yeast mortality, actively growing cultures were exposed to doxycycline for 1 hour with 30ºC incubation and then plated. The resultant drop in colony forming units (ESM Fig. S2b) shows that doxycycline at high concentrations, (>3 mg/mL) is lethal to actively growing cultures of this strain of yeast. This observation provides a potential explanation for why the reporter signal drops off at high levels of doxycycline, but comparison of these results to those in Figure 2 show that yeast in the BioPAD produce significant amounts of reporter up to concentrations of 10 mg/ml, indicating that they are less susceptible than yeast in suspension culture.
A remaining question is why the yeast in the BioPAD are more resistant to doxycycline than are yeast in suspension culture. One explanation might be that the hydrogel matrix sequesters or interferes with the doxycycline. We found that pre-incubation of doxycycline with hydrogel did not reduce the effect of the antibiotic solution (ESM Fig. S3). Previously, it has also been shown that glucose and α-lactoalbumin diffuse freely from 2% calcium alginate beads.[17] Thus, based on this evidence we find it unlikely that this problem is due to the hydrogel. It is interesting to speculate that the reduced doxycycline susceptibility in the BioPAD is similar to the microbial response seen in bacterial and fungal biofilms, a phenomenon that is not well understood.[18] These issues warrant future investigation.
To test the BioPAD using a field relevant sample, we measured its ability to identify the presence/absence of doxycycline in pharmaceutical dosage forms obtained from a local pharmacy. Doxycycline dosage forms (100 mg) used in this study were analyzed for purity by high performance liquid chromatography (HPLC), comparing the predominant peak with that from a doxycycline standard. The identity of this peak was confirmed to be doxycycline by an exact mass measurement from electrospray mass spectrometry (ESM Fig. S4). A pill was dissolved in approximately 100 mL of complete yeast media to comprise the test solution. With an eye towards BioPAD use in resource limited settings, testing was carried out with the more user-friendly lateral flow BioPAD, and test solution was assembled in a graduated baby bottle to avoid the use of laboratory measuring tools. 64 lateral flow test lanes were exposed to the doxycycline test solution and 56 to media only. Evaluation of the test results was carried out both visually and by ImageJ analysis to determine the predictive power of the BioPAD.
Test evaluations by visual and ImageJ analysis were found to be in agreement. A blind visual inspection, carried out by 2 readers comparing outcomes to standard images, identified 92% of samples containing doxycycline, and correctly identified 100% of samples containing no doxycycline (ESM Table S1) with only one sample disagreement, requiring a third reader as a tie-breaker. The second method, measuring reporter intensity using ImageJ software, found 95% of positive samples to have reporter intensities higher than samples not exposed to doxycycline. Intensity values from this second method were used to assemble a receiver operating characteristic (ROC) plot (Figure 3), able to measure the distinguishing power of a diagnostic. A ROC plot graphs the lateral flow BioPAD sensitivity (fraction of positive samples identified correctly) against 1-selectivity (fraction of false positives) for a series of possible thresholds that differentiate positive from negative outcomes. Ideal results in a ROC plot will show sensitivity approaching 100% before false positives are detected. Determining the area under the curve (AUC) is a common evaluation for the ROC plot with areas near .90 or greater indicating a useful diagnostic. In this case the AUC is .994 (n=120), using the Mann-Whitney U–test[19]. In a separate run, an analogous ROC plot produced an AUC of .87 (n=36) (ESM Fig. S5). aBoth of these measurements support the idea that the BioPAD is a useful indicator of the presence/absence of doxycycline at concentrations of 1 mg/mL.
Figure 3.
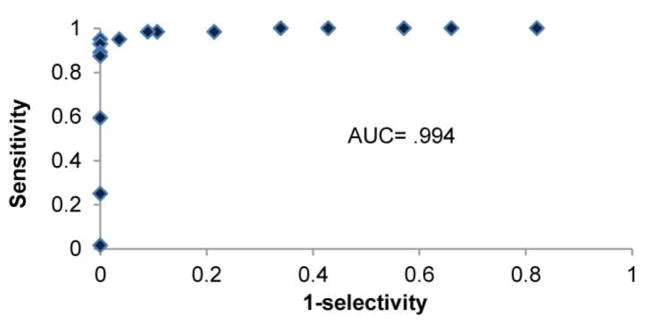
A receiver operating characteristic (ROC) plot was assembled reflecting the distinguishing power of the BioPAD for identifying the presence/absence of doxycycline at ~1mg/mL concentrations. The area under the curve (AUC) determined by Mann-Whitney is .994, indicating that the BioPAD is a useful tool for this analytical task
BioPADs stability in storage
One of the greatest advantages of yeast is their stability during storage[20]. Lateral flow BioPADs, wrapped in aluminum foil and sealed in a zipper-locked baggie containing desiccant, were stored in both warm and cold environments. Viability of the BioPAD was measured by comparing the mean intensity of reporter, measured in ImageJ, for test lanes exposed to 100 μg/mL doxycycline in media versus media only. BioPADs were considered viable when the average reporter intensity was determined to be significantly higher than the negative control (p<0.05) by the Student T-test. BioPADs kept at 4ºC were found to be responsive to analyte for at least 6 months (no later samples were available for testing) while those at −20ºC were viable for > 1 year (415 days with no later samples for testing) (ESM Fig. S6). BioPADs were found to remain viable following 12 days of storage in 54ºC conditions before losing activity, while those stored at 37ºC were responsive for at least 56 days with no further tests available. These results indicate that cool storage is preferred, but tests can remain reliable following exposure to warm or even hot temperatures.
CONCLUSION
A biologically-based Paper Analytical Device (BioPAD) was developed and is to our knowledge the first published example of a yeast whole-cell, paper-based biosensor. This research establishes the fabrication techniques and distinguishing power of this paper-based analytical device, composed of yeast, hydrogel and paper. Both stationary and lateral flow test designs allow multiple tests to be carried out on one paper device. BioPADs were able to indicate the presence of doxycycline in the range of 30 – 10,000 μg/mL with a peak in response around 100 μg/mL The present iteration of the BioPAD is more appropriate for qualitative than quantitative use, being able to discriminate between doxycycline concentrations within its detectable range from those outside of it. However, if the signal variability that is characteristic at the upper and lower ends of the responsive range could be reduced, perhaps through the use of a different reporter system, BioPADs have the potential to become more quantitative tools. In this study, the BioPAD tests proved to be an effective analytical tool, positively identifying the presence of active ingredient in doxycycline tablets with 92% and 95% success when interpreted by eye and computer image analysis, respectively. BioPAD viability, stored in aluminum foil and sealed in plastic zipper lock bags, was found to be at least 56 days at room temperature and at least 415 days when stored at 4ºC making them suitable for use in remote locations that require test transport and storage. While current testing requires a cell lysis step, too cumbersome for field testing, future work using fabrication methods presented here could be applied to other yeast biosensors. Given the current collection of genetically engineered yeast that exist[7, 21, 22], research in this vein could give rise to a series of yeast whole-cell, paper-based biosensor devices able to address a range of analytical questions that exist outside of the laboratory.
Supplementary Material
Acknowledgments
Galen Brown for acting as a third reader in BioPAD visual analysis. This work was supported by the following sources. Abigail Weaver is a fellow of the Chemistry-Biochemistry-Biology Interface (CBBI) Program at the University of Notre Dame, supported by training grant T32GM075762 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. Abigail is also a fellow of The Eck Institute for Global Health and the GLOBES program (an interdisciplinary graduate training program in environment and society, NSF award number 0504495) at the University of Notre Dame. Funding for some aspects of this work was provided by Notre Dame Advanced Diagnostics and Therapeutics.
References
- 1.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Diagnostics for the developing world: microfluidic paper-based analytical devices. Anal Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 2.Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew Chem Int Ed Engl. 2007;46:1318–20. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vella SJ, Beattie P, Cademartiri R, et al. Measuring markers of liver function using a micropatterned paper device designed for blood from a fingerstick. Anal Chem. 2012;84:2883–91. doi: 10.1021/ac203434x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver AA, Reiser H, Barstis T, et al. Paper analytical devices for fast field screening of Beta lactam antibiotics and antituberculosis pharmaceuticals. Anal Chem. 2013;85:6453–60. doi: 10.1021/ac400989p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W, Mo W, Shen D, et al. Yeast model uncovers dual roles of mitochondria in action of artemisinin. PLoS Genet. 2005;1:e36. doi: 10.1371/journal.pgen.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuhara H, Sor F. identification if two erthyomycin mutations in the mitochondrial gene coding for the large ribosomal RNA in yeast. 1982;10:6571–6577. doi: 10.1093/nar/10.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baronian K. The use of yeasts and moulds as sensing elements in biosensors. Biosens Bioelectron. 2004;19:953. doi: 10.1016/j.bios.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Raut N, O’Connor G, Pasini P, Daunert S. Engineered cells as biosensing systems in biomedical analysis. Anal Bioanal Chem. 2012;402:3147–59. doi: 10.1007/s00216-012-5756-6. [DOI] [PubMed] [Google Scholar]
- 9.Lagarde F, Jaffrezic-Renault N. Cell-based electrochemical biosensors for water quality assessment. Anal Bioanal Chem. 2011;400:947–64. doi: 10.1007/s00216-011-4816-7. [DOI] [PubMed] [Google Scholar]
- 10.Stocker J, Balluch D, Gsell M, et al. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ Sci Technol. 2003;37:4743–50. doi: 10.1021/es034258b. [DOI] [PubMed] [Google Scholar]
- 11.Struss A, Pasini P, Ensor CM, et al. Paper strip whole cell biosensors: a portable test for the semiquantitative detection of bacterial quorum signaling molecules. Anal Chem. 2010;82:4457–63. doi: 10.1021/ac100231a. [DOI] [PubMed] [Google Scholar]
- 12.Racek J. A yeast biosensor for glucose determination. Appl Microbiol Biotechnolopgy. 1991;45:473– 477. doi: 10.1002/ceat.270140404. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann M, Riedel K, Adler K, Kunze G. Amperometric measurement of copper ions with a deputy substrate using a novel Saccharomyces cerevisiae sensor. Biosens Bioelectron. 2000;15:211–219. doi: 10.1016/s0956-5663(00)00060-9. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura H, Suzuki K, Ishikuro H, et al. A new BOD estimation method employing a doublemediator system by ferricyanide and menadione using the eukaryote Saccharomyces cerevisiae. Talanta. 2007;72:210–216. doi: 10.1016/j.talanta.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Bellí G, Garí E, Piedrafita L, et al. An activator/repressor dual system allows tight tetracyclineregulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–7. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garí E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–48. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka H, Matsumura M, Veliky Ia. Diffusion characteristics of substrates in Ca-alginate gel beads. Biotechnol Bioeng. 1984;26:53–8. doi: 10.1002/bit.260260111. [DOI] [PubMed] [Google Scholar]
- 18.Van Acker H, Van Dijck P, Coenye T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014;22:326–33. doi: 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Mason S, Graham N. Areas beneath the relative operating characteristics (roc) and relative operating levels (rol) curves: Statistical significance and interpretation. Q J R. 2002:2145–2166. [Google Scholar]
- 20.Miyamoto-Shinohara Y, Imaizumi T, Sukenobe J, et al. Survival rate of microbes after freezedrying and long-term storage. Cryobiology. 2000;41:251–5. doi: 10.1006/cryo.2000.2282. [DOI] [PubMed] [Google Scholar]
- 21.Park M, Tsai S-L, Chen W. Microbial biosensors: engineered microorganisms as the sensing machinery. Sensors (Basel) 2013;13:5777–95. doi: 10.3390/s130505777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su L, Jia W, Hou C, Lei Y. Microbial biosensors: A review. Biosens Bioelectron. 2010;26:1788–1799. doi: 10.1016/j.bios.2010.09.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.