Abstract
Information on protein interactions (PIs) is valuable for biomedical research, but often lies buried in the scientific literature and cannot be readily retrieved. While much progress has been made over the years in extracting PIs from the literature using computational methods, there is a lack of free, public, user-friendly tools for the discovery of PIs. We developed PIMiner, an online tool for the extraction of PI relationships from PubMed-abstracts. Protein pairs and the words that describe their interactions are reported by PIMiner along with the interaction likelihood levels, so that new interactions can be readily detected within text. The option to extract only specific types of interactions is also provided. The PIMiner server can be accessed through a web browser or remotely through a client’s command line. PIMiner can process 50,000 PubMed abstracts in approximately seven minutes and is thus suitable for large scale processing of biological literature.
Keywords: Protein interactions, PIs, literature mining, biological textmining, systems biology, interactome mining, data mining, bioinformatics, complex networks
1 Introduction
Protein interactions (PIs), such as protein-protein interactions, provide important insights for many biochemical processes in the cell. Several important programs for predicting protein-protein interactions based on sequence or domains have been described (Chen et al., 2008; Dong, Zhou, and Liu, 2010; Li, Tan and Ng, 2006). However, the scientific literature offers a wealth of information regarding protein interactions and PI extraction from published articles has attracted much attention because of the importance of such interactions for research in biology and biomedicine. Manual curation of extracted PIs is highly time consuming, resource intensive, and is unable to keep pace with the ever rising number of publications. Thus, state-of-the-art PI extraction initiatives are designed to extract PIs using automated computational techniques, as well as a combination of automated methods and manual curation. Despite significant progress made in the development of automated PI extraction methods (Hoffmann and Valencia, 2004; Saetre, Kenji, and Tzujii, 2007; Hunter et al., 2008; Rebholz-Schuhmann et al., 2008; Chowdhary, Zhang, and Liu, 2009; Fundel, Küffner, and Zimmer, 2007; Jose, Vadivukarasi, and Devakumar, 2007), there has been a general lack of availability of ready-to-use, user-friendly and easily testable tools for extracting PIs from text (Jose, Vadivukarasi, and Devakumar, 2007; Kabiljo, Clegg, and Shepherd, 2009). In 2009, Kabiljo, Clegg, and Shephard analysed usability issues with several of these programs. Table 1 summarizes these results and complements them for the purposes of this study.
Table 1.
Comparison of state-of-the-art PI extraction programs (Kabiljo, Clegg, and Shepherd, 2009)
| PI programs | iHOP | AkanePPI | OpenDMAP | Protein Corral | Bayesian Networks Method | PIMiner |
|---|---|---|---|---|---|---|
| Source | Hoffmann and Valencia, 2005 | Saetre, Kenji, and Tsujii, 2008 | Hunter et al., 2008 | Rebholz-Schuhmann et al., 2008 | Chowdhary, Zhang, and Liu, 2009 | This Study |
| Webserver / webutility / standalone | Webserver and webservice | Standalone | Standalone | Webserver and webutility | Standalone | Webserver and webutility |
| Ease of installation, configuration and usage | Easy | Difficult requires installation of several, non-trivial components | Difficult configuration requires XML configuration file, not supplied | Easy webservice requires some basic coding | Difficult | Easy |
| Easily testable on different training/test data | Difficult does not accept user-submitted text | Difficult requires linguistic expertise | Difficult requires custom-written sample code from authors | Difficult does not allow pretagged text, considers only entities normalized to Uniprot | Difficult | Easy Module B designed specifically for this purpose |
| Allows user training data | No | Yes | Yes | No | No | Yes |
| Allows user raw text (test/query data) as input | No | Yes | Yes | Yes (in Whatizit version) | Yes | Yes |
| Allows user NER tagging | No | Yes | Yes | No | No | Yes |
| Easy integration with user programs | Yes | Yes | Yes | Yes | Yes | Yes |
| User specified interaction types | No | No | No | No | No | Yes |
Here we describe the PIMiner online application we have developed to address some of the above usability issues with an architecture that is flexible, user-friendly, and easy to test. The PIMiner tool is based on the PI extraction method we proposed in 2009 (Chowdhary, Zhang, and Liu, 2009). PIMiner significantly enhances our previous method by fulfilling the objective of easy use for routine PI extraction tasks in addition to several other improvements. Compared to the method described previously, PIMiner includes a more robust protein name library with the capacity to tag multiple-word names and has a larger interaction word list. PIMiner also provides an optional filter function to allow extraction of only specific interaction types; for example, one may wish to extract only phosphorylation or methylation types of interactions from the text, in which case there is an option to specify interaction type in the user interface. We have created a versatile, web-based, PI extraction tool, aimed to support research in biology and biomedical fields.
2 Methods
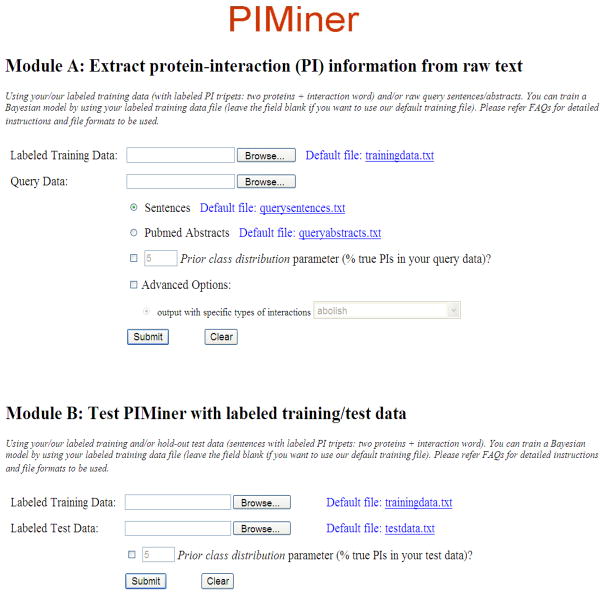
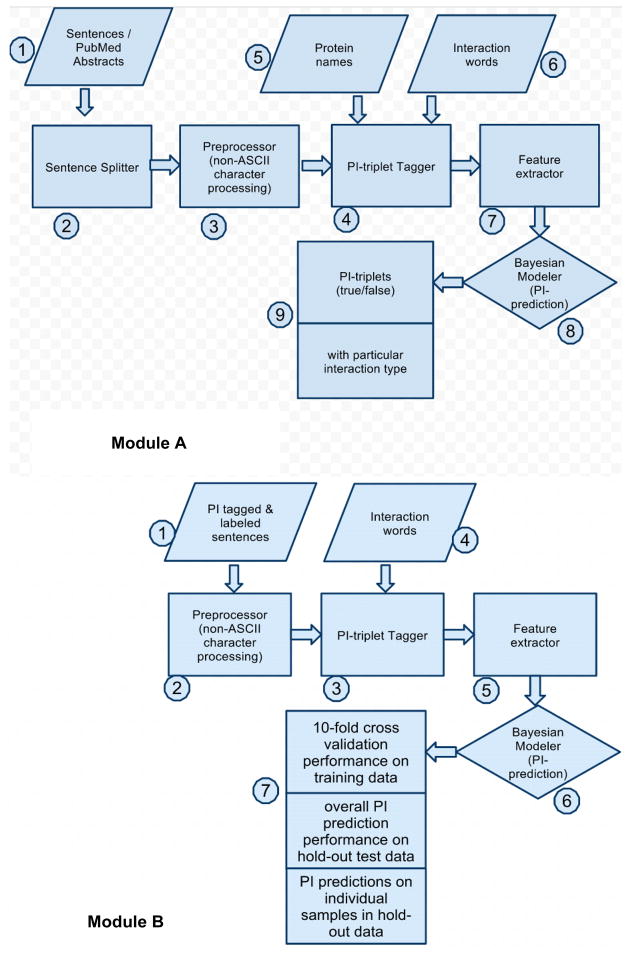
The PIMiner system consists of two modules, Module A and Module B (Figure 1). Module A allows for the extraction of PI information from raw text and Module B allows for testing of the PIMiner tool system with labelled training/test data (Figure 2).
Figure 1.
Web interface of PIMiner Modules A and B
Figure 2.
Workflow of PIMiner Modules A and B
2.1 Module A: PI Extraction
Module A is designed to extract interactions from raw query text (untagged/unlabelled text) in either sentence or PubMed abstract format. The workflow of Module A is shown in Figure 2. In this module, PubMed abstracts are first converted into individual sentences by the rule-based Splitter module. The processed sentences are then tagged for protein names and interaction words using the Tagger module. The tagger uses an exhaustive dictionary containing over eight million protein names and their variants. We generated the protein name dictionary by extracting data from various sources, including BioThesaurus (Liu et al., 2006), UniProtKB/Swiss-Prot database (UniProt Consortium 2010) and NCBI Entrez Gene database (Maglott et al., 2005). Extracting data from these sources not only provided more protein names than using BIOGrid (Stark et al., 2006), as done previously (Chowdhary, Zhang, and Liu, 2009), but also allowed for tagging of multiple-word names. The dictionary was cleaned by filtering out words that are not likely protein names using the Genia tagging program (Tsuruoka et al., 2005). Commonly occurring English words and one letter/digit acronyms/short-forms were also filtered out.
The interaction word list contains more than 2,000 unique terms, excluding variant forms that contain hyphens and those that represent American/British English language variations. Interaction terms describe the potential nature/type of the interaction between two interacting proteins in text. We generated our list of interaction words by combining various resources (Chowdhary, Zhang, and Liu, 2009) and also manually from the literature. An improvement in the interaction word list compared to the previously described method (Chowdhary, Zhang, and Liu, 2009) is the use of different syntactical forms of interaction words based on word endings without using the interaction words themselves.
Our protein name tagger is optimized for processing large volumes of text with linear/polynomial complexity in time. The tagger also attempts to detect variation in protein names by looking for certain types of domain-specific bag-of-words ahead of the detected protein name in the sentence. For example, the tagger will be able to detect protein ‘X receptor’ in a sentence even if protein ‘X receptor’ does not exist in the dictionary but protein ‘X’ does. The tagger handles case-sensitive variations of protein names by matching single-word protein names in a case-sensitive manner and multiple-word protein names in a case-insensitive manner. This is done to avoid matching of commonly occurring single non-protein words that are most frequently written in lower case. Case-insensitiveness is retained for matching protein names composed of multiple words because there is a much smaller chance of matching non-protein multiple-word concepts in text.
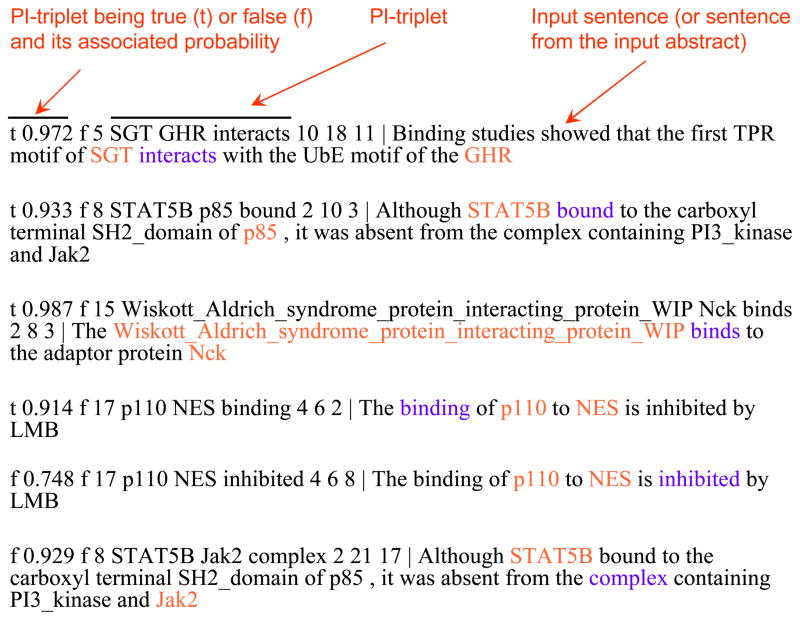
After tagging query sentences with protein names and interaction terms, the feature extractor module extracts feature vectors from the query sentences which are then passed to the trained PIMiner’s Bayesian modeller module. The Bayesian modeller module classifies the target interaction triplets (two target proteins + one interaction word) in the test sentences as true/false. Sample predictions are shown in Figure 3. For Bayesian model training, the feature vectors extracted from training sentences are utilized. The model is applied to classify target interactions in the query sentence file by assigning them labels of either true or false. Each triplet in the query sentence file is predicted with a probability value that indicates the likelihood of being true or false (Figure 3). More details about the Bayesian model training process are provided in our previous work (Chowdhary, Zhang, and Liu, 2009).
Figure 3.
Typical output from PIMiner. The PI triplet is shown highlighted in colour with red indicating the target protein pair and purple indicating the interaction word
PIMiner also outputs information about the type of interaction (e.g. phosphorylation, methylation and others) which characterizes the interaction between the two entities. We have implemented functions by which the system can either report all interaction types found in the query text, or the output can be restricted to specific types of interactions. This function may be very useful for researchers interested in only certain types of PIs.
2.2 Module B: Performance Evaluation
Module B is provided to allow for the performance evaluation of PIMiner. Module B accepts two input files, one that contains training sequences and another that contains test sentences where interaction triplets (two target proteins + one interaction word) are tagged and labelled as true or false. The workflow for Module B is shown in Figure 2. The feature extractor module extracts feature vectors from test sentences which are then passed to the trained PIMiner’s Bayesian modeller module as in Module A, which classifies the target interactions in the test sentences as true/false (see Figure 3 for sample predictions).
The output of Module B has three components related to i) performance of the system with a 10-fold cross validation conducted on the training data, ii) performance of the system on the test (hold-out) data, and iii) prediction results on individual samples in the test data. In addition to testing, Module B can also be used to extract interactions from sentences when protein names are already tagged by the user. This function may be helpful to the users who wish to use their own Named Entity Recognition (NER) tagger to pre-tag protein names in their dataset before using PIMiner. The user also has the option to use the default protein name tagger provided as part of Module A.
2.3 PIMiner Web Server
The PIMiner web tool can be accessed using an internet browser and also through a set of Perl programs that can be download and run locally. The downloadable programs also include a graphical interface, implemented in Perl-TK, to assist the user to input program parameters in a user-friendly manner. The downloadable version of PIMiner web tool is intended for “heavy duty” jobs.
The PIMiner server runs on an IBM HS22 blade which has 48 GB of RAM and 2 Quad core 2.93 GHz Nehalem Processors. The amount of resources allocated to the web server is currently 25GB of disk space, 8 GB of RAM and 1 core (2.93 GHz). Based on usage intensity, we may allocate more resources to the web server in the future. The web server is implemented using Apache, Perl, and Java programming languages.
3 Results
We evaluated our dictionary-based protein name tagger on the BioCreative task-1 (protein mention) test dataset (Hirschman et al., 2005) and the AIMed dataset (Bunescu and Mooney et al., 2006) and found that performance was satisfactory (Table 2). We evaluated our method for detection of known annotations of protein mentions in sentences in the BioCrative and AIMed datasets. We assessed both exact and partial match for the Biocreative dataset and exact match only for the AIMed dataset. Partial match was necessary because some of the annotations in the BioCreative test dataset do not point to specific proteins, rather to their semantic context in the sentence. For the AIMed dataset we considered only the maximal length protein name exact string match and ignored protein name substrings contained therein (e.g. if SP1 receptor was found in the text we considered SP1 receptor but not SP1). In contrast to the Biocreative dataset, the AIMed dataset tags only specific gene mentions and does not mention plural forms or family/general names that could possibly be normalized directly to their NCBI database IDs, and can therefore be considered more pure. The results of this analysis are shown in Table 2. We analysed PIMiner’s false positive predictions on the AIMed dataset and found that there were many protein names that were not tagged by AIMed. Examples include TNF appearing 23 times, 14-3-3 appearing 18 times, TNF receptor appearing 17 times, PDGF appearing 16 times, activin appearing 16 times, FGF appearing 15 times, and SH2 appearing 15 times. While these predictions may be considered true positive, for the purposes of evaluation we considered them false positive as per AIMed annotation. On analysing PIMiner’s false negative predictions on the AIMed dataset we observed that retinoblastoma was annotated as a true protein name which is not true since it is a name for retina cancer and also for a protein family.
Table 2.
Named entity recognition performance of protein name tagger in PIMiner
| Recall | Precision | F-measure | |
|---|---|---|---|
| On BioCreative-1 test data (Hirschman et al., 2005) | |||
| Partial match | 87.3 | 81.7 | 84.4 |
| Exact Match | 67.7 | 68.5 | 68.1 |
|
| |||
| On AIMed data (Bunescu and Mooney, 2006) | |||
| Exact Match | 79 | 68.8 | 73.6 |
4 Discussion
In this study we developed and introduced PIMiner, an online tool for the extraction of PIs from text. Development of PIMiner addressed several shortcomings related to effective, user-friendly PI extraction applications. Most PI extraction methods that have been proposed are either not available in a usable application form (Jose, Vadivukarasi, and Devakumar, 2007) or, if available, are not user-friendly (Kabiljo, Clegg, and Shepherd, 2009). Table 1 summarises specific issues with some of the state-of-the-art PI extraction systems available today based on discussions by Kabiljo, Clegg, and Shepherd (2009) and compares them to PIMiner. Due to these practical difficulties, recent progress in the development of PI extraction methods has not been able to truly benefit the potential end users of such systems. We have designed PIMiner with the goal of reducing some of these problems and providing researchers a tool that they can employ in a user-friendly manner for extracting PI information from PubMed text.
The PIMiner application is based on the PI extraction method we developed in 2009 (Chowdhary, Zhang, and Liu, 2009) with some additional features. For example, PIMiner uses a much larger protein name dictionary with over eight million names, including many gene names, compared to approximately 80,000 names used in our previous study. The protein name tagger in PIMiner is more sophisticated and is capable of tagging multiple-word protein names, while our previous method could handle only single-word protein names since because it was based on BioGRID reference data (Stark et al., 2006), which contained mostly single-word protein names. Our interaction word list is now of much larger size (> 2,000 interaction words) compared to what we used in our previous study (191 interaction words). Additionally, users can now input full PubMed abstracts to PIMiner in addition to the single sentence format that was handled by the previous version of the system (Chowdhary, Zhang, and Liu, 2009).
The PIMiner approach is different from our previous method in that PIMiner uses syntactical forms of the interaction words without using the interaction words themselves. For example, we categorize each word in our interaction word list into one of the eight predefined categories based on word endings and use these categories in our modelling. For example, the interaction words regulate, regulates, regulated, regulating, regulation, regulations, regulator and regulators would each belong to one of those eight categories. Thus, the number of model parameters to be learned in PIMiner is far fewer than in our earlier approach while maintaining similar prediction accuracy with an F-measure of 74.1% on the training data used in our earlier study (Chowdhary, Zhang, and Liu, 2009) based on 10-fold cross validation. This makes PIMiner more scalable to unseen data compared to our previous method. In contrast to our earlier method, PIMiner also provides the user the option to tune the model by taking into account the class distribution of the unseen data that might be different from that of the training data, thereby allowing the user to incorporate prior knowledge of the unseen query data in the modelling.
PIMiner is clearly improved compared to our previously described method (Chowdhary, Zhang, and Liu, 2009) and also outperforms other state-of-the-art PI extraction methods. In a head-to-head comparison of several PI extraction methods, Kabiljo, Cleff, and Shephard (2009) assessed the performance of both AkanePPI (Saetre, Kenji, and Tsujii, 2008) and OpenDMAP (Hunter et al., 2008) using the AIMed dataset. Recall, precision, and F-measure values were 74, 57.0, and 64.4, respectively, for AkanePPI and 9.1, 61, and 15.9, respectively, for OpenDMAP. PIMiner outperforms both PI extraction methods when tested using the same dataset with recall, precision, and F-measure values of 79, 68.8, and 73.6, respectively (Table 2). This is especially striking given that the AIMed dataset was used to develop the AkanePPI extraction system.
PIMiner provides two types of access to the remote user: i) browser access, and ii) command line access. Browser access is intended for “lightweight” jobs, while command line access is more suitable for “heavyweight” jobs. Command line access will allow users to run the PIMiner web utility remotely on their own computers and to save the results locally by default. It is possible for researchers to use PIMiner to build databases of protein-protein interactions and gene regulations using archived scientific literature. In addition, this utility can be used by researchers to extract PIs from papers that have been recently published or papers retrieved from PubMed using specific keywords, allowing them to obtain interactions that are absent in the general archived interaction databases. We have also provided functions that allow for consideration of only specific types of interactions. This allows researchers to target PIs of interest without getting lost in a large amount of unrelated interactions. PIMiner provides a generic framework for extracting interaction relationships of the type protein/protein and protein/gene in a user-friendly manner. In execution, PIMiner can process 50,000 PubMed abstracts in a little over seven minutes, making it suitable for processing large volumes of text in a reasonable period of time. We hope that the PIMiner service will provide a useful tool for researchers in the fields of biology and biomedicine.
The current version of PIMiner does not explicitly resolve the problem of polysemy, which is typical of a dictionary-based NER system. In addition, the current version does not attempt to detect PI cases where a protein might interact with itself. We plan to address these issues in the future versions of PIMiner. We also plan to extend this framework to extract interaction relationships in other related domains, such as small molecule-protein interactions and others. Additionally, we plan to adapt PIMiner so that it can be applied to the full text of papers.
Acknowledgments
Acknowledgements and funding
This study was supported in part by grant 1UL1RR025011 from the Clinical and Translational Science Award (CTSA) program of the National Center for Research Resources, National Institutes of Health. We thank Ryan Frahm for providing server support.
Biographies
Rajesh Chowdhary received his PhD in Computer Science from National University of Singapore. His research interests include gene regulation, systems biology, machine learning and biological text mining. He is Associate Research Scientist at Marshfield Clinic Research Foundation, USA.
Jinfeng Zhang is Assistant Professor at Department of Statistics, Florida State University, USA. His research interests include protein structure and function, systems biology, machine learning and biological text mining.
Sin Lam Tan is Systems Analyst at Marshfield Clinic Research Foundation, USA.
Daniel Osborne is student at Department of Statistics, Florida State University, USA.
Vladimir B. Bajic is Professor and Director, Computational Bioscience Research Center (CBRC), King Abdullah University of Science and Technology (KAUST), Kingdom of Saudi Arabia. His research interests include several areas of bioinformatics and computational biology.
Jun S. Liu is Professor at Department of Statistics, Harvard University. His research interests include statistics, bioinformatics and computational biology.
Footnotes
5 Availability and requirements
Project name: PIMiner web tool
Project home page: http://www.biotextminer.com/PPI/index.html
Operating system(s): Platform independent
Programming language (for running webutility): perl, perl-TK
Other requirements: Browsers IE 6, Firefox 3 or above versions
License: Webserver/webutility is free for use.
Any restrictions to use by non-academics: None
Contributor Information
Rajesh Chowdhary, Email: chowdhary.rajesh@mcrf.mfldclin.edu.
Jinfeng Zhang, Email: jinfeng@stat.fsu.edu.
Sin Lam Tan, Email: tan.sinlam@mcrf.mfldclin.edu.
Daniel Osborne, Email: dosborne@stat.fsu.edu.
Vladimir B. Bajic, Email: vladimir.bajic@kaust.edu.sa.
Jun S. Liu, Email: jliu@stat.harvard.edu.
References
- Bunescu R, Mooney R. Subsequence kernels for relation extraction. In: Weiss Y, Scholkopf B, Platt J, editors. Advances in Neural Information Processing Systems. Vol. 18. Cambridge, MA: MIT Press; 2006. pp. 171–178. [Google Scholar]
- Chen XW, Han B, Fang J, Haasl RJ. Large-scale protein-protein interaction prediction using novel kernel methods. Int J Data Mining and Bioinformatics. 2008;2(2):145–156. doi: 10.1504/ijdmb.2008.019095. [DOI] [PubMed] [Google Scholar]
- Chowdhary R, Zhang J, Liu JS. Bayesian inference of protein-protein interactions from biological literature. Bioinformatics. 2009;25(12):1536–1542. doi: 10.1093/bioinformatics/btp245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Zhou S, Liu X. Prediction of protein-protein interactions from primary sequences. Int J Data Mining and Bioinformatics. 2010;4(2):211–227. doi: 10.1504/ijdmb.2010.032151. [DOI] [PubMed] [Google Scholar]
- Fundel K, Küffner R, Zimmer R. RelEx – relation extraction using dependency parse trees. Bioinformatics. 2007;23(3):365–371. doi: 10.1093/bioinformatics/btl616. [DOI] [PubMed] [Google Scholar]
- Hirschman L, Yeh A, Blaschke C, Valencia A. Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinformatics. 2005;6(1):S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R, Valencia A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics. 2005;21(Suppl 2):ii252–ii258. doi: 10.1093/bioinformatics/bti1142. [DOI] [PubMed] [Google Scholar]
- Hunter L, Lu Z, Firby J, Baumgartner WA, Jr, Johnson HL, Ogren PV, Cohen KB. OpenDMAP: An open source, ontology-driven concept analysis engine, with application to capturing knowledge regarding protein transport, protein interactions and cell-type-specific gene expression. BMC Bioinformatics. 2008;9:78. doi: 10.1186/1471-2105-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose H, Vadivukarasi T, Devakumar J. Extraction of Protein Interaction Data: A Comparative Analysis of Methods in Use. EURASIP J Bioinform Syst Biol. 2007:53096. doi: 10.1155/2007/53096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabiljo R, Clegg AB, Shepherd AJ. A realistic assessment of methods for extracting gene/protein interactions from free text. BMC Bioinformatics. 2009;10:233. doi: 10.1186/1471-2105-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XL, Tan SH, Ng SK. Improving domain-based protein interaction prediction using biologically significant negative datasets. Int J Data Mining and Bioinformatics. 2006;1(2):138–149. doi: 10.1504/ijdmb.2006.010852. [DOI] [PubMed] [Google Scholar]
- Liu H, Hu ZZ, Zhang J, Wu C. BioThesaurus: a web-based thesaurus of protein and gene names. Bioinformatics. 2006;22(1):103–105. doi: 10.1093/bioinformatics/bti749. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebholz-Schuhmann D, Arregui M, Gaudan S, Kirsch H, Jimeno A. Text processing through web services: calling Whatiszit. Bioinformatics. 2008;24(2):296–298. doi: 10.1093/bioinformatics/btm557. [DOI] [PubMed] [Google Scholar]
- Saetre R, Kenji S, Tsujii J. In: Christopher JO, Baker SJ, editors. Syntactic features for protein-protein interaction extraction; Short Paper Proceedings of the 2nd International Symposium on Languages in Biology and Medicine; Singapore. 2007. pp. 6.1–6.14. [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruoka Y, Tateishi Y, Kim JD, Ohta T, McNaught J, Ananiadou S, Tsujii J. Developing a Robust Part-of-Speech Tagger for Biomedical Text. Advances in Informatics - 10th Panhellenic Conference on Informatics, LNCS; 2005. pp. 382–392. [Google Scholar]