Abstract
Nitrogen (N) fixation is fueling planktonic production in a multitude of aquatic environments. In meso- and poly-haline estuaries, however, the contribution of N by pelagic N2 fixation is believed to be insignificant due to the high input of N from land and the presumed absence of active N2-fixing organisms. Here we report N2 fixation rates, nifH gene composition and nifH gene transcript abundance for key diazotrophic groups over 1 year in two contrasting, temperate, estuarine systems: Roskilde Fjord (RF) and the Great Belt (GB) strait. Annual pelagic N2 fixation rates averaged 17 and 61 mmol N m−2 per year at the two sites, respectively. In RF, N2 fixation was mainly accompanied by transcripts related to heterotrophic (for example, Pseudomonas sp.) and photoheterotrophic bacteria (for example, unicellular diazotrophic cyanobacteria group A). In the GB, the first of two N2 fixation peaks coincided with a similar nifH-expressing community as in RF, whereas the second peak was synchronous with increased nifH expression by an array of diazotrophs, including heterotrophic organisms as well as the heterocystous cyanobacterium Anabaena. Thus, we show for the first time that significant planktonic N2 fixation takes place in mesohaline, temperate estuaries and that the importance of heterotrophic, photoheterotrophic and photosynthetic diazotrophs is clearly variable in space and time.
Introduction
Assimilation of reactive nitrogen (N) compounds and subsequent sedimentation of organic matter in aquatic environments may lead to N limitation of primary production (Howarth, 1988a). Fixation of atmospheric N2 accounts for more than 80% of the N input in some of these environments, thereby sustaining a significant fraction of the new production (Granhall and Lundgren, 1971; Horne and Galat, 1985). However, other N-limited aquatic systems do not display significant N2 fixation rates (Howarth et al., 1988b). In temperate estuaries, N is often limiting in summer due to the assimilation of reactive N by phytoplankton. As a consequence, the N:P ratio is below the Redfield ratio favoring N2 fixation (Boynton et al., 1982; Howarth et al., 1988b). Nonetheless, planktonic N2 fixation in temperate estuaries seems very low and is believed to be absent in estuaries exhibiting salinities >10 (Marino et al., 2002; Howarth and Marino, 2006; Conley et al., 2009). As an exception, the low-salinity (2–8) estuary of the Baltic Sea accommodates extensive summer blooms of heterocystous cyanobacteria (Stal et al., 2003). Yet, the high-sulfate concentrations that accompany more saline water may inhibit growth and N2 fixation by heterocystous cyanobacteria entering meso–polyhaline estuaries (Marino et al., 2002, 2003). The diazotrophic cyanobacteria currently believed to dominate oceanic N2 fixation, that is, members of the Trichodesmium genus, various cyanobacterial endosymbionts of diatoms, and unicellular diazotrophic cyanobacteria group A (UCYN-A) and B (Crocosphaera watsonii), have been found almost exclusively in tropical and subtropical waters, with growth and N2 fixation optima at temperatures >24 °C (Breitbarth et al., 2007; Church et al., 2008; Webb et al., 2009; Moisander et al., 2010). The UCYN-A group has, however, been observed in nifH cDNA clone libraries from waters as cold as 12 °C (Short and Zehr, 2007) and it has been associated with N2 fixation in temperate coastal waters (Rees et al., 2009; Mulholland et al., 2012). Yet, neither freshwater cyanobacterial diazotrophs nor oceanic cyanobacterial diazotrophs are thought to fix significant amounts of N in temperate estuaries.
New data on nitrogenase reductase (nifH) gene composition continue to expand the potential biome of marine N2 fixation both in terms of latitude and depth (Mehta et al., 2003; Farnelid et al., 2011; Fernandez et al., 2011; Hamersley et al., 2011) and diverse putative heterotrophic diazotrophs have been reported from temperate estuaries (for example, Affourtit et al., 2001; Jenkins et al., 2004; Farnelid et al., 2009), yet these organisms may thrive due to other traits than diazotrophy (Short and Zehr, 2007). They do, however, seem to fix N in estuarine sediments (Fulweiler et al., 2013) and heterotrophic nifH gene expression accompanies N2 fixation in the mesohaline deep waters of the Baltic Sea (Farnelid et al., 2013), indicating that these organisms could potentially be important diazotrophs in estuarine pelagic waters as well.
In this study, we investigated the temporal dynamics of N2 fixation in surface waters of two contrasting, temperate estuarine systems through monthly samples taken over a 1-year period. We determined the composition of present and active diazotrophs by Illumina amplicon sequencing of nifH genes and their RNA transcripts. Finally, we compared N2 fixation with a suite of environmental parameters and to the temporal dynamics of abundances of key groups of diazotrophs and their nifH transcription by quantitative PCR (qPCR) and reverse-transcription quantitative PCR (RT-qPCR).
Materials and methods
Sampling and environmental parameters
The two contrasting estuaries Roskilde Fjord (RF) and the Great Belt (GB) were sampled monthly throughout 2012. RF is a shallow (3-m mean depth), semi-enclosed and nutrient-rich estuary of 122 km2, which receives high-nutrient loads from its 1127 km2 drainage area (Flindt et al., 1997). The GB is part of the 37 333 km2 Kattegat and Belt Sea area (17-m mean depth), and is the main connection between the Baltic Sea and the North Sea and is therefore almost permanently stratified (Jørgensen et al., 2013). In GB, strong currents occur, changing between inflow of saline water from the north and inflow of less saline Baltic Sea water from the south.
Using 5-l free-flow bottles, surface water (1 m) was collected monthly from the stations in RF (55°42.00′N, 12°04.46′E, 4.8-m depth) and GB (55°30.27′N, 10°51.43′E, 35-m depth, Figure 1) in collaboration with the Danish Marine Monitoring Program. Samples were always obtained around noon (1000 to 1300 hours). In January, RF was inaccessible due to ice coverage and water was collected from a pier 1 km southeast of the station. Once on shore, aliquots for bacterial enumeration were fixed, bacterial production incubation initiated, and water for DNA and RNA extractions were filtered and preserved. The remaining analyses were conducted within 1–3 h of sampling.
Figure 1.
Map depicting the sampling locations. One sampling site was located in the shallow RF estuary and the other in the GB strait.
Nutrient concentrations were analyzed using a San++ Continuous Flow Analyzer (Skalar, Breda, The Netherlands) as previously described (Grasshoff, 1976). Detection limits were 0.3, 0.04 and 0.1 μmol N l−1 for NH4+, NO2− and NO3−, respectively. Chlorophyll a (Chl a) was measured fluorometrically (Holm-Hansen et al., 1965) and dissolved organic carbon (DOC) was measured using a TOC-Vcph analyzer (Shimadzu, Japan) according to Cauwet (1999). Bacterial production was estimated by 3H-thymidine incorporation (20 nmol l−1 final concentration, PerkinElmer, Waltham, MA, USA; Fuhrman and Azam, 1982) using microcentrifugation (Smith and Azam, 1992). Bacterial abundance was determined from samples fixed with glutaraldehyde (1% final concentration) using a FACSCanto II flowcytometer (BD Biosciences, Franklin Lakes, NJ, USA) according to Gasol and Del Giorgio (2000).
N2 fixation rate measurements
N2 fixation was measured by 15N-incorporation (Montoya et al., 1996) on all sampling dates, except for May where 15N2 gas was commercially unavailable. Enriched artificial sea water was used as a source of 15N rather than injecting 15N2 tracer gas directly into the samples, as this may underestimate N2 fixation (Mohr et al., 2010). The artificial sea water was prepared as previously described (Boström et al., 2007a). Following autoclaving, the hot artificial sea water was distributed into 50-ml borosilicate serum vials, which were sealed with butyl rubber stoppers and crimp sealed. In total, 1 ml 15N2 tracer gas (Campro Scientific, Veenendaal, The Netherlands; ⩾98%) was introduced into each vial using a gas-tight syringe and incubated horizontally for 24 h, at 150 r.p.m. rotation, which should ensure 90–100% tracer equilibration (Mohr et al., 2010). Sea water was transferred into 1.2-l polycarbonate bottles; filling the bottles three-fourth of the way up before adding 50 ml 15N2-enriched artificial sea water. Bottles were then filled with sea water until the formation of a meniscus and capped. Six replicate bottles were incubated for 24 h at in situ temperature ±1.5 °C; three replicates under in situ light conditions (Supplementary Table S1) and three replicates in darkness. Following incubation, 600 ml from each replicate was filtered onto pre-combusted (450 °C, 8 h) GF/F filters (Whatman, GE Healthcare, Little Chalfont, UK) and the remaining 600 ml from each replicate was filtered through a 10-μm polycarbonate filter (GE Water & Process Technologies, Trevose, PA, USA) and onto pre-combusted GF/F filters for differentiation of N2 fixation rates in the respective size fractions. Six controls were done at each sampling time point: two background δ15N controls, two un-amended controls filtered at the end of incubation and two amended controls filtered immediately after tracer addition. Filters were stored at −20 °C and dried at 60 °C before analysis at the Laboratory of Applied Physical Chemistry, Gent, Belgium on an isotope ratio mass spectrometer (PDZ Europa, Northwich, UK).
Extraction of nucleic acids and synthesis of cDNA
Water for DNA extraction was separated into size fractions. In total, 2 l of sea water was filtered onto a 10-μm polycarbonate filter (GE Water & Process Technologies) and 1 l of the filtrate was filtered onto a 0.22-μm Supor membrane filter (PALL Corporation, Port Washington, NY, USA). The filters were stored at −20 °C in 2-ml cryotubes containing 1 ml sucrose lysis buffer (400 mM NaCl, 750 mM sucrose, 20 mM EDTA, 50 mM Tris-HCL, pH 8). DNA was extracted using a phenol/chloroform-based protocol (Boström et al., 2004) and quantified (PicoGreen, Molecular Probes, Invitrogen, Eugene, OR, USA).
Water samples for RNA extraction were fixed immediately after sampling using stop solution (5% water-saturated phenol in absolute ethanol, Feike et al., 2012). Size fractionation was done as described for DNA samples, and filters were stored at −80 °C in 2-ml cryotubes containing 0.5 ml RNAlater (Ambion, Carlsbad, CA, USA). RNA was extracted (RNeasy kit; Qiagen, Hilden, Germany) using a slightly modified protocol (Boström et al., 2007b) and quantified (NanoDrop, 2000; Thermo Scientific, Waltham, MA, USA). cDNA was synthesized using TaqMan reverse-transcription reagents (Applied Biosystems, Foster City, CA, USA) and the nifH3 reverse primer (Zehr and McReynolds, 1989). Reverse transcriptase-free control reactions were included for the verification of complete DNA digestion during RNA extraction. Nested nifH PCR reactions with these were always negative.
nifH amplicon sequencing and sequence analysis
nifH amplicons were obtained from 56 samples, 28 from each station representing DNA and cDNA from the two size fractions in each of the 7 months: February, March, April, June, July, August and October. In total, 10 ng of DNA or cDNA corresponding to 4 ng RNA was added to Pure Taq Ready-To-Go PCR Beads (GE Healthcare) along with the nifH3 and nifH4 primers in a nested PCR approach (Zehr and Turner, 2001). Illumina indices (Supplementary Table S2) were added to amplicons in the second PCR round, which were done in triplicates for each sample. The triplicate PCR products were pooled, purified (AmPure, Agencourt Bioscience Corporation, Beverly, MA, USA), quantified (PicoGreen), and 30 ng from each sample were pooled and subjected to paired-end Illumina sequencing (MiSeq, Berlin Center for Genomics in Biodiversity Research, Berlin, Germany).
Sequence reads were de-multiplexed using CASAVA 1.8.2, assembled in Mothur 1.32.0 (Schloss et al., 2009), trimmed to 216 nucleotides and screened for ambiguous base calls. Poorly assembled sequences and sequences with frame shifts, that is, exhibiting poor alignment to the nifH seed sequences in the functional gene pipeline and repository database (http://fungene.cme.msu.edu/), were excluded. Chimeras were removed using UCHIME through Mothur. The sequences were clustered at 97% nucleotide similarity and representative sequences from the 84 most abundant operational taxonomic units (OTUs) were aligned in MEGA 5.05 (Tamura et al., 2011). The closest cultivated representatives were identified using the BLAST resource on NCBI and added to the alignment.
Inverse Simpson indices were used as a measure of alpha diversity, and subsampling down to 482 sequences per sample was chosen to accommodate the sample with the lowest number of sequences. Yue & Clayton measures of dissimilarity between samples and non-metrical multidimensional scaling coordinates were calculated in Mothur. Subsampling to 1200 sequences was applied and samples containing less sequences were excluded from the analysis. Non-metrical multidimensional scaling coordinates were plotted using KiNG through the make_3d_plots.py script in Qiime (Caporaso et al., 2010). The significance of the divergence between samples explained by size fraction, sampling station, N2 fixation rates or sample type (DNA or cDNA) was done using analysis of molecular variance. OTUs occurring in more than one cDNA sample from time points exhibiting high N2 fixation rates (>10 nmol N l−1 per day) were identified for each station using the Venn function in Mothur. Log10+1-transformed abundances of these OTUs were used to produce a heatmap using the CIMminer resource (http://discover.nci.nih.gov/cimminer/).
Sequences were uploaded to the Sequencing Read Archive database on NCBI (Accession number SRP038933).
Quantification of nifH genes and their expression
Five abundant OTUs (OTU0001, OTU0002, OTU0005, OTU0009 and OTU0014) were selected for quantification in DNA and RNA samples using qPCR and RT-qPCR. Primers and TaqMan probes were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/, Table 1). Specificities of the assays were verified using Primer-BLAST and by PCR tests against six available non-target DNA templates; the four non-target standards above, a Rhodopseudomonas nifH template (Farnelid et al., 2014), and a Clostridium-like Cluster III template (Bentzon-Tilia et al., 2014). The qPCR standards were synthesized and cloned into the pUC57 vector at GenScript (Piscataway, NJ, USA) and linearized by HindIII digestion. Reaction mixtures, thermal cycling settings, standard preparations and inhibition tests were done as previously described (Farnelid et al., 2013), supplementing the reaction mixtures with 1 mM MgCl2 and 0.2 vol/w % BSA. DNA and cDNA from the two size fractions were pooled according to the concentrations of DNA and RNA observed in the two size fractions after extraction. A total of 5 ng of DNA and cDNA corresponding to 4 ng RNA was added to the respective reactions. The obtained gene and transcript abundances were corrected for the PCR efficiency (Table 1).
Table 1. Primers and probes designed in this study for quantitative PCR and reverse-transcription quantitative PCR where R2 is the correlation coefficient of the standard curve and E is PCR efficiency.
| Target | Forward primer (5′-3′) | Reverse primer (5′-3′) | Probe (5′-3′) | R2 | E (%) |
|---|---|---|---|---|---|
| OTU0001 (Pseudomonas) | CAAGGCCCAGAACACCATCA | TTCGAGGAAGTTGATCGCGG | AAATGGCGGCGGAGGCCG | 0.932–0.989 | 97–104 |
| OTU0002 (UCYN-A) | AGCTGCAGAAAGAGGAACTGT | GGCTCAGGACCACCAGATTC | TCCTGGTTACAACAACGTTTTATGTG | 0.968–0.971 | 90–93 |
| OTU0005 (Anabaena) | TCCAAGGCTCAAACAACCGT | CAACCTACACCGGGTTCTGG | CCGCGACGTTAAATGCGTGGAATC | 0.926–0.935 | 95–106 |
| OTU0009 (Klebsiella) | ACCATTATGGAGATGGCCGC | GCCAATTTGCAGCACGTCTT | TCGGCTCGGTCGAGGACCTCGA | 0.971–0.972 | 92–100 |
| OTU0014 (Bradyrhizobium) | GATGCTTGGTGGATTGGCAC | ACAGAGTTCCGCTAAAGCCC | TGCGAGAAGAAGGCGAGGACGTCGAG | 0.949–0.995 | 92–93 |
Results
Environmental parameters
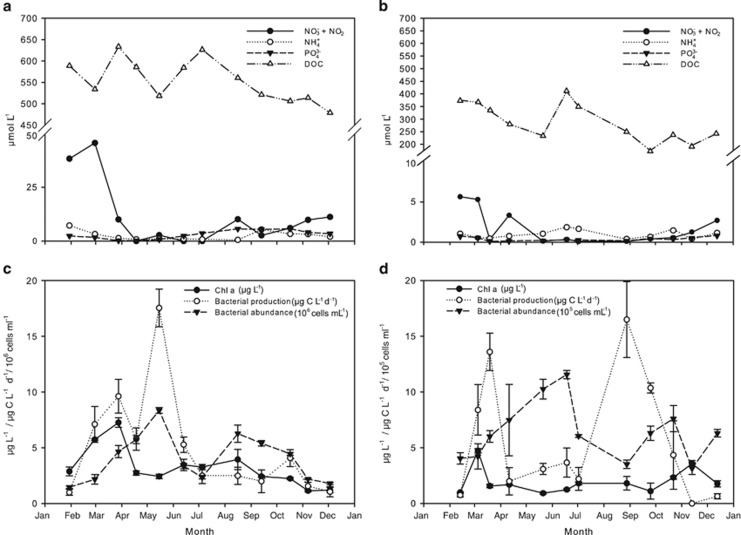
In RF, salinity and water temperature ranged from 12–15 and from −0.6 to 20 °C, respectively. In GB, salinities ranged from 11–24 with reduced values of 11 in February, 13–17 in June–August and 15 in October, suggesting extensive Baltic Sea water outflow in these periods. Water temperatures ranged from 0.4–17 °C in GB. In winter, the concentration of inorganic N was almost 10-fold higher in RF than in GB with combined NO2− and NO3− concentrations reaching values of 45.9 μmol N l−1 and 5.55 μmol N l−1, respectively (Figures 2a and b). NH4+ levels were 0.47–7.2 μmol N l−1 in RF and 0.34–1.9 μmol N l−1 in GB over the year (Figures 2a and b). Spring blooms appeared during February and March with Chl a concentrations reaching 7.2 μg l−1 in RF and 4.7 μg l−1 in GB (Figures 2c and d). Blooms lead to depletion of inorganic N, which increased again from September. PO43− concentrations displayed the same overall patterns as inorganic N (Figures 2a and b). The molecular N:P ratios exceeded the Redfield ratio in winter in RF, but decreased to <1 after the spring bloom with dissolved inorganic N concentrations <1 μmol l−1 in April, June and July. In GB, a N:P ratio of 25 was observed in June, but ratios were otherwise in the range of 2–10 and dissolved inorganic N concentrations were <1 μmol l−1 most of the year, collectively suggesting that N availability limits primary production in summer at both sites. The level of DOC was higher in RF (478–626 μmol l−1) than in GB (175–410 μmol l−1, Figure 2a and b). In RF, bacterial production increased from February to April and from May to June where it peaked at 18 μg C l−1 per day (Figure 2c). In GB, peaks were observed in March (14 μg C l−1 per day) and in August (16 μg C l−1 per day; Figure 2d). Bacterial abundances were highest in summer with RF exhibiting abundances about 10-fold higher than GB (Figures 2c and d).
Figure 2.
Seasonal changes in environmental data collected in RF (left panels) and the GB (right panels). Concentrations of inorganic nutrients and DOC for RF (a) and GB (b). (c) and (d) show concentrations of Chl a, bacterial production and bacterial abundance in RF and in GB, respectively. Note the difference in bacterial abundance units between stations. Error bars represent standard deviations.
N2 fixation
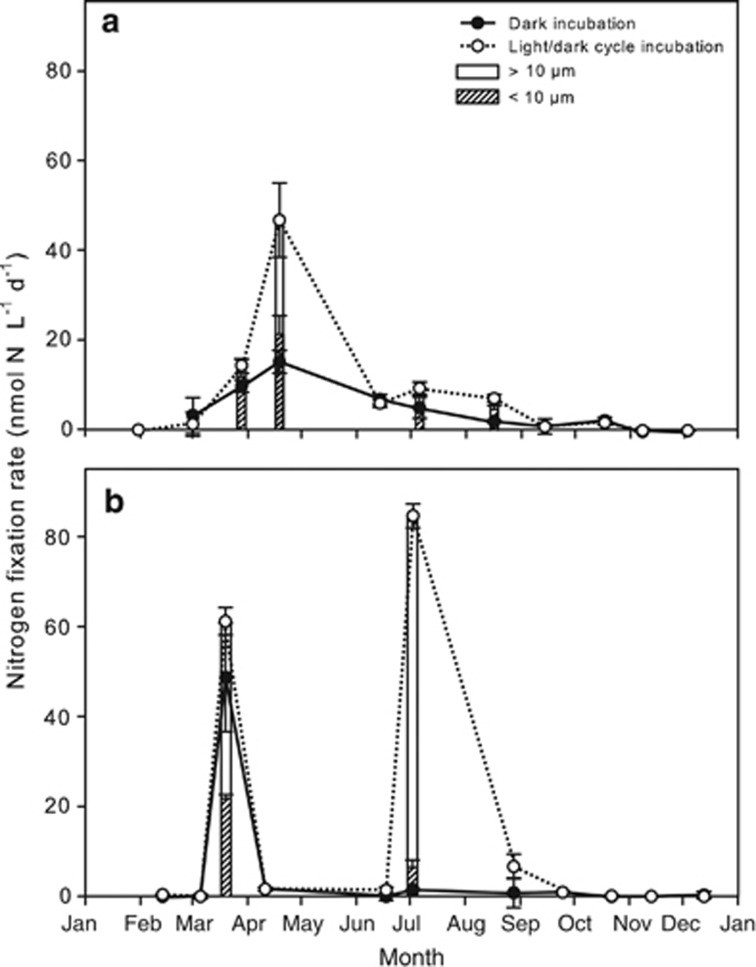
In RF, N2 fixation increased from the beginning of March with a pronounced N2 fixation peak in April at 47 nmol N l−1 per day (Figure 3a) coincided with the demise of the spring bloom and a 100-fold drop in inorganic N. Following the peak, N2 fixation stayed at 6.0–10 nmol N l−1 per day in June–August before decreasing in fall to 1.1–2.0 nmol N l−1 per day and being undetectable throughout the remainder of the year. N2 fixation rates in bottles incubated in darkness exhibited the same pattern as those exposed to a diurnal light cycle, but rates were lower. At the onset of the N2 fixation peak in late March, 76% of the fixed N was assimilated in the <10-μm size fraction. This changed to 46% in April and 61% in July and August. The annual areal input of fixed N at this shallow (4.8 m) station was 17 mmol N m−2 per year.
Figure 3.
N2 fixation measured throughout the year in RF (a) and the GB (b). Total N2 fixation in diurnal light cycle treatments is depicted as open circles, whereas closed circles represent the equivalent fixation in bottles incubated in complete darkness. N2 fixation data from the two different size fractions (>10 μm or <10 μm) are depicted as bars where available. Error bars indicate standard deviations.
In GB, N2 fixation in light was detectable from February to September with distinct peaks in March and July where N2 fixation rates reached 63 and 83 nmol N l−1 per day, respectively (Figure 3b). In the parallel dark incubations, a peak was also observed in March, but like for RF, the rates were lower than in the diurnal light cycle incubations. No peak was observed in the parallel dark incubations from July. The proportion of N fixed in the smaller size fraction was higher during the N2 fixation peak in March (33%) than in July (8%). The upper mixed layer was 13-m deep, and the integrated areal input of fixed N in this layer was 61 mmol N m−2 per year.
A negative correlation was observed between N2 fixation and NH4+ concentrations in RF (P=0.009, n=11, Supplementary Table S3), but not in GB. Here, N2 fixation was significantly negatively correlated with combined NO2− and NO3− (P=0.017, n=11, Supplementary Table S3). In RF, significant positive correlations were found between N2 fixation and DOC (P=0.005, n=11), bacterial production (P=0.015, n=11) and bacterial abundance (P=0.021, n=11; Supplementary Table S3). N2 fixation was not correlated with temperature or Chl a at either of the sampling sites.
Composition of the diazotrophic communities and transcript abundances
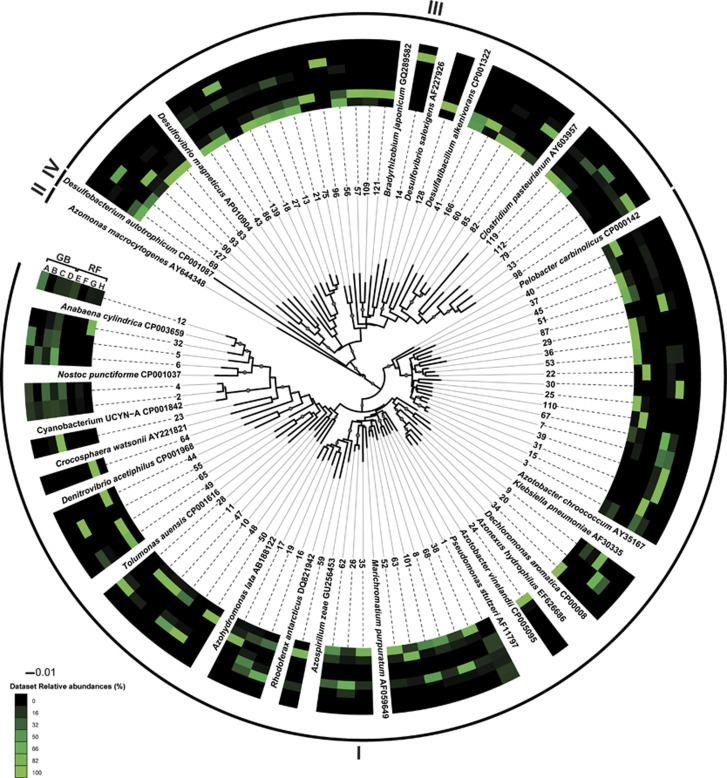
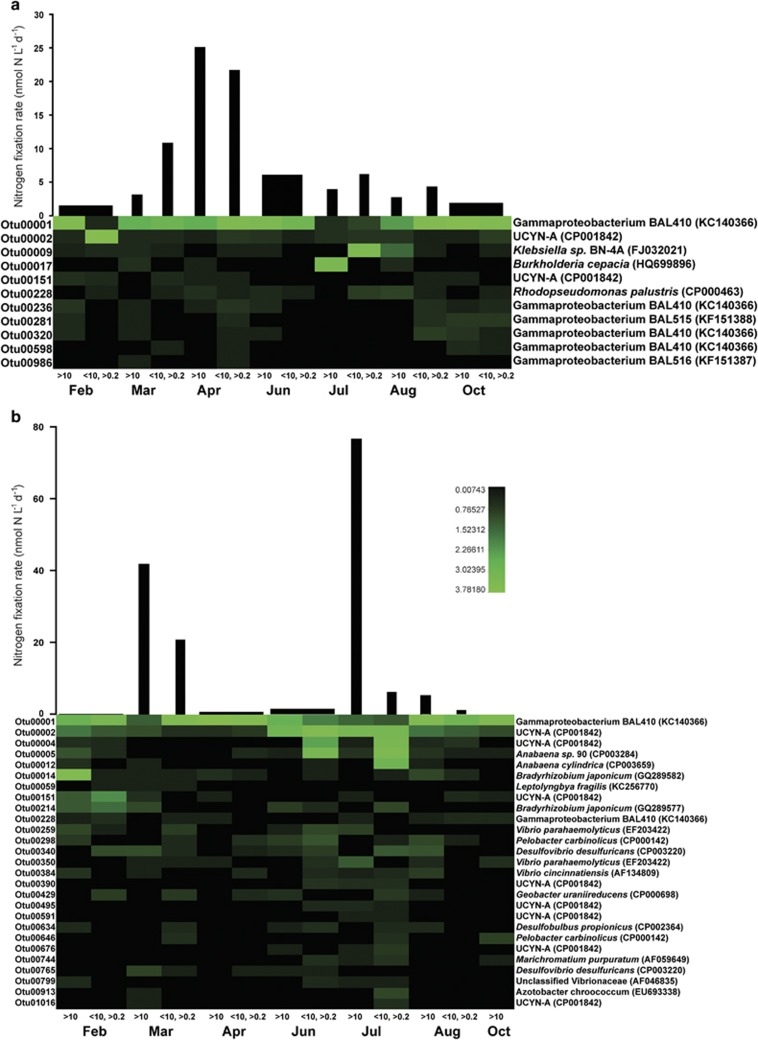
A total of 455 596 nifH sequences were obtained after initial quality check and assembly. Further evaluation of assembly quality and the occurrence of chimeric sequences reduced the number of sequences to a total of 287 494. Clustering the sequences at 97% similarity yielded 11 534 OTUs after elimination of singletons. The acquired sequences covered 87–99% of the nifH gene diversity in the samples. The 84 most abundant OTUs (⩾250 sequences each) accounted for >80% of all sequences and were exclusively affiliated with nifH clusters I and III (Figure 4). The most abundant OTU, OTU0001, was related to the gammaproteobacterium Pseudomonas stutzeri (cluster I; 99% nucleotide sequence similarity and 99% amino-acid sequence similarity) and accounted for 22% of all sequences. It was not recovered from all time points, but was present in both size fractions at both stations in DNA and RNA samples. The relative abundance of OTU0001 was highest in RNA samples (Figure 4). Eighteen OTUs were affiliated with the Geobacter/Pelobacter sub-group of Cluster I. These OTUs were recovered from both sampling sites, yet they all had highest relative abundances in DNA and were almost exclusively detected here. In total, 30 cluster III OTUs were detected at both stations. The majority of these exhibited a higher relative abundance in RF and most were exclusively recovered from this station. They showed higher relative abundance in DNA compared with RNA with OTU0014, OTU0090 and OTU0139 being the only three of the 30 cluster III OTUs exhibiting relative abundances of ⩾8% in RNA (Figure 4). Seven OTUs were affiliated with cluster I cyanobacteria (Figure 4). Most of these were related to the UCYN-A (16% of all sequences; 99% nucleotide sequence similarity and 99% amino-acid sequence similarity) and recovered from both stations, yet, exhibiting a higher relative abundance in GB (23%) compared with RF (5%). Similarly, the genus Anabaena showed a higher relative abundance in GB (8%) than in RF (1%). The fact that a large fraction of the diazotrophic community did not express the nifH gene at the seven selected time points was mirrored in the alpha diversity (Inverse Simpson). In DNA from RF, values ranged from 5.6–68 compared with 1.0–3.9 for RNA (Supplementary Figure S1). In GB, nifH diversity was higher in the RNA and lower in the DNA compared with RF.
Figure 4.
Neighbor-joining tree of representative sequences from the 84 most abundant OTUs (each representing ⩾250 sequences) resulting from the 97% nucleotide sequence similarity clustering. Each of the OTUs are designated a number chosen by Mothur. Bootstrap values ⩾50% are represented by size-proportional gray circles in the tree. The heatmap indicates the relative abundance of sequences associated with a given OTU in each of the following types of samples: GB, RNA, <10 μm (A), GB, RNA, >10 μm (B), GB, DNA, <10 μm (C), GB, DNA, >10 μm (D), RF, RNA, <10 μm (E), RF, RNA, >10 μm (F), RF, DNA, <10 μm (G), RF, DNA, >10 μm (H). The affiliations of the OTUs with the canonical nifH clusters (Chien and Zinder, 1996) is indicated by roman numerals.
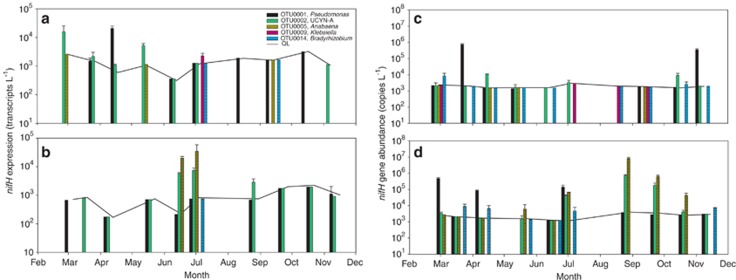
The molecular variance analysis showed a large discrepancy between the diazotrophic community present and the fraction of the community expressing nifH (P<0.001, n=52, Supplementary Figure S2a) and different community compositions at the two sites (P=0.004, n=52, Supplementary Figure S2b). To identify differences in the active diazotrophic communities, a core nifH-expressing microbiome (CExM) was defined for each station (i.e. OTUs recovered in more than one of the four samples per station exhibiting N2 fixation >10 nmol N l−1 per day; Supplementary Figure S3). In RF, the Pseudomonas-like OTU0001 dominated the CexM at most time points (Figure 5a). Most OTUs from the CexM were related to Pseudomonas-like gammaproteobacteria, yet their relative expression level seemed reduced during summer. To gain a quantitative measure of the nifH transcription of OTU0001, it was quantified by RT-qPCR (Table 1). Indeed, it was detectable in RNA at 7 of 10 dates from February to November (Figure 6) and transcripts were quantifiable in March (1.5 × 103 l−1) and April (2.1 × 104 l−1), coinciding with the N2 fixation peak (Figure 6a). Two OTUs similar to the UCYN-A group (OTU0002 and OTU0151) were also identified as belonging to the CexM and displayed roughly the same pattern as the Pseudomonas-like OTUs (Figure 5a). RT-qPCR quantification of the most abundant of these (OTU0002) showed highest activity in spring as well, with transcript abundances of 1.1 × 103–1.6 × 104 l−1 (Figure 6a). In July and August, the relative abundance of OTUs related to other proteobacteria increased in cDNA (Figure 5a). RT-qPCR data showed that transcripts of the Klebsiella-like OTU, OTU0009, was undetectable at all but one sampling date in July where it accounted for 2.3 × 103 transcripts per litre.
Figure 5.
The relative abundances of OTUs identified to be part of the core nifH-expressing microbiome over time in RF (a) and in the GB (b). The OTU-names are given to the left of the heatmap and the closest cultivated relative is indicated to the right of the heatmap. For reference, the volumetric N2 fixation rates showed in Figure 3 are given above the heatmaps. These rates are split into size fractions where available.
Figure 6.
Results from the reverse-transcription quantitative PCR (RT-qPCR) and quantitative PCR (qPCR) assays performed on RNA and DNA samples taken from February to November in RF and from March to November in the GB. RT-qPCR results from RF (a) and the GB (b). qPCR results from RF (c) and the GB (d). The quantification limit (QL) is defined as the number of gene copies and transcripts needed in situ to match the lowest standard; for example, 10 target transcripts in 4 ng of RNA. QL was calculated for all samples and is depicted as a solid black line. OTUs that were detectable, but not quantifiable, are depicted as bars without error bars at the QL line. Error bars indicate standard deviations.
In GB, a more complex CexM was present, representing 27 OTUs (Figure 5b, Supplementary Figure S2b). During the peak in N2 fixation in July, the relative proportion of nifH transcripts of the OTUs belonging to the CexM increased. These OTUs were related to the cyanobacteria UCYN-A and Anabaena, but also to several non-cyanobacterial genera. Four of the five OTUs that were included in the RT-qPCR assay were detectable in July, including OTUs related to Pseudomonas, UCYN-A, Anabaena and Bradyrhizobium. UCYN-A and Anabaena-like OTUs were, however, the only two quantifiable OTUs with 1.6 × 103 and 3.4 × 104 transcripts per litre, respectively (Figure 6b). Many of the same OTUs were detected during the N2 fixation peak in early spring, but a distinct change in the relative abundances of OTUs in the CexM was not observed at this time point. The most active OTUs, up to and during the spring peak, seemed to be the ones related to Pseudomonas, UCYN-A, Bradyrhizobium, Vibrio and Desulfovibrio (Figure 5b). In the RT-qPCR assay, OTUs related to Pseudomonas and UCYN-A were detected during spring but below the level of quantification (Figure 6b).
nifH gene abundances
In RF, the OTU related to Pseudomonas (OTU0001) reached highest abundance at the onset of the N2 fixation peak with 7.1 × 105 gene copies per litre (Figure 6c). It was not quantifiable in the summer, but increased in abundance again in autumn. During the N2 fixation peak in April, four of the five targeted OTUs were detectable by qPCR, but only the UCYN-A-like OTU0002 was quantifiable with 1.1 × 104 gene copies per litre (Figure 6c). The Klebsiella-like OTU (OTU0009) was detected in February and again from July–September exhibiting relatively low abundances (≈103 gene copies per litre).
As in RF, the Pseudomonas-like OTU reached highest abundances in March in GB just before the spring peak in N2 fixation with 4.8 × 105 gene copies per litre (Figure 6d). During this peak, all but OTU0009 was detectable (this OTU was never detected at this station). The Bradyrhizobium-like OTU0014 was, however, the only quantifiable OTU during the peak where it reached gene abundances of 9.0 × 103 gene copies per litre (Figure 6d). During the summer peak in N2 fixation, all OTUs, except for OTU0009, increased in abundance with OTU0001 reaching the highest abundance with 1.4 × 105 gene copies per litre (Figure 6d). The two cyanobacterial OTUs reached their highest abundance just after the N2 fixation peak in July with 7.6 × 105 and 8.1 × 106 gene copies per litre for OTU0002 (UCYN-A) and OTU0005 (Anabaena), respectively. These abundances decreased gradually following the peak (Figure 6d).
Discussion
N2 fixation in meso–polyhaline estuarine waters is believed to be negligible due to the high land runoff discharges of N and the presumed absence or inactivity of diazotrophic bacteria (Marino et al., 2002; Howarth and Marino, 2006; Conley et al., 2009). In contrast, we report substantial N2 fixation in surface waters of two contrasting temperate estuaries. In fact, the measured rates are among the highest reported for pelagic systems, reaching 83 nmol N2 l−1 per day in GB. This is higher than, for example, measurements from oceanic Trichodesmium blooms (Capone et al., 1997) and from productive, temperate coastal waters, for example, the North American east coast (Mulholland et al., 2012) and the western English Channel (Rees et al., 2009), but are similar to those measured in the Baltic Sea (Wasmund et al., 2005). Moreover, our molecular analyses revealed that the relative importance of the organisms responsible for the N2 fixation, heterotrophic, photoheterotrophic and heterocystous cyanobacteria, differed in space and time. This implies that heterocystous cyanobacteria are not always a pre-requisite for pelagic N2 fixation in temperate estuaries as previously assumed (Howarth et al., 1988b) and that significant N2 fixation takes place in these environments.
In both estuaries, the N2 fixation was accompanied by diverse diazotrophic bacteria, mainly related to (photo)heterotrophic bacteria, including the supposedly tropical to subtropical group of unicellular cyanobacteria, UCYN-A (Mazard et al., 2004; Moisander et al., 2010), which has recently been shown to be a metabolically reduced symbiont of a unicellular prymnesiophyte (Thompson et al., 2012). Interestingly, nifH transcripts related to this group have been reported from Chesapeake Bay at water temperatures as low as 12 °C (Short and Zehr, 2007), yet, here we measured nifH transcript abundances of >104 l−1 in RF in February at only 2.5 °C. Hence, our observations suggest that the geographic range of UCYN-A, conceivably of its eukaryotic host, and the associated N2 fixation extends to temperate regions, even in winter time.
The different physical properties of the two systems were reflected in community composition, with RF exhibiting a more diverse collection of diazotrophs. The cluster III diazotrophs, putative anaerobes, were particularly prevalent in the shallow, more heterotrophic RF, whereas almost all the cyanobacterial OTUs showed highest relative abundance in GB. Hence, we speculate that the surface community of RF was influenced by re-suspended sediment bacteria, whereas the GB community was influenced by the changing currents (see below).
Our data suggest that the genetic potential to fix N2 was, in many cases, not mirrored in the diversity of the genes transcribed; especially in the shallow RF in late fall and winter where the water column is well-mixed. Likewise for the temperate Chesapeake Bay, clone libraries indicated that only few phylotypes expressed nifH, and it was suggested that the distribution of indigenous diazotrophs is not always due to the selective advantage of diazotrophy per se (Zehr and Paerl, 2008). Yet, by applying high temporal sampling resolution and sequencing depth, we show that a significant fraction of the diverse heterotrophic diazotrophs present in such estuarine systems indeed occasionally expresses nifH. Cyanobacterial diazotrophs may exhibit diel patterns in nifH gene expression (for example, Shi et al., 2010). Sampling time can, therefore, have affected the relative proportions of transcripts from the respective functional types of diazotrophs in our data set. However, data on diurnal variation of nifH gene expression in heterotrophs is scarce (but see Church et al., 2005; Moisander et al., 2014); hence, at present, the consequences of the sampling time used here remains elusive. Future studies with increased diurnal sampling resolution would conceivably further advance our understanding of the explicit coupling between the key diazotrophs present and their nifH gene expression.
In RF, the CexM consisted exclusively of heterotrophic and photoheterotrophic diazotrophs. Their importance for N2 fixation was substantiated by the fact that significant N2 fixation was measured in dark incubations and in the <10-μm size fraction. N2 fixation was stimulated by light, but as photoheterotrophic groups like UCYN-A and Rhodopseudomonas-like organisms did express nifH at this site, light would indeed be expected to stimulate N2 fixation. Due to co-variation of environmental parameters over a season, correlation analyses should be interpreted with caution. Nevertheless, we see the positive correlations of N2 fixation with DOC, bacterial abundance and bacterial production as further support for the conclusion that heterotrophic organisms were the principal N2-fixing organisms in RF. In GB, the March peak in N2 fixation was accompanied by a CexM relatively similar to the one in RF, yet it was more diverse and qPCR of transcripts of the five predominant OTUs were below the level of quantification. OTUs related to UCYN-A and Pseudomonas were, however, detectable both by qPCR and RT-qPCR at the time of the spring peak in N2 fixation, and it is likely that these (photo)heterotrophic organisms played an important role at this time where currents moved polyhaline water (>18) in from the north. In fact, the Pseudomonas-like OTU reached the highest gene abundances (105–106 copies per litre) in spring at both stations. During the spring peaks, however, abundances of all quantified OTUs were generally low. Whether they were sufficient to account for the measured N2 fixation rates is difficult to determine, as empirical data on the N2 fixation potential of marine, heterotrophic diazotrophs and of the UCYN-A group are not currently available. However, adopting recently published assumptions about cell-specific N2 fixation rates in different groups of marine diazotrophs (Turk-Kubo et al., 2014), the five quantified OTUs could collectively account for N2 fixation rates in the range of 5–10 nmol N2 l−1 per day during the N2 fixation peak in RF. In contrast to the spring peaks, the summer N2 fixation peak was mainly associated with the >10-μm size fraction and a very low rate of N2 fixation in dark incubations, as would be expected for heterocystous cyanobacteria (Fay, 1976). Indeed, the nifH gene expression of Anabaena peaked at this time and nifH gene abundances from this group of organisms increased during the N2 fixation peak in summer, reaching the highest level observed in this study (8.1 × 106 copies per litre) just after the peak. It should, however, be noted that cyanobacteria, like the Anabaena genus, host multiple chromosomes per cell (Simon, 1977), and hence this high gene abundance does not translate directly into cell numbers. It is conceivable that the Anabaena-like OTUs in the Belt Sea area are advected from the Baltic Sea, since the summer N2 fixation peak coincided with a period of reduced salinity (13–17) presumably driven by a northerly current of Baltic Sea water. The salinity in GB should, however, swiftly constrain the activity of Anabaena (Marino et al., 2002; Moisander et al., 2002). Hence, the observation of nifH gene-expressing Anabaena in the Belt Sea area is surprising. Taken together, we conclude that heterotrophic and photoheterotrophic diazotrophs were responsible for N2 fixation in both estuaries following the spring bloom, while the heterocystous Anabaena was responsible for the pronounced peak in N2 fixation during summer in the GB.
Our study reports for the first time significant measured rates of pelagic N2 fixation in Danish coastal waters, even in two contrasting systems. Assuming that the sampled station is representative for the whole RF estuary, the annual N2 fixation input amounts to about 18 tons. In comparison, the combined RF/Isefjord system received 688 tons of N from land in 2011 (Pedersen et al., 2013). If RF receives half of this N load, pelagic N2 fixation corresponds to 5% of the N from land. It is worth noting though that part of the input from land will be bound as refractory-dissolved organic N (Jørgensen et al., 2013), and that the biological impact of N from N2 fixation may be relatively larger. It should be noted, though, that extrapolating N2 fixation measured at a single station to the basin scale is speculative, since the spatial variability in the RF estuary is not known. It is even more complicated to calculate the contribution from N2 fixation in GB due to the dynamic nature of this system. Nonetheless, the GB station is part of the 37 333 km2 Belt Sea and Kattegat area, which receives 63 000 tons N per year from land (Jørgensen et al., 2013), equal to 121 mmol N m−2 per year. In comparison, the mean rate of N2 fixation measured in this study was 61 mmol N m−2 per year, corresponding to 34 000 tons N per year for the Belt Sea and Kattegat area. Hence, it seems that in GB, pelagic N2 fixation could represent a substantial input. However, the rates are probably much lower in the northern and more saline Kattegat, and as the N fixation is mainly restricted to two days, any extrapolation is very uncertain. Nevertheless, it is roughly consistent with the annual input from N2 fixation of 11 000 tons N suggested from a recent modeling study (Jørgensen et al., 2013).
The present study highlights the need for more N2 fixation measurements to get confident measures of the contribution of pelagic N2 fixation to the systemic N budgets in the estuaries examined here. Taken together with the recent reports of significant N2 fixation in estuarine sediments (Fulweiler et al., 2013; Brown and Jenkins, 2014), the basin-scale implications of estuarine N2 fixation are likely considerable, suggesting that a reevaluation of the significance of N2 fixation in temperate estuaries and coastal waters is warranted. Moreover, the N2 fixation by heterotrophic, photoheterotrophic and heterocystous bacteria suggested from the present study highlights the importance of integrating all these three fundamentally different groups in future surveys of estuarine diazotrophy.
Acknowledgments
We thank Pia Moisander, Hanna Farnelid, and three anonymous reviewers for constructive comments improving the manuscript. We thank the Danish Nature Agency for including us in the sampling program. This work was supported by grants 09-066396 and 11-105450 from The Danish Council for Independent Research, Natural Sciences to LR and grant 067259 from the Strategic Research Council of Denmark to SM and HK-L.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Affourtit J, Zehr JP, Paerl HW. Distribution of nitrogen-fixing microorganisms along the Neuse River Estuary, North Carolina. Microb Ecol. 2001;41:114–123. doi: 10.1007/s002480000090. [DOI] [PubMed] [Google Scholar]
- Bentzon-Tilia M, Farnelid H, Jürgens K, Riemann L. Cultivation and isolation of N2-fixing bacteria from suboxic waters in the Baltic Sea. FEMS Microbiol Ecol. 2014;88:358–371. doi: 10.1111/1574-6941.12304. [DOI] [PubMed] [Google Scholar]
- Boström KH, Riemann L, Kühl M, Hagström Å. Isolation and gene quantification of heterotrophic N2-fixing bacterioplankton in the Baltic Sea. Environ Microbiol. 2007;9:152–164. doi: 10.1111/j.1462-2920.2006.01124.x. [DOI] [PubMed] [Google Scholar]
- Boström KH, Riemann L, Zweifel UL, Hagström Å. Nodularia sp. nifH gene transcripts in the Baltic Sea proper. J Plankton Res. 2007;29:391–399. [Google Scholar]
- Boström KH, Simu K, Hagström Å, Riemann L. Optimization of DNA extraction for quantitative marine bacterioplankton community analysis. Limnol Oceanogr-Meth. 2004;2:365–373. [Google Scholar]
- Boynton WR, Kemp WM, Keefe CW.1982A comparative analysis of nutrients and other factors influencing estuarine phytoplankton productionIn: Kennedy VS, (ed)Estuarine Comparisons Academic Press: New York; 69–90. [Google Scholar]
- Breitbarth E, Oschlies A, LaRoche J. Physiological constraints on the global distribution of Trichodesmium—effect of temperature on diazotrophy. Biogeosciences. 2007;4:53–61. [Google Scholar]
- Brown SM, Jenkins BD.2014Profiling gene expression to distinguish the likely active diazotrophs from a sea of genetic potential in marine sediments Environ Microbiole-pub ahead of print 25 March 2014doi: 10.1111/1462-2920.12403 [DOI] [PMC free article] [PubMed]
- Capone DG, Zehr JP, Paerl HW, Bergman B, Carpenter EJ. Trichodesmium, a globally significant marine cyanobacterium. Science. 1997;276:1221–1229. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauwet G.1999Determination of dissolved organic carbon (DOC) and nitrogen (DON) by high temperature combustionIn: Grasshoff K, Kremling K, Erhard M, (eds)Methods of Seawater Analyses Wiley-VCH; 407–420. [Google Scholar]
- Chien YT, Zinder SH. Cloning, functional organization, transcript studies, and phylogenetic analysis of the complete nitrogenase structural genes (nifHDK2) and associated genes in the archaeon Methanosarcina barkeri 227. J Bacteriol. 1996;178:143–148. doi: 10.1128/jb.178.1.143-148.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church MJ, Bjorkman KM, Karl DM, Saito MA, Zehr JP. Regional distributions of nitrogen-fixing bacteria in the Pacific Ocean. Limnol Oceanogr. 2008;53:63–77. [Google Scholar]
- Church MJ, Short CM, Jenkins BD, Karl DM, Zehr JP. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl Environ Microbiol. 2005;71:5362–5370. doi: 10.1128/AEM.71.9.5362-5370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley DJ, Paerl HW, Howarth RW, Boesch DF, Seitzinger SP, Havens KE, et al. ECOLOGY controlling eutrophication: nitrogen and phosphorus. Science. 2009;323:1014–1015. doi: 10.1126/science.1167755. [DOI] [PubMed] [Google Scholar]
- Farnelid H, Andersson AF, Bertilsson S, Abu Al-Soud W, Hansen LH, Sørensen S, et al. Nitrogenase gene amplicons from global marine surface waters are dominated by genes of non-cyanobacteria. PLoS One. 2011;6:e19223. doi: 10.1371/journal.pone.0019223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnelid H, Bentzon-Tilia M, Andersson AF, Bertilsson S, Jost G, Labrenz M, et al. Active nitrogen-fixing heterotrophic bacteria at and below the chemocline of the central Baltic Sea. ISME J. 2013;7:1413–1423. doi: 10.1038/ismej.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnelid H, Harder J, Bentzon-Tilia M, Riemann L.2014Isolation of heterotrophic diazotrophic bacteria from estuarine surface waters Environ Microbiole-pub ahead of print 16 December 2013doi: 10.1111/1462-2920.12335 [DOI] [PubMed]
- Farnelid H, Oberg T, Riemann L. Identity and dynamics of putative N2-fixing picoplankton in the Baltic Sea proper suggest complex patterns of regulation. Environ Microbiol Rep. 2009;1:145–154. doi: 10.1111/j.1758-2229.2009.00021.x. [DOI] [PubMed] [Google Scholar]
- Fay P. Factors influencing dark nitrogen fixation in a blue-green alga. Appl Environ Microbiol. 1976;31:376–379. doi: 10.1128/aem.31.3.376-379.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feike J, Jürgens K, Hollibaugh JT, Kruger S, Jost G, Labrenz M. Measuring unbiased metatranscriptomics in suboxic waters of the central Baltic Sea using a new in situ fixation system. ISME J. 2012;6:461–470. doi: 10.1038/ismej.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez C, Farias L, Ulloa O. Nitrogen fixation in denitrified marine waters. PLoS One. 2011;6:e20539. doi: 10.1371/journal.pone.0020539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flindt MR, Kamp-Nielsen L, Marques JC, Pardal MA, Bocci M, Bendoricchio G, et al. Description of the three shallow estuaries: Mondego River (Portugal), Roskilde Fjord (Denmark) and the Lagoon of Venice (Italy) Ecol Model. 1997;102:17–31. [Google Scholar]
- Fuhrman JA, Azam F. Thymidine Incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters–evaluation and field results. Mar Biol. 1982;66:109–120. [Google Scholar]
- Fulweiler RW, Brown SM, Nixon SW, Jenkins BD. Evidence and a conceptual model for the co-occurrence of nitrogen fixation and denitrification in heterotrophic marine sediments. Mar Ecol-Prog Ser. 2013;482:57–68. [Google Scholar]
- Gasol JM, Del Giorgio PA. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci Mar. 2000;64:197–224. [Google Scholar]
- Granhall U, Lundgren A. Nitrogen fixation in Lake Erken. Limnol Oceanogr. 1971;16:711–719. [Google Scholar]
- Grasshoff K. Methods of Seawater Analysis. Verlag Chemie: Weinheim; 1976. [Google Scholar]
- Hamersley MR, Turk KA, Leinweber A, Gruber N, Zehr JP, Gunderson T, et al. Nitrogen fixation within the water column associated with two hypoxic basins in the Southern California Bight. Aquat Microb Ecol. 2011;63:193–205. [Google Scholar]
- Holm-Hansen O, Lorenzen CJ, Holms RW, Strickland JDH. Fluorometric determination of chlorophyll. J Cons Int Explor Mer. 1965;30:3–15. [Google Scholar]
- Horne AJ, Galat DL. Nitrogen-fixation in an oligotrophic, saline desert lake—Pyramid Lake, Nevada. Limnol Oceanogr. 1985;30:1229–1239. [Google Scholar]
- Howarth RW. Nutrient limitation of net primary production in marine ecosystems. Annu Rev Ecol Syst. 1988;19:89–110. [Google Scholar]
- Howarth RW, Marino R. Nitrogen as the limiting nutrient for eutrophication in coastal marine ecosystems: evolving views over three decades. Limnol Oceanogr. 2006;51:364–376. [Google Scholar]
- Howarth RW, Marino R, Lane J. Nitrogen fixation in freshwater, estuarine, and marine ecosystems. 1. Rates and importance. Limnol Oceanogr. 1988;33:669–687. [Google Scholar]
- Jenkins BD, Steward GF, Short SM, Ward BB, Zehr JP. Fingerprinting diazotroph communities in the Chesapeake Bay by using a DNA macroarray. Appl Environ Microbiol. 2004;70:1767–1776. doi: 10.1128/AEM.70.3.1767-1776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen L, Markager S, Maar M.2013On the importance of quantifying bioavailable nitrogen instead of total nitrogen Biogeochemistrye-pub ahead of print 17 July 2013doi: 10.1007/s10533-013-9890-9 [DOI]
- Marino R, Chan F, Howarth RW, Pace M, Likens GE. Ecological and biogeochemical interactions constrain planktonic nitrogen fixation in estuaries. Ecosystems. 2002;5:719–725. [Google Scholar]
- Marino R, Howarth RW, Chan F, Cole JJ, Likens GE. Sulfate inhibition of molybdenum-dependent nitrogen fixation by planktonic cyanobacteria under seawater conditions: a non-reversible effect. Hydrobiologia. 2003;500:277–293. [Google Scholar]
- Mazard SL, Fuller NJ, Orcutt KM, Bridle O, Scanlan DJ. PCR analysis of the distribution of unicellular cyanobacterial diazotrophs in the Arabian Sea. Appl Environ Microbiol. 2004;70:7355–7364. doi: 10.1128/AEM.70.12.7355-7364.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta MP, Butterfield DA, Baross JA. Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca ridge. Appl Environ Microbiol. 2003;69:960–970. doi: 10.1128/AEM.69.2.960-970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr W, Grosskopf T, Wallace DWR, LaRoche J. Methodological underestimation of oceanic nitrogen fixation rates. PLoS One. 2010;5:e12583. doi: 10.1371/journal.pone.0012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisander PH, Beinart RA, Hewson I, White AE, Johnson KS, Carlson CA, et al. Unicellular cyanobacterial distributions broaden the oceanic N2 fixation domain. Science. 2010;327:1512–1514. doi: 10.1126/science.1185468. [DOI] [PubMed] [Google Scholar]
- Moisander PH, McClinton E, Paerl HW. Salinity effects on growth, photosynthetic parameters, and nitrogenase activity in estuarine planktonic cyanobacteria. Microb Ecol. 2002;43:432–442. doi: 10.1007/s00248-001-1044-2. [DOI] [PubMed] [Google Scholar]
- Moisander PH, Serros T, Paerl RW, Beinart RA, Zehr JP.2014Gammaproteobacterial diazotrophs and nifH gene expression in surface waters of the South Pacific Ocean ISME Je-pub ahead of print 10 April 2014doi: 10.1038/ismej.2014.49 [DOI] [PMC free article] [PubMed]
- Montoya JP, Voss M, Kahler P, Capone DG. A simple, high-precision, high-sensitivity tracer assay for N2 fixation. Appl Environ Microbiol. 1996;62:986–993. doi: 10.1128/aem.62.3.986-993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland MR, Bernhardt PW, Blanco-Garcia JL, Mannino A, Hyde K, Mondragon E, et al. Rates of dinitrogen fixation and the abundance of diazotrophs in North American coastal waters between Cape Hatteras and Georges Bank. Limnol Oceanogr. 2012;57:1067–1083. [Google Scholar]
- Pedersen TM, Sand-Jensen K, Markager S, Nielsen LS.2013Optical changes in a eutrophic estuary during reduced nutrient loadings Estuaries Coastse-pub ahead of print 4 December 2013doi: 10.1007/s12237-013-9732-y [DOI]
- Rees AP, Gilbert JA, Kelly-Gerreyn BA. Nitrogen fixation in the western English Channel (NE Atlantic Ocean) Mar Ecol-Prog Ser. 2009;374:7–12. [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Ilikchyan I, Rabouille S, Zehr JP. Genome-wide analysis of diel gene expression in the unicellular N2-fixing cyanobacterium Crocosphaera watsonii WH 8501. ISME J. 2010;4:621–632. doi: 10.1038/ismej.2009.148. [DOI] [PubMed] [Google Scholar]
- Short SM, Zehr JP. Nitrogenase gene expression in the Chesapeake Bay estuary. Environ Microbiol. 2007;9:1591–1596. doi: 10.1111/j.1462-2920.2007.01258.x. [DOI] [PubMed] [Google Scholar]
- Simon R. Macromolecular composition of spores from the filamentous cyanobacterium Anabaena cylindrica. J Bacteriol. 1977;129:1154–1155. doi: 10.1128/jb.129.2.1154-1155.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DC, Azam F. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs. 1992;6:107–114. [Google Scholar]
- Stal LJ, Albertano P, Bergman B, von Brockel K, Gallon JR, Hayes PK, et al. BASIC: Baltic Sea cyanobacteria. An investigation of the structure and dynamics of water blooms of cyanobacteria in the Baltic Sea—responses to a changing environment. Cont Shelf Res. 2003;23:1695–1714. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AW, Foster RA, Krupke A, Carter BJ, Musat N, Vaulot D, et al. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science. 2012;337:1546–1550. doi: 10.1126/science.1222700. [DOI] [PubMed] [Google Scholar]
- Turk-Kubo KA, Karamchandani M, Capone DG, Zehr JP.2014The paradox of marine heterotrophic nitrogen fixation: abundances of heterotrophic diazotrophs do not account for nitrogen fixation rates in the Eastern Tropical South Pacific Environ Microbiole-pub ahead of print 7 January 2014doi: 10.1111/1462-2920.12346 [DOI] [PubMed]
- Wasmund N, Nausch G, Schneider B, Nagel K, Voss M. Comparison of nitrogen fixation rates determined with different methods: a study in the Baltic Proper. Mar Ecol-Prog Ser. 2005;297:23–31. [Google Scholar]
- Webb EA, Ehrenreich IM, Brown SL, Valois FW, Waterbury JB. Phenotypic and genotypic characterization of multiple strains of the diazotrophic cyanobacterium, Crocosphaera watsonii, isolated from the open ocean. Environ Microbiol. 2009;11:338–348. doi: 10.1111/j.1462-2920.2008.01771.x. [DOI] [PubMed] [Google Scholar]
- Zehr JP, McReynolds LA. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1989;55:2522–2526. doi: 10.1128/aem.55.10.2522-2526.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr JP, Paerl HW.2008Molecular ecological aspects of nitrogen fixation in the marine environmentIn: Kirchman DL, (ed)Microbial Ecology of the Oceans John Whiley & Sons: Hoboken; 481–525. [Google Scholar]
- Zehr JP, Turner PJ.2001Nitrogen fixation: nitrogenase genes and gene expressionIn: Paul JH, (ed)Methods in Microbiology Academic Press: New York; 271–285. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.