Abstract
Eukaryotic microbes are key ecosystem drivers; however, we have little theory and few data elucidating the processes influencing their observed population patterns. Here we provide an in-depth quantitative analysis of population separation and similarity in the yeast Saccharomyces cerevisiae with the aim of providing a more detailed account of the population processes occurring in microbes. Over 10 000 individual isolates were collected from native plants, vineyards and spontaneous ferments of fruit from six major regions spanning 1000 km across New Zealand. From these, hundreds of S. cerevisiae genotypes were obtained, and using a suite of analytical methods we provide comprehensive quantitative estimates for both population structure and rates of gene flow or migration. No genetic differentiation was detected within geographic regions, even between populations inhabiting native forests and vineyards. We do, however, reveal a picture of national population structure at scales above ∼100 km with distinctive populations in the more remote Nelson and Central Otago regions primarily contributing to this. In addition, differential degrees of connectivity between regional populations are observed and correlate with the movement of fruit by the New Zealand wine industry. This suggests some anthropogenic influence on these observed population patterns.
Introduction
Research to date attempting to elucidate the patterns and processes involved in shaping natural populations has largely focused on readily observed macroorganisms but comparatively little work has been conducted on microbial species (Anderson and Kohn, 1998; Martiny et al., 2006; Prosser et al., 2007). Because of their large population sizes, and ease of transfer, one might expect microbial populations to be well mixed (Finlay, 2002); however, there is increasing evidence showing that many are not homogeneous but structured (Taylor et al., 2006; Whitaker and Banfield, 2006; Hanson et al., 2012). Most microbial ecology studies have focused on bacteria, but eukaryotic microbes, which undergo sex (with recombination), also have key ecosystem roles (Green et al., 2008; Van Der Heijden et al., 2008). It is not clear whether the population patterns estimated for eukaryotic ‘macrobes' generally hold for eukaryotic microbes (Hartl and Clark, 1997; Anderson and Kohn, 1998; Halkett et al., 2005; Taylor et al., 2006; Prosser et al., 2007; Tsai et al., 2008).
A metapopulation comprises a number of spatially separated populations of the same species that interact to some extent. To date all studies examining microbial populations have simply examined whether population structure is evident or not (Aa et al., 2006; Achtman, 2008; Liti et al., 2009; Goddard et al., 2010; Anderson and Shearer, 2011; Härnström et al., 2011; Gayevskiy and Goddard, 2012; Wang et al., 2012). Merely defining microbial populations as either structured or homogeneous is highly unlikely to reflect the true biological situation. A more accurate approach is not only to assess the degree to which populations are structured, but also connected by gene flow, and crucially go onto quantify these processes; however, to the best of our knowledge, there are no previous studies that have used a unified framework to do this. Here we analyse the natural population of Saccharomyces cerevisiae in New Zealand (NZ), and in doing so take the first steps towards quantifying microbial population structure and similarity.
S. cerevisiae, a budding yeast, has been closely associated with humans since the dawn of civilisation because of its fermentative capabilities, and has come to be of significant commercial importance in the production of bread, wine, beer and other alcoholic beverages (McGovern et al., 1996; Pretorius, 2000; Cavalieri et al., 2003; Chambers and Pretorius, 2010). S. cerevisiae is also a classic model organism for research into cell biology, genetics and increasingly ecology and evolution (Chambers and Pretorius, 2010; Dujon, 2010; Gray and Goddard, 2012; Hittinger, 2013; Hyma and Fay, 2013). Recent studies have revealed a large genetic diversity within S. cerevisiae, and there is good evidence for population structure at intercontinental scales (Fay and Benavides, 2005; Schuller et al., 2005; Aa et al., 2006; Lopandic et al., 2008; Liti et al., 2009; Goddard et al., 2010; Mercado et al., 2011; Di Maio et al., 2012; Wang et al., 2012). Similar inferences have been made at finer scales with reports that some genotypes were unique to different geographic locations in Austria, although many were also ubiquitous across regions (Lopandic et al., 2008). In addition, Bayesian inference shows genetic differentiation between populations spanning hundreds of kilometres in NZ (Gayevskiy and Goddard, 2012). While the scales of these studies differ, they all commonly report the presence of hybrid or mosaic strains indicative of some levels of connectivity between populations via gene flow.
Global-scale analyses have suggested that ecological function may define population structure to a greater extent than geographic origin (Fay and Benavides, 2005; Legras et al., 2007). Strains associated with wine appear somewhat distinct from those isolated from distilling, bread making, fermented milk, rice wine, ale and lager, with geographic origin only explaining 28% of variability (Legras et al., 2007). Furthermore, whole-genome analyses of a limited number of strains suggest specific S. cerevisiae populations associated with vineyards, sake and related ferments, although some of these clusters are confounded with geographic origin (Liti et al., 2009; Schacherer et al., 2009). In contrast, a recent rigorous genome-wide population study has provided evidence of gene flow across small distances (<17 km) between distinct populations inhabiting vineyards and oak trees, showing connectivity between ecological niches at small scales (Hyma and Fay, 2013).
Despite these excellent efforts to date, most studies have been drawn from widely dispersed isolates often from different ecological niches, with relatively small sample sizes from any one discrete population (Fay and Benavides, 2005; Aa et al., 2006; Legras et al., 2007; Liti et al., 2009; Schacherer et al., 2009; Wang et al., 2012) and have thus not afforded adequate power to quantify ‘ecological scale' population processes such as gene flow. Previous work using both microsatellite and RAD-seq analyses shows that a distinct S. cerevisiae population resides in NZ, suggesting that this population is not subject to rampant inward international gene flow (Goddard et al., 2010; Cromie et al., 2013). Therefore, in addition to its geographic isolation, the NZ S. cerevisiae population appears relatively self-contained and thus provides a good population to study the processes we are interested in. Here we analyse close to a thousand S. cerevisiae isolates from four niches across six regions spanning over 1000 km. We quantify both the degree to which this population is structured and go on to quantify the extent to which the various regional populations are connected by gene flow in one of the most comprehensive studies of a microbial metapopulation to date. Lastly, this study sheds light on the connection between farmed (managed) and native ecosystems by examining the relationship between microbial populations residing in vineyards and native NZ forest.
Materials and methods
Sample collection and processing
Six to seven Vitis vinifera var. Sauvignon Blanc vineyards were selected from each of Hawke's Bay, Martinborough, Nelson, Awatere Valley, Wairau Valley and Central Otago in NZ (Figure 1). Approximately 5 g of soil were aseptically taken from each of these 37 vineyards between 1 and 4 weeks before harvest in mid-March 2011. Ten litres of juice derived from the same vineyards were collected from commercial settling tanks (one vineyard provided juice samples from two pressing tiers, resulting in a total of 38 juice samples). Soil and fruit samples were taken from six native NZ plants located in non-managed native bush reserves within each region (Supplementary Table S2), ranging from 0.1 to 50 km from the vineyard sites, totalling 72 native samples (36 soil and 36 fruit). S. cerevisiae is rare in niches other than in actively fermenting fruit, so equivalent selective culturing methods were employed for all samples to control for the effects of high sugar and ethanol (Mortimer and Polsinelli, 1999; Pretorius, 2000; Xufre et al., 2006; Goddard, 2008; Taylor et al., 2014). An enrichment method emulating fermenting selection pressures was employed for all 147 environmental samples (Mortimer and Polsinelli, 1999; Serjeant et al., 2008). Samples were submerged in 10 ml SelMed media (1% yeast extract, 2% peptone, 10% glucose and 5% ethanol) for six days; 500 μl was then transferred to 10 ml fresh SelMed for four additional days, and then dilutions plated onto YPD (1% yeast extract, 2% peptone, 2% glucose) with 50 μg ml−1 chloramphenicol to retard bacterial growth. All incubation was at 28 °C. Up to 94 colonies were taken from each sample and stored in 15% glycerol at −80 °C. A total of 7144 individuals were isolated from environmental samples. A natural enrichment of the juice samples was performed by allowing them to ferment spontaneously at 15 °C. In all, 100 ml was concentrated by centrifugation after 21 days, and plated on YPD with 50 μg ml−1 chloramphenicol. Again 94 colonies were isolated from each ferment sample totalling 3572 individuals. All niches were thus evenly sampled and in total 10 716 individuals were collected.
Figure 1.
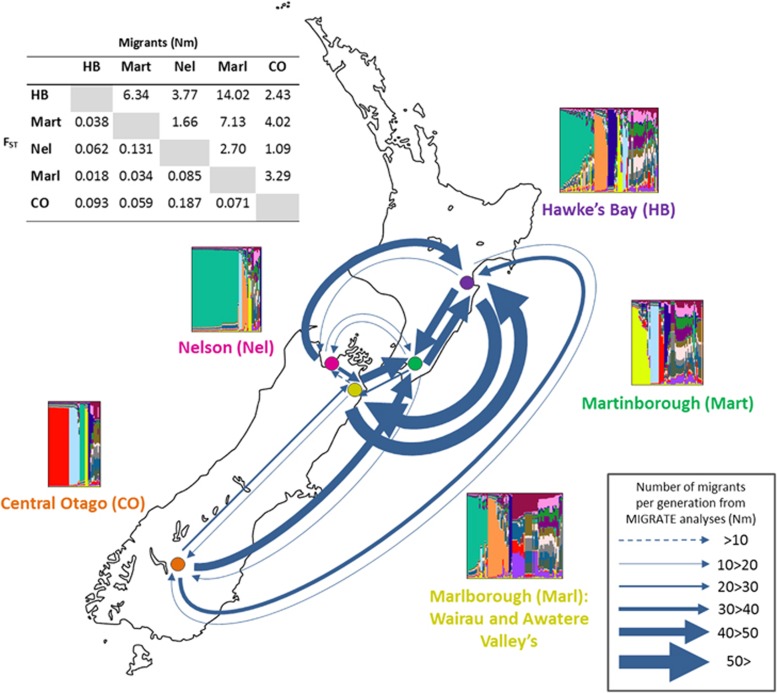
The location of NZ regions and analyses of population structure and connectivity. Plots of the ancestry profiles are shown beside each region: each vertical line represents an individual with the different colours showing the proportion of ancestry of each individual to each of the 16 inferred populations. Arrows connecting different regions show directional migration rates as calculated in MIGRATE with the width of the arrows representing the number of migrants per generation as indicated in the scale. Absolute numbers can be found in Supplementary Table S4. The table reports pairwise FST values below the diagonal and the number of migrants per generation (Nm) as calculated from FST above the diagonal. All FST values are significant (P<0.01).
Molecular methods
Genomic DNA was extracted from colonies with 15 μl of 1.25 mg ml−1 Zymolyase solution dissolved in 1.2 M sorbitol and 0.1 M KH2PO4 at pH 7.2 and treated with EMA to bind unwanted DNA fragments (Rueckert and Morgan, 2007). We employed a multiplex PCR reaction to distinguish S. cerevisiae, and this also identifies S. uvarum (de Melo Pereira et al., 2010). DNA from eight S. cerevisiae colonies from each sample were initially amplified and scored at 10 unlinked loci as described by Richards et al. (2009) using capillary electrophoresis on an ABI3130XL (Applied Biosystems, Life Technologies, Mulgrave, VIC, Australia). If all eight initial isolates were genotypically identical then no further genotyping was performed for that sample; however, if more than one genotype was recovered, another eight were genotyped until either no new genotypes were seen, or all isolates from the sample had been genotyped. A number of control samples were submitted for the calculation of error rates per allele and per locus as described by Pompanon et al. (2005). To further ascertain the reliability of microsatellite loci amplification and scoring, we analysed an additional 96-well control plate replicating the same strain for DNA extraction, PCR amplification and genotyping.
Data analyses
A ±1-bp error in size calling from run to run variation and plus-A effects was observed and loci were binned accordingly using Genemapper (Version 4). F-statistics, migration estimates (Nm values) and Mantel tests were performed with GenAlEx (Genetic Analyses in Excel) version 6.5 (Peakall and Smouse, 2006; Peakall and Smouse, 2012). Estimates of population diversity were calculated by rarefaction (which controls for unequal sample sizes) using EstimateS (Colwell, 2006). The maximum likelihood outcrossing rates were estimated in Mathematica 7 following the method used by Johnson et al. (2004) that estimates the proportion of matings between spores from the same meiotic event (that is, that are asci mates), and those from independent meiotic events (code available at http://goddardlab.auckland.ac.nz/data-and-code/). Allelic richness was estimated using rarefaction with HP rare, again controlling for unequal sample sizes, based on the lowest number of 94 observed alleles among sampled populations (Kalinowski, 2005).
Population structure was evaluated using the Bayesian clustering method implemented in InStruct, which does not assume Hardy–Weinberg Equilibrium, accounts for inbreeding, and makes no a priori assumptions about the sampling location of the genotypes (Gao et al., 2007). This method estimates the most likely number of populations and assigns genotypes to these probabilistically. Admixture was allowed and the proportion of each genotype's ancestry in each inferred population was estimated. Three chains of one million MCMC iterations with a burn-in of 10 000 were run for K=1–25. Convergence of the MCMC chain was confirmed using the Gelman–Rubin statistic (Gelman and Rubin, 1992). Analyses of the resulting ancestry profiles evaluating and quantifying the contribution of niche and geographic region to population structure was conducted with ObStruct (Gayevskiy et al., 2014).
Directional migration rates were quantified using the Bayesian coalescent approach implemented in MIGRATE that assumes constant population sizes, random mating, a constant mutation rate and that populations are connected only through migration, not population divergence (Beerli and Felsenstein, 2001; Beerli, 2006, 2009; Beerli and Palczewski, 2010). Mutation-scaled population sizes (θ) were calculated using the number of sampled alleles (Haasl and Payseur, 2010). We employed a Brownian motion allele mutation model with starting estimates of the mutation-scaled migration rate derived from FST calculations to estimate all possible migration routes. Chains of one million steps with a burn-in of 50 000 were run with 10 replicates, sampling every 100 steps (Beerli, 2009). The analysis was run in parallel on the NeSI pan cluster at the University of Auckland.
Results
S. cerevisiae presence, abundance and genetic diversity
PCR analyses revealed that 3900 (36%) of the 10 716 isolates were S. cerevisiae. Of the 3780 isolates from spontaneous ferments, 2210 (56%) were S. cerevisiae and 1570 (40%) S. uvarum, revealing the co-existence of a sister Saccharomyces species in this niche. Here we do not pursue the population genetics of S. uvarum. S. cerevisiae was detected in 13 of the 37 vineyard soils, and four and one of the 36 native soil and fruit samples, respectively. The breakdown of samples that yielded S. cerevisiae is shown in Supplementary Tables S1 and S2.
From control samples, two loci (YOR267C and YBR240C) amplified unreliably and were removed from all analyses. Overall the mean error rates per allele and locus were ±4.08% and 4.35%, respectively. In total 850 individuals were genotyped, with 681 isolates from spontaneous ferments, 130 from vineyard soil, 31 from native soil and 8 from native fruits. Identical genotypes within the same sample were collapsed to conservatively account for clonal expansion during enrichment and fermentation meaning the data set was compressed to 380 genotype profiles. Just 11 genotypes matched commercially available wine strains commonly used in NZ (Richards et al., 2009) and were removed from further analyses. This resulted in a final data set comprising 369 microsatellite profiles (Supplementary Data set S1). Interestingly, no genotypes matched a genetically and ecologically diverse set of international strains (Liti et al., 2009) genotyped using the same method (Richards et al., 2009; Goddard et al., 2010).
For the entire data set, a large allelic diversity was detected at all loci. YFR028C and YML091C had the greatest diversity with 25 and 30 alleles, respectively, and all other loci had between 11 and 16 alleles. Overall, 295 different genotypes were recovered and only 38 of these were identified in more than one sample. On average samples that yielded S. cerevisiae contained 4.7 unique genotypes, although most alleles were shared between populations (Supplementary Data set S1). Rarefaction analyses (Chao, 1987; Colwell, 2006) estimate that these genotypes were sampled from an underlying NZ population containing ∼1700 different genotypes (with 95% confidence limits of 1159–2486).
Testing for ecological drivers of population structure
Only four different genotypes were derived from native fruits and soil, and 21 from vineyard soil (Supplementary S1 and S2). This translates to low statistical power to test how the niche of isolation affects population structure. Despite this, observations of identical genotypes between niches within, but not between, regions are striking. For example, genotypes recovered from native soil and fruit in the Martinborough region (Waiohine Gorge) were identical to spontaneous ferment isolates recovered 20 km away in Martinborough vineyards but were not found in other NZ regions; three of the genotypes isolated from vineyard soil in the Wairau Valley were identical to isolates from spontaneous ferments sourced in the same region, with one of these vineyard soil genotypes being identical to an isolate from the spontaneous ferment from the same vineyard. FST values between environmental samples (from native soil and fruit and vineyard soil) and spontaneous ferment samples within regions are extremely low (<0.005) and insignificant (P>0.33) with the exception of Martinborough (FST=0.046, P=0.003); however, this FST value is classed as only representing ‘low' differentiation (Wright, 1978). There is complete overlap of isolates deriving from all niches in a principal component analysis of genetic distances between genotypes (Supplementary Figure S1) and no significant population differentiation between niches within regions using InStruct and subsequent ObStruct analyses (P>0.119, Supplementary Figure S2). Together, this provides no substantial evidence of an effect of niche on population structure within regions. Some of the S. cerevisiae genotypes contributing to spontaneous ferments may have derived from wineries, as opposed to the ‘environment' (Bokulich et al., 2013). As all of these wineries reside within the same geographic regions the fruit was collected from, these potentially winery derived genotypes form part of the local population we wish to study. Thus, individuals from various niches within regions comprise homogenous populations, and so we combined all genotypes from different niches within regions to form regional populations for further analyses.
Testing for geographic drivers of population structure
There was significant genetic differentiation, as estimated by pairwise FST values, between populations deriving from all six regions (P<0.01), with the exception of those between the Wairau and Awatere Valley's (FST=0.001, P=0.310). These two valleys comprise the wider Marlborough region and were thus combined to represent one population residing in Marlborough. The subsequent pairwise FST values between regions are shown in Figure 1. A low albeit significant correlation was observed between genetic and geographic distance (Mantel Test: R2=0.181, P<0.001). Population diversity, as estimated by rarefaction analyses to control for uneven numbers of genotypes, differs by as much as threefold between regions (Table 1). Hawke's Bay and Marlborough harbour the greatest diversity, whereas Nelson and Central Otago the least. Allelic richness across regions is comparable to estimates within one s.d. of each other (Table 1). All eight loci in all regions are significantly out of Hardy–Weinberg equilibrium (P<0.001), and show strong signals for inbreeding; however, outcrossing rates are significantly above zero within each region (Table 1).
Table 1. Summary of the populations isolated from each region.
| Region | Number of individualsa | Number of different genotypes | Estimated total number of genotypesb | Mean allelic richnessc | Inbreeding Coefficient FIS | Outcrossing rated | Region-specific genotypes |
|---|---|---|---|---|---|---|---|
| Hawke's Bay | 94 | 87 | 482: 272–932 | 10.72±4.86 | 0.6483 | 0.12: 0.09–0.16 | 79/87: 91% |
| Martinborough | 68 | 56 | 291: 150–647 | 11.02±4.29 | 0.6067 | 0.11: 0.08–0.16 | 52/56: 93% |
| Nelson | 66 | 48 | 131: 82–252 | 7.29±1.83 | 0.2620 | 0.42: 0.32–0.54 | 44/48: 92% |
| Marlborough | 91 | 76 | 415: 138–1610 | 10.47±3.64 | 0.5331 | 0.18: 0.14–0.24 | 70/76: 92% |
| Central Otago | 50 | 44 | 144: 84–293 | 8.29±2.84 | 0.5869 | 0.14: 0.09–0.20 | 39/44: 89% |
The number of individuals included in the final analysis after the collapsing of identical individuals because of clonal expansion.
The estimated number of genotypes in the population by rarefaction with 95% upper and lower rarefaction limit calculated using EstimateS (Colwell, 2006).
Calculated for each locus independently using HP Rare and based on 94 genes. The mean±1 s.d. are reported.
The average proportion of spores mating from independent tetrads per meiotic generation surrounded by 2 log likelihood support limits (equivalent to a 95% confidence interval), calculated following Johnson et al. (2004).
Quantifying geographic population structure
InStruct analyses (Gao et al., 2007) indicate the optimal number of populations, given the data are 16. Examination of the ancestry profile plots (Rosenberg, 2004) resulting from this analysis are indicative of population structure by region to some degree (for example, the blocks of green, red and yellow in Nelson, Central Otago and Martinborough, respectively) and are in agreement with the magnitude of the pairwise FST estimates (Figure 1). Subsequent ObStruct analyses revealed that the inferred population structure is significantly correlated with geographic location (R2=0.16, P<0.0001), and this explains about one-sixth of the genetic variability observed. Individuals from the Nelson and Central Otago regions contributed the greatest signal to overall population structure with significant decreases in the R2 values observed when these are removed (R2=Δ−0.05 and Δ−0.02, respectively). The R2 value remained constant when data from Martinborough were removed but increased when Hawke's Bay and Marlborough data were independently removed (R2=Δ+0.03 for both). Increases in R2 suggest individuals from these regions add noise to any signal for structure (that is, have homogenised not localised populations). Further, canonical discriminant analysis shows that ∼80% of the variation in ancestry profiles can be represented with the first and second axes, suggesting that most of the variation can be visualised in these graphical representations of the data (Supplementary Figure S3). Ancestry profiles from Central Otago and Nelson cluster the most discretely in these plots, recapitulating that populations from these regions provide the strongest signals for differentiation. Pairwise comparisons between regions all significantly differ (P<0.001 or P=0.06 between Hawke's Bay and Marlborough), but the R2 values vary from 0.02 to 0.23 (Supplementary Table S3).
Quantifying population connectivity and migration
Pairwise estimates of migration between the regions (Nm values) using classic methods derived from FST values (Hartl and Clark, 1997) suggest that Hawke's Bay and Marlborough are the most connected, closely followed by Marlborough and Martinborough, and Hawke's Bay and Martinborough (Figure 1). Nelson and Central Otago share the lowest number of migrants with an estimate of just one per generation (Figure 1). MIGRATE analyses showed an acceptance ratio for each parameter ranging from 0.38 to 0.65, and an effective sample size of approximately two million, suggesting that the chain length was sufficient. The autocorrelation between parameters and the prior was high and estimated to be around 0.96 overall, indicating a lack of information in data. This is reflected in the high confidence intervals surrounding the estimates (Supplementary Table S4). However, consistent patterns between multiple runs were evident, allowing meaningful estimates of gene flow between regions to be made. Inferred mean rates of movement between regions span an order of magnitude ranging from 6 to 63 migrants per generation (Figure 1 and Supplementary Table S4), and show differential inward and outward movements for some regions. Correlating with the classic Nm estimates, and the analyses of population structure, Nelson and Central Otago show greatest isolation with twofold greater rates of outward than inward migration, and show an average of just 51 inward migrants per generation, 3.2-fold less than the overall average inward migration rate of 164 migrants per generation for all other regions (Figure 1 and Supplementary Table S4). Conversely, Marlborough and the Hawke's Bay, which harbours some of the least distinctive and most diverse populations, experience some of the greatest inward migration rates at an average of 171 migrants per generation, 1.4-fold more than the average inward migration rate (Supplementary Table S4). In line with the low FST estimates, a high degree of individuals with shared ancestry from InStruct, and a large proportion of admixed individuals (Figure 1), Marlborough and the Hawke's Bay are the most connected regions, and experience an average of twofold more migration between these regions than the average overall migration rate. The extent of migration between regions does not correlate with geographic location (P>0.21) showing the difference in the extent of gene flow is not simply a function of distance.
Discussion
We have very few models attempting to generally describe the population biology of microbes. Accurate quantification of short-term population level processes is necessary to understand the likely longer-term evolutionary trajectories of populations (Smadja and Butlin, 2011; Gray and Goddard, 2012), as well as how microbes may interact with other members of the community (Ruxton et al., 2014). We have attempted to make a significant step forward: rather than simply describing this S. cerevisiae population as either structured or not, here we paint a more biologically realistic picture by quantifying the role that geography has in defining structure, and go on to provide quantitative estimates of gene flow between populations residing in different regions.
S. cerevisiae has clearly been isolated many times from managed vineyard ecosystems and ferments of fruit (Lopandic et al., 2008; Liti et al., 2009; Schacherer et al., 2009; Goddard et al., 2010; Gayevskiy and Goddard, 2012; Bokulich et al., 2014). This species is also well reported from native niches in the northern hemisphere (Sniegowski et al., 2002; Wang et al., 2012; Hyma and Fay, 2013), and in the Southern hemisphere has been isolated from exotic Quercus species in NZ, and from Nothofagus in Patagonia (Zhang et al., 2010; Libkind et al., 2011). Here we provide the first report of S. cerevisiae from multiple native tree species in the South Pacific region. Overall, this NZ S. cerevisiae metapopulation displays large genetic variance, compounding evidence that NZ harbours a large and diverse population of this species (Goddard et al., 2010; Gayevskiy and Goddard, 2012; Cromie et al., 2013). Within regions, which typically encompass a radius of under 100 km, there is no compelling evidence for genetic differentiation between niches within managed ecosystems nor more strikingly between managed and native ecosystems. The lack of genetic differentiation between managed and native ecosystems seen here does not permit us to determine whether vineyards or native forests are the sources or sinks of these populations, just that they are connected. Thus, there appears to be a free flow of individuals between these various niches at subregional scales, supporting previous reports from NZ and the United States of America (Goddard et al., 2010; Hyma and Fay, 2013). The inference of little differentiation between niches at regional scales is in contrast to previous reports showing differentiation between isolates from various ecological niches at global scales (Fay and Benavides, 2005; Legras et al., 2007). One explanation for this is the extent of sample effort within any one population. The studies, including this one, reporting a minor effect of niche examined a large number of individuals from specific more localised populations, and in some sense evaluate ‘ecological scale' processes: it may be that some strains are less well adapted to various niches and that selection will eventually result in their removal. Studies evaluating strains from different geographic and ecological sources only include a handful of strains from any one specific population and unfortunately tend to confound geographic location with niche, but conclude that niche has a stronger role; in some sense these studies might examine populations where selection has possibly had more time to operate. Perhaps, the drivers of population structure differ at different scales? Lastly, it might be that NZ has relatively recently been colonised by only one of the inferred lineages of S. cerevisiae, and this has radiated to all niches. This would also provide a signal for the lack of differentiation between niches. Estimates of the rates of global flux for S. cerevisiae would help disentangle these possibilities.
Whereas populations appear homogeneous within regions, analyses provide compelling evidence for various degrees of genetic differentiation between populations inhabiting major NZ regions. This differentiation is not absolute and there is also a degree of connectivity between regions. This is in line with a previous smaller-scale study with this species that reported both differentiation and connection between regions in the North Island of NZ (Gayevskiy and Goddard, 2012), and this is also echoed at global scales (Liti et al., 2009; Wang et al., 2012). Here all analyses, both classic and more sophisticated Bayesian approaches, converge on the same conclusion. FST, Bayesian, ancestry profile and migration analyses show that the populations residing in Nelson and Central Otago are the most distinct and experience the least inward migration. Conversely, Marlborough and Hawke's Bay have smaller pairwise FST values and Bayesian, ancestry profile and migration analyses show that these regions are the most mixed and connected. Marlborough and Hawke's Bay experience the most inward migration at approximately three times that into Nelson and Central Otago. This is consistent with the higher genetic diversity observed in these regions and implies that they accumulate genetic diversity from around the country.
S. cerevisiae cells and spores are sessile; however, there are a variety of possible vectors that may move this unicellular eukaryote around. S. cerevisiae has been shown to be associated with both wasps and bees and has long been known to be associated with fruit flies (Reuter et al., 2007; Goddard et al., 2010; Stefanini et al., 2012). Recent work provides evidence that certain volatiles released by S. cerevisiae attract Drosophila, and this enhances the likelihood of movement, and potentially facilitates a mutualism between these species (Buser et al., 2014). These insect species easily move over regional scales, and so presumably have some part in the homogenisation of S. cerevisiae within regions. Insects less likely move S. cerevisiae over hundreds of kilometres between regions, although S. cerevisiae may also be associated with birds that can easily cover these distances (Francesca et al., 2012). Humans are also obvious vectors. Indeed, the patterns of separation, and rates of migration in and out the various regions shown here are nicely in line with the flow of fruit and equipment because of the actions of the NZ wine industry. Marlborough and the Hawke's Bay are the two largest viticultural and winemaking regions in the country, and fruit from other regions is often transferred to them, mirroring the inferred migration of S. cerevisiae into these regions. This national ‘ecological' scale picture complements and mirrors the global ‘evolutionary' scale picture revealed for this species: that this is a genetically diverse species that shows some degree of structure and connectivity, and these patterns are consistent with human-influenced dispersal (Fay and Benavides, 2005; Legras et al., 2007).
Although the above interpretation fits nicely with the population patterns observed here, it is important to consider alternate explanations. The connections between populations could instead be indicative of recent divergence events. The NZ wine industry is very young in evolutionary terms and it is possible that S. cerevisiae was introduced to these regions via the introduction of vines and winery equipment such as barrels (Goddard et al., 2010). The patterns observed in this analysis could be explained by the large wine-producing regions of Hawke's Bay and Marlborough being the source of variation and the outlying regions resulting from founder events with subsequent population expansion and divergence (Hartl and Clark, 1997). The method of migration analysis employed here assumes that population divergence has not occurred, and only invokes migration to explain any similarity in genetic diversity between populations (Beerli, 2009). One issue with a divergence (as opposed to migration) explanation is that source populations must exist before the populations they are proposed to have founded. Whereas Hawke's Bay is one of the oldest wine-producing regions in NZ, Marlborough is one of the youngest having only been established around 1970. Thus, the divergence hypothesis fits less well, given the vast diversity and admixture observed in the recently established Marlborough region. In addition, whilst it appears that the NZ S. cerevisiae population is reasonably internationally distinct, these patterns of differentiation may also be explained by the inward migration of genotypes from offshore. These explanations are not mutually exclusive and it is likely that population divergence from founding populations is occurring alongside inevitable national and international migration of strains because of the vast movement of fruit, equipment and people by the wine industry.
The demonstration that certain regions have ‘signature' microbial populations is of relevance to the wine industry. It is often suggested that certain wines reflect their geographic origin, and this is encapsulated in the concept of terroir (Bokulich et al., 2014). Classically, this was thought to largely result from the interaction between specific Vitis vinifera varieties and the local soils, geography and climate; however, there is limited but increasing evidence showing that the microbes that influence vine growth, fermentation and wine style (as S. cerevisiae does) also exhibit regional differentiation (Gayevskiy and Goddard, 2012; Bokulich et al., 2014; Taylor et al., 2014), as we again demonstrate here. Thus, these data further support the concept that there could be a microbial aspect to terroir. Metabolic profiling of regionally defined genotypes is necessary to determine whether the genetic differentiation demonstrated here translates to phenotypes that are relevant to wine, and thus whether microbes contribute to terroir in a predictable and consistent way.
Here we provide a more advanced insight into the population biology of a well-established model microbial eukaryote that has also been biotechnologically harnessed by humans since the dawn of civilisation. We take a significant step towards quantifying these processes by providing the first estimates for metapopulation separation and similarity. We reveal S. cerevisiae population differentiation in NZ at scales over 100 km, with the most signal provided by the more remote regions, but no differentiation within regions, even between populations inhabiting native forests and vineyards. We also show differential migration of this species between regions, and postulate that this may be due, at least in part, to human influence. By quantifying the magnitude of these forces in microbes we begin to provide one crucial aspect of an inclusive framework attempting to more fully integrate ecological and evolutionary processes.
Acknowledgments
We thank Amisfeild, Ata Rangi, Churton, Coal Pit, Constellation, Delegats, Domain Road, Frey Vineyard, Huia, Misha's Vineyard, Mt Difficulty, Mt Riley, Neudorf, Palliser, Pernod Ricard, Rippon, Seifried, Seresin, Te Kairanga, Te Whare Ra, Tohu, Trinity Hill, Villa Maria and Vita Brevis for access to their land and providing samples. Soon Lee, Peter Morrison-Whittle and Velimir Gayevskiy assisted with sample collection and processing. We also acknowledge the contribution of the NeSI high-performance computing facilities at the University of Auckland and the staff, particularly Jordi Blasco, at NeSI and Centre for eResearch. New Zealand's national facilities are provided by the New Zealand eScience Infrastructure (NeSI) and funded jointly by NeSI's collaborator institutions and through the Ministry of Business, Innovation and Employment's Infrastructure programme. This work was funded by a University of Auckland doctoral scholarship to SK and grants to MG from the New Zealand Ministry of Business, Innovation and Employment, New Zealand Winegrowers and Plant and Food Research.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aa E, Townsend JP, Adams RI, Nielsen KM, Taylor JW. Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6:702–715. doi: 10.1111/j.1567-1364.2006.00059.x. [DOI] [PubMed] [Google Scholar]
- Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- Anderson JB, Kohn LM. Genotyping, gene genealogies and genomics bring fungal population genetics above ground. Trends Ecol Evol. 1998;13:444–449. doi: 10.1016/s0169-5347(98)01462-1. [DOI] [PubMed] [Google Scholar]
- Anderson JL, Shearer CA. Population genetics of the aquatic fungus Tetracladium marchalianum over space and time. PLoS One. 2011;6:e15908. doi: 10.1371/journal.pone.0015908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics. 2006;22:341–345. doi: 10.1093/bioinformatics/bti803. [DOI] [PubMed] [Google Scholar]
- Beerli P.2009How to use MIGRATE or why are Markov chain Monte Carlo programs difficult to useBertorelle G, Bruford MW, Hauffe HC, Rizzoli A, Vernesi C (eds), Population Genetics for Animal Conservation Cambridge University Press: New York, USA; 42–79. [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Palczewski M. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics. 2010;185:313–326. doi: 10.1534/genetics.109.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Ohta M, Richardson PM, Mills DA. Monitoring seasonal changes in winery-resident microbiota. PLoS One. 2013;8:e66437. doi: 10.1371/journal.pone.0066437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Thorngate JH, Richardson PM, Mills DA. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci USA. 2014;111:E139–E148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser CC, Newcomb RD, Gaskett AC, Goddard MR.2014Niche construction initiates the evolution of mutualistic interaction Ecol Lets(in press). [DOI] [PubMed]
- Cavalieri D, McGovern PE, Hartl DL, Mortimer R, Polsinelli M. Evidence for S. cerevisiae fermentation in ancient wine. J Mol Evol. 2003;57 (Suppl 1:S226–S232. doi: 10.1007/s00239-003-0031-2. [DOI] [PubMed] [Google Scholar]
- Chambers PJ, Pretorius IS. Fermenting knowledge: the history of winemaking, science and yeast research. EMBO Rep. 2010;11:914–920. doi: 10.1038/embor.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. Estimating the population size for capture - recapture data with unequal catchability. Biometrics. 1987;43:783–791. [PubMed] [Google Scholar]
- Colwell RK.2006. EstimateS: Statistical estimation of species richness and shared species from samples www.purl.oclc.ord/estimates .
- Cromie GA, Hyma KE, Ludlow CL, Garmendia-Torres C, Gilbert TL, May P, et al. Genomic sequence diversity and population structure of Saccharomyces cerevisiae assessed by RAD-seq. G3 (Bethesda) 2013;3:2163–2171. doi: 10.1534/g3.113.007492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo Pereira GV, Ramos CL, Galvão C, Souza Dias E, Schwan RF. Use of specific PCR primers to identify three important industrial species of Saccharomyces genus: Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces pastorianus. Lett Appl Microbiol. 2010;51:131–137. doi: 10.1111/j.1472-765X.2010.02868.x. [DOI] [PubMed] [Google Scholar]
- Di Maio S, Polizzotto G, Di Gangi E, Foresta G, Genna G, Verzera A, et al. Biodiversity of indigenous Saccharomyces populations from old wineries of south-eastern Sicily (Italy): preservation and economic potential. PLoS One. 2012;7:e30428. doi: 10.1371/journal.pone.0030428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Yeast evolutionary genomics. Nat Rev Genet. 2010;11:512–524. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005;1:0066–0071. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Francesca N, Canale DE, Settanni L, Moschetti G. Dissemination of wine-related yeasts by migratory birds. Environ Microbiol Rep. 2012;4:105–112. doi: 10.1111/j.1758-2229.2011.00310.x. [DOI] [PubMed] [Google Scholar]
- Gao H, Williamson S, Bustamante CD. A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics. 2007;176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayevskiy V, Goddard MR. Geographic delineations of yeast communities and populations associated with vines and wines in New Zealand. ISME J. 2012;6:1281–1290. doi: 10.1038/ismej.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayevskiy V, Klaere S, Knight S, Goddard MR. ObStruct: a method to objectively analyse factors driving population structure using bayesian ancestry profiles. PLoS One. 2014;9:e85196. doi: 10.1371/journal.pone.0085196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Stat Sci. 1992;7:457–472. [Google Scholar]
- Goddard MR. Quantifying the complexities of Saccharomyces cerevisiae's ecosystem engineering via fermentation. Ecology. 2008;89:2077–2082. doi: 10.1890/07-2060.1. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Anfang N, Tang R, Gardner RC, Jun C. A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ Microbiol. 2010;12:63–73. doi: 10.1111/j.1462-2920.2009.02035.x. [DOI] [PubMed] [Google Scholar]
- Gray JC, Goddard MR. Gene-flow between niches facilitates local adaptation in sexual populations. Ecol Lett. 2012;15:955–962. doi: 10.1111/j.1461-0248.2012.01814.x. [DOI] [PubMed] [Google Scholar]
- Green JL, Bohannan BJM, Whitaker RJ. Microbial biogeography: from taxonomy to traits. Science. 2008;320:1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- Haasl RJ, Payseur BA. The number of alleles at a microsatellite defines the allele frequency spectrum and facilitates fast accurate estimation of θ. Mol Biol Evol. 2010;27:2702–2715. doi: 10.1093/molbev/msq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkett F, Simon J-C, Balloux F. Tackling the population genetics of clonal and partially clonal organisms. Trends Ecol Evol. 2005;20:194–201. doi: 10.1016/j.tree.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol. 2012;10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- Härnström K, Ellegaard M, Andersen TJ, Godhe A. Hundred years of genetic structure in a sediment revived diatom population. Proc Natl Acad Sci USA. 2011;108:4252–4257. doi: 10.1073/pnas.1013528108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG.1997Principles of Population Genetics3rd ednSinauer Associates, Inc.: Sunderland, MA, USA [Google Scholar]
- Hittinger CT. Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 2013;29:309–317. doi: 10.1016/j.tig.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Hyma KE, Fay JC. Mixing of vineyard and oak-tree ecotypes of Saccharomyces cerevisiae in North American vineyards. Mol Ecol. 2013;22:2917–2930. doi: 10.1111/mec.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LJ, Koufopanou V, Goddard MR, Hetherington R, Schäfer SM, Burt A. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics. 2004;166:43–52. doi: 10.1534/genetics.166.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST. HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes. 2005;5:187–189. [Google Scholar]
- Legras JL, Merdinoglu D, Cornuet JM, Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Libkind D, Hittinger CT, Valeŕio E, Gonca̧lves C, Dover J, Johnston M, et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci USA. 2011;108:14539–14544. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopandic K, Tiefenbrunner W, Gangl H, Mandl K, Berger S, Leitner G, et al. Molecular profiling of yeasts isolated during spontaneous fermentations of Austrian wines. FEMS Yeast Res. 2008;8:1063–1075. doi: 10.1111/j.1567-1364.2008.00385.x. [DOI] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- McGovern PE, Voigt MM, Glusker DL, Exner LJ. Neolithic resinated wine. Nature. 1996;381:480–481. [Google Scholar]
- Mercado L, Sturm ME, Rojo MC, Ciklic I, Martínez C, Combina M. Biodiversity of Saccharomyces cerevisiae populations in Malbec vineyards from the ‘Zona Alta del Río Mendoza' region in Argentina. Int J Food Microbiol. 2011;151:319–326. doi: 10.1016/j.ijfoodmicro.2011.09.026. [DOI] [PubMed] [Google Scholar]
- Mortimer R, Polsinelli M. On the origins of wine yeast. Res Microbiol. 1999;150:199–204. doi: 10.1016/s0923-2508(99)80036-9. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse P. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research – an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompanon F, Bonin A, Bellemain E, Taberlet P. Genotyping errors: causes, consequences and solutions. Nat Rev Genet. 2005;6:847–859. doi: 10.1038/nrg1707. [DOI] [PubMed] [Google Scholar]
- Pretorius IS. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Prosser JI, Bohannan BJM, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, et al. The role of ecological theory in microbial ecology. Nat Rev Microbiol. 2007;5:384–392. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- Reuter M, Bell G, Greig D. Increased outbreeding in yeast in response to dispersal by an insect vector. Curr Biol. 2007;17:R81–R83. doi: 10.1016/j.cub.2006.11.059. [DOI] [PubMed] [Google Scholar]
- Richards KD, Goddard MR, Gardner RC. A database of microsatellite genotypes for Saccharomyces cerevisiae. Antonie van Leeuwenhoek. 2009;96:355–359. doi: 10.1007/s10482-009-9346-3. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Rueckert A, Morgan HW. Removal of contaminating DNA from polymerase chain reaction using ethidium monoazide. J Microbiol Methods. 2007;68:596–600. doi: 10.1016/j.mimet.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Ruxton GD, Wilkinson DM, Schaefer HM, Sherratt TN. Why fruit rots: theoretical support for Janzen's theory of microbe–macrobe competition. Proc R Soc B Biol Sci. 2014;281:20133320. doi: 10.1098/rspb.2013.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller D, Alves H, Dequin S, Casal M. Ecological survey of Saccharomyces cerevisiae strains from vineyards in the Vinho Verde Region of Portugal. FEMS Microbiol Ecol. 2005;51:167–177. doi: 10.1016/j.femsec.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Serjeant K, Tang R, Anfang N, Beggs JR, Goddard MR. Yeasts associated with the New Zealand Nothofagus honeydew system. N Z J Ecol. 2008;32:209–213. [Google Scholar]
- Smadja CM, Butlin RK. A framework for comparing processes of speciation in the presence of gene flow. Mol Ecol. 2011;20:5123–5140. doi: 10.1111/j.1365-294X.2011.05350.x. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Dombrowski PG, Fingerman E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Stefanini I, Dapporto L, Legras J-L, Calabretta A, Di Paola M, De Filippo C, et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc Natl Acad Sci USA. 2012;109:13398–13403. doi: 10.1073/pnas.1208362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philos Trans R Soc Lond B Biol Sci. 2006;361:1947–1963. doi: 10.1098/rstb.2006.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MW, Tsai P, Anfang N, Ross HA, Goddard MR.2014Pyrosequencing reveals regional differences in fruit-associated fungal communities Environ Microbioldoi: 10.1111/1462-2920.12456 [DOI] [PMC free article] [PubMed]
- Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc Natl Acad Sci USA. 2008;105:4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Heijden MGA, Bardgett RD, Van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. 2008;11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- Wang Q-M, Liu W-Q, Liti G, Wang S-A, Bai F-Y. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol Ecol. 2012;21:5404–5417. doi: 10.1111/j.1365-294X.2012.05732.x. [DOI] [PubMed] [Google Scholar]
- Whitaker RJ, Banfield JF. Population genomics in natural microbial communities. Trends Ecol Evol. 2006;21:508–516. doi: 10.1016/j.tree.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Wright S.Ed.. (1978Variability within and among Natural Populations. Evolution and the Genetics of Populations University of Chicago Press: Chicago [Google Scholar]
- Xufre A, Albergaria H, Inácio J, Spencer-Martins I, Gírio F. Application of fluorescence in situ hybridisation (FISH) to the analysis of yeast population dynamics in winery and laboratory grape must fermentations. Int J Food Microbiol. 2006;108:376–384. doi: 10.1016/j.ijfoodmicro.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Zhang H, Skelton A, Gardner RC, Goddard MR. Saccharomyces paradoxus and Saccharomyces cerevisiae reside on oak trees in New Zealand: evidence for migration from Europe and interspecies hybrids. FEMS Yeast Res. 2010;10:941–947. doi: 10.1111/j.1567-1364.2010.00681.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.