Abstract
The bottom of the Red Sea harbors over 25 deep hypersaline anoxic basins that are geochemically distinct and characterized by vertical gradients of extreme physicochemical conditions. Because of strong changes in density, particulate and microbial debris get entrapped in the brine-seawater interface (BSI), resulting in increased dissolved organic carbon, reduced dissolved oxygen toward the brines and enhanced microbial activities in the BSI. These features coupled with the deep-sea prevalence of ammonia-oxidizing archaea (AOA) in the global ocean make the BSI a suitable environment for studying the osmotic adaptations and ecology of these important players in the marine nitrogen cycle. Using phylogenomic-based approaches, we show that the local archaeal community of five different BSI habitats (with up to 18.2% salinity) is composed mostly of a single, highly abundant Nitrosopumilus-like phylotype that is phylogenetically distinct from the bathypelagic thaumarchaea; ammonia-oxidizing bacteria were absent. The composite genome of this novel Nitrosopumilus-like subpopulation (RSA3) co-assembled from multiple single-cell amplified genomes (SAGs) from one such BSI habitat further revealed that it shares ∼54% of its predicted genomic inventory with sequenced Nitrosopumilus species. RSA3 also carries several, albeit variable gene sets that further illuminate the phylogenetic diversity and metabolic plasticity of this genus. Specifically, it encodes for a putative proline-glutamate ‘switch' with a potential role in osmotolerance and indirect impact on carbon and energy flows. Metagenomic fragment recruitment analyses against the composite RSA3 genome, Nitrosopumilus maritimus, and SAGs of mesopelagic thaumarchaea also reiterate the divergence of the BSI genotypes from other AOA.
Introduction
The Red Sea is home to over 25 brines (salinity>25%) occurring along its central axis at great depths beneath the sea (1.4–2.0 km below sea surface), and enclosed in seafloor depressions (Karbe, 1987; Antunes et al., 2011), also known as deep hypersaline anoxic basins (DHABs; van der Wielen et al., 2005). DHABs are also found in the Mediterranean Sea and the Gulf of Mexico (Joye et al., 2009; Antunes et al., 2011; and references therein). Generally, individual brines contain dense mixtures of gases and heavy metals (Hartmann et al., 1998; Schmidt et al., 2003), are characterized by geochemically and spatially distinct layers (Antunes et al., 2011) and some are still hydrothermally active (Swift et al., 2012).
A common theme among all these DHABs is that the transition from the overlying seawater to the brine–commonly referred to as the brine-seawater interface (BSI), forms a particularly interesting environment, which is characterized by steep gradients of temperature, salinity, pH and dissolved oxygen (Eder et al., 2001; Sass et al., 2001; Daffonchio et al., 2006; Borin et al., 2009; Swift et al., 2012). Here also, the concentration of nutrients (organic carbon, NH4+ and NO3–) and minerals are a few orders of magnitude higher than in the overlying seawater (Ryan et al., 1969; Anschutz et al., 1999; Bougouffa et al., 2013). Moreover, the BSI is a microbial ‘hotspot' harboring dense microbial populations, which are also metabolically more active than the adjacent layers (Karbe, 1987; Daffonchio et al., 2006; Yakimov et al., 2007; Bougouffa et al., 2013).
Molecular-based evidences and geochemical data gathered in all brine pools studied to date document that sulfur cycling and aerobic oxidation of methane are the main energy-generating processes in the BSI habitat (Faber et al., 1998; Eder et al., 2002; Schmidt et al., 2003; Daffonchio et al., 2006; Yakimov et al., 2007; Borin et al., 2009; Joye et al., 2009; La Cono et al., 2011; Ferrer et al., 2012; Bougouffa et al., 2013). Although ammonia-oxidizing archaea (AOA) typically predominate the deep-sea realm of the global ocean (Stahl and de la Torre, 2012; and references therein), data from some Mediterranean Sea brines suggest that Archaea are less predominant in the BSI environment (∼10% of all prokaryotes; Daffonchio et al., 2006; Yakimov et al., 2007; Borin et al., 2009). This implies that the conditions in the transition layer probably act as a major constraint on the success of AOA.
Previous studies have also shown that increasing salinity leads to loss of diversity among ammonia-oxidizing prokaryotes in various aquatic environments (Bernhard et al., 2005; Erguder et al., 2009; Biller et al., 2012). Because the crenarchaeotal membrane lipid signature of surface sediments in the brine-bearing northern Red Sea area deviates from that in the open ocean (Trommer et al., 2009), it is also presumed that the thaumarchaeotal subpopulations in the bathypelagic habitats of these areas represent salt-adapted lineages. Prompted by our limited knowledge of thaumarchaeal diversity and an understanding of how putative AOA in the BSI may thrive under high salinity, we hypothesized that osmotic stress may have constrained the success of most bathypelagic lineages, thus selecting for a highly diverged AOA subpopulation with distinct genetic attributes. Accordingly, we employed phylogenomic-based approaches combining pyrotag amplicon-sequencing, phylogenetics, single-cell genomics (Stepanauskas, 2012) and fragment recruitment analyses to both assess and compare the AOA diversity in the BSI of several geochemically distinct brines of the Red Sea, and to elucidate their unique genetic attributes for halotolerance.
Materials and methods
Four hundred and fifty-four pyrosequencing, single-cell genomics and metagenomic analyses were conducted on microbial biomass captured from BSI samples collected from various deep hypersaline anoxic brines of the Red Sea during the 3rd WHOI-KAUST sampling expedition on board the R/V Aegaeo in October/November 2011 (Table 1; Supplementary Figure 1). Temperature, salinity, oxygen and pressure were measured on-line using a conductivity–temperature–depth unit (Sea-Bird Electronic, Bellevue, WA, USA). The elemental composition and concentrations of nutrients in the BSI and brine samples were determined using the commercial service provided by GEOMAR Helmholtz Centre for Ocean Research, Kiel (Germany). DNA extracts of three samples (200, 700 and 1500 m) previously collected from the water column overlying Atlantis II Deep in central Red Sea (21°20.76′N, 38°04.68′E; October 2008; Ngugi and Stingl, 2012) were also included for phylogenetic community comparison with that of the BSI. Pyrotag sequencing was done using 16S rRNA gene primers for the V3–V6 (B343F/B1099rc; Liu et al., 2007) and V6–V9 (A934F/U1492R; Lane, 1991; Teske and Sorensen, 2008) regions for bacteria and archaea, respectively. Cell sorting and single-cell genomics were conducted at the Single Cell Genomics Center, Bigelow Laboratory for Ocean Sciences (East Boothbay, ME, USA) as described in Rinke et al. (2014). Comparative genome analysis was performed using the phylogenomic platform EDGAR (Blom et al., 2009), which uses an automatically calculated Blast score ratio value cutoff for identifying conserved and unique gene sets among genome pairs based on bi-directional best blast hit analyses. For this analysis, we used the complete genome of Nitrosopumilus maritimus SCM1 as reference (Walker et al., 2010). Unless mentioned otherwise, the gene locus IDs of the reference genome will be used to provide gene context throughout the text. Full details of materials and methods are provided as Supplementary Information.
Table 1. The physical and geochemical parameters in the BSI layer and brine bodies of five deep hypersaline anoxic brines from the Red Sea.
| Parameters | Atlantis II Deep (S-W basin)a | Discovery Deepa | Erba Deepa | Kebrit Deepa | Nereus Deep (E-W basin)a | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
BSI |
UCL1 |
UCL2 |
LCL1 |
Brine |
BSI |
Brine |
BSI |
Brine |
BSI |
Brine |
BSI |
Brine |
| Depth (m) | 1998 | 2006 | 2025 | 2048 | 2062 | 2038 | 2141 | 2381 | 2392 | 1467 | 1490 | 2432 | 2445 |
| Salinity (%) | 5.6 | 8.6 | 11.2 | 15.4 | 26.2 | 13.8 | 26.0 | 9.8 | 18.1 | 18.2 | 26.0 | 15.4 | 22.4 |
| Temp. (°C) | 31.8 | 46.0 | 52.1 | 65.0 | 67.8 | 35.8 | 44.8 | 23.1 | 28.2 | 21.9 | 23.4 | 27.9 | 30.1 |
| pH | 6.5 | 5.8 | 5.7 | 5.2 | 5.1 | 6.6 | 6.4 | 7.7 | 7.2 | 5.7 | 5.2 | 7.7 | 5.5 |
| Oxygen (μM) | 2.2 | 0.5 | 0.5 | 0.5 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 4.6 | 2.3 | 0.3 | 2.8 |
| Nitrate (μM)b | 55.3 | 37.9 | 46.1 | 21.9 | 32.7 | 30.1 | 43.9 | 29.0 | 55.5 | 28.4 | 44.1 | 24.2 | 59.1 |
| Ammonia (μM) | 2.0 | 107.0 | 174.0 | 378.0 | 806.0 | 289.0 | 852.0 | 190.0 | 691.0 | 2017.0 | 2657.0 | 650.0 | 1116.0 |
| Urea (μM) | 1.7 | 1.5 | 1.3 | 1.0 | < 0.1 | 1.3 | 1.5 | 1.4 | 1.6 | < 0.1 | 1.1 | 2.4 | < 0.1 |
| Sulfate (mM) | 32.5 | 30.2 | 28.7 | 25.2 | 8.3 | 26.7 | 8.4 | 38.6 | 41.6 | 31.1 | 27.7 | 22.5 | 10.5 |
| H2S (μM) | –c | – | – | – | – | – | – | – | – | 149.8 | 526.7 | – | – |
| Mn (mM) | 0.1 | 0.8 | 1.2 | 1.8 | 1.7 | 0.5 | 1.1 | 0.2 | 0.6 | 0.1 | 0.2 | 0.9 | 1.0 |
| Fe (μM) | 0.5 | 0.8 | 24.5 | 70.5 | 1528.0 | 0.6 | 0.1 | < 0.1 | 31.8 | 40.4 | 1.5 | 0.5 | 27.2 |
| Chloride (M) | 0.8 | 1.5 | 2.0 | 3.0 | 5.2 | 2.5 | 5.1 | 1.6 | 3.3 | 3.4 | 5.0 | 2.9 | 4.2 |
| Bromide (mM) | 1.0 | 1.1 | 1.2 | 1.3 | 1.5 | 1.3 | 1.5 | 1.1 | 1.4 | 2.5 | 3.0 | 2.4 | 1.5 |
| Ca (mM) | 17.3 | 38.8 | 52.3 | 81.6 | 142.2 | 64.0 | 134.1 | 21.0 | 32.7 | 49.0 | 53.6 | 140.0 | 224.7 |
| B (mM) | 0.5 | 0.6 | 0.6 | 0.7 | 1.0 | 0.7 | 1.0 | 0.6 | 0.7 | 2.1 | 2.5 | 0.8 | 0.8 |
| Li (μM) | 49.6 | 126.0 | 174.9 | 286.0 | 522.7 | 231.3 | 431.9 | 42.9 | 68.8 | 360.7 | 406.6 | 248.9 | 186.9 |
| Na (M) | 0.7 | 1.4 | 1.8 | 2.9 | 4.9 | 2.3 | 4.7 | 1.5 | 3.2 | 3.2 | 4.8 | 2.5 | 3.5 |
| K (mM) | 13.2 | 20.8 | 26.3 | 40.4 | 63.2 | 31.4 | 60.8 | 17.1 | 34.5 | 26.5 | 35.5 | 52.8 | 71.4 |
| Mg (mM) | 61.8 | 55.9 | 51.6 | 44.6 | 29.1 | 49.5 | 33.0 | 61.1 | 71.4 | 104.3 | 118.7 | 67.3 | 67.4 |
| Sr (μM) | 121.5 | 190.7 | 236.2 | 338.4 | 596.8 | 284.6 | 599.7 | 176.9 | 265.8 | 631.6 | 685.2 | 476.5 | 712.2 |
| Si (mM) | 0.1 | 0.4 | 0.5 | 0.7 | 1.0 | 0.3 | 0.1 | 0.1 | 0.2 | 0.4 | 0.6 | 0.1 | 0.1 |
| Ba (μM) | 1.7 | 6.2 | 9.7 | 1.2 | 1.9 | 1.7 | 5.6 | 5.0 | 1.2 | 0.8 | 1.0 | 0.2 | 3.7 |
Abbreviations: BSI, brine-seawater interface; LCL, lower convective layer; Temp., temperature; UCL, upper convective layer.
Sampling locations (Lat./Long.): Atlantis II (21° 20.76'/38° 04.68'); discovery (21° 16.98'/38°) nereus (23° 11.53'/37° 25.09'). More details in Supplementary Figure 1.
Nitrite was below detection limit (0.1 μM).
Below detection limit (∼1 μM).
Results and Discussion
Physicochemical conditions in the BSI
Ammonia, and not nitrate, dominates the inorganic nitrogen pool of all layers of the brines (Table 1; Supplementary Figure 1), ranging between 2 μM and 2 mM in the BSI and 0.8–2.7 mM in the brine body. Nitrate concentrations ranged from 22 to 59 μM, being highest in the brine, whereas urea and nitrite were extremely low (<0.1–2 μM). Owing to the reported sensitivity of AOA to high ammonia concentrations (Prosser and Nicol, 2012), the high ammonium levels in the BSI should favor ammonia-oxidizing bacteria (AOB) over AOB, but AOB are completely absent in the BSI environment (Yakimov et al., 2007; see below), suggesting that differences in ammonia uptake kinetics (Martens-Habbena et al., 2009) may not solely explain niche differentiation of AOA and AOB in these habitats. In contrast, dissolved oxygen is present at relatively low concentrations in the BSI compared with the deep-seawater mass in the Red Sea (∼1–5 μM vs ∼90 μM; Ngugi and Stingl, 2012; Table 1), but is within the minimal reported concentration at which AOA are still capable of nitrification (∼2–10 μM O2; Martens-Habbena et al., 2009).
The total iron content in the BSI (∼0.01–70 μM) is much higher than in the deep-seawater (<0.1 μM; Danielsson et al., 1980), whereas manganese is very high (up to 1.7 mM) and seems to rise with increasing salinity of the brine. Owing to their high concentration (Table 1), affinity and possible precipitation with other elements (Krom and Berner, 1980), these two ions may exacerbate the already limited availability of some essential nutrients in the BSI, for example, phosphate (<1 μM; Karbe, 1987; Bougouffa et al., 2013). Sulfate concentrations broadly decreased from normal-seawater levels (∼30 mM) to about 10 mM in the brine, indicating sulfate reduction in the anoxic brine layers. Only Kebrit Deep had remarkably high sulfide concentrations––both in the BSI (150 μM H2S) and the brine (527 μM H2S), which based on isotopic data, are proposed to be mostly of abiotic origin ( Blum and Puchelt, 1991; Hartmann et al., 1998).
Microbial community structure in the BSI
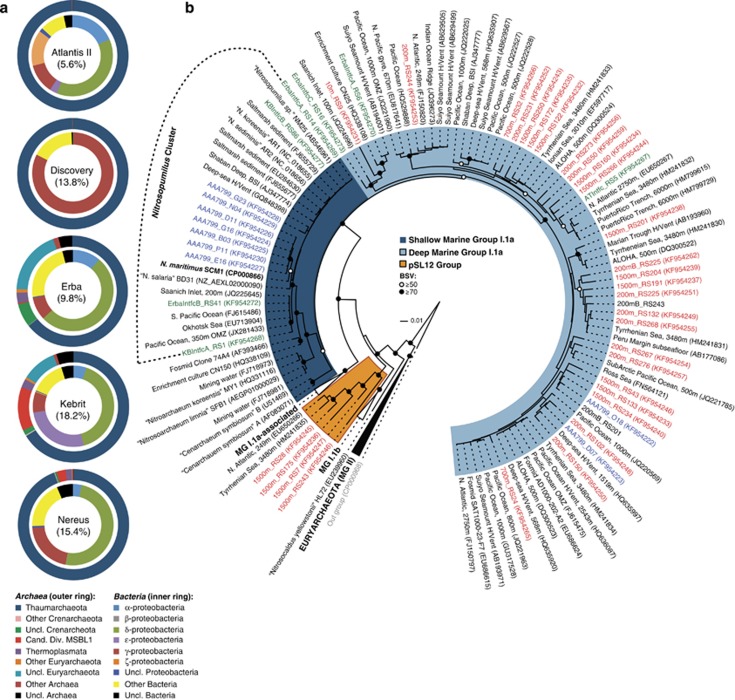
These variable conditions and the high salinity (5.6–18.2% (w/v)) of the BSI layer in all five DHABs studied here should impact the microbial community structure of the BSI. Based on 454 amplicon data (details in Supplementary Material), we found that members of the phylum Thaumarchaeota predominated the archaeal community of the BSI (64–99%) irrespective of the sample (Figure 1a), whereas affiliates of the domain Bacteria were diverse, brine specific and predominated by the phylum Proteobacteria (72–98%), similar to Mediterranean Sea DHABs (Borin et al., 2009; La Cono et al., 2011). Like in earlier studies (Yakimov et al., 2007), none of the canonical AOB were found (that is, Nitrosomonas, Nitrosospira and Nitrosococcus; Kowalchuk and Stephen, 2001) even after our extensive sequencing effort (93–99% Good's estimated coverage). Complementary blast-based analyses for homologs of bacterial ammonia monooxygenase genes in metagenomic data sets (Supplementary Table 8) corroborated the absence of AOB in the BSI (data not shown). Based on the 454 data, we also observed that sequences belonging to a putative nitrite-oxidizing Nitrospina-like cluster accounted up to one-third of the bacterial fraction in Atlantis II Deep but only marginal (∼2%) in other brines including samples from the overlying water column (data not shown). Collectively, this means that these AOA are probably the sole ammonia oxidizers in the BSI environment, and also hints for a potential metabolic interaction between Nitrospina-like bacteria and thaumarchaea in Atlantis II Deep.
Figure 1.
Microbial community structuring in the brine-seawater interface (BSI) of five brine pools (a) and the phylogeny of thaumarchaea (b) in the BSI and the overlying seawater. Classification of 454 reads is based on the Greengenes taxonomy, whereas the phylogenetic tree was inferred using the neighbor-joining approach from nearly full-length 16S rRNA gene sequences from the overlying seawater (in red), BSI (in green); only representative sequences (at 97% identity; Supplementary Figure 2B) are shown. Sequences from single-cell amplified genomes are shown in blue (more details in supporting materials). The percentages in brackets denote the salinity of the sample. Phylogenetic lineage definitions follow Francis et al. (2005).
Thaumarchaeal diversity and phylogeny in the BSI
The thaumarchaeal population in the BSI layer of all five studied brine pools exhibits a high degree of conservation, as a single highly abundant and BSI-specific phylotype is dominating. Based on 16S rRNA gene analyses, this phylotype accounts for >98% of all sequences (Supplementary Figure 2A). To gain further insight on the phylogeny of this ubiquitous phylotype, we sequenced nearly full-length 16S rRNA genes from three of the BSIs together with those from the water column overlying Atlantis II Deep. Again, clustering of all these clonal sequences (identity at 97%) proved that most of the thaumarchaeal sequences in the BSI habitat belonged to a single AOA phylotype, which was absent from the overlying water column (200–1500 m; Supplementary Figure 2B).
Interestingly, phylogenetic analysis of representatives of the above sequence clusters further indicated that the predominant BSI-specific AOA belong to the Shallow Marine Group I (SMGI) clade (Francis et al., 2005) and not the Deep Marine Group I (DMGI) clade or even the pSL12 lineage (Figure 1b). Their 16S rRNA gene sequences are ∼99% identical to the type strain N. maritimus SCM1 (Könneke et al., 2005) and four other recently published enrichment cultures (Matsutani et al., 2011; Mosier et al., 2012; Park et al., 2012b). Analogous analyses using functional gene markers encoding for the alpha subunits of the ammonia monooxygenase and 4-hydroxybutyryl-CoA dehydratase enzymes also corroborate the membership of the BSI thaumarchaea within the genus Nitrosopumilus (Supplementary Figure 3), which is more representative of environmental sequences from marine sediments rather than the open-ocean environment (Francis et al., 2005). Overall, these results provide strong evidence for niche partitioning among the AOA in the water column and brine pools of the Red Sea. Moreover, a large number of environmental sequences that cluster with the Nitrosopumilus cluster were retrieved from habitats with a broad salinity range (5–33% (w/v)), or replete with ammonium (0.1–2 mM) and sulfate (0.1–701 mM; Yakimov et al., 2007; Nelson et al., 2009; Swan et al., 2010; Park et al., 2010; La Cono et al., 2011), which implies a broader halotolerance and greater tolerance for ammonium in some species of this genus.
Single-cell genome features
To explore the special genetic attributes of the Nitrosopumilus species residing in the BSI environment, we subsequently employed single-cell genomics (Stepanauskas, 2012) to assess the unique adaptations of putative AOA in the Atlantis-BSI. Here, the genomes of nine individual cells were reconstructed from single sorted cells, out of which seven belonged to the SMG1 clade, whereas two affiliated with the DMGI clade (Figure 1b). Only single-cell amplified genomes (SAGs) with an assembly size greater than 0.4 Mbp were considered for further analyses (Supplementary Tables 1 and 2). The assembled SAGs ranged in sizes of 0.43–0.52 Mbp (DMGI clade) and 1.0–1.42 Mbp (SMGI clade). The overall features of the draft genomes of our SMGI clade SAGs were similar to the reference N. maritimus SCM1 genome (Walker et al. 2010; Supplementary Table 1) including GC content, the fraction of genes predicted functional (40–75%) or orthologous to SCM1 (∼60%). Based on the occurrence of a set of 801 conserved single-copy genes found in seven published MGI.1a thaumarchaea (∼5–84.4% Supplementary Table 3), we estimate that the genome size of the AOA subpopulation represented by these SAGs ranges from 1.0 Mbp (DMGI clade) to 1.7 Mbp (SMGI clade).
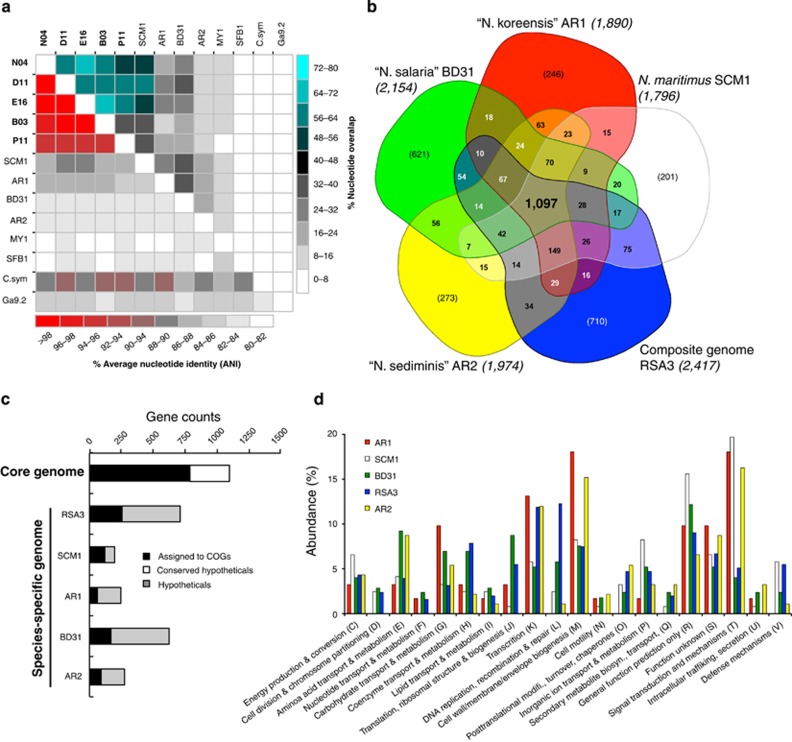
Further, analyses comparing the average nucleotide identity (ANI) of the overlapping nucleotide bases (Teeling et al., 2004) between individual SAGs and to published Nitrosopumilus genomes, showed that any of our SAG pairs within the SMGI clade had ∼50–70% of their assembled bases overlapping with ANIs of greater than 96%, but were on average less than 90% identical to reference strains (Figure 2a). The draft DMGI clade SAGs were, however, less identical to each other (∼37% genome overlap; 86% ANI) and also to representative genomes within the SMGI clade (∼60% genome overlap; 70% ANI), which includes five of our SAGs (Supplementary Table 1). Altogether, this highlights the strong variability and possible divergence of our SAGs to the currently sequenced Nitrosopumilus genomes and also their uncultured deep-sea counterparts. In view of the operational ANI of ∼95%, which has become a standard for species circumscription (Konstantinidis and Tiedje, 2005), we conclude that the population represented by our SAGs in the SMGI clade constitutes a novel species of the genus Nitrosopumilus, whereas those in the DMGI clade represents an uncharacterized lineage. Because of the high representation and the unprecedented occurrence of SMGI clade-like thaumarchaea in BSI habitats examined here, we subsequently focused our comparative genome analysis on the related groups of SAGs.
Figure 2.
Comparison of all Nitrosopumilus genomes based on (a) whole-genome alignment of the assembled bases, (b) reciprocal blast analysis of their predicted protein-coding genes, (c) functional assignment of cluster of orthologous genes (COGs) in the core and species-specific genomes and (d) relative abundance of COG categories in their variable genomes. (a) Highlights the relatedness of our SAGs (in bold) to each other than to published genomes, whereas b–d inventories differences in the proteome of the co-assembled composite Nitrosopumilus-like genome RSA3 to four publicly available Nitrosopumilus genomes. (b) The Nitrosopumilus core genome with 1097 genes (in bold), the species-specific gene sets (in bracket) and the flexible genes present in some but not all genomes. The total predicted protein-coding genes in each genome are indicated in brackets next to their names.
Taking into account the length of the assembly (>1 Mbp) coupled to both the proportion of genomic overlap (>63%) and ANI of greater than 97% exhibited by E16, D11 and N04 (Figure 2a), and their high degree of gene order conservation (synteny) of more than 57% (Supplementary Table 4) as well as negligible phylogenetic distance as inferred from multiple gene sets (Supplementary Figure S4), we selected these three SAGs for combined assembly. The co-assembled genome thus represents a composite genome of the subpopulation specific to this environment (5.6% (w/v); Atlantis II Deep), which is designated here as composite genome Nitrosopumilus-like RSA3 to reflect its membership within the genus Nitrosopumilus and the fact that it is a co-assembly of three Red Sea SAGs (that is, RSA3). The composite RSA3 genome provides a greater increase of sequenced data at 1.85 Mbp from 109 contigs (N50=41 609 bp) with an estimated completeness of 91% (Supplementary Tables 1 and 3). Also like other recently published Nitrosopumilus genomes, the co-assembled genome shares up to 60% of its proteome with N. maritimus (Supplementary Table 1). Unless otherwise mentioned, this composite genome will be referred throughout as RSA3, whereas reference to its protein-coding genes will be denoted with ‘SCCG RSA3' followed by a specific gene locus ID.
The pan- and core genome of the genus Nitrosopumilus
In order to place the RSA3 genome into perspective, we estimated both the total set of non-redundant genes in all Nitrosopumilus species (that is, the pan-genome) and the subset that is shared by all species (that is, the core genome). The pan-genome of all five Nitrosopumilus genomes comprises of 4094 genes with a conserved core of 1097 genes (Figure 2b). When the calculations were done to include the pan-genome (3149 genes) of all five SAGs within the SMGI clade instead of the co-assembled RSA3 genome, the core genome remained almost the same at 1129 genes. Together with the fact that 62–67% of the protein-coding genes from RSA3 are conserved in their order relative to the three SAGs used for the co-assembly (E16, D11 and N03), this also means that the composite genome well captures the core content of the local AOA subpopulation despite probable MDA amplification biases. Nonlinear prediction models based on decay functions (Tettelin et al., 2005) further estimate the Nitrosopumilus core genome size to plateau at around 1020 genes (Supplementary Figure 5), which is equivalent to 42–57% of their predicted proteomes. This degree of genome conservation follows the general trend of other marine bacterioplankton with similar levels of intraspecific divergence (∼3% 16 S rRNA gene divergence; Kettler et al., 2007; Oda et al., 2008; Grote et al., 2012), and compares well to recent estimates for separate MGI lineages (1057 genes; Alonso-Sáez et al., 2012). Still, the high count of singleton genes indicates both the openness of the Nitrosopumilus pan-genome (Supplementary Figure 5), and the likelihood of finding more unique genes in subsequent genomes.
Core and variable gene inventory
Most genes in the core genome (72%) matched numerous functional categories in the Clusters of Orthologous Groups database (Figure 2c), comprising those encoding enzymes for several central metabolic pathways synonymous with members of the phylum Thaumarchaeota (Supplementary Table 5). These include the 3-hydroxypropionate/4-hydroxybutyrate carbon fixation pathway (Berg et al., 2007), the incomplete tri-carboxylic acid cycle (Hallam et al., 2006b), ammonia oxidation (Walker et al., 2010), assimilatory sulfate reduction and the CDVABC- and the ftsZ-based cell division systems (Pelve et al., 2011), as well as housekeeping functions such as protein synthesis, nucleic and amino-acid metabolism, and transport functions (Supplementary Tables 5 and 6). However, one-third of the core genes encode for hypothetical proteins (Figure 2c), which indicates that they are conserved in all Nitrosopumilus genomes, therefore implying that many facets of the biology of this genus still remain unresolved.
We also found 201–710 genes that are unique to each genome (that is, the species-specific genome), which could serve for species-specific adaptations and provide means for functional differentiation among members of this genus (Figure 2b). These gene sets equate from as little as 11% (N. maritimus SCM1) to a maximum of 29% (RSA3) of their predicted protein-coding genes, and again include many that cannot be assigned to Clusters of Orthologous Groups, especially for four of the genomes (RSA3, AR1, AR2 and BD31), where they account between 64 and 75% of their total unique genes (Figure 2c). The high proportion of hypothetical proteins in the variable genome implies that there might be several critical, albeit unknown, mechanisms of niche adaptation.
For those genes with functional predictions, many pertain to Clusters of Orthologous Groups related to ‘cell wall/membrane/envelope biogenesis' (AR1 and AR2), ‘signal transduction mechanisms' (SCM1 and AR1), ‘transcription' (RSA3 and AR2) or ‘general functions' (SCM1 and BD31; Figure 3d). There is also a large fraction of genes in the unique genome of RSA3 (12%) encoding for ‘repair, replication and recombination' enzymes (for example, methylases and recombinases; Supplementary Table 7), presumably as a consequence of the destabilization effect of salts on biomolecules like DNA and proteins (Ziemienowicz et al., 2011), which also increases homologous recombination events in some microorganisms (Boyandin et al., 2000), preferentially in (halophilic) Archaea than in bacteria (Naor et al., 2012).
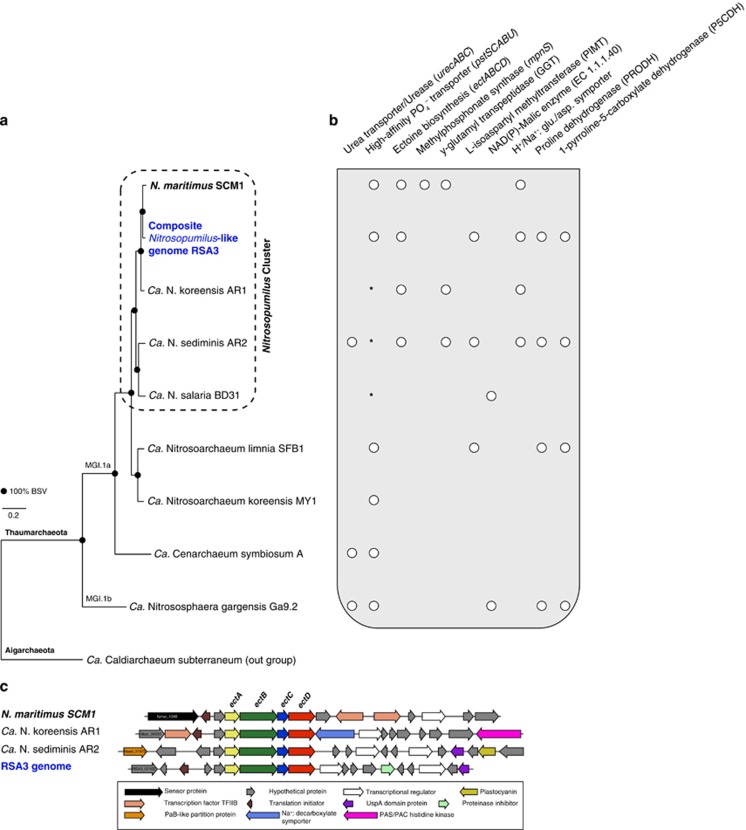
Figure 3.
Core gene-based phylogeny (a) and inventory of select metabolic features of the thaumarchaeal variable proteome (b and c). (a) A maximum-likelihood tree of 301 concatenated genes, (b) depicts the presence (shown as white dots) or absence of genes in sequenced thaumarchaeal genomes, whereas (c) emphasizes the gene-neighborhood differences around the ectABCD operon involved in ectoine biosynthesis between Nitrosopumilus species. Asterisk (*), low-affinity phosphate transporters; ectA, L-2,4-diaminobutyrate Nγ-acetyltransferase; ectB, L-2,4-diaminobutyrate acetyltransferase; ectC, ectoine synthase; ectD, ectoine hydroxylase.
Functional plasticity in the genus Nitrosopumilus
Although members of the Nitrosopumilus cluster are very close at the phylogenomic level (amino-acid identity 86–93% relative to SCMI; Figure 3a) and at the 16S rRNA gene level (up to 99% identical), we found many, albeit species-specific metabolic differences that provide evidence for functional differentiation among species of these ubiquitous marine Archaea (Figure 3b; Supplementary Table 7). Some of these metabolic traits relate to, and corroborate initial genome reports and activity measurements including: the potential for urea utilization (for example, AR2; Park et al., 2012a), the capacity to synthesize methlyphosphonate (for example, only in SCM1; Metcalf et al., 2012), alternative routes for phosphoenolpyruvate biosynthesis using the NAD(P)-dependent malic enzyme and the potential inability to synthesize ectoine (for example, BD31; Mosier et al., 2012; Spang et al., 2012), as well as the presence of putative high-affinity phosphate uptake systems (Hallam et al., 2006a; Walker et al., 2010; Mosier et al., 2012; Park et al., 2012b).
By including the composite genome of RSA3 in these analyses, we further show that the putative AOA subpopulation in the Atlantis-BSI harbors and shares most but not all of these metabolic facets with the type species of this genus. For example, RSA3 and SCM1 are the only members of their genus harboring a putative high-affinity phosphate-specific transport (Pst) system (pstSCABU; Nmar_0479/0481–0484; Figure 3b), which might be correlated with the extremely low concentration of dissolved inorganic phosphate in the Atlantis-BSI (<1 μM; Bougouffa et al., 2013); the other species are predicted to have the low-affinity phosphate inorganic transporter. The Pst system occurs also in non-marine thaumarchaea (Kim et al., 2011; Blainey et al., 2011; Spang et al., 2012; Figure 3b), suggesting that phosphorus limitation is not necessarily confined to marine Thaumarchaeota.
Other functional differences present in the pan genome of thaumarchaea from the BSI relate to the potential use of γ-glutamyl peptides as a source of sulfur-containing amino acids from the catabolism of glutathione (γ-L-glutamyl-L-cysteinyl-glycine; Meister and Anderson, 1983). For example, both RSA3 and BD31 lack the gene encoding for a γ-glutamyl transpeptidase (Nmar_1004; Figure 3b), which releases cysteine by catalyzing the breakage of the gamma linkage in glutathione. However, the genome of RSA3 (and AR2) harbors a gene encoding for L-isoaspartyl methyltransferase (SCCG-RSA3_00440; Figure 3b), which recognizes altered aspartyl residues in proteins and polypeptides and methylates them to normal L-aspartyl residues (Young et al., 2001). Owing to the potential exposure of cells to heavy metals, oxides (for example, Fe2+ and HS–) and radicals in the brine or sediments, this enzyme could help reduce damage to cellular biomolecules.
Osmotolerance
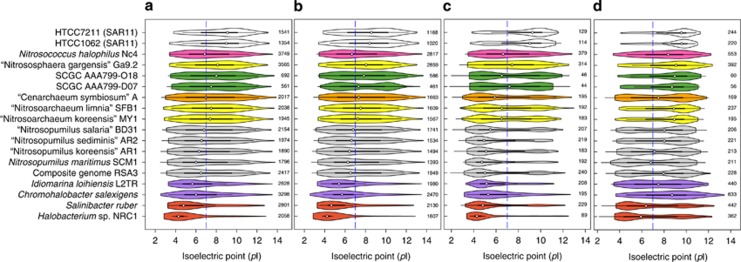
Osmotic stress is considered as one of the key factors restricting the success of ammonia oxidizers (Oren, 2011), with salinity being implicated as a major driving force for niche adaptation among aquatic AOA and AOB (Bernhard et al., 2005; Erguder et al., 2009; Biller et al., 2012). Most extreme halophiles (for example, Halobacterium) adapt to high salinity by increasing their intracellular salt concentrations (Oren et al., 2002). Consequently, the proteins of halophilic organisms tend to have acidic residues on their surface and show optimal activity and stability at high salinity (Paul et al., 2008; Oren, 2013). In contrast, moderate halophiles, which grow at 5–20% NaCl (Kanekar et al., 2012), keep their intracellular ionic concentrations at low levels and use organic solutes to produce the necessary osmotic balance without dramatically altering the structure of their proteome (Oren, 2011). By comparing the distribution of genes based on the isoelectric point (pI) of the encoded proteins (Figure 4), we found that for all Nitrosopumilus species, the median pI of the overall proteome or only those of predicted proteins with no integral membrane domains were similar to each other (∼6.5) but significantly higher (Kruskal–Wallis test, P<0.001) than in exemplary moderate halophiles (∼5.7). However, proteins of both groups of organisms with a single predicted integral membrane domain showed a remarkably similar unimodal distribution (median pI of ∼5.1; Figure 4c). Also, both had a significantly lower pI (P<0.001) compared with other AOA lineages including our mesopelagic SAGs (∼6.9), and the most abundant planktonic bacteria of the SAR11 clade (∼9.1). We estimate that these proteins account for 17–25% of the predicted proteome of Nitrosopumilus species, with about 10% containing signal peptides. The highly acidic nature of only these proteins and not of integral membrane proteins with two or more trans-membrane helices (Figure 4d) altogether implies that the osmotolerance of Nitrosopumilus species (and other moderate halophiles) partly depends on the adaptation of peptides located peripheral to the cell membrane.
Figure 4.
Violin plots showing the distribution in the isoelectric point of predicted proteomes among members of the genus Nitrosopumilus (in gray), AOA from soil/sediment (in yellow), host-associated AOA (in orange), mesopelagic AOA (in green), an obligately halophilic ammonia-oxidizing bacterium (in magenta) and typical planktonic bacteria (in white) relative to extreme (in red) and moderate (in purple) halophiles. The four panels show data for the overall predicted proteomes (a), as well as protein-coding genes without any trans-membrane domains (b), with a single trans-membrane domain (c), and those with two or more trans-membrane domains (d). The total gene counts for each data are shown on the right side of each panel. The dashed blue line demarcates a pI of 7.0, whereas ringed dots show the median pI.
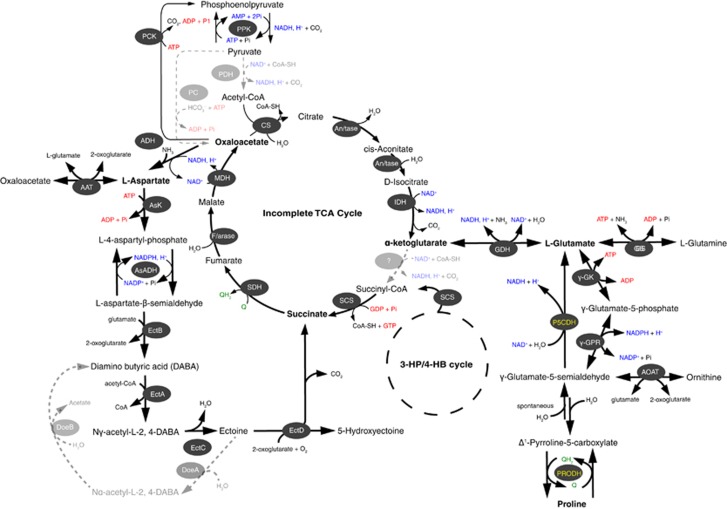
The presence of genes putatively encoding for enzymes involved in ectoine and hydroxyectoine production in marine Nitrosopumilus species (ectABCD; Nmar_1343–1346; Figure 3b) further suggests that they additionally rely on ectoine for osmoprotection. The same strategy is probably used by the most halotolerant, ammonia oxidizer known to date, Nitrosococcus halophilus, which grows with up to 94 g NaCl l–1 (Koops et al., 1990). ‘Ca. Nitrosopumilus salaria' BD31 is the only species in this genus lacking these genes, presumably because it was isolated from brackish sediments in the San Francisco Bay estuary with variable salinity (Mosier et al., 2012; Figure 3c). Subsequent analyses of several mesopelagic thaumarchaeal SAGs from a variety of marine environments present in the Integrated Microbial Genome database (https://img.jgi.doe.gov/; Rinke et al., 2013; Luo et al., 2014) indicate that these deep-water phylogenetic clades lack ectoine biosynthesis genes. This implies that this biochemical pathway is probably exclusive to marine Nitrosopumilus species given also the highly conserved organization of their ectABCD operon (Figure 3c), thus supporting the idea of salinity as a driver for the niche speciation of marine AOA (Erguder et al., 2009; Biller et al., 2012). Also, the fact that the accessory gene cassette co-transcribed with the ectABCD unit is variable among different Nitrosopumilus species, further suggests some degree of flexibility on the levels of ectoine produced. Although the mechanism of action remains to be ascertained, it is possible that the proteinase inhibitor l1 Kazal (SCCG-RSA3_02089) on the operon of RSA3 serves to regulate ectoine production or the enzymatic cascade leading to ectoine. The linkage of this operon also to a universal stress response protein (UspA), implies that it may also be responsive to other stress cues. Moreover, the potential to convert ectoine further to hydroxyectoine, which is produced in some organisms as a response to heat stress (Pastor et al., 2010), might provide thermo-protective properties for AOA residing in the Atlantis II Deep (32–68 °C; Table 1).
The presence of numerous transport systems in the Nitrosopumilus core genome that potentially take up glutamate (via a putative sodium-dependent glutamate/aspartate symporter; Nmar_1093; Figure 3b) or glycerol (using major intrinsic channel proteins; Nmar_0489/0561) also implies the possibility of using exogenously supplied osmolytes. Glutamate is possibly also replenished internally through the reversible reactions of glutamate dehydrogenase (Nmar_1312), glutamine synthetase (Nmar_1771/1790) or via the ornithine–glutamate reaction cascade (Figure 5) catalyzed by γ-acetylyglutamate kinase (Nmar_1290), N-acetyl-γ-glutamyl phosphate reductases (Nmar_1289) and acetylornithine aminotransferase (Nmar_1291).
Figure 5.
Metabolic scheme of ectoine, glutamate and proline synthesis and catabolism inferred from the Nitrosopumilus genomes. Enzymes for each pathway are represented as oval shapes and color-coded according to their occurrence in all species (white font), or only in RSA3 and AR2 (yellow font). Gray dashed arrows highlight pathways that are absent in all Nitrosopumilus species. AAT, aspartate aminotransferase; ADH, aspartate dehydrogenase; AOAT, acetylornithine aminotransferase; AsK, aspartate kinase; AsADH, aspartyl-semialdehyde dehydrogenase; GDH, glutamate dehydrogenase; GS, glutamine synthetase; γ-GK, acetylyglutamate kinase; γ-GPR, N-acetyl-γ-glutamyl phosphate reductase; EctA, L-2,4-diaminobutyrate acetyltransferase; EctB, L-2,4-diaminobutyrate transaminase; EctC, ectoine synthase; EctD, ectoine hydrolase; DoeA, ectoine hydrolase; DoeB, Nα-acetly-L-2,4-diaminobutyrate deacetylase; P5CDH, 1-pyrroline-5-carboxylate dehydrogenase; PRODH, proline dehydrogenase. Other enzymes shown are for pyruvate (de)carboxylation, the Tri-carboxylic acid (TCA) cycle and 3-hydroxypropionate/4-hydroxybutyrate (3-HP/4-HB) cycle including: An/tase, aconitase; CS, citrate synthase; F/arase, Fumarase; IDH, isocitrate dehydrogenase; MDH, malate dehydrogenase; PPK, phosphoenolpyruvate (PEP) kinase; PCK, PEP carboxykinase; PC, pyruvate carboxylase; PDH, pyruvate dehydrogenase; SCS, succinyl-CoA synthetase; SDH, succinate dehydrogenase.
Interestingly, we found also that only RSA3 and AR2 carry the two key enzymes mediating the catabolism of proline to glutamate (Figures 3b and 5), namely 1-pyrroline-5-carboxylate dehydrogenase (SCCG-RSA3_02256) and a putative proline dehydrogenase (PRODH; SCCG-RSA3_02257/02258). This suggests the potential for osmolyte switching (Martin et al., 2001; Saum and Muller, 2007), which might offer these AOA a greater osmotolerance in a halocline environment like the BSI. PRODH genes also occur in two other thaumarchaea (Figure 3b), and altogether cluster in a phylogenetic branch of mono-functional PRODHs (Supplementary Figure 6), having an average sequence identity of ∼35% to characterized peptides from Thermus thermophilus and Bacillus subtilis.
Bioenergetic considerations
At increasing salinities, cells require extra energy to synthesize or take up solutes and to extrude sodium ions, and this energetic liability has often been used to explain the observed decline in metabolic diversity with increasing salinity (Oren, 2011). Thus, metabolic processes from which less energy is gained might be precluded at higher salinities given the high cost of synthesizing compatible solutes (Oren, 2011). For example, the energy gained from the aerobic oxidation of ammonia or nitrite (ΔG0=–45.8 and –37.1 kJ per mol e– transferred, respectively) is relatively lower compared with sulfide oxidation to sulfate (–99.6 kJ). As such, the low-energy yield of nitrification may preclude this process at high salt concentrations, which may partially explain the reduced biomass of AOA and the higher density and phylogenetic diversity of sulfur oxidizers in the BSI layer (Yakimov et al., 2007, 2013).
Although the upper salinity limit for autotrophic ammonia oxidation is still uncertain, nitrification appears to be constrained at salt concentrations exceeding 100 g l–1 (Oren, 2011). The obligately halotolerant AOB Nitrosococcus halophilus Nc4, grows optimally at 41 g NaCl l–1 (maximum at 94 g NaCl l–1) having been isolated from a salt lagoon (off Sardinia, Mediterranean Sea; Koops et al., 1990). This salinity range is within the values in the BSI habitats studied here (47–199 g NaCl l–1; Table 1). However, the lack of AOB in the ammonium-rich BSI layer (Yakimov et al., 2007; this study) suggests that AOA are more salt tolerant that AOB.
Comparative genome analyses suggest that most marine Nitrosopumilus species are capable of producing ectoine and hydroxyectoine (Hallam, et al., 2006a; Walker et al., 2010; this study). Energetically, the production of such low-molecular-weight compounds (3–5 carbon units) could be advantageous for two reasons. They are relatively inexpensive to synthesize compared with large solutes and have no significant negative effects on cytoplasmic constituents. Oren (1999), for example, estimated that the cost for synthesizing ectoine (50 ATPs per mol ectoine) by autotrophs using the Calvin cycle to fix CO2 was twice lower that needed to produce sucrose or trehalose. Ectoine is probably also used strictly for osmoprotection as putative genes encoding enzymes for the degradation of ectoine namely ectoine hydrolase and Nα-acetyl-L-2,4-diaminobutyric acid deacetylase are absent in all thaumarchaeal genomes (Figure 5). Although there is no data of whether ectoine is excreted by thaumarchaea, the potentially one-way-only biosynthesis pathway implies that there is no ectoine turnover; therefore, very little energy would be invested subsequent to its production.
16S rRNA gene-based analyses indicate that thaumarchaea are part of the metabolically active prokaryotic community of the BSI (Yakimov et al., 2007), where the capacity to fix dissolved inorganic CO2 was also higher than in the bathypelagic waters (Yakimov et al., 2007; 2013). The high dissolved inorganic CO2-incorporating activity coincidentally matched profiles of amoA-like mRNA gene transcripts (Yakimov et al., 2007), suggesting that most of the AOA are autotrophic. Recent studies however, indicate that not all AOA are strictly chemoautotrophic as previously assumed (Mussmann et al., 2011; Tourna et al., 2011; Stahl and de la Torre, 2012; Seyler et al., 2014), as they are capable of using a variety of alternative sources of energy (for example, urea) and carbon (for example, acetate, pyruvate and 2-oxoglutarate). They also efficiently take up amino acids from their environment (Ouverney and Fuhrman, 2000; Teira et al., 2006; Varela et al., 2008), including aspartate––the precursor for ectoine biosynthesis. Hence, the co-assimilation of such organic substrates might potentially leverage the cost for combating salt stress by lowering the energy invested for the (de novo) synthesis of intermediates in the pathway series leading to ectoine and the glutamate family of osmoprotectants (glutamine and proline; Figure 5). Several central intermediates of their incomplete tri-carboxylic acid cycle are also renewable from reactions involving glutamate dehydrogenase (Nmar_1312), ectoine hydroxylase (Nmar_1343) and aspartate aminotransferase (Nmar_0546), which suggests a potential anaplerotic contribution of osmolytes to the carbon and energy flow of some (halotolerant) AOA.
Genotypic variants in the BSI environment
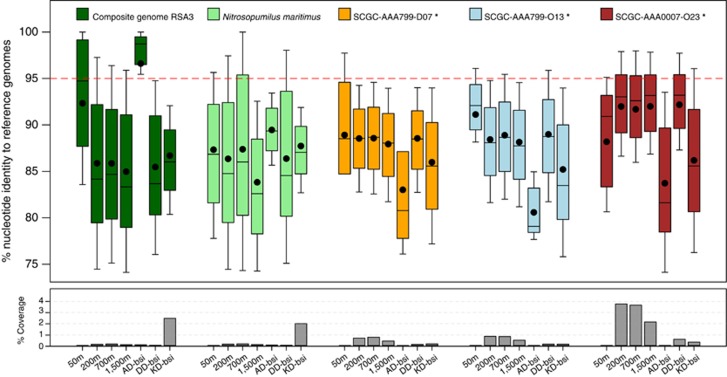
To better capture the thaumarchaeal population structure in distinct BSI habitats, we employed fragment recruitment analyses (Rusch et al., 2007) to recruit homologs of the assembled/reference genomes from several metagenomic data sets (Figure 6; Supplementary Table 8). Here, the genomes representing the SMGI clade (that is, RSA3 and SCM1; Walker et al., 2010; this study) and the DMG1 clade (that is, SCGC-AAA799-D07; SCGC-AAA799O18; SCGC-AAA0007-O23; Rinke et al., 2013; this study) were used as queries (more details in Supplementary Materials). Although fragments recruited using RSA3 genomes gave a high ANI of 97±5.0% (that is, in the Atlantis-BSI), we found no differences in the coverage of recruited reads against RSA3 and SCM1 genomes (<0.6% of total reads length) in all metagenomic data sets, despite our relaxed 70–100% nucleotide identity threshold. The exception was the Kebrit-BSI (18.2% salinity), where coverage was relatively high (2.0–2.4%) and ANI was around 87±4.5% (Figure 6), suggesting the presence of a diverged but highly abundant thaumarchaeal population in Kebrit Deep closely related to RSA3 (and SCM1). The abundance of these thaumarchaea is similar in range (∼26% of the total prokaryotic 16S rRNA genes) to those found in the Atlantis II Deep water column (200–1500 m; 10–26%) but higher than in the most BSI samples (∼10% Supplementary Table 9). This increase is not unexpected as the highly sulfidic (and suboxic) setting of Kebrit Deep is likely to favor thaumarchaea due to the inhibitory effect of sulfide and S-containing compounds on AOB activities (or Bacteria in general; Erguder et al., 2009).
Figure 6.
Fragment recruitment of metagenomic data from the Atlantis II Deep water column and the brine-seawater interfaces (BSI) of three brine pools in the Red Sea against the composite RSA3 genome and four others including the type species (N. maritimus SCM1) and three SAGs representing mesopelagic AOA (asterisked). The upper panel shows the distribution of recruited reads at a relaxed nucleotide identity of 70–100%. Bar plots are color-coded according to the reference genome used, whereas the average nucleotide identity of recruited reads in each metagenome is indicated by a black dot. The red dotted line demarcates the operational 95% identity level for similar genotypes as the reference. The lower panel shows the coverage of recruited fragments (per Mbp of data). AD, Atlantis II Deep; DD, Discovery Deep; KD, Kebrit Deep. See Supplementary Table 8 and Supplementary Methods for more details.
In the overlying water column, however, the reference DMGI clade SAGs gave better coverage (2.1–3.6%) and ANI (92±4.5%) than the SMGI clade representatives (Figure 6; Supplementary Table 8), which reinforces our previous observations that the bathypelagic AOA in the Red Sea are different from those in the BSI layer. The high average identity of recruited reads (∼97%) coupled with the low coverage (<1%) and diversity (1 operational taxonomic unit) of thaumarchaeal 16S rRNA gene sequences in the Atlantis-BSI, implies that it is probably the sole AOA residing in this habitat. The high ANI of recruited fragments from the 50-m sample above Atlantis II Deep (92±7.4%) against the RSA3 genome but low values at intervening water column depths (86±7.5% Figure 6), also suggests strong evolutionary connections between AOA in the euphotic zone and in the BSI layer.
Conclusion and perspectives
The BSI is a unique environment and a ‘hotspot' for many chemolithoautotrophs. Yet very little is known about how the conditions in this transition layer impact the microbiology of ammonia-oxidizing archaea (AOA), which globally predominate the oceanic deep-sea realm. Previous studies have shown that thaumarchaea are active and predominate the archaeal community in the BSI layer of Mediterranean Sea brines (Daffonchio et al., 2006; Yakimov et al., 2007; Borin et al., 2009). The question that has lingered since then is whether the population represented by these organisms are evolutionary related to those in the bathypelagic water mass. Here, we not only confirmed their abundance but also showed that the thaumarchaea residing in the BSI of Red Sea's brines represent novel species that are highly diverged from their bathypelagic counterparts, and that they instead share ancestry with shallow (epipelagic) marine clades. The existence of unique genotypes both in the overlying water column and the BSI of geochemically distinct brines implies that these populations cannot effectively compete in the same ecological space, which reinforces the concept of ecotype differentiation among marine Thaumarchaeota.
The synergistic effects of high salinity, low dissolved oxygen, increased sulfide and heavy metal concentrations––as is the case in many hypersaline environments––would be prohibitive for the growth of most planktonic aerobes, including AOA. However, the ability to synthesize, take up and potentially also to switch osmolytes (ectoine, glutamate and proline) could be crucial for the adaptation of AOA in saline systems. Although the tri-carboxylic acid cycle is incomplete in marine AOA (Walker et al., 2010), the potential metabolic interconnection of intermediates of this pathway with the biosynthesis of osmolytes further hints that the production of these osmoprotectants might be energetically less costly, and could potentially contribute to the carbon flow of some (halotolerant) thaumarchaea thus enabling AOA to thrive in environments with a broad salinity range.
Data access, sequence deposition and accession numbers
All 454 pyrosequencing data for 16S rRNA gene amplicons have been deposited in the NCBI Sequence Read Archive under study accession number SRP034153. Sanger sequences have been deposited in GenBank under accession numbers: KF954222–KF954277 (16S rRNA genes), KF954278–KF954311 (4-hbd) and KF954312–KF954414 (amoA genes), whereas the SAGs are under Bio-project numbers PRJNA248330, PRJNA248331, PRJNA248333, PRJNA248336–PRJNA248339 and PRJNA248555.
Acknowledgments
We are grateful for financial support from the Saudi Aramco (Bio-CCC RGC/3/1051-01) and the SEDCO Research Excellence Award program. We also thank the 3rd WHOI-KAUST Red Sea 2011 Cruise team, the captain and the crew on board the RV Aegeo, and the Coastal and Marine Resources Core Lab (CMRC) for their excellent technical assistance; the Genomics Core Lab Facility for sequencing efforts; and Dr Till Bayer for helpful bio-informatics discussions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alonso-Sáez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL, et al. Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci USA. 2012;109:17989–17994. doi: 10.1073/pnas.1201914109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anschutz P, Blanc G, Chatin F, Geiller M, Pierret M-C. Hydrographic changes during 20 years in the brine-filled basins of the Red Sea. Deep Sea Res Part IOceanogr Res Pap. 1999;46:1779–1792. [Google Scholar]
- Antunes A, Ngugi DK, Stingl U. Microbiology of the Red Sea (and other) deep-sea anoxic brine lakes. Environ Microbiol Rep. 2011;3:416–433. doi: 10.1111/j.1758-2229.2011.00264.x. [DOI] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- Bernhard AE, Donn T, Giblin AE, Stahl DA. Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ Microbiol. 2005;7:1289–1297. doi: 10.1111/j.1462-2920.2005.00808.x. [DOI] [PubMed] [Google Scholar]
- Biller SJ, Mosier AC, Wells GF, Francis CA. Global biodiversity of aquatic ammonia-oxidizing archaea is partitioned by habitat. Front Microbiol. 2012;3:252. doi: 10.3389/fmicb.2012.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom J, Albaum SP, Doppmeier D, Pühler A, Vorhölter F-J, Zakrzewski M, et al. EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics. 2009;10:154. doi: 10.1186/1471-2105-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum N, Puchelt H. Sedimentary-hosted polymetallic massive sulfide deposits of the Kebrit and Shaban Deeps, Red Sea. Mineralium Deposita. 1991;26:217–227. [Google Scholar]
- Borin S, Brusetti L, Mapelli F, D'Auria G, Brusa T, Marzorati M, et al. Sulfur cycling and methanogenesis primarily drive microbial colonization of the highly sulfidic Urania deep hypersaline basin. Proc Natl Acad Sci USA. 2009;106:9151–9156. doi: 10.1073/pnas.0811984106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougouffa S, Yang JK, Lee OO, Wang Y, Batang Z, Al-Suwailem A, et al. Distinctive microbial community structure in highly stratified deep-sea brine water columns. Appl Environ Microbiol. 2013;79:3425–3437. doi: 10.1128/AEM.00254-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyandin AN, Lobova TI, Krylova TY, Kargatova TV, Popova LY, Pechurkin NS. Effect of salinity on the adaptive capacity of recombinant strains of Escherichia coli and Bacillus subtilis. Microbiology. 2000;69:196–199. [PubMed] [Google Scholar]
- Daffonchio D, Borin S, Brusa T, Brusetti L, van der Wielen PWJJ, Bolhuis H, et al. Stratified prokaryote network in the oxic-anoxic transition of a deep-sea halocline. Nature. 2006;440:203–207. doi: 10.1038/nature04418. [DOI] [PubMed] [Google Scholar]
- Danielsson L-G, Dyrssen D, Granéli A. Chemical investigations of Atlantis II and discovery brines in the Red Sea. Geochimica et Cosmochimica Acta. 1980;44:2051–2065. [Google Scholar]
- Eder W, Jahnke LL, Schmidt M, Huber R. Microbial diversity of the brine-seawater interface of the Kebrit Deep, Red Sea, studied via 16S rRNA gene sequences and cultivation methods. Appl Environ Microbiol. 2001;67:3077–3085. doi: 10.1128/AEM.67.7.3077-3085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder W, Schmidt M, Koch M, Garbe Schönberg D, Huber R. Prokaryotic phylogenetic diversity and corresponding geochemical data of the brine–seawater interface of the Shaban Deep, Red Sea. Environ Microbiol. 2002;4:758–763. doi: 10.1046/j.1462-2920.2002.00351.x. [DOI] [PubMed] [Google Scholar]
- Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing Archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- Faber E, Botz R, Poggenburg J, Schmidt M, Stoffers P, Hartmann M. Methane in Red Sea brines. Org Geochem. 1998;29:363–379. [Google Scholar]
- Ferrer M, Werner J, Chernikova TN, Bargiela R, Fernández L, La Cono V, et al. Unveiling microbial life in the new deep-sea hypersaline Lake Thetis. Part II: a metagenomic study. Environ Microbiol. 2012;14:268–281. doi: 10.1111/j.1462-2920.2011.02634.x. [DOI] [PubMed] [Google Scholar]
- Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the oceans. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ, et al. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. MBio. 2012;3:e00252–12. doi: 10.1128/mBio.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:520–536. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M, Scholten JC, Stoffers P, Wehner F. Hydrographic structure of brine-filled deeps in the Red Sea—new results from the Shaban, Kebrit, Atlantis II, and Discovery Deep. Marine Geol. 1998;144:311–330. [Google Scholar]
- Joye SB, Samarkin VA, Orcutt BN, MacDonald IR, Hinrichs K-U, Elvert M, et al. Metabolic variability in seafloor brines revealed by carbon and sulphur dynamics. Nat Geosci. 2009;2:349–354. [Google Scholar]
- Kanekar PP, Kanekar SP, Kelkar AS, Dhakephalkar PK.2012Halophiles – taxonomy, diversity, physiology, and applicationsIn: Satyanaraya T, Johri BN, Prakash A (eds)Microorganisms in Environmental Management: Microbes and Environment Springer: Dordrecht; 1–34. [Google Scholar]
- Karbe L.1987Hot brines and the deep-sea environmentIn: Edwards A, Head MS (eds)Red Sea Key Environments Series: Exeter, Great Britain; 70–89. [Google Scholar]
- Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BK, Jung M-Y, Yu DS, Park S-J, Oh TK, Rhee S-K, et al. Genome sequence of an ammonia-oxidizing soil archaeon, ‘Candidatus Nitrosoarchaeum koreensis' MY1. J Bacteriol. 2011;193:5539–5540. doi: 10.1128/JB.05717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koops HP, Böttcher B, Möller UC, Pommerening-Roser A, Stehr G. Description of a new species of Nitrosococcus. Arch Microbiol. 1990;154:244–248. [Google Scholar]
- Kowalchuk GA, Stephen JR. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu Rev Microbiol. 2001;55:485–529. doi: 10.1146/annurev.micro.55.1.485. [DOI] [PubMed] [Google Scholar]
- Könneke M, Bernhard AE, la Torre de JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Krom MD, Berner RA. Adsorption of phosphate in anoxic marine sediments. Limnol Oceanogr. 1980;25:797–806. [Google Scholar]
- La Cono V, Smedile F, Bortoluzzi G, Arcadi E, Maimone G, Messina E, et al. Unveiling microbial life in new deep-sea hypersaline Lake Thetis. Part I: Prokaryotes and environmental settings. Environ Microbiol. 2011;13:2250–2268. doi: 10.1111/j.1462-2920.2011.02478.x. [DOI] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M (eds)Nucleic Acid Techniques in Bacterial Systematics New York; 115–175. [Google Scholar]
- Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Tolar BB, Swan BK, Zhang CL, Stepanauskas R, Ann Moran M, et al. Single-cell genomics shedding light on marine Thaumarchaeota diversification. ISME J. 2014;8:732–736. doi: 10.1038/ismej.2013.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, la Torre de JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- Martin DD, Ciulla RA, Robinson PM, Roberts MF. Switching osmolyte strategies: response of Methanococcus thermolithotrophicus to changes in external NaCl. Biochim Biophys Acta. 2001;1524:1–10. doi: 10.1016/s0304-4165(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Matsutani N, Nakagawa T, Nakamura K, Takahashi R, Yoshihara K, Tokuyama T. Enrichment of a novel marine ammonia-oxidizing archaeon obtained from sand of an Eelgrass zone. Microbes Environ. 2011;26:23–29. doi: 10.1264/jsme2.me10156. [DOI] [PubMed] [Google Scholar]
- Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- Metcalf WW, Griffin BM, Cicchillo RM, Gao J, Janga SC, Cooke HA, et al. Synthesis of methylphosphonic acid by marine microbes: a source for methane in the aerobic ocean. Science. 2012;337:1104–1107. doi: 10.1126/science.1219875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosier AC, Allen EE, Kim M, Ferriera S, Francis CA. Genome sequence of ‘Candidatus Nitrosopumilus salaria' BD31, an ammonia-oxidizing archaeon from the San Francisco Bay Estuary. J Bacteriol. 2012;194:2121–2122. doi: 10.1128/JB.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussmann M, Brito I, Pitcher A, Damsté JSS, Hatzenpichler R, Richter A, et al. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci USA. 2011;108:16771–16776. doi: 10.1073/pnas.1106427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naor A, Lapierre P, Mevarech M, Papke RT, Gophna U. Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr Biol. 2012;22:1444–1448. doi: 10.1016/j.cub.2012.05.056. [DOI] [PubMed] [Google Scholar]
- Nelson KA, Moin NS, Bernhard AE. Archaeal diversity and the prevalence of Crenarchaeota in salt marsh sediments. Appl Environ Microbiol. 2009;75:4211–4215. doi: 10.1128/AEM.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngugi DK, Stingl U. Combined analyses of the ITS Loci and the corresponding 16S rRNA genes reveal high micro- and macrodiversity of SAR11 populations in the Red Sea. PLoS One. 2012;7:e50274. doi: 10.1371/journal.pone.0050274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Larimer FW, Chain PSG, Malfatti S, Shin MV, Vergez LM, et al. Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. Proc Natl Acad Sci USA. 2008;105:18543–18548. doi: 10.1073/pnas.0809160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev. 1999;63:334–348. doi: 10.1128/mmbr.63.2.334-348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. Thermodynamic limits to microbial life at high salt concentrations. Environ Microbiol. 2011;13:1908–1923. doi: 10.1111/j.1462-2920.2010.02365.x. [DOI] [PubMed] [Google Scholar]
- Oren A. Life at high salt concentrations, intracellular KCl concentrations, and acidic proteomes. Front in Microbiol. 2013;4:1–6. doi: 10.3389/fmicb.2013.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A, Heldal M, Norland S, Galinski EA. Intracellular ion and organic solute concentrations of the extremely halophilic bacterium Salinibacter ruber. Extremophiles. 2002;6:491–498. doi: 10.1007/s00792-002-0286-3. [DOI] [PubMed] [Google Scholar]
- Ouverney CC, Fuhrman JA. Marine planktonic archaea take up amino acids. Appl Environ Microbiol. 2000;66:4829–4833. doi: 10.1128/aem.66.11.4829-4833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B-J, Park S-J, Yoon D-N, Schouten S, Damste JSS, Rhee S-K. Cultivation of autotrophic ammonia-oxidizing Archaea from marine sediments in coculture with sulfur-oxidizing Bacteria. Appl Environ Microbiol. 2010;76:7575–7587. doi: 10.1128/AEM.01478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-J, Kim J-G, Jung M-Y, Kim S-J, Cha I-T, Ghai R, et al. Draft genome sequence of an ammonia-oxidizing archaeon,‘Candidatus Nitrosopumilus sediminis' AR2, from Svalbard in the Arctic Circle. J Bacteriol. 2012;194:6948–6949. doi: 10.1128/JB.01869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-J, Kim J-G, Jung M-Y, Kim S-J, Cha I-T, Kwon K, et al. Draft genome sequence of an ammonia-oxidizing archaeon, ‘Candidatus Nitrosopumilus koreensis' AR1, from marine sediment. J Bacteriol. 2012;194:6940–6941. doi: 10.1128/JB.01857-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor JM, Salvador M, Argandona M, Bernal V, Reina-Bueno M, Csonka LN, et al. Ectoines in cell stress protection: Uses and biotechnogical production. Biotechnol Adv. 2010;28:782–801. doi: 10.1016/j.biotechadv.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Paul S, Bag SK, Das S, Harvill ET, Dutta C. Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 2008;9:R70. doi: 10.1186/gb-2008-9-4-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelve EA, Lindås A-C, Martens-Habbena W, la Torre de JR, Stahl DA, Bernander R. Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus. Mol Microbiol. 2011;82:555–566. doi: 10.1111/j.1365-2958.2011.07834.x. [DOI] [PubMed] [Google Scholar]
- Prosser JI, Nicol GW. Archaeal and bacterial ammonia-oxidizers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 2012;20:523–531. doi: 10.1016/j.tim.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Rinke C, Lee J, Nath N, Goudeau D, Thompson B, Poulton N, et al. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat Protocol. 2014;9:1038–1048. doi: 10.1038/nprot.2014.067. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan WBF, Thorndike EM, Ewing M, Ross DA.1969Suspended matter in the Red Sea brines and its detection by light scatteringIn: Degens E, Ross DA (eds)Hot Brines and Recent Heavy Metal Deposits in the Red Sea Springer: New York, NY; 153–157. [Google Scholar]
- Sass AM, Sass H, Coolen MJ, Cypionka H, Overmann J. Microbial communities in the chemocline of a hypersaline deep-sea basin (Urania basin, Mediterranean Sea) Appl Environ Microbiol. 2001;67:5392–5402. doi: 10.1128/AEM.67.12.5392-5402.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saum SH, Muller V. Salinity-dependent switching of osmolyte strategies in a moderately halophilic bacterium: glutamate induces proline biosynthesis in Halobacillus halophilus. J Bacteriol. 2007;189:6968–6975. doi: 10.1128/JB.00775-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Botz R, Faber E, Schmitt M, Poggenburg J, Garbe-Schönberg D, et al. High-resolution methane profiles across anoxic brine–seawater boundaries in the Atlantis-II, Discovery, and Kebrit Deeps (Red Sea) Chem Geol. 2003;200:359–375. [Google Scholar]
- Seyler LM, McGuiness LM, Kerkhof LJ. Crenarchaeal heterotrophy in salt marsh sediments. ISME J. 2014;8:1534–1543. doi: 10.1038/ismej.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Poehlein A, Offre P, Zumbrägel S, Haider S, Rychlik N, et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012;14:3122–3145. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- Stahl DA, de la Torre JR. Physiology and diversity of ammonia-oxidizing Archaea. Annu Rev Microbiol. 2012;66:83–101. doi: 10.1146/annurev-micro-092611-150128. [DOI] [PubMed] [Google Scholar]
- Stepanauskas R. Single cell genomics: an individual look at microbes. Curr Opin Microbiol. 2012;15:613–620. doi: 10.1016/j.mib.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Swan BK, Ehrhardt CJ, Reifel KM, Moreno LI, Valentine DL. Archaeal and Bacterial communities respond differently to environmental gradients in anoxic sediments of a California Hypersaline Lake, the Salton Sea. Appl Environ Microbiol. 2010;76:757–768. doi: 10.1128/AEM.02409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift SA, Bower AS, Schmitt RW. Vertical, horizontal, and temporal changes in temperature in the Atlantis II and Discovery hot brine pools, Red Sea. Deep Sea Res Part I Oceanogr Res Pap. 2012;64:118–128. [Google Scholar]
- Teeling H, Meyerdierks A, Bauer M, Amann R, Glöckner FO. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ Microbiol. 2004;6:938–947. doi: 10.1111/j.1462-2920.2004.00624.x. [DOI] [PubMed] [Google Scholar]
- Teira E, Van Aken H, Veth C, Herndl GJ. Archaeal uptake of enantiomeric amino acids in the meso- and bathypelagic waters of the North Atlantic. Limnol Oceanogr. 2006;51:60–69. [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome'. Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske A, Sorensen KB. Uncultured Archaea in deep marine subsurface sediments: have we caught them all. ISME J. 2008;2:3–18. doi: 10.1038/ismej.2007.90. [DOI] [PubMed] [Google Scholar]
- Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trommer G, Siccha M, van der Meer MTJ, Schouten S, Damsté JSS, Schulz H, et al. Distribution of Crenarchaeota tetraether membrane lipids in surface sediments from the Red Sea. Organic Geochem. 2009;40:724–731. [Google Scholar]
- van der Wielen PWJJP BolhuisHH, Borin SS, Daffonchio DD, Corselli CC, Giuliano LL, et al. The enigma of prokaryotic life in deep hypersaline anoxic basins. Science. 2005;307:121–123. doi: 10.1126/science.1103569. [DOI] [PubMed] [Google Scholar]
- Varela MM, van Aken HM, Sintes E, Herndl GJ. Latitudinal trends of Crenarchaeota and Bacteria in the meso- and bathypelagic water masses of the Eastern North Atlantic. Environ Microbiol. 2008;10:110–124. doi: 10.1111/j.1462-2920.2007.01437.x. [DOI] [PubMed] [Google Scholar]
- Walker CB, la Torre de JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimov MM, La Cono V, Denaro R, D'Auria G, Decembrini F, Timmis KN, et al. Primary producing prokaryotic communities of brine, interface and seawater above the halocline of deep anoxic lake L'Atalante, Eastern Mediterranean Sea. ISME J. 2007;1:743–755. doi: 10.1038/ismej.2007.83. [DOI] [PubMed] [Google Scholar]
- Yakimov MM, La Cono V, Slepak VZ, La Spada G, Arcadi E, Messina E, et al. 2013Microbial life in the Lake Medee, the largest deep-sea salt-saturated formation Sci Rep 33554 . . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AL, Carter WG, Doyle HA, Mamula MJ, Aswad DW. Structural integrity of histone H2B in vivo requires the activity of protein L-isoaspartateO-methyltransferase, a putative protein repair enzyme. J Biol Chem. 2001;276:37161–37165. doi: 10.1074/jbc.M106682200. [DOI] [PubMed] [Google Scholar]
- Ziemienowicz A, Rahavi SMR, Kovalchuk I. The stimulatory effect of CaCl2, NaCl and NH4NO3 salts on the ssDNA-binding activity of RecA depends on nucleotide cofactor and buffer pH. BMB Rep. 2011;44:341–346. doi: 10.5483/BMBRep.2011.44.5.341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.