Abstract
The resilience of microbial communities to press disturbances and whether ecosystem function is governed by microbial composition or by the environment have not been empirically tested. To address these issues, a whole-ecosystem manipulation was performed in a full-scale activated sludge wastewater treatment plant. The parameter solids retention time (SRT) was used to manipulate microbial composition, which started at 30 days, then decreased to 12 and 3 days, before operation was restored to starting conditions (30-day SRT). Activated sludge samples were collected throughout the 313-day time series in parallel with bioreactor performance (‘ecosystem function'). Bacterial small subunit (SSU) rRNA genes were surveyed from sludge samples resulting in a sequence library of >417 000 SSU rRNA genes. A shift in community composition was observed for 12- and 3-day SRTs. The composition was altered such that r-strategists were enriched in the system during the 3-day SRT, whereas K-strategists were only present at SRTs⩾12 days. This shift corresponded to loss of ecosystem functions (nitrification, denitrification and biological phosphorus removal) for SRTs⩽12 days. Upon return to a 30-day SRT, complete recovery of the bioreactor performance was observed after 54 days despite an incomplete recovery of bacterial diversity. In addition, a different, yet phylogenetically related, community with fewer of its original rare members displaced the pre-disturbance community. Our results support the hypothesis that microbial ecosystems harbor functionally redundant phylotypes with regard to general ecosystem functions (carbon oxidation, nitrification, denitrification and phosphorus accumulation). However, the impacts of decreased rare phylotype membership on ecosystem stability and micropollutant removal remain unknown.
Introduction
Mixed-culture microbial assemblages present in biological wastewater treatment systems, such as activated sludge, are the most important component of modern sanitation systems used in domestic and industrial wastewater treatment (Tchobanoglous et al., 2003; Seviour and Nielsen, 2010). These assemblages are organized into planktonic biofilms or flocs that degrade complex organic matter, attenuate toxic compounds and transform inorganic nutrients in wastewater. In addition to the importance of activated sludge in modern sanitation, these systems are uniquely suited for whole-ecosystem scale experimentation due to the tight controls on chemical, physical and biological processes (Daims et al., 2006). As a model system for microbial ecology, research on activated sludge has greatly informed our understanding of microbe–microbe interactions (Wagner ,2002; Daims et al., 2006; Maixner et al., 2006), undiscovered biochemistry (Strous et al., 2006; Maixner et al., 2008; Lücker et al., 2010; He and McMahon, 2011; Sorokin et al., 2012) and community assembly theory (Sloan et al., 2007; Ofiteru et al., 2010). However, little work has focused on the ecological consequences of press (that is, prolonged) disturbances on microbial diversity and the extent of compositional versus environmental effects on ecosystem function (Allison and Martiny, 2008). Most community ecology studies in activated sludge systems have been limited to microbial diversity surveys of full-scale municipal treatment plants that are not open to experimental manipulation (Wells et al., 2009, 2011; Yang et al., 2011; Ye et al., 2011; Zhang et al., 2012; Saunders et al., 2013). Thus, a large knowledge gap exists with regard to activated sludge microbial community temporal variability, responses to species-selection pressures and resilience to disturbances.
Environmental factors, such as pH and temperature, are known to influence specific functions (for example, nitrification) in activated sludge bioreactors. However, system performance and microbial diversity are most influenced by operating conditions (for example, organic loading rate and feed composition) and reactor configuration (for example, batch-fed versus continuous; Pholchan et al., 2010). Pholchan et al. (2010) found that microbial diversity and performance increased under a batch-fed configuration, but diversity itself could not be systematically manipulated, which indicates a knowledge gap regarding the control of both the membership and structure of engineered microbial communities. The effects of other operational parameters on microbial diversity such as solids retention time (SRT), which represents the average time that microorganisms reside in a bioreactor, have yet to be investigated thoroughly and under realistic conditions (Saikaly et al., 2005; Tan et al., 2008). For example, real wastewater, rather than synthetic feed solutions, should be used to account for mechanisms of spatial dynamics such as colonization (Leibold et al., 2004), and reactor volume must be large enough to (1) ensure that biomass collection does not act as a confounding variable on SRT and (2) operation should be at a relevant scale for community ecology and environmental engineering (Carpenter, 1996).
It is plausible that SRT can be used to control both diversity and composition of microorganisms because it is related to the specific biomass growth rate (Tchobanoglous et al., 2003) and would likely influence the abundance of bacterial, archaeal and eucaryal clades within the community. The manipulation of SRT also implies that microorganisms can be selected along a continuum of life-history strategies, which forms the bases of r–K selection theory (Reznick et al., 2002). Only organisms that have doubling times less than a corresponding SRT will be capable of growing quickly enough in the system to avoid being washed out. Thus, there is a trade-off between maximum growth rate and resource-use efficiency: (1) Fixed volume bioreactors operated at high SRTs will be highly saturated with organisms (that is, density-dependent growth) that are capable of efficiently utilizing scarce resources (‘K-strategists') (2) Low SRTs will enrich for fast-growing organisms that are adapted for high resource utilization (‘r-strategists') and typically dominate unstable environments where disturbances have recently occurred (Pianka, 1970).
To explore the consequences of a press disturbance in the context of ecosystem function and microbial diversity, we sequentially reduced the SRT from 30 days, to 12 days and 3 days, before operation was restored to a 30-day SRT, over a timecourse of 313 days. This study was performed in a full-scale activated sludge wastewater treatment plant (sequencing batch membrane bioreactor configuration, as described by Vuono et al., (2013)) in order to tightly control environmental conditions such as pH, dissolved oxygen concentrations, biomass concentrations and food:microorganism (F:M) ratio. We hypothesized that both microbial community structure and membership would shift with changes in SRT, and the shift would reflect a selective enrichment of microorganisms along a life-history continuum (r versus K-strategists). We further hypothesized that if microbial communities are resilient to press disturbances, we should observe secondary succession of a community comprising pioneering species to a climax community with the same diversity prior to the disturbance. Because diversity can be viewed from a spectrum of viewpoints, we used Hill numbers to measure and compare diversity through time and across treatments (Hill, 1973; Jost, 2006; Chao et al., 2010; Leinster and Cobbold, 2012). In addition, we hypothesized that if community composition was a major factor that governs ecosystem function, the same community should return after the disturbance and perform the same functions. In this study, a culture-independent approach was used to monitor bacterial diversity with barcoded amplicon sequences of the bacterial SSU rRNA gene. In parallel, a wide range of metadata was collected as operational, performance and water quality parameters to identify break points for ecosystem function gain and loss.
Materials and methods
Sampling, amplification and sequencing
All biological samples for each time point observation were collected in triplicate. Activated sludge samples were collected from a ∼75 l/min sludge recirculation line between sequencing batch reactor and membrane biological reactor tanks (for details on reactor operation, see Supplementary Information Materials and methods). Samples were stored at −20 °C prior to processing. DNA was extracted within 1 month of sampling using MoBio PowerBiofilm DNA extraction kit following manufacturer's protocol with a 1 minute bead-beating step for cellular disruption. Barcoded SSU rRNA gene primers 515f-927r were incorporated with adapter sequences for the GSFLX-Titanium platform of the Roche 454 Pyrosequencing technology (for details, see Supplementary Information Materials and methods).
SSU rRNA processing pipeline and quality control
SSU rRNA gene amplicons generated from pyrosequencing were binned by barcode and quality filtered using the ‘split_libraries.py' script in the Quantitative Insights Into Microbial Ecology (QIIME v1.5-dev) software (Caporaso et al., 2010). Sequences with errors in the barcode or primer, shorter than 400 nt, longer than 460 nt, with a quality score <50, homopolymer run greater than 6 nt and sequences that contained ambiguous base calls were discarded from downstream analysis. Flowgrams for remaining sequences were denoised using DeNoiser version 1.3.0-dev by Reeder and Knight (2010). Chimeric sequences were identified using UCHIME under reference mode and de novo mode (Edgar et al., 2011) (for details, see Supplementary Information Materials and methods). The remaining 672 521 sequences were processed in Mothur as outlined by Schloss et al., (2009) Schloss SOP version data 15 February2013 (for details, see Supplementary Information Materials and methods). Singletons were discarded from downstream analysis prior to diversity calculations. A phylogenetic tree was constructed from the filtered alignment using FastTree. Unweighted and weighted UniFrac (WU) distance matrices were calculated from the phylogenetic tree along with Morisita–Horn (MH) distances. Beta diversity metrics were derived from a rarefied OTU table from the sample with lowest sequencing depth (for details on diversity calculations and statistical analysis, see Supplementary Information Materials and methods). These sequence data have been submitted to MG-RAST database under the metagenomic ID 9726 (static link http://metagenomics.anl.gov/linkin.cgi?project=9726).
Results
Analysis of ecosystem function
To provide context for the biological response to the SRT disturbance, we measured a wide range of operational and environmental parameters obtained through a Supervisory Control and Data Acquisition (SCADA) system (>1.0 × 108 measurements) (Supplementary Figure S1). In addition, we characterized ecosystem processes by measuring removal efficiencies of chemical oxygen demand (COD), total nitrogen (TN), ammonia, nitrate, nitrite and total phosphorus (TP). Throughout the investigation, COD removal was consistently >90% and was not significantly different between pre-disturbance and the 3-day SRT (P=0.123, Wilcoxon signed-rank test). Mean±s.d. of pre- and post-disturbance COD removal were 96.3±0.9% and 95.6±0.8%, respectively, and were not significantly different (P=0.102, Wilcoxon signed-rank test). TN and TP removal were also not significantly different between pre- and post-disturbance (TN: P=0.069, TP: P=0.79, Wilcoxon signed-rank test). TN removal exceeded 90% on day 214, whereas TP removal exceeded 95% on day 236. Removal efficiencies are summarized in Supplementary Table S1. All general ecosystem functions (that is, COD, TN and TP removal) recovered within 54 days after the end of the 3-day SRT, or maximum disturbance state (MDS) (for details, see Supplementary Information Materials and methods).
The transition from a 12-day SRT to 3-day SRT resulted in a major shift in the nitrogen cycle within the treatment plant (Supplementary Figure S2). The complete loss of nitrification was observed within 2 days of starting the 3-day SRT (day 162). Correspondingly, effluent ammonia concentrations gradually increased and peaked at 39.2 mg NH3-N/L (day 182). After biomass wasting ceased (day 178), ammonia and nitrite oxidation gradually increased and recovered by day 205 and 214, respectively. Heterotrophic denitrification efficiency also recovered rapidly by day 219, as indicated by the lack of nitrite and nitrate accumulation in the treated effluent.
Effects of SRT on bacterial diversity
We monitored bacterial communities along the experimental time-course by collecting activated sludge samples (n=81) and sequencing the V4-V5 hypervariable region of the bacterial SSU rRNA gene. A total of 417 515 high quality amplicon reads were obtained at an average read depth of 5155 sequences/sample, with sequence statistics of 1925/10 059/4947/1829 (min/max/median/s.d., respectively) and a total of 2478 operational taxonomic units (OTUs; 97% sequence similarity by average-neighbor method). To accurately compare bacterial diversities across the different SRT treatments, we calculated Hill numbers based on OTU sequence counts (Figure 1c; Equation 1).
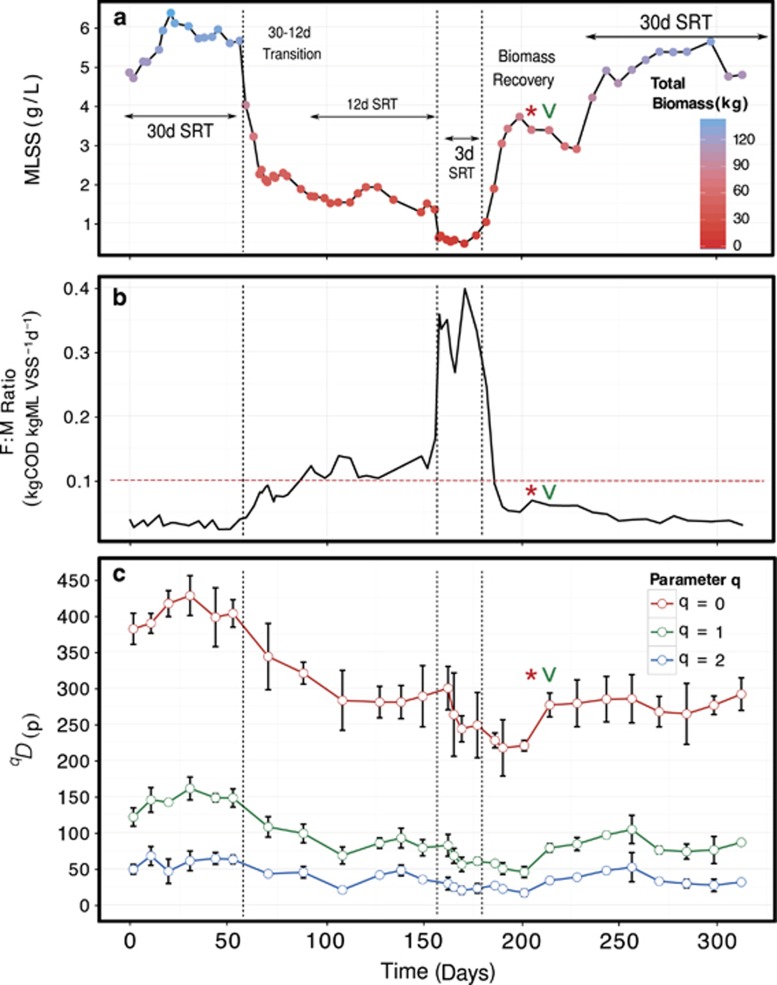
Figure 1.
Mixed liquor suspended solids (MLSS) concentration (gl−1) within the sequencing batch membrane bioreactor with corresponding reactor SRTs, transitions and biomass recover periods as a function of time (a). Total Biomass (kg) is also indicated as new membranes were installed on day 225 and bioreactor volume was reduced; food:microorganism (F:M) ratio as a function of time (b). Red horizontal dashed line indicates the F:M ratio threshold (<0.1 kgCOD kgMLVSS−1 d−1) used in engineering design for SBR reactors (Tchobanoglous et al., 2003); Three orders of diversity q=0, 1 and 2 as a function of time and corresponding treatment conditions (c). Calculations were made after rarefying to an equal number of reads for all samples to control for unequal sampling effort. Error bars indicate 95% confidence interval approximated from a t-distribution. Red ‘asterisk' and green ‘arrow' indicate when nitrogen removal and phosphorus removal recovered in the system, respectively.
As an ecological parameter, the term ‘diversity' is more inclusive than simply species richness and includes aspects of community evenness. Furthermore, estimating the total number of microbial species (as ‘OTUs') in a microbial community, even if asymptotic estimators such as Chao1 are used, cannot be accomplished from sample data alone (Haegeman et al., 2013). Generalized diversity metrics such as Hill numbers facilitate our interpretation of diversity by maintaining a mathematical ‘doubling property', producing ecologically intuitive quantities that allow for confidence in both ratios and percentage changes. Thus, Hill numbers can inform our understanding of the magnitude of change between two communities, which is often more relevant for biological interpretation (Jost, 2006). Hill numbers are calculated using the following equation (Hill, 1973). Let 0⩽q<∞, such that
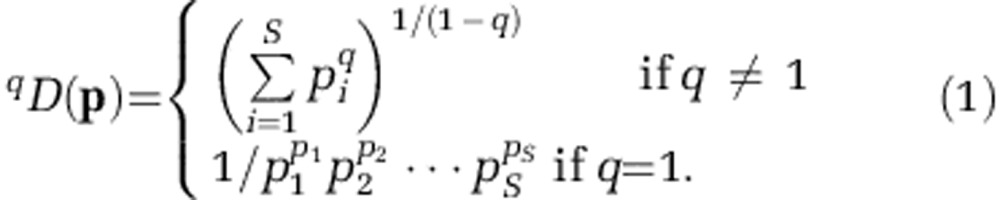 |
where S is the number of species sampled, pi is the species frequency of the ith species and the parameter q, or sensitivity parameter, is equal to the ‘order' of diversity that applies weight to common or rare species. 0D is equivalent to species richness, 1D is equal to exp(Shannon entropy (H)=-∑piln pi), and 2D is equivalent to 1/Simpson concentration (Hill, 1973; Jost, 2006). Together, 1⩽q⩽2 provides a unified framework for diversity measurements, which can be interpreted as the effective number of species (ENS), or a community of S equally abundant species.
During our time-series, the sequential decrease of SRT from 30 to 3 days had a large negative effect on taxonomic diversity. Our results from estimating species richness (0D) indicate the large range of variation within time-point replicates (Figure 1c). These results are consistent with the finding of Haegman et al. (2013), who demonstrate the large biases involved in species richness estimations and the interpretation of results from values of q<1. However, at q⩾1, we see the true impact of the SRT press disturbance on the activated sludge community. Our calculations for 1D, which weigh each OTU exactly by its frequency, indicate a 57% reduction of diversity from a pre-disturbance 30-day SRT to a 3-day SRT (mean±s.d.: 145.3±12.8 and 62.5±6.0 ENS, respectively). During the biomass recovery phase 1D decreased further (52±5.7 ENS) and moderately recovered by the post-disturbance recovery 30-day SRT (86.5±12.6 ENS, 59.5% of the pre-disturbance ENS). At 2D, where dominant species are disproportionally emphasized by the diversity calculation, we see diversity trends similar to 1D. Between pre-disturbance 30-day SRT and press disturbance 3-day SRT; we observed an even greater decline in diversity (60.6%) compared with 1D, which indicates a less-even community than during the MDS. Post-disturbance diversity recovered to 63.4% of the original starting conditions, with corresponding means of 59.5±9.0 and 37.7±10.3 ENS for pre- and post-disturbance, respectively.
To ascertain if diversities at each SRT treatment were statistically different, we modeled the series as an autoregressive (1) process in order to account for autocorrelation (for details, see Supplementary Information Materials and methods). For both 1D and 2D, we tested the null hypothesis that mean values were equal across SRT treatments: μ30d-pre=μ12d=μ3d=μ30-post. For both cases, diversities were statistically different at the 5% level (q=1: P=0.0053, likelihood ratio statistic 12.73; q=2: P<0.001, likelihood ratio statistic 19.61; distributed X32). When comparing pre- and post-disturbance diversities only (μ30d-pre=μ30-post), 1D was significantly different (P=0.0064, likelihood ratio statistic of 7.42, distributed X12), whereas 2D was not (P=0.0746, likelihood ratio statistic of 3.18, distributed X12). These results suggest that the system recovered to a greater extent with regard to common OTUs (2D), but was significantly less diverse in its pool of rare OTUs (1D).
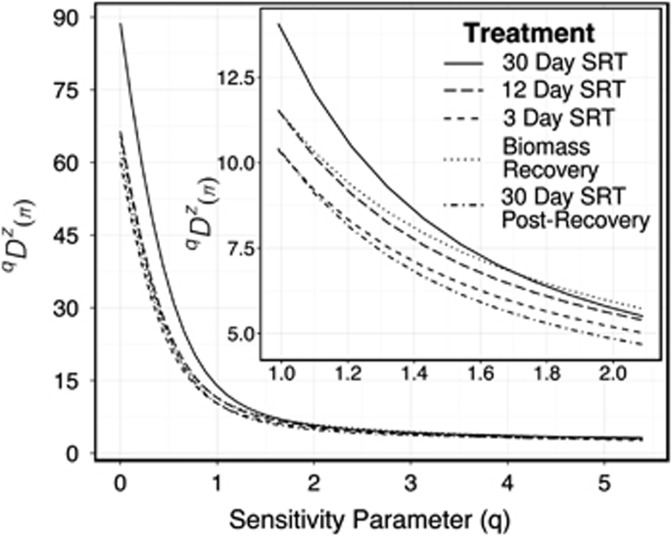
To verify that the observed estimates of taxonomic diversity for orders 1 and 2 were congruent (because OTU-based measures are arbitrary and can be affected by OTU-picking methods (Schloss, 2010; Schloss and Westcott, 2011; Sun et al., 2011)), we plotted phylogenetic diversity (Faith, 1992) based on Hill numbers (Chao et al., 2010) as described by Leinster and Cobbold (2012) and Armitage et al. (2012); (Figure 2). In Figure 2, a series of effective numbers using mean qDZ(π) values for each SRT treatment are plotted versus q, where π is the relative abundance of ‘historical species' (for detailed calculation, see Supplementary Information Materials and methods). Figure 2 (inset) displays the qDZ(π) values between 1⩽q⩽2. Crossover points for each treatment indicate transitions between communities that differ in their diversities of rare to common taxa. The pre-disturbance (30-day SRT; solid line) community is more diverse than all treatments, except at q=1.72 when the solid line crosses below the profile for biomass recovery (dotted line). As q approaches 2, the biomass recovery community is the most diverse with regard to common taxa (that is, it is the most even). However, the post-disturbance SRT (dotted-dashed line) is the least diverse, as the profile lies below all other treatment profiles. At q≈1, pre-disturbance ENS is 30.3% more diverse than post-disturbance and at q≈2, pre-disturbance ENS is 16.9% more diverse than post-disturbance. These results indicate the incomplete recovery of phylogenetic diversity post-disturbance with regard to rare taxa.
Figure 2.
Rarefied phylogenetic diversity profile, qDZ(π), for activated sludge microbial communities, binned by SRT treatment. As parameter q increases, rare taxa are weighted less and q becomes a measure of evenness. Values of 1<q<2 are shown in inset box.
Effects of SRT on community composition
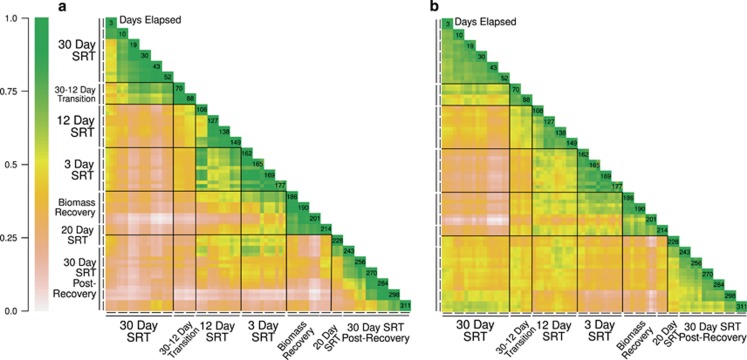
Pairwise ecological dissimilarity metrics of composition and relatedness were used to measure changes in community composition across all SRTs. We hypothesized that because ecosystem function recovered by 54 days after the MDS, similarity should be observed between pre- and post-disturbance communities. Given that rare community members (1D) recovered to a lesser degree than common members (2D) (Figures 1c and 2), we chose the MH and WU indices to characterize the recovery of common community members. Rather than perform an Eigen-decomposition of the MH and WU distance matrices to be visualized and condensed into ordination space, we chose to visualize the distance matrices directly (Figure 3). The MH index, which measures compositional similarity of abundant OTUs, showed greater similarity within each SRT (values approaching unity as blocks of green along diagonal), whereas the largest shifts in similarity were between SRTs (Figure 3a). Next, analysis of similarity (ANOSIM) and permutational multivariate analysis of variance (ADONIS) were used to test if bacterial community composition (with transition categories removed) within the same SRT and between SRTs were statistically different. Results suggest a strong distinction between treatments (RANOSIM=0.86, P<0.001; R2ADONIS=0.75, P<0.001). After the system was returned to a 30-day SRT (lower right, Figure 3a), time-decay was observed, which indicates that the community was still undergoing dynamic succession with addition and replacement of new community members. Similar trends were observed after compositionally based dissimilarity metrics were used, which include both abundance weighted (for example, Bray-Curtis) and presence/absence (unweighted UniFrac) metrics (Supplementary Figure S3).
Figure 3.
Heatmaps displaying pairwise MH distances (a) and WU distances (b) for each sample date and corresponding SRTs. Values along unity indicate days elapsed from the start of the study. Values close to 1 (green-yellow) indicate high similarity, whereas values close to 0 (brown-white) indicate dissimilarity. Opposing side of each matrix is omitted for clarity.
Investigation of phylogenetic relatedness (WU, Figure 3b) revealed similar patterns to MH between SRTs (value approaching unity along diagonal). However, in contrast to MH, phylogenetic similarity during the post-recovery 30-day SRT remained relatively high through time. Also, during the biomass recovery period (WU, Figure 3b) a clear bifurcation was observed after day 201 where pairwise similarities among pre- (30-day SRT) and post-disturbance increased and were at their maximum values (0.73 on day 311). These results suggest that phylogenetically related organisms reestablish in the post-disturbance community. Here, reestablish is defined as either recolonization (immigration from a source community) or regrowth by organisms (for example, dormant) within the bioreactor that were not completely washed out. These results are also corroborated using ordination techniques, as the trajectory of the bacterial communities along the principal coordinate (PCo) 1 axis return to their original starting conditions WU (Supplementary Figure S4). The distinction of community similarity within, rather than between SRTs, however, remained high (RANOSIM=0.92, P<0.001; R2ADONIS=0.68, P<0.001) despite the observed trends of similarity (that is, return of PCo scores along PCo1 axis).
Bacterial dynamics/response during selective enrichment of r–K-strategists
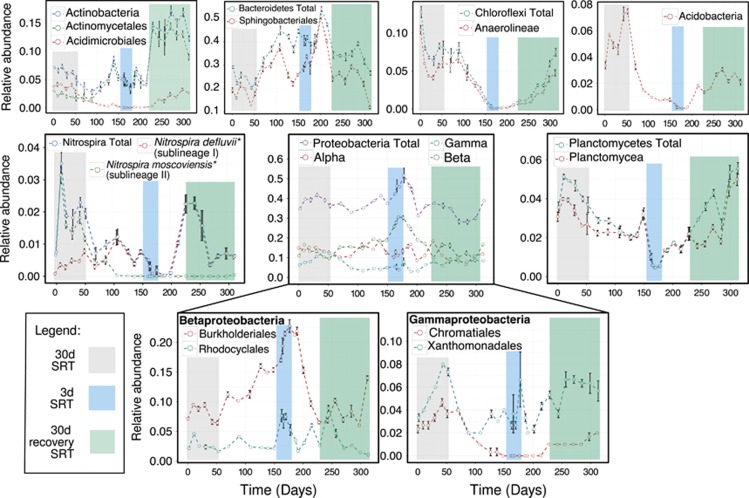
Taxonomic assignments were mapped onto OTUs to measure the relative shifts in abundance of specific bacterial clades. The abundance of major bacterial phyla are plotted versus time in Figure 4. Here, dynamic fluctuations of higher bacterial taxonomic ranks are seen both through time and across SRTs. The most notable shifts in relative abundance occurred during the MDS (3-day SRT). Several bacterial phyla, such as Acidobacteria, Planctomycetes, Chloroflexi and Nitrospira declined in abundance, which indicates that these phyla were washed out of the system as SRT decreased. Because Nitrospira is associated with the oxidation of nitrite, these results explain the decline in nitrogen removal efficiency and the accumulation of nitrite in the treated effluent (Supplementary Figure S2). After the MDS, ammonia and nitrite concentrations in the treated effluent were below detection limit on days 205 and 219, respectively. These results correspond to the increase of ammonia-oxidizing bacteria and nitrite-oxidizing bacteria in the activated sludge. Although these results are not surprising, because nitrogen removal efficiency has long been known to improve with longer SRTs (typically>5 days), prior studies have not demonstrated the complete washout and recovery of such functionally important organisms. For example, two sublineages of known nitrite-oxidizing bacteria within the phylum Nitrospira were present before the initial transition to a 12-day SRT: sublineage II, whose cultured representative is Nitrospira moscoviensis, was present at greater relative abundance (1.5–2%) than its sublineage I counterpart (‘Candidatus' Nitrospira defluvii) (∼0.5%). However, by day 90 (during the 30–12-day SRT transition) sublineage I displaced sublineage II and persisted as the only Nitrospira OTU to recover from the MDS.
Figure 4.
Relative abundance of major bacterial phyla as function of time and corresponding SRTs: from left to right is 30-day SRT, 3-Day SRT and post-recovery 30-day SRT. Error bars represent the standard error of the sample mean. *Cultured representatives of each Nitrospira sublineage detected.
The relative abundance of organisms at higher taxonomic ranks such as Alpha- and Gamma-proteobacteria remained constant in response to SRT operational transitions, which indicates generalism (that is, mildly fluctuating dynamics and persistent occurrence through time). However, at lower taxonomic ranks a differential response was observed. For example, taxa of the order Actinomycetales within the Actinobacteria persisted at a relatively consistent level through the MDS but more than doubled in abundance after the MDS. The majority of the increase in abundance within Actinomycetales (48.1%) was attributed to sequences associated with the genus Tetrasphaera (OTU 1153), known to include phosphate-accumulating organisms. However, taxa within Acidimicrobiales declined to extinction during the MDS and reestablished thereafter. Similar patterns were observed within Alphaproteobacteria (Supplementary Figure S5) and Gammaproteobacteria. In the latter, taxa within the order Xanthomonadales did not appear to be greatly affected by the MDS (indicating generalism), whereas phylogenetically related taxa of Chromatiales were pushed to temporal extinction but reestablished thereafter. These results highlight the variability and diversity of life-history strategies, even among phylogenetically related groups.
Orders within two phyla displayed a positive response to the MDS, indicating the selection of r-strategists. Within Betaproteobacteria, taxa within the order Burkholderiales increased to a maximum of 23% of the total community during the MDS. Interestingly, ammonia-oxidizing bacteria within the Betaproteobacteria were pressed to temporal extinction (Supplementary Figure S6). Members of Rhodocyclales also increased in relative abundance ( from ∼2.5% to ∼7%); however, post-disturbance abundance fluctuated in a comparable range. Members of the phylum Bacteroidetes increased in abundance precisely as the system transitioned to a 12-day SRT. The relative abundance of taxa within the order Sphingobacteriales increased by 20% above the 12- and 3-day SRT levels during the biomass recovery period. The most prominent taxa were affiliated with members of the family Saprospiraceae, which are known to be epiphytic protein hydrolyzers commonly attached to filamentous bacteria (Xia et al., 2008).
The phenotypic effects associated with rRNA gene copy number have been shown to positively correlate with the rate at which phylogenetically diverse bacteria respond to resource availability (Klappenbach et al., 2001). Thus, the translational power or number of rRNA gene operons for abundant OTUs should be inversely related to SRT. The number of rRNA gene operons from the most abundant OTUs was estimated at various SRTs with the Ribosomal RNA Database (Lee et al., 2009). The two most abundant OTUs during the 3-day SRT MDS were members of Betaproteobacteria order Comamonadaceae (OTU 418) and Bacteroidetes order Sphingobacteriales (OTU 1200). Together, OTU 418 and 1200 matched at 100% bootstrap confidence at the family-level in the Ribosomal RNA Database with upper estimates of rRNA gene operon copy number of 5 and 6 and means of 2.8 and 3.0 at order and family levels, respectively. During the biomass recovery phase, the two most abundant OTUs, which were also of orders Comamonadaceae and Sphingobacteriales (OTU 1453 and 1, respectively), had upper rRNA gene operon copy number estimates of 5 and 6, respectively. K-selected organisms found in our sequence libraries, such as Nitrospira sp., Nitrosomonas sp., and taxa phylogenetically associated with Planctomycetes and Chloroflexi all contained 1 rRNA gene operon and at the family-level had average estimates of 1.75. K-selected organisms were present only at SRTs ⩾12-days, whereas r-selected organisms were abundant during all SRTs. The decline in abundance of K-selected organisms during the MDS validates that SRT is an appropriate parameter to enrich for a variety of organisms that display trade-offs between maximum growth rate and resource-use efficiency (that is, life-history strategy).
Discussion
Microbial diversity is not resilient to all types of disturbance
In this study, it is demonstrated that (1) the operational parameter SRT can selectively enrich for organisms based on life-history strategy (that is, selection of r–K-strategists); (2) in contrast to studies that show resilience of microbial diversity to pulse-disturbances (Shade et al., 2012), microbial communities in activated sludge did not fully recover to their original state after the press disturbance and (3) the recovery of microbial composition is not a prerequisite for the recovery of general ecosystem functions. Concordantly, our results support the functional redundancy hypothesis for microbial ecosystems (Allison and Martiny, 2008) and indicate that after a major disturbance, new microbial communities reassemble to perform the same ecosystem functions. Thus, environmental conditions are the main driver in selecting for specific ecosystem processes. A major issue in microbial ecology has been to elucidate compositional versus environmental effects on ecosystem processes. Allison and Martiny (2008) suggest that this could be accomplished through manipulation of microbial composition while controlling the abiotic environment. We chose SRT as the most appropriate parameter to address this question because of its capacity to select for organisms over a range of life-history strategies. The manipulation of SRT is also analogous to macro-scale ecological processes where diversity in a climax community is reduced by disturbance followed by an interspecific competition and secondary succession of early colonizers (Horn, 1974). Indeed, over 55% reduction in diversity was observed during the MDS and early colonizers that were phylogenetically affiliated with bacterial orders; Comamonadaceae and Sphingobacteriales outcompeted late colonizers, such as ammonia-oxidizing bacteria, nitrite-oxidizing bacteria, Planctomycetes, Chloroflexi and populations of Actinobacteria and Gammaproteobacteria. Our results show that although some phylogenetically related organisms reestablish after the disturbance, community composition was significantly different and ∼40% less diverse (1D) with rare membership comprising the majority of diversity loss.
Despite the incomplete recovery of diversity and composition, general bioreactor functions (that is, carbon oxidation, nitrification/denitrification and phosphorus accumulation) recovered relatively quickly (54 days). These results indicate functional redundancy of the post-disturbance community. It is important to note that although complete recovery of diversity was not observed during the study period, it is possible that rare taxa may have returned if the study period were extended. Furthermore, we have not addressed how diversity loss in the rare species pool may affect ecosystem stability in response to future disturbances, as rare and dormant organisms have an important role in maintaining diversity (Jones and Lennon, 2010). In addition, rare community members are likely responsible for other metabolically specialized functions such as biotransformation and degradation of organic micropollutants (Helbling et al., 2012; Johnson et al., 2012). Additional studies should evaluate the impact of disturbance on micropollutant removal coupled with surveys of associated functional genes. Finally, the mechanisms by which microbial ecosystems can be repopulated after disturbance, either through recolonization (that is, immigration) or regrowth, must also be evaluated in order to inform our understanding of diversity maintenance and enhance recovery of microbial ecosystems after disturbance.
Improved methods of comparing microbial diversity
A common approach in microbial community ecology studies is to assess ecosystem resilience and recovery through comparison of univariate metrics and visualization of community data in ordination space (Fierer et al., 2008; Costello et al., 2010; Dethlefsen and Relman, 2010; Caporaso et al., 2011; Werner et al., 2011; Zhao et al., 2012; Shade et al., 2013). These methods often oversimplify and condense complex community data into a single datum point, which cannot be partitioned into independent alpha- and beta- components (Jost, 2007). In the current study, we expand the suite of measurements to include a variety of ecosystem processes and demonstrate the utility of using Hill numbers to more comprehensively measure and compare microbial diversity. For example, if conclusions regarding community resilience were based solely on WU PCoA results (Supplementary Figure S3), one could falsely conclude that the microbial community completely recovered. However, the use of Hill numbers expands our understanding of diversity by revealing the opposite conclusion: microbial diversity was not resilient because diversity across all orders of q did not recover to the pre-disturbance state. We contend that Hill numbers, calculated both taxonomically (that is, OTU-based) and phylogenetically (that is, using diversity profiles), are the most informative metrics for comparing microbial diversity, and that these metrics should permeate into other fields of microbial ecology such as clinical microbiology associated with the massive DNA sequence studies of the Human Microbiome Project (H.M.P. Consortium, 2012).
Prediction of ecosystem health based on indicator species
In this study, the activated sludge metacommunity is temporally partitioned by selecting for organisms based on a range of life-history strategies (that is, r versus K-strategists). Fierer et al. (2007) found that certain bacterial phyla in soil could be partitioned into r–K categories using both experimental soil columns and meta-analysis, measured by the net carbon mineralization rate. The relative abundance of Acidobacteria negatively correlated with net carbon mineralization rate (r2=0.26, P<0.001), whereas Bacteroidetes and the Beta-class of the phylum Proteobacteria were positively correlated with net carbon mineralization rates (r2=0.34, P<0.001 and r2=0.35, P<0.001, respectively) (Fierer et al., 2007). Our study yields similar results, albeit through an alternative selection method: adjustment of SRT in a wastewater treatment system. We demonstrate that more ecosystem processes (nitrification/denitrification and phosphate-accumulating organisms activity) are present at higher SRTs when K-strategists are detected. At lower SRTs (3 days), K-strategists were absent and the only remaining ecosystem process, among the performance parameters that were measured, was carbon oxidation. As more genomes are sequenced and added to the Ribosomal RNA Database and other databases, future studies can enumerate the number of K-strategists in their sequence libraries to gauge ecosystem health and successional stage.
Merging ecology and environmental engineering
Activated sludge bioreactors are a model system for microbial ecology, but they also serve a major role in water quality and public health protection and as a renewable source of freshwater in urban centers with scarce water supplies. Knowledge and integration of both disciplines can improve existing technologies in which engineered biological systems are optimized for specific purposes (McMahon et al., 2007). Such systems will improve upon our current strategies for water supply and treatment: current ‘linear' methods of water supply (that is, inter-basin water transfers, use, and discharge) are not sustainable because as urban centers grow larger, regional water scarcity becomes more prevalent and water supply may be compromised owing to climate change (Daigger, 2009). Thus, new paradigms of urban water management must treat wastewater as a resource, rather than a liability, by planning for water reuse and nutrient reclamation. For example, the concept of ‘tailored' nutrient management (Vuono et al., 2013) aims to support this paradigm by recovering both water and nutrients by blending high nutrient streams (for example, anaerobic digester centrate) with reclaimed water or through flexible treatment plant operation. In the latter, greater knowledge of ecosystem resilience, robustness and stability within wastewater treatment systems in response to changing operating conditions will enable engineers and city water planners to sustainably manage freshwater resources through water and nutrient reclamation.
Acknowledgments
We are most grateful to Terry Reid, Lloyd Johnson and Aqua-Aerobic Systems, for their generous support. The material presented is based in part upon work supported by the National Science Foundation under Cooperative Agreement EEC-1028968. We thank Dave Armitage, Chuck Pepe-Ranney, Nicholas Gotelli, Holger Daims, Pat Schloss, Chuck Robertson, Kirk Harris, Ashley Shade, Robert Almstrand, Lee Stanish, Cathy Lozupone and Greg Caporaso for thoughtful discussions, help with data interpretation and bioinformatics suggestions. We additionally thank Ryan Holloway, Dean Heil and John McEncroe for their analytical support on this project.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage DW, Gallagher KL, Youngblut ND, Buckley DH, Zinder SH. Millimeter-scale patterns of phylogenetic and trait diversity in a salt marsh microbial mat. Front Microbiol. 2012;3:293. doi: 10.3389/fmicb.2012.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high- throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter SR. Microcosm experiments have limited relevance for community and ecosystem ecology. Ecology. 1996;77:677–680. [Google Scholar]
- Chao A, Chiu C-H, Jost L. Phylogenetic diversity measures based on Hill numbers. Philos Trans R Soc Lond B Biol Sci. 2010;365:3599–3609. doi: 10.1098/rstb.2010.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- H.M.P. Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Gordon JI, Secor SM, Knight R. Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J. 2010;4:1375–1385. doi: 10.1038/ismej.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigger GT. Evolving urban water and residuals management paradigms: water reclamation and reuse, decentralization, and resource recovery. Water Environ Res. 2009;81:809–823. doi: 10.2175/106143009x425898. [DOI] [PubMed] [Google Scholar]
- Daims H, Taylor MW, Wagner M. Wastewater treatment: a model system for microbial ecology. Trends Biotechnol. 2006;24:483–489. doi: 10.1016/j.tibtech.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Relman D. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci. 2010;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman B, Hamelin J, Moriarty J, Neal P, Dushoff J, Weitz JS. Robust estimation of microbial diversity in theory and in practice. ISME J. 2013;7:1092–1101. doi: 10.1038/ismej.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, McMahon KD. ‘Candidatus Accumulibacter' gene expression in response to dynamic EBPR conditions. ISME J. 2011;5:329–340. doi: 10.1038/ismej.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbling DE, Johnson DR, Honti M, Fenner K. Micropollutant biotransformation kinetics associate with WWTP process parameters and microbial community characteristics. Environ Sci Technol. 2012;46:10579–10588. doi: 10.1021/es3019012. [DOI] [PubMed] [Google Scholar]
- Hill MO. Diversity and Evenness: A Unifying Notation and Its Consequences. Ecology. 1973;54:427–432. [Google Scholar]
- Horn HS. The ecology of secondary succession. Annu Rev Ecol Syst. 1974;5:25–37. [Google Scholar]
- Johnson DR, Goldschmidt F, Lilja EE, Ackermann M. Metabolic specialization and the assembly of microbial communities. ISME J. 2012;6:1985–1991. doi: 10.1038/ismej.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci. 2010;107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. Entropy and diversity. Oikos. 2006;113:363–375. [Google Scholar]
- Jost L. Paritioning diversity into independent alpha and beta components. Ecology. 2007;88:2427–2439. doi: 10.1890/06-1736.1. [DOI] [PubMed] [Google Scholar]
- Klappenbach Ja, Saxman PR, Cole JR, Schmidt TM. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ZM-P, Bussema C, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37:D489–D493. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, et al. The metacommunity concept: a framework for multi-scale community ecology. Ecology Letters. 2004;7:601–613. [Google Scholar]
- Leinster T, Cobbold Ca. Measuring diversity: the importance of species similarity. Ecology. 2012;93:477–489. doi: 10.1890/10-2402.1. [DOI] [PubMed] [Google Scholar]
- Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, et al. A Nitrospira metagenomeilluminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci. 2010;107:13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maixner F, Noguera DR, Anneser B, Stoecker, Wegl G, Wagner M, et al. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol. 2006;8:1487–1495. doi: 10.1111/j.1462-2920.2006.01033.x. [DOI] [PubMed] [Google Scholar]
- Maixner F, Wagner M, Lücker S, Pelletier E, Schmitz-Esser S, Hace K, et al. Environmental genomics reveals a functional chlorite dismutase in the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”. Environ Microbiol. 2008;10:3043–3056. doi: 10.1111/j.1462-2920.2008.01646.x. [DOI] [PubMed] [Google Scholar]
- McMahon KD, Martin HG, Hugenholtz P. Integrating ecology into biotechnology. Curr Opin Biotechnol. 2007;18:287–292. doi: 10.1016/j.copbio.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Ofiteru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis C, et al. Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci USA. 2010;107:15345–15350. doi: 10.1073/pnas.1000604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pholchan MK, Baptista JDC, Davenport RJ, Curtis TP. Systematic study of the effect of operating variables on reactor performance and microbial diversity in laboratory-scale activated sludge reactors. Water Res. 2010;44:1341–1352. doi: 10.1016/j.watres.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Pianka E. On r- and K- selection. Am Nat. 1970;102:592–597. [Google Scholar]
- Reeder J, Knight R. Rapid denoising of pyrosequencing amplicon data: exploiting the rank-abundance distribution. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick D, Bryant M, Bashey F. r - and K -Selection Revisited: the role of population regulation in life-history evolution. Ecology. 2002;83:1509–1520. [Google Scholar]
- Saikaly PE, Stroot PG, Oerther DB. Use of 16S rRNA Gene Terminal Restriction Fragment Analysis To Assess the Impact of Solids Retention Time on the Bacterial Diversity of Activated Sludge. Appl Environ Microbiol. 2005;71:5814–5822. doi: 10.1128/AEM.71.10.5814-5822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Larsen P, Nielsen PH. Comparison of nutrient-removing microbial communities in activated sludge from full-scale MBRs and conventional plants. Water Sci Technol. 2013;68:366–371. doi: 10.2166/wst.2013.183. [DOI] [PubMed] [Google Scholar]
- Schloss PD. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput Biol. 2010;6:e1000844. doi: 10.1371/journal.pcbi.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol. 2011;77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seviour R, Nielsen PH.2010Microbial Ecology of Activated Sludge1st ednIWA Publishing: London, UK [Google Scholar]
- Shade A, Mcmanus PS, Handelsman J. Unexpected diversity during community succession in the apple flower microbiome. MBio. 2013;4:1–12. doi: 10.1128/mBio.00602-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Read JS, Youngblut ND, Fierer N, Knight R, Kratz TK, et al. Lake microbial communities are resilient after a whole-ecosystem disturbance. ISME J. 2012;6:2153–2167. doi: 10.1038/ismej.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan WT, Woodcock S, Lunn M, Head IM, Curtis TP. Modeling taxa-abundance distributions in microbial communities using environmental sequence data. Microb Ecol. 2007;53:443–455. doi: 10.1007/s00248-006-9141-x. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Lücker S, Vejmelkova D, Kostrikina Na, Kleerebezem R, Rijpstra WIC, et al. Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 2012;6:2245–2256. doi: 10.1038/ismej.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, et al. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature. 2006;440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- Sun Y, Yunpeng C, Huse SM, Knight R, Farmerie WG, Wang X, et al. A large-scale benchmark study of existing algorithms for taxonomy-independent microbial community analysis. Brief Bioinform. 2011;13:107–121. doi: 10.1093/bib/bbr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TW, Ng HY, Ong SL. Effect of mean cell residence time on the performance and microbial diversity of pre-denitrification submerged membrane bioreactors. Chemosphere. 2008;70:387–396. doi: 10.1016/j.chemosphere.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Tchobanoglous G, Burton F, Stensel D.2003Wastewater Engineering; Treatment and Reuse4th ednMcGraw-Hill Inc.: New York [Google Scholar]
- Vuono D, Henkel J, Benecke J, Cath TY, Reid T, Johnson L, et al. Flexible hybrid membrane treatment systems for tailored nutrient management: a new paradigm in urban wastewater treatment. J Memb Sci. 2013;446:34–41. [Google Scholar]
- Wagner M. Bacterial community composition and function in sewage treatment systems. Curr Opin Biotechnol. 2002;13:218–227. doi: 10.1016/s0958-1669(02)00315-4. [DOI] [PubMed] [Google Scholar]
- Wells GF, Park H-D, Eggleston B, Francis CA, Criddle CS. Fine-scale bacterial community dynamics and the taxa-time relationship within a full-scale activated sludge bioreactor. Water Res. 2011;45:5476–5488. doi: 10.1016/j.watres.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Wells GF, Park H-D, Yeung C-H, Eggleston B, Francis CA, Criddle CS. Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ Microbiol. 2009;11:2310–2328. doi: 10.1111/j.1462-2920.2009.01958.x. [DOI] [PubMed] [Google Scholar]
- Werner JJ, Knights D, Garcia ML, Scalfone NB, Smith S, Yarasheski K, et al. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci. 2011;108:4158–4163. doi: 10.1073/pnas.1015676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Kong Y, Thomsen TR, Halkjaer Nielsen P. Identification and ecophysiological characterization of epiphytic protein-hydrolyzing saprospiraceae (“Candidatus Epiflobacter” spp.) in activated sludge. Appl Environ Microbiol. 2008;74:2229–2238. doi: 10.1128/AEM.02502-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Zhang W, Liu R, Li Q, Li B, Wang S, et al. Phylogenetic diversity and metabolic potential of activated sludge microbial communities in full-scale wastewater treatment plants. Environ Sci Technol. 2011;45:7408–7415. doi: 10.1021/es2010545. [DOI] [PubMed] [Google Scholar]
- Ye L, Shao M-F, Zhang T, Tong AHY, Lok S. Analysis of the bacterial community in a laboratory-scale nitrification reactor and a wastewater treatment plant by 454-pyrosequencing. Water Res. 2011;45:4390–4398. doi: 10.1016/j.watres.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Zhang T, Shao MF, Ye L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012;6:1137–1147. doi: 10.1038/ismej.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci. 2012;109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.