Abstract
Five distinct cryptic lineages (clades I–V) have recently been recognized in the Escherichia genus. The five clades encompass strains that are phenotypically and taxonomically indistinguishable from Escherichia coli sensu stricto; however, scant data are available on their ecology, virulence and pathogenic properties. In this study 20 cryptic E. coli strains isolated from marine sediments were investigated to gain insights into their virulence characteristics and genetic traits. The ability to adhere to intestinal cells was highest among clade V strains, which also harbored the genes involved in gut colonization as well as the genes (pduC and eut operon) typically found in environmentally adapted E. coli strains. The pduC gene was significantly associated with clade V. Multilocus sequence typing of three representative clade V isolates revealed new sequence types (STs) and showed that the strains shared two allelic loci (adk 51 and recA 37). Our findings suggest that cryptic Escherichia lineages are common in coastal marine sediments and that this habitat may be suitable for their growth and persistence outside the host. On the other hand, detection in clade V strains of a gene repertoire and adhesion properties similar to those of intestinal pathogenic strains could indicate their potential virulence. It could be argued that there is a dual nature of cryptic clade V strains, where the ability to survive and persist in a secondary habitat does not involve the loss of the host-associated lifestyle. Clade V could be a group of closely related, environmentally adapted E. coli strains.
Introduction
Escherichia coli is one of the best known bacterial species and the most important model organism in genetic and molecular biology studies (Tenaillon et al., 2010). It is the predominant commensal aerobic organism in the gut microbiota of vertebrates, but the species also includes intra- and extra-intestinal pathogenic strains. Transmission of E. coli pathogenic strains is primarily through the fecal–oral route. This often requires transient passage through the secondary habitats such as soil, water and sediments, where E. coli must survive environmental stressors (Bergholz et al., 2011). Transit through water and sediments explains the global use of the species as an indicator of fecal pollution in water bodies. However, it has recently been demonstrated that E. coli is not only a transient organism in secondary habitats, but perhaps part of their natural microflora. Indeed, E. coli can grow and reach high densities outside mammalian hosts under favorable environmental conditions, also in the absence of fecal inputs (Power et al., 2005; Byappanahalli et al., 2006; Walk et al., 2007).
Environmentally adapted E. coli strains are phenotypically and taxonomically indistinguishable from E. coli sensu stricto. It has recently been recognized that some belong to five distinct cryptic lineages, Escherichia clades I–V (CI–CV; Walk et al., 2009; Clermont et al., 2011). Phylogenetic analysis showed that only CI should be considered to be related to E. coli species, whereas CII–CV are genetically divergent (Walk et al., 2009). Data on the prevalence and distribution of strains belonging to cryptic clades are still limited, but CV strains are the most abundant and have been recovered from aquatic environment more frequently than from fecal samples (Walk et al., 2009; Clermont et al., 2011). A recent study of over 200 commensal E. coli isolates from human and animal fecal samples reported the absence of cryptic strains (Lescat et al., 2013), supporting the emerging hypothesis that their habitat may be outside the host. The few studies assessing the genetic diversity of environmental E. coli strains have found different gene sets and pathways between environmental and enteric strains (Luo et al., 2011; Oh et al., 2012). The presence in aquatic environments of Escherichia populations that are not easily distinguishable from E. coli on the basis of biochemical profiling is likely to compromise its use as an indicator organism of fecal contamination and water pollution (Whitman and Nevers, 2003), because naturalized E. coli might interfere with enumeration of fecal coliforms (Clermont et al., 2011; Luo et al., 2011). Furthermore, the significance of Escherichia clades for human health in aquatic ecosystems remains to be elucidated.
In this work, the pathogenic potential of cryptic Escherichia strains from a set of 138 E. coli isolates, collected in marine sediments along the Adriatic coast (Luna et al., 2010; Vignaroli et al., 2013), was investigated by evaluating their adhesion and invasion properties in cell culture models. The genetic traits involved in gut colonization or associated with environmentally adapted strains were also assessed. Selected isolates were typed by multilocus sequence typing (MLST).
Materials and methods
Bacterial strains: phylogenetic grouping and clade assignment
A collection of 138 E. coli strains isolated from coastal marine sediments collected by our group in the Adriatic Sea (Luna et al., 2010; Vignaroli et al., 2013) and tested for their antibiotic resistance and virulence traits (Luna et al., 2010; Vignaroli et al., 2012, 2013) were screened to establish whether they could be assigned to one of the five cryptic clades.
Their phylogenetic grouping had been assessed by amplification of the chuA and yjaA genes and the DNA fragment TSPE4.C2 according to Clermont et al. (2000). The presence/absence of these DNA fragments led the strains to be assigned to phylogenetic groups A, B1, B2 or D (Luna et al., 2010; Vignaroli et al., 2013).
In this study, strains were attributed to cryptic lineages I, II, III, IV or V by a new PCR method based on aes and chuA allele-specific amplifications developed by Clermont et al. (2011).
PCR detection of virulence and metabolic genes
The virulence genes typical of enteroaggregative E. coli pathotype, that is, a gene encoding dispersin protein (aap), aggregative-adherence probe-associated protein (aatA) and fimbrial transcriptional activator (aggR), were sought using primers, PCR conditions and amplification protocols described in Sarantuya et al. (2004) and Thomazini et al. (2011). Screening for the capsule synthesis genes (kpsMTII, kpsMTIII) was performed using the PCR protocols and primers reported by Johnson and Stell (2000).
The presence of genes that are frequently associated with gut colonization (genes of the fucose operon and for maltose/maltodextrin transport and glycogen metabolism; Jones et al., 2008; Luo et al., 2011; Oh et al., 2012) and of genes that are commonly carried by the environmentally adapted strains (genes involved in propanediol and ethanolamine metabolism and in lysozyme production; Luo et al., 2011; Oh et al., 2012) was determined by single PCR assays. The gene list and primer pairs used (synthesized by Sigma-Genosys, Milan, Italy) are reported in Supplementary Table S1. Primer design and PCR conditions were developed in this study. Each PCR assay was performed in a 50-μl final reaction volume containing 1X buffer, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate (Euroclone, Celbio, Milan, Italy), 0.2 or 0.5 μM of each primer pair according to the gene tested (Supplementary Table S1), 1U of Hot Rescue Taq DNA polymerase (Diatheva, Fano, Italy) and 5 μl of DNA template obtained from crude lysates of bacterial cultures (1 ml) grown overnight in brain heart infusion broth (Oxoid, Basingstoke, UK). Crude lysates were obtained using the procedure described by Hynes et al. (1992), modified by reducing incubation times (20 min at 37 °C and 30 min at 60 °C). The amplification program was as follows: 1 cycle of 10 min at 95 °C, followed by 30 cycles of 30 s at 95 °C, 30 s at an annealing temperature specific for each primer pair (see Supplementary Table S1), 30 s at 72 °C and a final extension step of 7 min at 72 °C. Reference strains E. coli ATCC 35150 and K-12 C600, or E. coli strains from our laboratory collection, testing positive for the virulence/metabolic genes examined, were used as positive controls after sequencing of all PCR products.
Cell lines
Caco-2 (ATCC HTB37) and Intestine 407 (Int407, ATCC CCL-6) cells were used as intestinal cell models. Cells were routinely cultured in 50 ml (25 cm2) plastic tissue culture flasks (CELLSTAR, Greiner Bio-One, Frickenhausen, Germany) in an atmosphere containing 5% CO2 at 37 °C in modified Eagle's medium (Lonza Bio Whittaker, Verviers, Belgium) supplemented with 1% (v/v) L-glutamine, 1% (v/v) non-essential amino acids and 10% (v/v) fetal calf serum (all were purchased from Gibco, Grand Island, NY, USA), with a change of culture medium every 48 h.
Adhesion and invasion assays
For adhesion and invasion assays, confluent cell monolayers were trypsinized and resuspended in culture medium; 2 ml of the cell suspension was then seeded in six-well microplates and incubated for 5–6 days under the conditions described above until they reached semiconfluence. After overnight growth in Luria-Bertani broth (Oxoid), bacterial cultures were diluted (1:100) and incubated in the same medium at 37 °C in a shaking bath. After reaching an optical density of 0.1 at 625 nm (OD625), corresponding to 107–108 CFU ml−1, bacterial cells were washed once with phosphate-buffered saline (PBS) and resuspended in modified Eagle's medium without serum. Intestinal monolayers were washed in PBS and 2 ml of the bacterial suspension (at OD625=0.1) was added to each well and the microplate was incubated for 2 h at 37 °C in 5% CO2.
To evaluate adherent bacteria, infected monolayers were washed three times with PBS and lysed with 0.1% Triton X-100. Aliquots of the appropriate dilutions of the lysate were plated in brain heart infusion agar and incubated overnight at 37 °C to count total adherent bacteria (CFU ml−1). To determine viable intracellular bacteria, infected monolayers were washed three times with PBS and covered with 2 ml modified Eagle's medium supplemented with gentamicin (100 μg ml−1), to kill extracellular bacteria. After 2 h at 37 °C in 5% CO2, monolayers were washed and lysed as described above; viable internalized bacteria were counted by plating the lysates on brain heart infusion agar. The number of adherent and invasive bacteria was expressed as a percentage of the initial inoculum. The adhesion index (AI) was calculated as the ratio of adherent bacteria (%) of the tested strain to adherent bacteria (%) of the positive control strain. E. coli ATCC 35150 and K-12 C600 were the positive and negative control strains, respectively.
For microscopic observation, a suspension of 107 bacteria per ml was added to the tissue culture and incubated for 2 h at 37 °C in 5% CO2. After three washes with PBS, cells were fixed with methanol for 10 min, stained with 20% Giemsa for 20 min (Darfeuille-Michaud et al., 1998) and examined with a Leica DMRB microscope (Leica Microsystems, Wetzlar, Germany) using the X100 oil-immersion objective.
MLST
MLST was performed as described previously (Vignaroli et al., 2012, 2013) using sequence analysis of internal fragments of the seven housekeeping genes adk, fumC, gyrB, icd, mdh, purA and recA, according to the protocol reported on the E. coli MLST website (http://mlst.ucc.ie/mlst/mlst/dbs/Ecoli/documents).
eBURSTv3 software, available at http://eburst.mlst.net, was used to compare our MLST data with the MLST database and to represent results as an eBURST diagram.
Statistical analysis
Fisher's test was used to evaluate whether the association between assignment to CV and presence of the genes commonly found in environmental strains (pduC, yegX and eut) was significant. Significance was set at a P-value <0.05.
Results
Assignment to the cryptic clades
Of the 138 strains of our collection, 20 (14.5%) were found to be cryptic members of the Escherichia genus. Application of the typing method recently developed by Clermont et al. (2011) enabled them to be assigned to one of the five cryptic clades as follows: 18 strains belonged to CV, 1 to CIII and 1 to CIV.
Detection of virulence and metabolic genes
The frequency of the virulence and metabolic genes in the 20 cryptic Escherichia strains is reported in Supplementary Table S2. PCR data demonstrated that none of the strains carried the typical enteroaggregative E. coli pathotype virulence genes (aap, aatA and aggR); nonetheless, 60% were positive for the astA gene, which encodes EAST1 enterotoxin, as described previously (Vignaroli et al., 2012, 2013). Moreover, 70% of strains were positive for the kpsMT genes, specific for group II capsules. None of the strains carried group III capsule genes. The virulence gene profile, including the additional genes (aer, fyuA and iroN) previously found in each cryptic strain (Luna et al., 2010; Vignaroli et al., 2012, 2013), is reported in Table 1.
Table 1. Virulence and metabolic genes carried by the 20 cryptic clade strains.
| Strain | Clade | Virulence gene | fuc operon | glgA | mal genes | aggR | aap | aat | pduC | yegX | eut operon |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FE5E1 | V | / | K, U | + | F, P, Q | − | − | − | + | − | A, B, E, L, P, S |
| FE5E3 | V | astA, kpsMTII | K, R, U | + | F, P, Q | − | − | − | + | − | A, B, E, L, P, S |
| FE5E7 | III | astA, kpsMTII | / | + | F, P, Q | − | − | − | + | + | A, B, E, L, P, S |
| PE10i1 | V | kpsMTII | K, R, U | + | F, P, Q | − | − | − | + | − | A, B, E, L, P, S |
| PE11i3 | V | kpsMTII | P, K, R, U | + | F, P, Q | − | − | − | + | + | A, B, E, L, P, S |
| PE11E1 | V | astA, fyuA | P, K, R, U | + | F, P, Q | − | − | − | + | + | A, B, E, L, P, S |
| PE11E2 | V | astA, kpsMTII | P, K, R, U | + | F, P, Q | − | − | − | + | − | A, B, E, L, P, S |
| CF12i2 | V | astA | P, K, R, U | + | F, P, Q | − | − | − | + | + | A, B, E, L, P, S |
| CF12i9 | V | astA, kpsMTII | P, K, R, U | + | F, P, Q | − | − | − | + | − | A, B, E, L, P, S |
| CF12i12 | V | astA, kpsMTII | P, K, R, U | + | F, P, Q | − | − | − | + | − | A, B, E, L, P, S |
| PE9i1 | V | kpsMTII | K, R, U | + | F, P, Q | − | − | − | + | − | A, B, E, L, P, S |
| PE9i5 | V | kpsMTII | P, K, R, U | + | F, P, Q | − | − | − | + | + | A, B, E, L, P, S |
| PE9i15 | V | astA, kpsMTII | P, K, R, U | + | F, P, Q | − | − | − | + | + | A, B, E, L, P, S |
| PE9i21 | V | / | P, K, R, U | + | F, P, Q | − | − | − | + | + | A, B, E, L, P, S |
| PE9i22 | V | astA, kpsMTII | P, K, R, U | + | F, P, Q | − | − | − | + | − | A, B, E, L, P, S |
| PE9i36 | IV | iutA, iroN, kpsMTII | / | + | F, P, Q | − | − | − | − | + | A, B, E, L, P, S |
| PR15E9 | V | astA | P, K, R, U | + | F, P, Q | − | − | − | + | − | A, B, E, L, P, S |
| PR15i10 | V | astA, fyuA | P, K, R, U | + | F, P, Q | − | − | − | − | + | A, B, E, L, P, S |
| PR16E3 | V | astA, fyuA, aer, iroN | K, R, U | + | F, P, Q | − | − | − | + | − | A, B, E, L, P, S |
| PR17E3 | V | kpsMTII | K, R, U | + | F, P, Q | − | − | − | + | + | A, B, E, L, P, S |
Most of the isolates (60–100%) carried genes for fucose, maltose, amylomaltase and maltodextrin metabolism (Table 1, Supplementary Table S2). The CIII strain and the CIV strain both lacked the fuc operon genes (fucPKUR).
All 20 Escherichia cryptic strains harbored the eutABELPS genes of the eut operon, which is involved in ethanolamine degradation. The yegX gene, encoding a muramidase, was detected in the CIII and CIV strains and in 8/18 CV strains. However, as the control strains (enterohemorrhagic ATCC 35150 and commensal K-12 C600) also carried eut and yegX, the statistical analysis was not informative.
All but two strains (the single CIV strain and CV E. coli PR15i10) carried the pduC gene, encoding a propanediol dehydratase; both control strains lacked this gene. To establish whether it was specific to CV isolates, 22 additional E. coli strains from our collection, representing different phylogroups but belonging to none of the five cryptic clades, were also subjected to pduC gene screening. We found that 15/22 strains were negative and Fisher's exact test showed a significant association between the pduC gene and CV (P=0.0001).
The virulence and metabolic genes carried by the 20 cryptic strains are listed in Table 1.
Adhesion and invasion
Eight cryptic strains (6/18 CV strains, the CIII strain and the CIV strain), which were found by PCR to differ in their content in the genes tested, were selected for adhesion and invasion assays using Caco-2 and Int 407 cell lines.
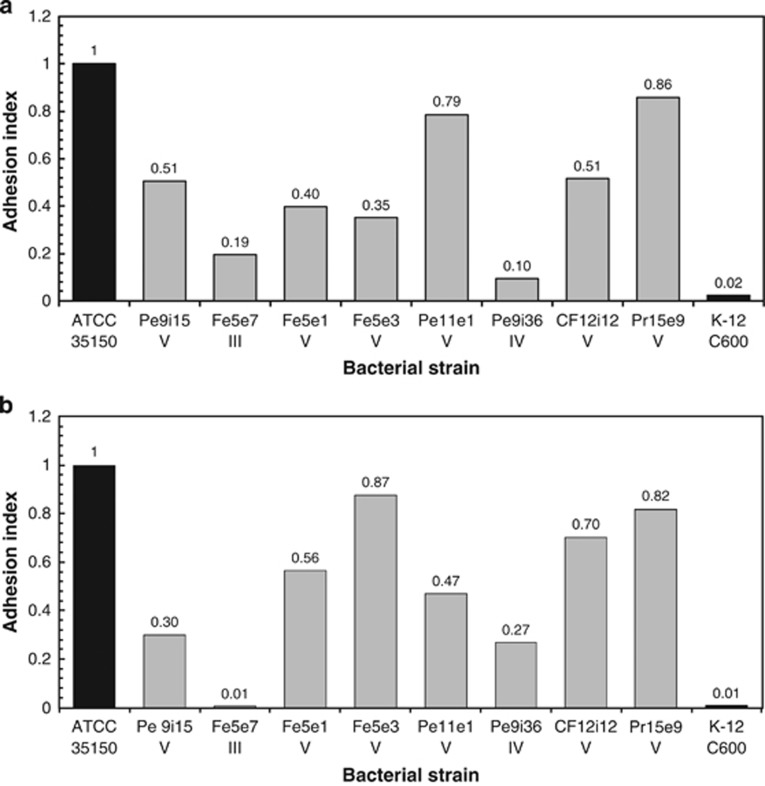
The CV strains (FE5E1, FE5E3, PE11E1, CF12i12, PE9i15 and PR15E9) showed a good AI with both cell lines, whereas the CIII strain (FE5E7) and the CIV strain (PE9i36) had very low AI (Figures 1a and b).
Figure 1.
Adhesion of eight cryptic E. coli strains to Caco-2 (a) and Int 407 (b) cells. The adhesion index is a dimensionless value ranging from 0 (no adhesion) to 1 (maximum adhesion, equal to that observed in the positive control E. coli ATCC 35150).
In the assays with Caco-2 cells (Figure 1a), two CV strains (PE11E1 and PR15E9) showed AI (0.79 and 0.86, respectively) close to those of the positive control E. coli ATCC 35150 (AI=1). The AI of the other six CV strains ranged from 0.35 to 0.51. In tests with Int 407 cells (Figure 1b), CV strain PR15E9 showed a similar AI as in the experiment with Caco-2 cells, whereas the AI of CV strains CF12i12 and FE5E3 were higher. The CIII strain exhibited the lowest AI (0.01).
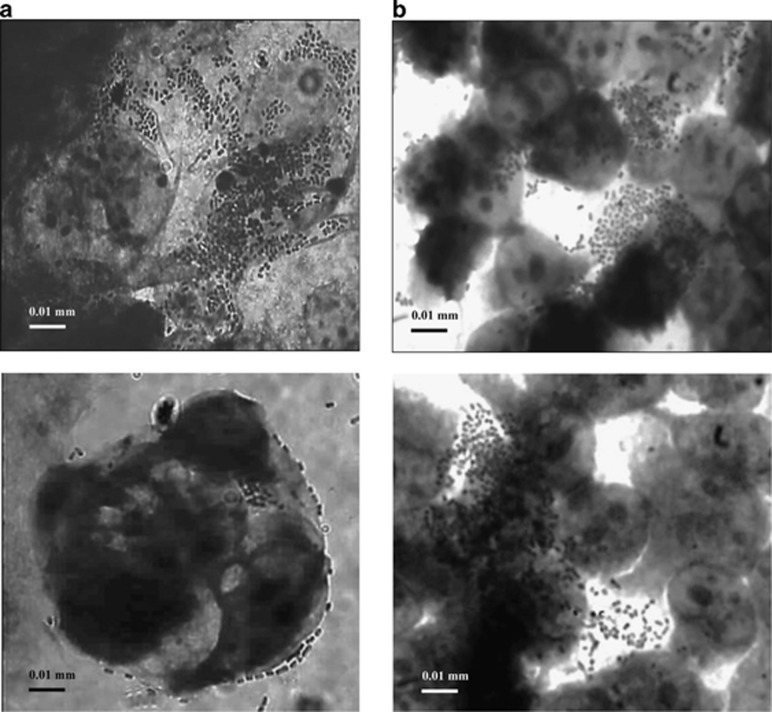
No strain exhibited the ability to invade either of the intestinal cell line. By microscopic analysis, we found that the strains displayed a localized pattern of adherence to both the intestinal cell lines, with microcolony formation (Figures 2a and b).
Figure 2.
Localized pattern of adherence of clade V strain PE9i15 to Caco-2 cells (a) and of clade V strain FE5E3 to Int 407 cells (b). Standard light microscopy of Giemsa-stained cells.
MLST analysis
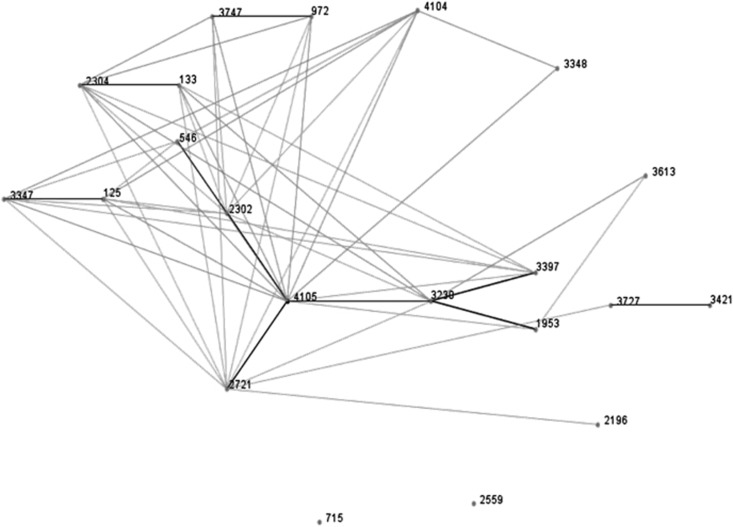
Three CV strains PE11E1, PE9i15 and PR15E9, the CIII strain FE5E7 and the CIV strain PE9i36 underwent ST determination. Their allelic combinations are reported in Table 2. The CIII strain, which had a low AI and lacked the fuc operon genes, was assigned to ST2371. The other four strains exhibited new allelic profiles and were deposited as new STs. A new fumC allele was documented in one of the CV adherent isolates, E. coli PE9i15, CV strain PR15E9 showed a new combination of known alleles and CV strain PE11E1 displayed a new icd allele. The CIV strain exhibited 5/7 new alleles. It is worth noting that adk 51 and recA 37 were documented in all three CV strains. All available STs (n=21) containing adk 51 and recA 37 in the MLST database (see Supplementary Table S3) were then compared with our strains using the eBURST algorithm. In the descent patterns provided by the eBURST diagram (Figure 3) ST4105, assigned to CV E. coli PR15E9, seems to be the founder of a clonal complex encompassing most of the selected STs as single and double locus variants of the founder.
Table 2. Multilocus sequence typing results showing the allelic combinations of the cryptic strains typed in this study.
| Strain | Clade | adk | fumC | gyrB | icd | mdh | purA | recA | Sequence type |
|---|---|---|---|---|---|---|---|---|---|
| FE5E7 | III | 276 | 356 | 279 | 304 | 242 | 235 | 210 | ST2371 |
| PE9i36 | IV | 356 | 531 | 387 | 441 | 56 | 322 | 63 | ST4103 |
| PE11E1 | V | 51 | 48 | 45 | 442 | 34 | 279 | 37 | ST4104 |
| PE9i15 | V | 51 | 468 | 220 | 266 | 235 | 42 | 37 | ST3613 |
| PR15E9 | V | 51 | 48 | 45 | 266 | 34 | 42 | 37 | ST4105 |
Figure 3.
e-BURST diagram showing the putative founder ST4105 (E. coli PR15E9) and the patterns of descent. The single and double locus variants of the 21 sequence types analyzed are connected by black and gray lines, respectively.
Discussion
It is well established that E. coli strains can survive and persist in soil and aquatic environments (Byappanahalli et al., 2006; Walk et al., 2007; Walk et al., 2009; Bergholz et al., 2011). Such strains are phenotypically indistinguishable but genetically divergent from commensal E. coli and have been assigned to the five novel cryptic clades, CI–CV (Walk et al., 2009). As data on the distribution, genetic diversity and virulence of cryptic strains are still limited (Gordon et al., 2002; Walk et al., 2007; Clermont et al., 2011; Luo et al., 2011), this study was designed to assess the virulence features, genetic traits and phylogenetic relationships of cryptic strains isolated from coastal marine sediments.
Of the 138 E. coli strains isolated from marine sediment (Luna et al., 2010; Vignaroli et al., 2013), 20 (14.5%) were found to belong to a cryptic clade. Most (85%) were susceptible to all the antimicrobials tested in our previous studies (Vignaroli et al., 2012, 2013), also in line with the findings reported by Ingle et al. (2011): these data are consistent with the hypothesis of the non-intestinal origin of cryptic clades. The sporadic recovery of strains from clade II–V from vertebrate hosts suggests a poor ability of these strains to compete for gastrointestinal tract colonization. Conversely, their features probably favor their persistence in the environment (water, soil and sediment), where these strains are believed to pose a low-level threat to public health (Walk et al., 2009; Ingle et al., 2011; Luo et al., 2011). Different gene sets and pathways may facilitate E. coli survival in the human gastrointestinal tract and the environment; in particular, the genes related to the transport and utilization of nutrients (N-acetylglucosamine, gluconate, 5-C and 6-C sugars such as fucose), which are abundant in the gastrointestinal tract, may be crucial for gut colonization and survival (Chang et al., 2004; Luo et al. 2011; Oh et al., 2012). Furthermore, other nutrients available in the mammalian gut, such as maltose, maltodextrins and glycogen, seem to play an important role not only in E. coli colonization but also in the maintenance of rapid growth rates in the host (Jones et al., 2008).
Our experiments showed that all CV strains possessed two to four fuc genes of the fucose operon, whereas the CIII and CIV strains lacked them completely. These findings are consistent with the total absence of the fuc operon in other clade III (TW09231, TW09276) and clade IV (TW14182, TW11588, H605) Escherichia strains (Luo et al., 2011), whose genomes are available on http://www.broadinstitute.org and http://www.patricbrc.org. Genes for glycogen, maltose, amylomaltase and maltodextrin metabolism were carried by all 20 strains. It is worth noting that the control enterohemorrhagic strain E. coli ATCC 35150 also carried these genes. As our cryptic strains showed a gene content similar to that of control strains, they may have maintained the typical genetic traits of enterobacteria. Clearly, further assays are required to test strains' gene expression and their ability to grow in the presence of the various sugar substrates.
The 20 cryptic strains were also positive for several genes of the eut operon, either key (eutABE) and accessory (eutLSP) genes involved in the aerobic utilization of ethanolamine, a compound also found in cell membranes. In the absence of other nutrients, bacteria such as E. coli can use it as the sole source of carbon and nitrogen via the enzyme ethanolamine ammonia-lyase (encoded by eutAB; Kofoid et al., 1999). Although ethanolamine metabolism has been related to E. coli environmental survival by Luo et al. (2011), the relationship between the ability to use ethanolamine and bacterial pathogenesis has been highlighted by others (Kofoid et al., 1999; Garsin, 2010).
Within the environment-specific gene set, yegX (for lysozyme production) and pduC (for propanediol utilization) seem to be crucial for survival in the environment (Luo et al., 2011). However, detection of yegX in 10 cryptic strains and in the two control strains (pathogenic E. coli ATCC 35150 and commensal E. coli K-12 C600) rules out its use as a marker of environmental adaptation. In contrast, pduC was significantly associated with CV and may be a more valuable marker of environmentally adapted isolates.
The 20 cryptic strains harbored a small number of virulence genes and did not share a common virulence profile. The presence of the astA gene associated with the absence of the aggR regulon, seen in most of our strains, has also been described in several commensal and pathogenic E. coli isolates (Ménard and Dubreuil, 2002) that have been designated atypical enteroaggregative E. coli due to the unclear role of EAST1 (Kaper et al., 2004; Croxen and Finlay, 2010). Our findings agree with those of Ingle et al. (2011), who reported a high frequency of some virulence factors (astA, aer, kpsE) in cryptic isolates, particularly CV strains, even though these strains were avirulent, in a mouse model of extra-intestinal infection.
Given the presence in our strains of genetic traits similar to those of intestinal isolates, we tested their in vitro ability to adhere to human enterocytes. All the CV strains used in these experiments displayed good adhesion to at least one of the intestinal cell lines (Caco-2, Int 407). Neither the CIII nor the CIV strain carried the fuc genes, suggesting that the different AI among clades could be related to their genetic background. The adhesion patterns observed in the CV strains further demonstrate their ability to interact with the human host and do not allow excluding their pathogenicity to humans.
The MLST data support the view that cryptic clades are more abundant in the environment and bird gut than in the human gastrointestinal tract (Walk et al., 2009; Lescat et al., 2013). The CIII strain belonged to ST2371, previously described in an avian E. coli strain (E. coli MLST database), whereas the CIV and CV strains were assigned to new STs. Interestingly, the three CV strains, though belonging to different STs, shared adk 51 and recA 37; in addition, CV strains PE11E1 and PR15E9 shared three more alleles (fumC 48, gyrB 45 and mdh 34), and PE9i15 and PR15E9 (CV strains) shared two more alleles (icd 266 and purA 42). Despite their different origin from the sediments sampled at different times and sites, all strains showed a common genetic background.
Querying the MLST database disclosed that 18 E. coli isolates assigned to different STs shared adk 51, fumC 48 and recA 37 with our CV strains. Moreover, all 18 strains displayed allelic combinations that are very similar to those found in our strains. Most were environmental or animal isolates and three were described as CV strains (E. coli Z205, RL325/96, Walk et al. (2009) and E. coli E1118, Luo et al. (2011)). This ST data set was used to explore the genotypic correlation among these strains and our CV isolates by eBURST analysis. The approach produced a diagram where these STs are linked (clonal complex) and where our CV PR15E9 strain (ST4105) emerges as the putative founder, differing from most of the other STs at a single or double locus. These findings support the hypothesis that, despite their wide geographic distribution in different environments, CV isolates are a distinct group of closely related non-human strains.
The present study provides new insights on the ecology and evolution of cryptic Escherichia lineages in the sedimentary habitat and suggests that they could potentially confound water quality assays, leading to misinterpretation of the risks of exposure to fecal bacteria (Rijal et al., 2011). Current practices of water quality monitoring could be improved with new molecular methods aimed to identify and distinguish E. coli from Escherichia cryptic clades. These methods should be based on discriminating genetic targets and our data point out pduC gene as a putative useful biomarker for CV strains, even if further evidences are required.
It may be concluded that Escherichia cryptic clades are also common in coastal marine sediments and that this habitat may be suitable for their growth/persistence outside the host; such ability probably depends on a gene repertoire favoring survival in the environment. However, they also exhibit genetic traits and adhesion properties similar to those of intestinal pathogenic strains, indicating potential virulence. It could be argued a dual nature of cryptic CV, where the ability to survive and persist in a secondary habitat does not involve the loss of the host-associated lifestyle.
Acknowledgments
We are grateful to Prof Anna Pianetti (University of Urbino) for supplying both the intestinal cell lines used in this work.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Bergholz PW, Noar JD, Buckley DH. Environmental patterns are imposed on the population structure of Escherichia coli after fecal deposition. Appl Environ Microbiol. 2011;77:211–219. doi: 10.1128/AEM.01880-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli MN, Whitman RL, Shively DA, Ting WT, Tseng CC, Nevers MB. Seasonal persistence and population characteristics of Escherichia coli and enterococci in deep backshore sand of two freshwater beaches. J Water Health. 2006;4:313–320. doi: 10.2166/wh.2006.518. [DOI] [PubMed] [Google Scholar]
- Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O, Gordon DM, Brisse S, Walk ST, Denamur E. Characterization of the cryptic Escherichia lineages: rapid identification and prevalence. Environ Microbiol. 2011;13:2468–2477. doi: 10.1111/j.1462-2920.2011.02519.x. [DOI] [PubMed] [Google Scholar]
- Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol. 2010;8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DM, Bauer S, Johnson JR. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology. 2002;148:1513–1522. doi: 10.1099/00221287-148-5-1513. [DOI] [PubMed] [Google Scholar]
- Hynes WL, Ferretti JJ, Gilmore MS, Segarra RA. PCR amplification of streptococcal DNA using crude cell lysates. FEMS Microbiol Lett. 1992;94:139–142. doi: 10.1016/0378-1097(92)90597-h. [DOI] [PubMed] [Google Scholar]
- Ingle DJ, Clermont O, Skurnik D, Denamur E, Walk ST, Gordon DM. Biofilm formation by and thermal niche and virulence characteristics of Escherichia spp. Appl Environm Microbiol. 2011;77:2695–2700. doi: 10.1128/AEM.02401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains of patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- Jones SA, Jorgensen M, Chowdhury FZ, Rodgers R, Hartline J, Leatham MP, et al. Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect Immun. 2008;76:2531–2540. doi: 10.1128/IAI.00096-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescat M, Clermont O, Woerther PL, Glodt J, Dion S, Skurnik D, et al. Commensal Escherichia coli strains in Guiana reveal a high genetic diversity with host-dependant population structure. Environ Microbiol Rep. 2013;5:49–57. doi: 10.1111/j.1758-2229.2012.00374.x. [DOI] [PubMed] [Google Scholar]
- Luna GM, Vignaroli C, Rinaldi C, Pusceddu A, Nicoletti L, Gabellini M, et al. Extraintestinal Escherichia coli carrying virulence genes in coastal marine sediments. Appl Environ Microbiol. 2010;76:5659–5668. doi: 10.1128/AEM.03138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci USA. 2011;108:7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard LP, Dubreuil JD. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): a new toxin with an old twist. Crit Rev Microbiol. 2002;28:43–60. doi: 10.1080/1040-840291046687. [DOI] [PubMed] [Google Scholar]
- Oh S, Buddenborg S, Yoder-Himes DR, Tiedje JM, Konstantinidis KT. Genomic diversity of Escherichia isolates from diverse habitats. PLoS One. 2012;7:e47005. doi: 10.1371/journal.pone.0047005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power ML, Littlefield-Wyer J, Gordon DM, Veal DA, Slade MB. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ Microbiol. 2005;7:631–640. doi: 10.1111/j.1462-2920.2005.00729.x. [DOI] [PubMed] [Google Scholar]
- Rijal G, Tolson JK, Petropoulou C, Granato TC, Glymph A, Gerba C, et al. Microbial risk assessment for recreational use of the Chicago area waterway system. J Water Health. 2011;9:169–186. doi: 10.2166/wh.2010.020. [DOI] [PubMed] [Google Scholar]
- Sarantuya J, Nishi J, Wakimoto N, Erden S, Nataro JP, Sheikh J, et al. Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in mongolian children. J Clin Microbiol. 2004;42:133–139. doi: 10.1128/JCM.42.1.133-139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- Thomazini CM, Samegima DA, Rodrigues MA, Victoria CR, Rodrigues J. High prevalence of aggregative adherent Escherichia coli strains in the mucosa-associated microbiota of patients with inflammatory bowel diseases. Int J Med Microbiol. 2011;301:475–479. doi: 10.1016/j.ijmm.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Vignaroli C, Luna GM, Rinaldi C, Di Cesare A, Danovaro R, Biavasco F. New sequence types and multidrug resistance among pathogenic Escherichia coli isolates from coastal marine sediments. Appl Environ Microbiol. 2012;78:3916–3922. doi: 10.1128/AEM.07820-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignaroli C, Luna GM, Pasquaroli S, Di Cesare A, Petruzzella R, Paroncini P, et al. Epidemic Escherichia coli ST131 and Enterococcus faecium ST17 in coastal marine sediments from an Italian beach. Environ Sci Technol. 2013;47:13772–13780. doi: 10.1021/es4019139. [DOI] [PubMed] [Google Scholar]
- Walk ST, Alm EW, Calhoun LM, Mladonicky JM, Whittam TS. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ Microbiol. 2007;9:2274–2288. doi: 10.1111/j.1462-2920.2007.01341.x. [DOI] [PubMed] [Google Scholar]
- Walk ST, Alm EW, Gordon DM, Ram JL, Toranzos GA, Tiedje JM, et al. Cryptic lineages of the genus Escherichia. Appl Environ Microbiol. 2009;75:6534–6544. doi: 10.1128/AEM.01262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman RL, Nevers MB. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Appl Environ Microbiol. 2003;69:5555–5562. doi: 10.1128/AEM.69.9.5555-5562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.