Background: PRODH and P5CDH from Thermus thermophilus are monofunctional enzymes in proline catabolism.
Results: Steady-state kinetics and intermediate trapping data show the PRODH and P5CDH reactions are coupled by a channeling step.
Conclusion: Substrate channeling in monofunctional enzymes is achieved via weak interactions.
Significance: Evidence for substrate channeling between monofunctional proline catabolic enzymes is shown and confirms the Rosetta Stone hypothesis.
Keywords: Amino Acid, Bacterial Metabolism, Dehydrogenase, Enzyme Kinetics, Enzyme Mechanism, Flavoprotein, Proline, Substrate Channeling
Abstract
Proline dehydrogenase (PRODH) and Δ1-pyrroline-5-carboxylate (P5C) dehydrogenase (P5CDH) catalyze the four-electron oxidation of proline to glutamate via the intermediates P5C and l-glutamate-γ-semialdehyde (GSA). In Gram-negative bacteria, PRODH and P5CDH are fused together in the bifunctional enzyme proline utilization A (PutA) whereas in other organisms PRODH and P5CDH are expressed as separate monofunctional enzymes. Substrate channeling has previously been shown for bifunctional PutAs, but whether the monofunctional enzymes utilize an analogous channeling mechanism has not been examined. Here, we report the first evidence of substrate channeling in a PRODH-P5CDH two-enzyme pair. Kinetic data for the coupled reaction of PRODH and P5CDH from Thermus thermophilus are consistent with a substrate channeling mechanism, as the approach to steady-state formation of NADH does not fit a non-channeling two-enzyme model. Furthermore, inactive P5CDH and PRODH mutants inhibit NADH production and increase trapping of the P5C intermediate in coupled assays of wild-type PRODH-P5CDH enzyme pairs, indicating that the mutants disrupt PRODH-P5CDH channeling interactions. A dissociation constant of 3 μm was estimated for a putative PRODH-P5CDH complex by surface plasmon resonance (SPR). Interestingly, P5CDH binding to PRODH was only observed when PRODH was immobilized with the top face of its (βα)8 barrel exposed. Using the known x-ray crystal structures of PRODH and P5CDH from T. thermophilus, a model was built for a proposed PRODH-P5CDH enzyme channeling complex. The structural model predicts that the core channeling pathway of bifunctional PutA enzymes is conserved in monofunctional PRODH-P5CDH enzyme pairs.
Introduction
The oxidative conversion of proline to glutamate, known as the proline catabolic pathway, involves two enzymes, a flavin-dependent proline dehydrogenase (PRODH)2 and a NAD+-dependent Δ1-pyrroline-5-carboxylate (P5C) dehydrogenase (P5CDH) (Fig. 1). The intervening step involves the hydrolysis of P5C to l-glutamate-γ-semialdehyde (GSA). The proline catabolic pathway helps organisms respond to changes in the nutritional environment by initiating the breakdown of proline as a source for nitrogen, carbon, and energy (1–6). In most organisms, PRODH and P5CDH are expressed as separate enzymes. In Gram-negative bacteria, however, PRODH and P5CDH are fused into one polypeptide known as proline utilization A (PutA) (7).
FIGURE 1.
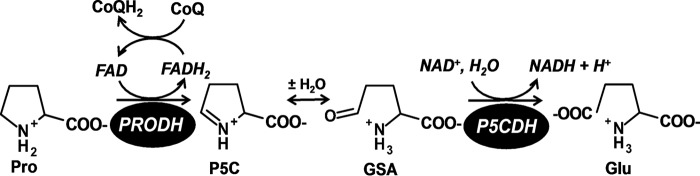
Reactions catalyzed by PRODH and P5CDH.
A unique aspect of proline catabolism is the discovery of a substrate channeling mechanism for P5C/GSA transfer between the PRODH and P5CDH domains of PutA. Crystal structures of PutA enzymes from Bradyrhizobium japonicum (8) and Geobacter sulfurreducens (9) revealed that the two active sites are separated by a linear distance of 40–50 Å and connected by a curved tunnel that traverses ∼70 Å. Kinetic data for these enzymes are consistent with a substrate channeling mechanism (8, 9). Blockage of the tunnel in B. japonicum PutA by mutagenesis abrogates channeling, which indicates that the tunnel functions as the conduit for the intermediate P5C/GSA (10). Furthermore, a substrate channeling mechanism is supported by kinetic studies of PutA from Escherichia coli (11) and Salmonella typhimurium (12) indicating that a general feature of PutA enzymes is direct transfer of P5C between the PRODH and P5CDH active sites.
A potential rationale for channeling P5C/GSA may be to limit reactions with other molecules and regulate its signaling activity. P5C/GSA is appreciably labile and has been shown to react with metabolites (13, 14), inhibit enzymes (15–18), and affect signaling pathways (19–22). High levels of P5C/GSA were shown to generate inactive adducts with pyridoxal phosphate, leading to lower amounts of functional vitamin B6 in patients (13). P5C has also been shown to act as a signaling molecule in eukaryotes resulting in apoptosis (19, 20). Very recently, P5C was proposed to act as a signaling metabolite linking proline metabolism with lipid oxidation in Caenorhabditis elegans (22). A more compelling reason for channeling P5C/GSA, however, may be to avoid futile cycling (11, 12, 23, 24). Cycling between proline catabolism and proline biosynthesis would be energetically costly for the cell. Thus, substrate channeling may be a necessary mechanism for maintaining efficient proline catabolic flux.
A major question in proline catabolism is whether PRODH and P5CDH enzymes that are not covalently linked utilize a substrate channeling mechanism similar to PutA. According to the Rosetta Stone hypothesis, which predicts protein-protein interactions based on gene fusion events (25, 26), individual PRODH and P5CDH enzymes are expected to interact, possibly in a manner analogous to PutA. To date, however, no detailed studies have been performed to test substrate channeling between monofunctional PRODH and P5CDH enzymes.
To address this gap, we chose to characterize the coupled reaction of PRODH and P5CDH enzymes from Thermus thermophilus. The x-ray crystal structures of T. thermophilus PRODH (TtPRODH) and P5CDH (TtP5CDH) have been solved providing a solid framework for investigating substrate channeling with these enzymes (27, 28). TtPRODH has a (βα)8 barrel structure which is conserved in the PRODH domain of PutA (8, 9, 29). TtP5CDH shares the fundamental aldehyde dehydrogenase (ALDH) dimeric structure of the ALDH superfamily (28), which is found in PutA and P5CDH from eukaryotes (30, 31). Thus, TtPRODH and TtP5CDH have domain structures that are conserved from prokaryotes to higher eukaryotes.
Here, we present a variety of kinetic strategies for evaluating substrate channeling between TtPRODH and TtP5CDH. The sum of the kinetic evidence supports a substrate channeling mechanism for the TtPRODH-TtP5CDH coupled reaction. TtPRODH and TtP5CDH interactions were also explored by surface plasmon resonance and used to help model a putative TtPRODH-TtP5CDH bienzyme complex. The structural model suggests that TtPRODH-TtP5CDH forms a channeling pathway that is conserved with covalently linked PRODH-P5CDH domains in PutA.
EXPERIMENTAL PROCEDURES
Materials
All chemicals and buffers were obtained from Sigma-Aldrich or Thermo Fisher Scientific Inc. unless otherwise noted. (DL)-P5C (50/50 mixture) was synthesized by the method of Williams and Frank and stored in 1 m HCl at 4 °C and was neutralized the day of experiments on ice by titrating with 6 m NaOH (32). Assays containing exogenously added (DL)-P5C contained ∼150 mm NaCl from the pH neutralization. N-(Biotinoyl)-N′-(iodoacetyl)ethylenediamine (BIAM) and dimethyl sulfoxide were obtained from Molecular Probes. The streptavidin (SA) sensor chip and PD-10 desalting columns were purchased from GE Healthcare. Active site and surface mutants for this study were generated using the GeneTailor mutagenesis kit (Invitrogen). All mutations were confirmed by DNA sequencing. All experiments were conducted in Nanopure water.
Purification of Enzymes
TtPRODH wild-type and mutants (R288M/R289M, S9C, and S88C), TtP5CDH wild-type and mutant C322A, and Deinococcus radiodurans P5CDH (DrP5CDH) mutant C325A were expressed from a pKA8H expression vector while the Saccharomyces cerevisiae P5CDH (Put2p) mutant C351A was expressed from a pET14b expression vector (pET14b-PUT2). All enzymes were expressed and purified with a N-terminal His tag as previously described in E. coli BL21(DE3) pLysS cells (27, 31, 33). Assays with non-His tag P5CDH enzymes confirmed that the His tag does not affect the Kirsch assays used for testing substrate channeling. The final concentration of purified protein was determined using the bicinchoninic acid method (Thermo Scientific Pierce) using bovine serum albumin (BSA) as a standard and is expressed as concentration of monomer unless stated otherwise (34).
PRODH and P5CDH Kinetic Assays
TtPRODH activity was measured at 25 °C using Coenzyme Q1 (CoQ1) as the terminal electron acceptor. Kinetic parameters for proline and CoQ1 were determined in assays using TtPRODH (0.5 μm) in 50 mm phosphate buffer (pH 8.0) and varying proline (0–100 mm) at a fixed concentration of CoQ1 (150 μm) or varying CoQ1 (0–300 μm) at a fixed proline concentration (150 mm). Reaction progress curves were monitored by following reduction of CoQ1 at 275 nm (ϵ = 13700 m−1 cm−1) using a 0.15 cm path length on a Hi-Tech Scientific SF-61DX2 stopped-flow instrument (TgK Scientific) at 25 °C (11). The kinetic parameters Km and kcat were estimated by fitting initial velocities to the Michaelis-Menten equation (35, 36).
P5CDH activity was measured by monitoring NADH formation at 340 nm (ϵ340 nm = 6200 m−1cm−1) at 25 °C. Assays were performed using 0.5 μm P5CDH, 0.2 mm NAD+, 50 mm potassium phosphate (pH 7.5), 25 mm NaCl and varying l-P5C (1–300 μm). The concentration of l-P5C is considered to be half the total (DL)-P5C concentration (11, 12). The kinetic parameters Km and kcat were estimated by fitting initial velocities to the Michaelis-Menten equation (35, 36). The inhibition of TtP5CDH by proline was determined by varying l-P5C (1–300 μm) at different fixed concentrations of proline (0–15 mm). Initial velocities were fit to a competitive inhibition equation (Equation 1) using SigmaPlot 12 Enzyme Kinetics Module (Systat Inc.), where v is P5CDH velocity (μm/s), ET is P5CDH concentration (μm), kcat (s−1) and Km (μm) are the Michaelis-Menten constants for P5CDH, [S] is l-P5C concentration (μm), [I] is the proline concentration (μm) and KI is the competitive inhibition constant for l-proline.
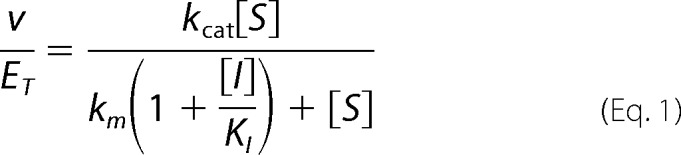 |
Coupled TtPRODH-TtP5CDH Activity Assays
The optimal ratio of TtPRODH to TtP5CDH was determined first by varying TtPRODH (0.1 μm to 10 μm) and holding TtP5CDH fixed at 0.5 μm. Assays were performed with proline (1 mm), CoQ1 (0.1 mm), and NAD+ (0.2 mm) in 50 mm potassium phosphate (pH 7.5) and 25 mm NaCl. Equal volumes of substrate and enzyme solutions were mixed on a SF-61DX2 stopped-flow spectrophotometer and the reaction was monitored by following NAD+ reduction at 340 nm (6200 cm−1 m−1). The non-channeling PRODH-P5CDH reaction was simulated as previously described (37) using a free diffusion two-enzyme model (see Equation 2).
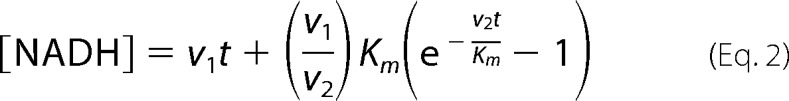 |
In the above equation, v1 is the experimentally determined rate of PRODH activity under the specified assay conditions with 1 mm proline, t is time in seconds (s), Km (43 μm l-P5C) and v2 (0.26 μm/s) are the steady-state Michaelis-Menten constants Km and V, respectively, for TtP5CDH. The transient time (τ) to reach steady-state formation of NADH is equivalent to Km/v2.
The strategy described by Geck and Kirsch (38) was used to test substrate channeling between TtPRODH and TtP5CDH. The coupled TtPRODH-TtP5CDH assay (channeling assay) was performed at increasing concentrations of inactive mutants of TtPRODH (R288M/R289M), TtP5CDH (C322A), DrP5CDH (C325A), and Put2p (C351A). The lack of catalytic activity in the TtPRODH mutant R288M/R289M and the P5CDH mutants was confirmed by the assays described above. BSA was used as negative control in the channeling assays.
For the channeling assays, a 1:1 mixture of TtPRODH (0.5 μm) and TtP5CDH (0.5 μm) was prepared in 50 mm potassium phosphate (pH 7.5) and 25 mm NaCl. The effect of inactive P5CDH on TtPRODH-TtP5CDH coupled activity was tested by adding 1–25 μm (monomer) of the different inactive P5CDH mutants (TtP5CDH 322A, DrP5CDH 325A, and Put2p 351A) to the TtPRODH:TtP5CDH mixture. The effect of inactive TtPRODH was performed similarly using 1 to 25 μm TtPRODH R288M/R289M. The ratio of inactive mutant (PRODH or P5CDH) to the corresponding wild-type enzyme thus ranged from 0–50-fold in the assays. A separate substrate solution of proline (2 mm), CoQ1 (0.2 mm), and NAD+ (0.4 mm) was prepared in 50 mm potassium phosphate (pH 7.5) and 25 mm NaCl. Equal volumes of enzyme and substrate solutions were mixed on a SF-61DX2 stopped-flow spectrophotometer and the reaction was monitored by following NAD+ reduction at 340 nm (6200 m−1 cm−1). The percent coupled TtPRODH-TtP5CDH activity is the ratio of NADH formation with and without inactive mutants.
P5C Trapping Assays
To trap the P5C intermediate, TtPRODH-TtP5CDH coupled assays were performed in the presence of o-aminobenzaldehyde (o-AB), which forms a dihydroquinazolinium compound with P5C that can be monitored at 443 nm (ϵ340 nm = 2900 m−1cm−1) (39). Coupled TtPRODH-TtP5CDH activity assays were performed as described above except in the presence of 1 mm o-AB. The maximum amount of P5C trapped by o-AB was determined in assays without NAD+. The effect of inactive mutants TtP5CDH C322A and Put2p C351A on P5C trapping was determined by assays using increasing concentrations of the inactive P5CDH mutants (0–37.5 μm, monomer). The ratio of inactive P5CDH mutant to wild-type TtP5CDH ranged from 0 to 75. Assays were performed in a 96 well plate (200 μl total assay volume) and initiated by addition of a substrate mixture with final concentrations after mixing of 0.5 μm TtPRODH, 0.5 μm TtP5CDH, 0–37.5 μm inactive P5CDH mutant, 1 mm proline, 0.2 mm NAD+, 0.2 mm CoQ1, and 1 mm o-AB in 50 mm potassium phosphate buffer (pH 7.5, 25 mm NaCl). P5C-o-AB complex formation was followed at 443 nm for 20 min on a Powerwave XS 96-well plate reader (Biotek).
SPR Analysis of TtPRODH-TtP5CDH Interactions
Cysteine residues were introduced on opposite sides of TtPRODH at surface residues Ser-9 and Ala-88. TtPRODH mutants S9C and A88C were then purified as described above. Purified TtPRODH mutants S8C and A88C required the presence of the reducing agent Tris(3-hydroxypropyl)phosphine to prevent precipitation during purification. The specific activities of TtPRODH mutants S9C and A88C relative to wild-type TtPRODH were 74 and 60%, respectively. Purified TtPRODH mutants S8C and A88C were labeled with biotin at the incorporated cysteine residue using the thiol-reactive BIAM reagent. A 5 mm stock of BIAM was prepared by dissolving in dimethyl sulfoxide. Under anaerobic conditions, purified TtPRODH mutants S9C and A88C and BIAM were mixed at final concentrations of 51 μm (TtPRODH) and 715 μm (BIAM). The labeling reaction was allowed to proceed overnight at 4 °C until the next morning when 25 mm β-mercaptoethanol was added to quench unreacted BIAM. Biotin-labeled TtPRODH was then separated from quenched BIAM by passing the reaction mixture through a PD-10 desalting column. The concentration of total protein was measured using the Pierce 660 nm Protein Assay Reagent (Pierce). After the biotinylation procedure, the specific activity of both mutants dropped to 44% relative to wild-type TtPRODH. Biotin labeling of TtPRODH was confirmed by Western blot analysis using streptavidin-conjugated horseradish peroxidase (Pierce). ECL prime reagents (GE Healthcare) were used for detection and visualization of biotin-labeled TtPRODH.
The interaction between TtPRODH and TtP5CDH was studied using a BIAcore 3000 (BIACORE AB, Uppsala, Sweden) instrument maintained in the Nanomaterials Characterization Core Facility at the University of Nebraska Medical Center. A streptavidin (SA) sensor chip was first washed with HBS-EP buffer (10 mm HEPES, pH 7.4, 0.15 m NaCl, 3.4 mm EDTA, and 0.05% surfactant P20) for 6 min (20 μl/min flow rate). Prior to immobilization, the SA sensor chip was activated with three consecutive 40 μl injections of activating buffer (1 m NaCl and 50 mm NaOH). Biotinylated TtPRODH mutants S9C and A88C were then diluted in HBS-EP to a final concentration of 250 μm and injected at a flow rate of 5 μl/min onto cells 2 (TtPRODH S9C) and 4 (TtPRODH A88C). Sensor chip flow cells 1 and 3 were not exposed to TtPRODH mutants S9C and A88C and were used as reference cells. After completing the injection of biotinylated TtPRODH mutants S9C and A88C, the total response units (RU) were ∼1900 RU and ∼2600 RU, respectively. Thus, similar amounts of TtPRODH mutants S9C and A88C were coated on the SA chip. After washing the coated flow cells, TtP5CDH (1 μm) was injected onto both surfaces.
The binding of TtP5CDH to immobilized TtPRODH S9C and A88C was tested using TtP5CDH, DrP5CDH and BSA as analytes. The analytes were equilibrated in HBS-EP buffer and diluted to a concentration of 1 μm immediately prior to injection. Each protein analyte was then injected onto flow cells 1–4 for 2 min followed by a 5 min dissociation phase at a flow rate of 30 μl/min.
The kinetics of TtP5CDH binding to immobilized TtPRODH A88C were estimated by injecting increasing concentrations of TtP5CDH (0.5–7.5 μm, monomer) pre-equilibrated in HBS-EP buffer. From lowest to highest concentration, wild-type TtP5CDH was injected for 2 min (30 μl/min) followed by a 5 min dissociation phase at a flow rate of 30 μl/min with HBS-EP buffer. The SA-TtPRODH binding surface was regenerated using 90 μl of 2 m NaCl (30 μl/min) and equilibrated with HBS-EP buffer (30 μl/min, 5 min) after each injection of TtP5CDH. The resulting sensorgrams were globally fitted to 1:1 Langmuir binding model using BIA-evaluation software 4.1. The experiments were carried out at 25 °C. All buffers were filtered and degassed prior to injection on SA sensor chip. Signals from the reference flow cells were subtracted from the sensorgrams.
RESULTS
Inhibition of TtP5CDH by Proline
X-ray crystal structures of TtP5CDH (28) have shown that both glutamate (PDB ID:2BHQ) and proline (PDB ID:2J40) bind to the active site of TtP5CDH with identical orientations of the carboxylate group resulting in favorable hydrogen bonding interactions with Ser-321 and backbone amide nitrogen atoms. Proline inhibition of TtP5CDH, however, has not been characterized. Thus, prior to examining the coupled TtPRODH-TtP5CDH reaction, proline inhibition of the TtP5CDH reaction was determined.
The steady-state kinetic parameters determined for TtP5CDH were Km of 43 ± 1 μm l-P5C and kcat of 0.52 ± 0.03 s−1. When varying proline, the decrease in P5CDH activity fit well to a competitive inhibition model with an inhibition constant (KI) of 3.9 mm proline (Fig. 2A). A double reciprocal plot of 1/v versus 1/[S] with varying proline concentrations is also shown (Fig. 2B) and is consistent with proline as a competitive inhibitor of P5CDH with respect to P5C/GSA.
FIGURE 2.
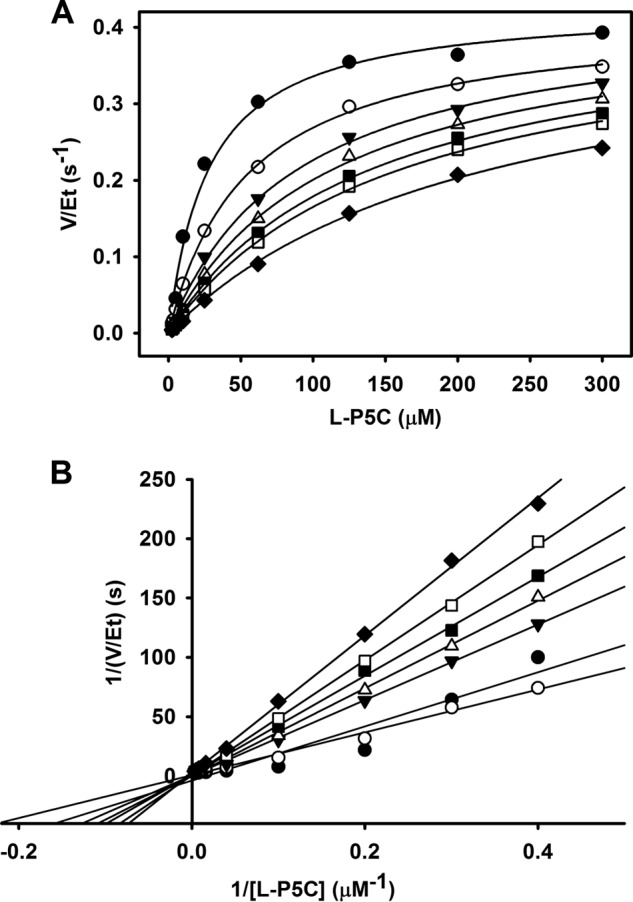
Inhibition pattern of TtP5CDH with l-proline. A, non-linear least squares fit to the competitive inhibition model (Eq. 1) with 0 (●), 2 (○), 4 (▾), 6 (▵), 8 (■), 10 (□), and 15 mm (♦) proline and varied P5C. Best fit parameters were kcat = 0.43 ± 0.02 s−1, Km = 27.7 ± 4.3 μm, and KI (pro) = 3.9 ± 0.7 mm proline. B, double reciprocal plot of the same data in panel A showing competitive inhibition pattern.
Transient Time of Coupled TtPRODH-TtP5CDH Reaction
The ratio of TtPRODH:TtP5CDH was initially optimized to ensure that the coupled TtPRODH-TtP5CDH reaction was not limited by the concentration of either enzyme and to prevent surplus formation of P5C by excess TtPRODH. To avoid proline inhibition of TtP5CDH, we characterized coupled TtPRODH-TtP5CDH activity using 1 mm proline which is < KI(pro) (3.9 mm). Fig. 3 shows the dependence of NADH formation on the concentration of TtPRODH while keeping the substrates and TtP5CDH fixed. With TtP5CDH fixed at 0.5 μm, the NADH formation rate plateaus above 1 μm TtPRODH. Thus, an equimolar mixture of 0.5 μm TtPRODH and 0.5 μm TtP5CDH was considered to be appropriate for testing substrate channeling.
FIGURE 3.
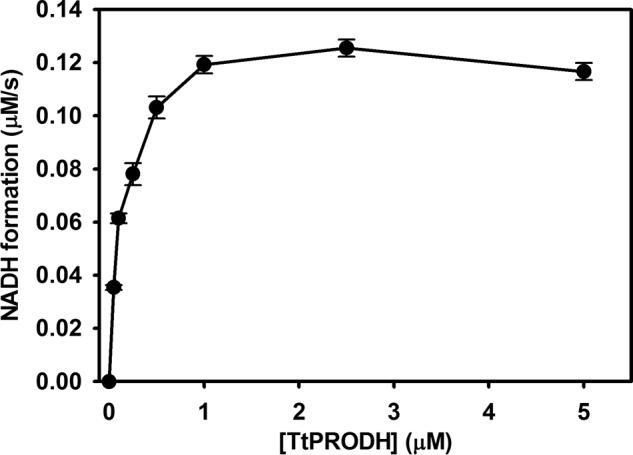
Dependence of the NADH formation rate of P5CDH on TtPRODH concentration. The reaction mixture contained 0.5 μm TtP5CDH, 1 mm proline, 0.2 mm NAD+, and 0.1 mm CoQ1 with TtPRODH varied from 0.1 to 10 μm. Assays were performed in 50 mm potassium phosphate (pH 7.5, 25 mm NaCl), and NADH formation was monitored at 340 nm. Data points are the mean ± S.D. from three assays.
The reaction progress curve of NADH formation using an equimolar mixture of TtPRODH-TtP5CDH (0.5 μm:0.5 μm) was next examined and compared with that of a diffusion-limited two-enzyme reaction model (Eq. 2). An experimental transient time of 38.5 s was observed for an equimolar mixture of TtPRODH-TtP5CDH prior to achieving a steady-state velocity of 0.08 μm NADH s−1 (Fig. 4). The transient time for a non-channeling TtPRODH-TtP5CDH coupled reaction was then simulated using the kinetic constants of the individual reactions. The steady-state kinetic parameters determined for TtPRODH were Km of 8.2 ± 0.3 mm proline and 162 ± 8 μm CoQ1 with an overall kcat of 17.0 ± 0.4 s−1. Under the assay conditions for the coupled reaction, TtPRODH generates P5C/GSA at 0.08 μm/s (v1). The predicted transient time before reaching a linear rate of NADH formation is 164.2 s for a non-channeling TtPRODH-TtP5CDH coupled reaction. Thus, although the simulated steady-state rate of NADH formation is identical to the experimental rate, the transient time is 4-fold longer than that observed experimentally. The shorter lag time for the experimental reaction is suggestive of substrate channeling between TtPRODH and TtP5CDH.
FIGURE 4.
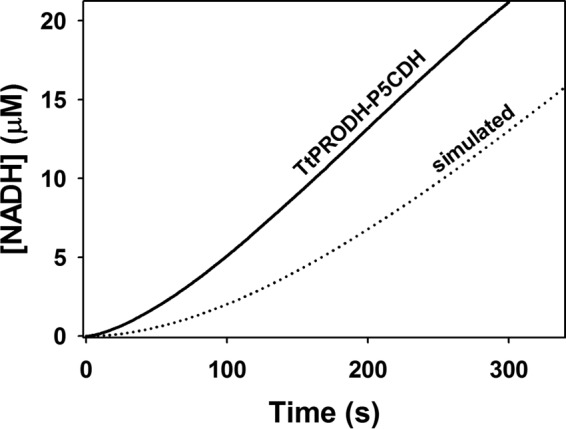
Experimental and simulated reaction progress curves of coupled TtPRODH and TtP5CDH activity. The solid line is the steady-state formation of NADH followed at 340 nm in an assay containing an equimolar mixture of TtPRODH (0.5 μm) and TtP5CDH (0.5 μm), 1 mm proline, 0.2 mm NAD+, and 0.1 mm CoQ1 (pH 7.5). The dotted line is simulated NADH formation from a non-channeling free diffusion model (Eq. 2) using experimentally determined values of v1 (0.08 μm/s), Km2 (43 μm), and v2 (0.26 μm/s).
One explanation for the shorter lag time of the TtPRODH-TtP5CDH-coupled reaction, however, could be allosteric activation of one or both enzymes (40). Upon adding up to 50-fold excess TtP5CDH, however, we observed no increase in TtPRODH activity. Likewise, no activation of TtP5CDH activity was observed with 50-fold excess of the TtPRODH mutant R288M/R289M. Thus, we found no evidence of allosteric activation between TtPRODH and TtP5CDH.
P5C Trapping Shows Evidence of Channeling
If the intermediate P5C/GSA is channeled between TtPRODH and TtP5CDH, then the amount of intermediate in the bulk solvent should be sensitive to TtP5CDH activity, which requires NAD+. The ability of excess o-AB (1 mm), which traps P5C as a dihydroquinazolinium compound that is detected at 443 nm, was examined using an equimolar mixture of TtPRODH and TtP5CDH in assays with and without NAD+. Without NAD+, the dihydroquinazolinium (P5C-o-AB complex) is formed, consistent with P5C being released into bulk solvent (Fig. 5A). In the presence of NAD+ (0.2 mm), o-AB trapping of P5C is significantly reduced indicating that a large fraction of P5C is not solvent accessible during the TtPRODH-TtP5CDH coupled reaction. In these assays, the apparent fraction of P5C that is channeled is 80%. Trapping assays performed with the TtP5CDH C322A inactive mutant showed no change in P5C trapping with NAD+.
FIGURE 5.
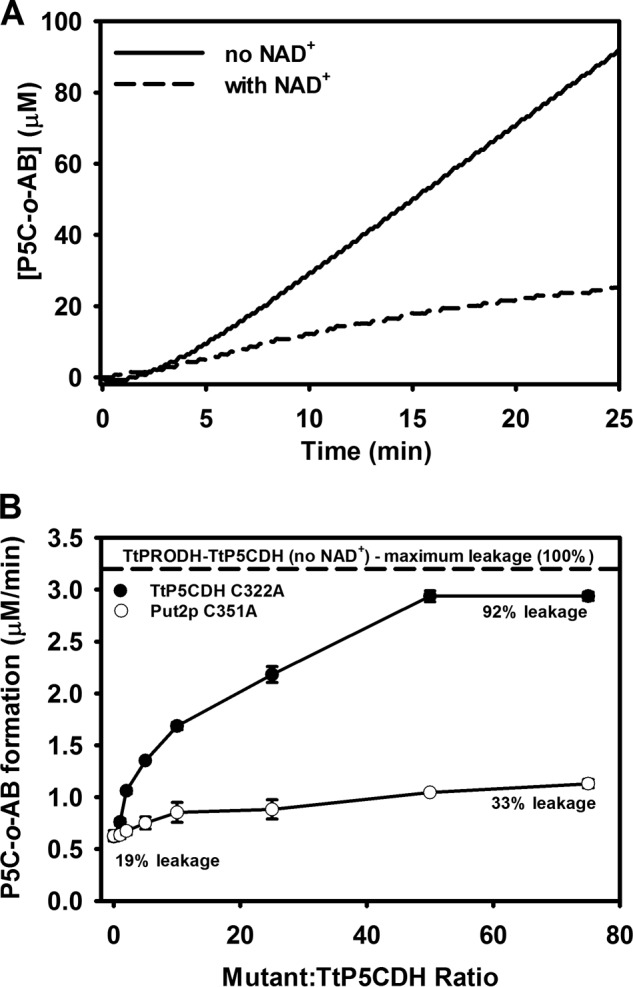
Trapping of P5C intermediate shows evidence of substrate channeling. A, solid line is P5C-o-AB production from an equimolar mixture of TtPRODH (0.5 μm) and TtP5CDH (0.5 μm) with 1 mm proline and 0.2 mm CoQ1. The dotted line is the same reaction mixture but with 0.2 mm NAD+. Assays were performed in 50 mm potassium phosphate (pH 7.5) and 1 mm o-AB. Complex formation of P5C-o-AB is followed at 443 nm. B, steady-state P5C-o-AB formation rate from an equimolar mixture of TtPRODH (0.5 μm) and TtP5CDH (0.5 μm) as a function of increased ratio of inactive TtP5CDH C322A (filled circles) or Put2p C351A mutants (open circles) (mutant/wild-type TtP5CDH). Assays were performed in 50 mm potassium phosphate (pH 7.5) containing 1 mm proline, 0.2 mm NAD+, 0.2 mm CoQ1, and 1 mm o-AB. Percent leakage was estimated by dividing the rate of P5C-o-AB formation by the maximum rate of P5C-o-AB formation determined by assays without NAD+ (dotted line).
A second approach for testing P5C release into bulk solvent was developed using a strategy devised by Geck and Kirsch (38). In this strategy, inactive mutants are used as competitors of the putative channeling complex. For these assays, an inactive mutant of TtP5CDH was generated by mutating Cys-322, which is the essential active site nucleophile. If the coupled TtPRODH-TtP5CDH reaction involves a channeling complex, the inactive TtP5CDH C322A mutant is anticipated to increase P5C trapping by disrupting wild-type TtPRODH-TtP5CDH interactions. Fig. 5B shows that the amount of P5C trapped by o-AB in a TtPRODH-TtP5CDH coupled reaction is about 19% of the maximum possible. Thus, the leakage of P5C during the coupled TtPRODH-TtP5CDH reaction is estimated to be ∼19%. Addition of the inactive TtP5CDH C322A mutant shows a concentration dependent increase in trapped P5C with 92% leakage estimated at 50-fold excess of the C322A mutant relative to wild-type TtP5CDH. This result indicates the C322A mutant causes a nearly 5-fold increase in P5C release into bulk solvent consistent with disrupting a TtPRODH-TtP5CDH channeling complex. We also tested the effect of an inactive P5CDH from another organism. Fig. 5B shows that an inactive P5CDH mutant from yeast S. cerevisiae, Put2p C351A, had a much a smaller effect with an increase of only 33% in P5C trapping. Thus, the effect appears to be specific for the TtP5CDH C322A mutant.
Inactive Mutants Disrupt TtPRODH-TtP5CDH-coupled Reaction
Substrate channeling in the TtPRODH-TtP5CDH coupled reaction was next tested using the original strategy of Geck and Kirsch. In these assays, the effects of inactive TtP5CDH and TtPRODH mutants on the overall coupled reaction rate were determined. The inactive TtPRODH mutant R288M/R289M was generated by replacing two catalytic arginine residues (Arg-288 and Arg-289) that are essential in PRODH and PutA for proline binding via ionic interactions with the carboxylate moiety of proline (9, 30, 41, 42).
Fig. 6 shows that the inactive TtP5CDH C322A mutant significantly decreased the rate of the PRODH-P5CDH coupled reaction in a concentration dependent manner (Fig. 6). The rate of NADH formation decreased by 60% in the presence of 50-fold excess of TtP5CDH C322A (25 μm) relative to wild-type TtP5CDH (0.5 μm). To test the specificity of this effect, inactive P5CDH from other organisms were used in identical assays. An inactive mutant of P5CDH from D. radiodurans (DrP5CDH C325A) was added up to 50-fold excess relative to TtP5CDH. The maximum decrease in PRODH-P5CDH activity was 16% at a 10-fold excess of DrP5CDH C325A mutant. Also, no decrease in activity was observed with 50-fold excess of the Put2p inactive mutant C351A or 50-fold excess of BSA in the PRODH-P5CDH coupled assay (data not shown). Thus, the observed decrease in the PRODH-P5CDH activity appears to be specific for the TtP5CDH C322A mutant.
FIGURE 6.
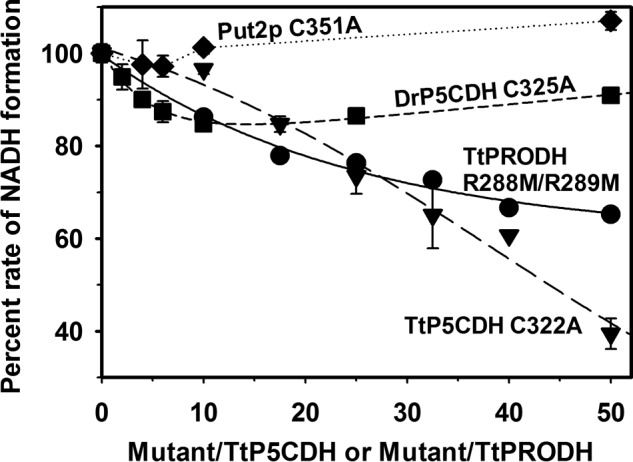
Test for substrate channeling. Percent rate of NADH formation using an equimolar mixture of TtPRODH (0.5 μm) and TtP5CDH (0.5 μm) is plotted as a function of increased ratio of inactive mutants of P5CDH (Mutant/wild-type TtP5CDH) or PRODH (mutant/wild-type TtPRODH). Shown are mutants TtP5CDH C322A (triangles), TtPRODH R288M/R289M (circles), DrP5CDH C325A (squares), and Put2p C351A (diamonds). Assays were performed in 50 mm potassium phosphate (pH 7.5) with 1 mm proline, 0.2 mm NAD+, and 0.1 mm CoQ1. Percent NADH formation rate is the rate of the initial velocity progress curve without mutant divided by the rate in the presence of mutant enzyme. The concentration of inactive mutants was increased from 0 to 25 μm. Data points are the mean ± S.D. from three assays.
Adding the TtPRODH inactive mutant R288M/R289M to the PRODH-P5CDH reaction also decreased activity but to a lesser extent. A 50-fold excess of TtPRODH R288M/R289M (25 μm) relative to wild-type TtPRODH (0.5 μm) resulted in a 35% decrease in the rate of NADH formation (Fig. 6).
Evidence for TtPRODH-TtP5CDH Binding
Prompted by the kinetic evidence of a substrate channeling mechanism for the coupled TtPRODH-TtP5CDH reaction, potential binding interactions between TtPRODH and TtP5CDH were investigated by SPR. For the SPR analysis, TtPRODH was immobilized on a BIAcore streptavidin (SA) sensor chip in two different orientations. TtPRODH lacks cysteine residues making it convenient to specifically label engineered cysteine residues. Surface residues Ser-9 and Ala-88 were chosen for cysteine replacement. Ser-9 and Ala-88 are near the top and bottom of the PRODH β8α8 barrel, respectively, enabling TtPRODH to be immobilized in two different orientations (Fig. 7A). Immobilization of TtPRODH S9C should leave the bottom face of the PRODH β8α8 barrel exposed to solvent (Fig. 7A) whereas the top face of the PRODH β8α8 barrel should be solvent accessible with TtPRODH A88C.
FIGURE 7.
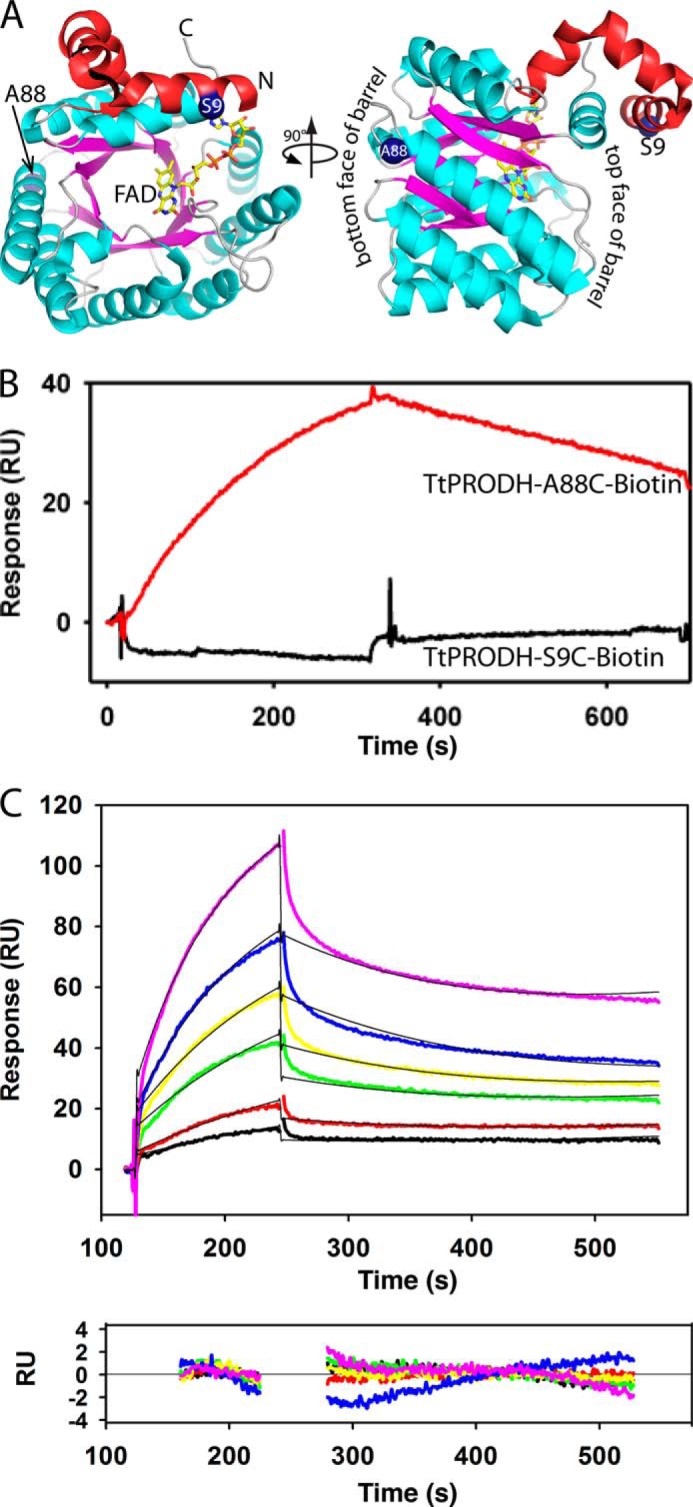
TtP5CDH binds to immobilized TtPRODH. A, structure of TtPRODH (PDB ID 2G37) showing the sites of immobilization for SPR analysis (blue spheres, Ser-9 and Ala-88). The N-terminal domain is colored red and the (βα)8 barrel is in cyan/magenta. B, SPR analysis of TtP5CDH binding to TtPRODH immobilized in two different orientations via S9C and A88C. Sensorgrams show binding response of TtP5CDH (1 μm) injected onto surface with immobilized TtPRODH S9C (black curve) or A88C (red curve) at a flow rate of 30 μl/min in HEPES-EP buffer (pH 7.4) at 25 °C. The association and dissociation phases were 300 s each. C, SPR kinetic analysis of TtP5CDH binding to immobilized TtPRODH A88C in HEPES-EP buffer (pH 7.4) at 25 °C. Sensorgrams are of increasing TtP5CDH concentrations of 0.5 μm (black), 1 μm (red), 2 μm (green), 3 μm (yellow), 5 μm (blue), and 7.5 μm (pink). The association phase is the injection of TtP5CDH at 30 μl/min for 120 s and the dissociation phase is HEPES-EP buffer (pH 7.4) at 30 μl/min for 300 s. The data were fit by global analysis to a 1:1 Langmuir binding isotherm using BIAevaluation 4.1 software to yield a KD of 3 μm. Bottom panel shows the residuals from the fitting analysis (chi-square value 0.58). Signals from the control surface have been subtracted.
Fig. 7B shows that upon injecting TtP5CDH, a binding response was observed with immobilized TtPRODH A88C but not with immobilized TtPRODH S9C. This result suggests that TtP5CDH distinguishes between the two orientations of TtPRODH and specifically recognizes the configuration of immobilized TtPRODH A88C. It also indicates that the immobilized orientation of TtPRODH S9C conceals a surface on TtPRODH that is critical for TtP5CDH binding. No binding response was observed for DrP5CDH or BSA with the immobilized TtPRODH mutants (data not shown) further indicating a specific binding event between immobilized TtPRODH A88C and TtP5CDH.
The binding kinetics of TtP5CDH to immobilized TtPRODH A88C were then evaluated (Fig. 7C). SPR analysis shows TtP5CDH binds to TtPRODH A88C with a dissociation constant (KD) of 3 μm. The estimated kon and koff rate constants for TtP5CDH binding were 1.49 × 103 m−1s−1 and 4.51 × 10−3 s−1, respectively.
DISCUSSION
The channeling of metabolites is generally perceived to enhance metabolic flux thereby benefiting cellular fitness (43–45). Metabolic channeling has been proposed for various pathways such as fatty acid β-oxidation (46), tricarboxylic acid cycle (47), and purine biosynthesis (48, 49). Generating kinetic and structural evidence for channeling between two enzymes of consecutive steps in a pathway, however, is challenging. As a simple two-enzyme system, the proline catabolic pathway affords the opportunity to investigate mechanisms of substrate channeling and their importance in vivo. In addition, the two enzymes of proline catabolism are either free or fused together (i.e. PutA) enabling the comparison of two biological strategies for handling the same metabolite. The Rosetta Stone hypothesis predicts that if two enzyme pairs are fused together in one organism, then in other systems in which they are free, they will interact (25, 26).
The first observation of substrate channeling for the PRODH-P5CDH reaction was reported in PutA from Salmonella typhimurium by Surber and Maloy who concluded that the oxidation of proline to glutamate involves a leaky channel (12). In recent years, kinetic and structural data of PutA enzymes from B. japonicum and G. sulfurreducens have provided unprecedented molecular details of P5C/GSA channeling including the three-dimensional space and electronic environment of the intervening cavity between PRODH and P5CDH, potential exit and entry sites for water, and replacement of residues lining the cavity with bulkier side chains that block substrate channeling. In addition, a novel hysteretic mechanism was found for PutA from E. coli suggesting a channeling step in the overall PRODH-P5CDH reaction becomes activated in the first few catalytic turnovers. From these aforementioned studies, substrate channeling as a mechanism for the two-step conversion of proline into glutamate is now well established in PutA enzymes.
In this study, we present the first evidence of substrate channeling for a PRODH-P5CDH pair that are not covalently linked. Rate measurements for the coupled TtPRODH-TtP5CDH enzyme pair demonstrates that steady-state formation of NADH is achieved faster than the theoretical approach to steady-state by two individual enzymes. Although such rapid product formation can be explained by allosteric activation by one enzyme on the other (40), our results show that this is not the case for TtPRODH and TtP5CDH. The observed lag time is greater than that observed for PutA. The lag times reported for PutA enzymes thus far range from no apparent lag to 23 s for E. coli PutA (8, 9, 11). The longer lag time of the TtPRODH-TtP5CDH pair most likely reflects the dynamic nature of a potential channeling pathway between two free enzymes.
The experiments utilizing the catalytically inactive TtP5CDH C322A mutant were very useful for testing substrate channeling in the TtPRODH-TtPCDH reaction. The inactive TtP5CDH mutant clearly disrupted the TtPRODH-TtP5CDH coupled reaction as observed by decreased NADH formation and increased trapping of the P5C intermediate. As noted by Yadid et al., assays involving trapping of the intermediate provide a rigorous test for channeling, and kinetic evidence for protection of a reactive intermediate implies a protein-protein complex (50). Thus, our kinetics data provide strong evidence that the TtPRODH-TtP5CDH reaction employs a channeling mechanism and imply a protein-protein complex.
It appears that TtPRODH and TtP5CDH form a weak transient complex. Such complexes are characterized by a KD in the micromolar range (51) and can be challenging to study using traditional biophysical methods. We found no evidence of a stable complex using gel filtration chromatography (data not shown) but were able to estimate a KD of ∼3 μm for TtPRODH-TtPP5CDH using SPR. These results are consistent with TtPRODH and TtP5CDH forming a weak transient complex. Thus, although PRODH and P5CDH from T. thermophilus are not covalently linked as in PutA and do not form a tightly bound complex, substrate channeling is still achieved via dynamic interactions. We note that there is precedent for weak transient enzyme-enzyme complexes in other metabolic pathways, such as purine biosynthesis (48), the aspartate pathway of amino acid biosynthesis (52), branched chain amino acid catabolism (53), and the tricarboxylic acid cycle (47). In particular, the KD of 3 μm that we measured for TtPRODH-TtP5CDH is very similar to that of 2.8 μm measured in branched chain amino acid catabolism (53).
The advantage of a dynamic channeling system versus a fixed system such as PutA, may be the ability to regulate P5C/GSA levels and utilize P5C as a metabolite signaling molecule (19–22) or drive the proline-P5C redox cycle (54–56). If the consecutive enzymes were fixed together, the potential to regulate P5C would be limited. In organisms with free PRODH and P5CDH, additional regulation of the proline catabolic pathway is possible due to dynamic interactions. Thus, a stable and perfect channeling enzyme pair would not necessarily be a benefit for the cell.
Crystal structures of PutA (8, 9) provide a basis for building testable models of the TtPRODH-TtP5CDH complex. TtPRODH and TtP5CDH have the same folds as the respective domains of PutA. Therefore, a simple model of the channeling complex was made by superimposing a TtP5CDH dimer and two copies of the TtPRODH β8α8 barrel onto a dimer of B. japonicum PutA (PDB 3HAZ) (Fig. 8A). In this model, the top face of the TtPRODH β8α8 barrel forms the interface with TtP5CDH (Fig. 8B). This prediction is consistent with the SPR data showing that TtP5CDH specifically binds to the top face of the TtPRODH barrel but not to the bottom face (Fig. 7B). The size of the interface is also reasonable. According to a trend observed by Nooren and Thornton, a transient heterodimer with a KD ∼3 μm can be expected to have an interface contact area of ∼500 Å2 (57). From our model, we calculated a contact area of 700 Å2 for the binding interface between TtPRODH and TtP5CDH (58), which is consistent with this trend. TtP5CDH also forms a trimer-of-dimers hexamer at > 1 mg/ml (>17 μm monomer). Application of the 3-fold symmetry operator of the TtP5CDH hexamer to the model in Fig. 8A generates a dodecamer model (Fig. 8C). Notice that in this model, the NAD+ binding site is visible from the exterior indicating that the NADH and glutamate can exit into the bulk solvent rather than into the hexameric core region. Whether such a large assembly is present at the relatively low protein concentrations used in our assays (0.5 μm) is unknown, but if so, the population is likely low. These models will be tested by site-directed mutagenesis of the predicted interfaces.
FIGURE 8.
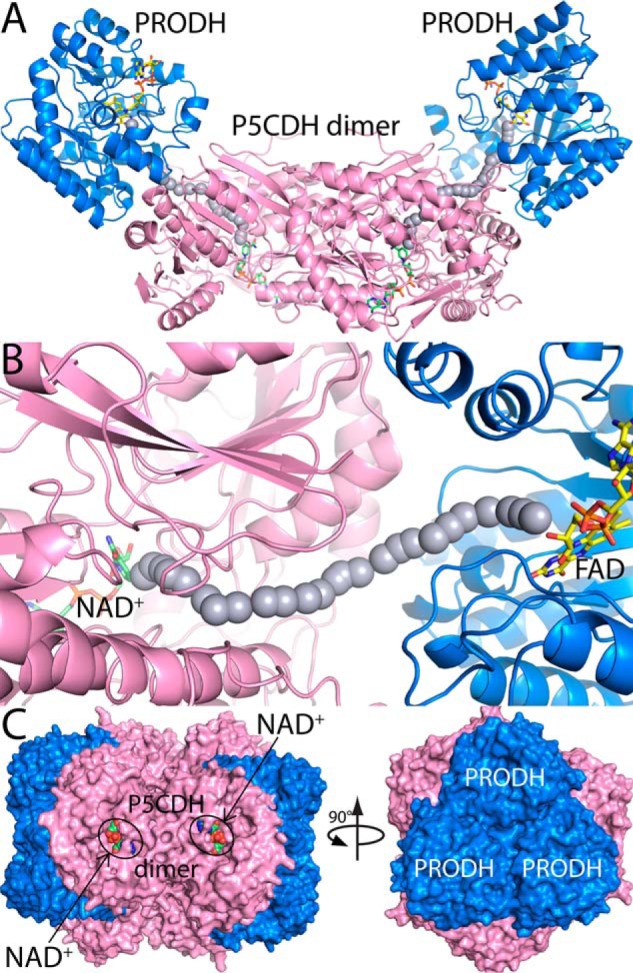
Models of TtPRODH-TtP5CDH complexes. A, model of a tetramer containing a P5CDH dimer (pink) and two copies of TtPRODH (blue). FAD and NAD+ are drawn as yellow and green sticks, respectively. The gray spheres represent the path between the PRODH and P5CDH active sites. This model was built by superimposing a TtP5CDH dimer (PDB 2BHQ) and two copies of TtPRODH (PDB 2G37) onto a dimer of BjPutA (PDB 3HAZ). B, close-up view of the modeled PRODH-P5CDH interface. C, model of a dodecamer generated by applying 3-fold symmetry to the tetramer in panel A. Each tetramer has a different color. NAD+ bound to P5CDH is shown in green spheres.
In summary, the results here show that the two-step reaction catalyzed by PRODH and P5CDH from T. thermophilus involves a substrate channeling mechanism. This is the first evidence of substrate channeling between PRODH and P5CDH enzymes that are not fused together. This study also provides a useful approach for examining substrate channeling in PRODH and P5CDH enzymes from other organisms as well as other enzyme pairs that catalyze consecutive reactions.
Acknowledgment
We thank Dr. Chantey R. Morris from the University of Nebraska Medical Center Nanomaterials Characterization Core Facility for technical assistance with SPR.
This work was supported, in whole or in part, by National Institutes of Health Grants GM065546 and P30GM103335 and is a contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act.
- PRODH
- proline dehydrogenase
- BIAM
- N-(biotinoyl)-N′-(iodoacetyl)ethylenediamine
- CoQ1
- ubiquinone-1
- DrP5CDH
- Δ1-pyrroline-5-carboxylate dehydrogenase from D. radiodurans
- GSA
- l-glutamate-γ-semialdehyde
- o-AB
- o-aminobenzaldehyde
- P5C
- Δ1-pyrroline-5-carboxylate
- P5CDH
- Δ1-pyrroline-5-carboxylate dehydrogenase
- PutA
- proline utilization A
- Put2p
- Δ1-pyrroline-5-carboxylate dehydrogenase from S. cerevisiae
- SPR
- surface plasmon resonance
- TtPRODH
- proline dehydrogenase from T. thermophilus
- TtP5CDH
- Δ1-pyrroline-5-carboxylate dehydrogenase from T. thermophilus.
REFERENCES
- 1. Phang J. M., Pandhare J., Liu Y. (2008) The metabolism of proline as microenvironmental stress substrate. J. Nutr. 138, 2008S–2015S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takagi H. (2008) Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications. Appl. Microbiol. Biotechnol. 81, 211–223 [DOI] [PubMed] [Google Scholar]
- 3. Krishnan N., Doster A. R., Duhamel G. E., Becker D. F. (2008) Characterization of a Helicobacter hepaticus putA mutant strain in host colonization and oxidative stress. Infect. Immun. 76, 3037–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Curtis J., Shearer G., Kohl D. H. (2004) Bacteriod proline catabolism affects N2 fixation rate of drought-stressed soybeans. Plant Physiol. 136, 3313–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kohl D. H., Schubert K. R., Carter M. B., Hagedorn C. H., Shearer G. (1988) Proline metabolism in N2-fixing root nodules: Energy transfer and regulation of purine synthesis. Proc. Natl. Acad. Sci. U.S.A. 85, 2036–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lamour N., Rivière L., Coustou V., Coombs G. H., Barrett M. P., Bringaud F. (2005) Proline metabolism in procyclic Trypanosoma brucei is down-regulated in the presence of glucose. J. Biol. Chem. 280, 11902–11910 [DOI] [PubMed] [Google Scholar]
- 7. Tanner J. J. (2008) Structural biology of proline catabolism. Amino Acids. 35, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Srivastava D., Schuermann J. P., White T. A., Krishnan N., Sanyal N., Hura G. L., Tan A., Henzl M. T., Becker D. F., Tanner J. J. (2010) Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. U.S.A. 107, 2878–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singh H., Arentson B. W., Becker D. F., Tanner J. J. (2014) Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site. Proc. Natl. Acad. Sci. U.S.A. 111, 3389–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arentson B. W., Luo M., Pemberton T. A., Tanner J. J., Becker D. F. (2014) Kinetic and structural characterization of tunnel-perturbing mutants in Bradyrhizobium japonicum proline utilization A. Biochemistry 53, 5150–5161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moxley M. A., Sanyal N., Krishnan N., Tanner J. J., Becker D. F. (2014) Evidence for hysteretic substrate channeling in the proline dehydrogenase and Delta1-pyrroline-5-carboxylate dehydrogenase coupled reaction of proline utilization A (PutA). J. Biol. Chem. 289, 3639–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Surber M. W., Maloy S. (1998) The PutA protein of Salmonella typhimurium catalyzes the two steps of proline degradation via a leaky channel. Arch. Biochem. Biophys. 354, 281–287 [DOI] [PubMed] [Google Scholar]
- 13. Farrant R. D., Walker V., Mills G. A., Mellor J. M., Langley G. J. (2001) Pyridoxal phosphate de-activation by pyrroline-5-carboxylic acid. Increased risk of vitamin B6 deficiency and seizures in hyperprolinemia type II. J. Biol. Chem. 276, 15107–15116 [DOI] [PubMed] [Google Scholar]
- 14. Berney M., Weimar M. R., Heikal A., Cook G. M. (2012) Regulation of proline metabolism in mycobacteria and its role in carbon metabolism under hypoxia. Mol. Microbiol. 84, 664–681 [DOI] [PubMed] [Google Scholar]
- 15. Bearne S. L., Wolfenden R. (1995) Glutamate γ-semialdehyde as a natural transition state analogue inhibitor of Escherichia coli glucosamine-6-phosphate synthase. Biochemistry 34, 11515–11520 [DOI] [PubMed] [Google Scholar]
- 16. Bearne S. L., Hekmat O., Macdonnell J. E. (2001) Inhibition of Escherichia coli CTP synthase by glutamate γ-semialdehyde and the role of the allosteric effector GTP in glutamine hydrolysis. Biochem. J. 356, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thoden J. B., Huang X., Raushel F. M., Holden H. M. (1999) The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway. Biochemistry 38, 16158–16166 [DOI] [PubMed] [Google Scholar]
- 18. Nishimura A., Nasuno R., Takagi H. (2012) The proline metabolism intermediate Delta1-pyrroline-5-carboxylate directly inhibits the mitochondrial respiration in budding yeast. FEBS Lett. 586, 2411–2416 [DOI] [PubMed] [Google Scholar]
- 19. Nomura M., Takagi H. (2004) Role of the yeast acetyltransferase Mpr1 in oxidative stress: regulation of oxygen reactive species caused by a toxic proline catabolism intermediate. Proc. Natl. Acad. Sci. U.S.A. 101, 12616–12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maxwell S. A., Davis G. E. (2000) Differential gene expression in p53-mediated apoptosis-resistant vs. apoptosis-sensitive tumor cell lines. Proc. Natl. Acad. Sci. U.S.A. 97, 13009–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pang S., Curran S. P. (2014) Adaptive capacity to bacterial diet modulates aging in C. elegans. Cell Metab. 19, 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pang S., Lynn D. A., Lo J. Y., Paek J., Curran S. P. (2014) SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat. Commun. 5, 5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ekena K., Maloy S. (1990) Regulation of proline utilization in Salmonella typhimurium: how do cells avoid a futile cycle? Mol. Gen. Genet. 220, 492–494 [DOI] [PubMed] [Google Scholar]
- 24. Moses S., Sinner T., Zaprasis A., Stöveken N., Hoffmann T., Belitsky B. R., Sonenshein A. L., Bremer E. (2012) Proline utilization by Bacillus subtilis: uptake and catabolism. J. Bacteriol. 194, 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marcotte E. M., Pellegrini M., Ng H. L., Rice D. W., Yeates T. O., Eisenberg D. (1999) Detecting protein function and protein-protein interactions from genome sequences. Science. 285, 751–753 [DOI] [PubMed] [Google Scholar]
- 26. Enright A. J., Iliopoulos I., Kyrpides N. C., Ouzounis C. A. (1999) Protein interaction maps for complete genomes based on gene fusion events. Nature 402, 86–90 [DOI] [PubMed] [Google Scholar]
- 27. White T. A., Krishnan N., Becker D. F., Tanner J. J. (2007) Structure and kinetics of monofunctional proline dehydrogenase from Thermus thermophilus. J. Biol. Chem. 282, 14316–14327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Inagaki E., Ohshima N., Takahashi H., Kuroishi C., Yokoyama S., Tahirov T. H. (2006) Crystal structure of Thermus thermophilus δ1-pyrroline-5-carboxylate dehydrogenase. J. Mol. Biol. 362, 490–501 [DOI] [PubMed] [Google Scholar]
- 29. Lee Y. H., Nadaraia S., Gu D., Becker D. F., Tanner J. J. (2003) Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein. Nat. Struct. Biol. 10, 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luo M., Arentson B. W., Srivastava D., Becker D. F., Tanner J. J. (2012) Crystal structures and kinetics of monofunctional proline dehydrogenase provide insight into substrate recognition and conformational changes associated with flavin reduction and product release. Biochemistry 51, 10099–10108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pemberton T. A., Srivastava D., Sanyal N., Henzl M. T., Becker D. F., Tanner J. J. (2014) Structural studies of yeast δ(1)-pyrroline-5-carboxylate dehydrogenase (ALDH4A1): active site flexibility and oligomeric state. Biochemistry 53, 1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Williams I., Frank L. (1975) Improved chemical synthesis and enzymatic assay of delta-1-pyrroline-5-carboxylic acid. Anal. Biochem. 64, 85–97 [DOI] [PubMed] [Google Scholar]
- 33. Luo M., Singh R. K., Tanner J. J. (2013) Structural determinants of oligomerization of δ(1)-pyrroline-5-carboxylate dehydrogenase: identification of a hexamerization hot spot. J. Mol. Biol. 425, 3106–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 35. Michaelis L., Menten M. L. (1913) Die Kinetik der Invertinwirkung. Biochim. Z. 49, 333–369 [Google Scholar]
- 36. Michaelis L., Menten M. L., Johnson K. A., Goody R. S. (2011) The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry 50, 8264–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meek T. D., Garvey E. P., Santi D. V. (1985) Purification and characterization of the bifunctional thymidylate synthetase-dihydrofolate reductase from methotrexate-resistant Leishmania tropica. Biochemistry 24, 678–686 [DOI] [PubMed] [Google Scholar]
- 38. Geck M. K., Kirsch J. F. (1999) A novel, definitive test for substrate channeling illustrated with the aspartate aminotransferase/malate dehydrogenase system. Biochemistry 38, 8032–8037 [DOI] [PubMed] [Google Scholar]
- 39. Mezl V. A., Knox W. E. (1976) Properties and analysis of a stable derivative of pyrroline-5-carboxylic acid for use in metabolic studies. Anal. Biochem. 74, 430–440 [DOI] [PubMed] [Google Scholar]
- 40. Baker P., Carere J., Seah S. Y. (2012) Substrate specificity, substrate channeling, and allostery in BphJ: an acylating aldehyde dehydrogenase associated with the pyruvate aldolase BphI. Biochemistry 51, 4558–4567 [DOI] [PubMed] [Google Scholar]
- 41. Zhang M., White T. A., Schuermann J. P., Baban B. A., Becker D. F., Tanner J. J. (2004) Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors. Biochemistry 43, 12539–12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ostrander E. L., Larson J. D., Schuermann J. P., Tanner J. J. (2009) A conserved active site tyrosine residue of proline dehydrogenase helps enforce the preference for proline over hydroxyproline as the substrate. Biochemistry 48, 951–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang X., Holden H. M., Raushel F. M. (2001) Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu. Rev. Biochem. 70, 149–180 [DOI] [PubMed] [Google Scholar]
- 44. Srere P. A., Ovadi J. (1990) Enzyme-enzyme interactions and their metabolic role. FEBS Lett. 268, 360–364 [DOI] [PubMed] [Google Scholar]
- 45. Ovadi J. (1991) Physiological significance of metabolic channelling. J. Theor. Biol. 152, 1–22 [PubMed] [Google Scholar]
- 46. Ishikawa M., Tsuchiya D., Oyama T., Tsunaka Y., Morikawa K. (2004) Structural basis for channelling mechanism of a fatty acid beta-oxidation multienzyme complex. EMBO J. 23, 2745–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meyer F. M., Gerwig J., Hammer E., Herzberg C., Commichau F. M., Völker U., Stülke J. (2011) Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon. Metab. Eng. 13, 18–27 [DOI] [PubMed] [Google Scholar]
- 48. Rudolph J., Stubbe J. (1995) Investigation of the mechanism of phosphoribosylamine transfer from glutamine phosphoribosylpyrophosphate amidotransferase to glycinamide ribonucleotide synthetase. Biochemistry 34, 2241–2250 [DOI] [PubMed] [Google Scholar]
- 49. Zhao H., French J. B., Fang Y., Benkovic S. J. (2013) The purinosome, a multi-protein complex involved in the de novo biosynthesis of purines in humans. Chem. Commun. 49, 4444–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yadid I., Rudolph J., Hlouchova K., Copley S. D. (2013) Sequestration of a highly reactive intermediate in an evolving pathway for degradation of pentachlorophenol. Proc. Natl. Acad. Sci. U.S.A. 110, E2182–E2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perkins J. R., Diboun I., Dessailly B. H., Lees J. G., Orengo C. (2010) Transient protein-protein interactions: structural, functional, and network properties. Structure 18, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 52. James C. L., Viola R. E. (2002) Production and characterization of bifunctional enzymes. Substrate channeling in the aspartate pathway. Biochemistry 41, 3726–3731 [DOI] [PubMed] [Google Scholar]
- 53. Islam M. M., Wallin R., Wynn R. M., Conway M., Fujii H., Mobley J. A., Chuang D. T., Hutson S. M. (2007) A novel branched-chain amino acid metabolon. Protein-protein interactions in a supramolecular complex. J. Biol. Chem. 282, 11893–11903 [DOI] [PubMed] [Google Scholar]
- 54. Monteoliva M. I., Rizzi Y. S., Cecchini N. M., Hajirezaei M. R., Alvarez M. E. (2014) Context of action of proline dehydrogenase (ProDH) in the Hypersensitive Response of Arabidopsis. BMC Plant Biol. 14, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miller G., Honig A., Stein H., Suzuki N., Mittler R., Zilberstein A. (2009) Unraveling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J. Biol. Chem. 284, 26482–26492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu W., Phang J. M. (2012) Proline dehydrogenase (oxidase) in cancer. Biofactors. 38, 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nooren I. M., Thornton J. M. (2003) Structural characterisation and functional significance of transient protein-protein interactions. J. Mol. Biol. 325, 991–1018 [DOI] [PubMed] [Google Scholar]
- 58. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]


