Background: Hedgehog acyltransferase (Hhat) transfers palmitate onto Sonic Hedgehog, which is required for efficient signaling.
Results: Hhat membrane topology was determined using in silico and empirical methods.
Conclusion: Hhat contains 10 transmembrane domains and 2 re-entrant loops.
Significance: This analysis provides a framework for understanding the mechanism of action of Hhat and other membrane bound O-acyltransferase enzymes with protein substrates.
Keywords: Membrane Bilayer, Membrane Protein, Protein Palmitoylation, Sonic Hedgehog (SHH), Transmembrane Domain
Abstract
Hedgehog acyltransferase (Hhat) is a multipass transmembrane enzyme that mediates the covalent attachment of the 16-carbon fatty acid palmitate to the N-terminal cysteine of Sonic Hedgehog (Shh). Palmitoylation of Shh by Hhat is critical for short and long range signaling. Knowledge of the topological organization of Hhat transmembrane helices would enhance our understanding of Hhat-mediated Shh palmitoylation. Bioinformatics analysis of transmembrane domains within human Hhat using 10 different algorithms resulted in highly consistent predictions in the C-terminal, but not in the N-terminal, region of Hhat. To empirically determine the topology of Hhat, we designed and exploited Hhat constructs containing either terminal or 12 different internal epitope tags. We used selective permeabilization coupled with immunofluorescence as well as a protease protection assay to demonstrate that Hhat contains 10 transmembrane domains and 2 re-entrant loops. The invariant His and highly conserved Asp residues within the membrane-bound O-acyltransferase (MBOAT) homology domain are segregated on opposite sides of the endoplasmic reticulum membrane. The localization of His-379 on the lumenal membrane surface is consistent with a role for this invariant residue in catalysis. Analysis of the activity and stability of the Hhat constructs revealed that the C-terminal MBOAT domain is especially sensitive to manipulation. Moreover, there was remarkable similarity in the overall topological organization of Hhat and ghrelin O-acyltransferase, another MBOAT family member. Knowledge of the topological organization of Hhat could serve as an important tool for further design of selective Hhat inhibitors.
Introduction
Hedgehog acyltransferase (Hhat)2 catalyzes the covalent attachment of the 16-carbon fatty acid palmitate to the N-terminal cysteine of Sonic Hedgehog (Shh) (1), a modification that is critical for signaling activity (2–6). Shh signaling is required for proper embryogenesis and tissue development, and defects in Shh signaling result in abnormal development of the neural tube, gastrointestinal tract, and limbs (7). In adults, aberrant Shh signaling promotes the initiation and progression of various tumors including gastrointestinal, pancreatic, and prostate cancers (8). In addition, Shh signaling plays a role in maintaining cancer stem cells and in mediating resistance to cancer therapies (9). Furthermore, recent studies elucidated the importance of Hhat activity in mediating the proliferation of pancreatic and lung squamous cell carcinomas (10–13). These studies established Hhat as an attractive drug target, and small molecule inhibitors of Hhat are currently under development (14).
Hhat is a multipass transmembrane protein that belongs to the membrane-bound O-acyltransferase (MBOAT) family (15). The MBOAT family is characterized by a conserved homology domain with an invariant His residue (His-379 in Hhat) surrounded by hydrophobic residues and a highly conserved Asp/Asn residue (Asp-339 in Hhat) surrounded by moderately hydrophobic residues. Most MBOAT enzymes transfer fatty acids and other lipids onto hydroxyl groups of membrane bound lipids. Hhat and two other MBOAT members (Porcupine and ghrelin O-acyltransferase) are unique in that they catalyze the transfer of fatty acids onto secreted proteins. Porcupine transfers a monounsaturated 16-carbon fatty acid onto the Wnt family of ligands (16), and ghrelin O-acyltransferase (GOAT) transfers an 8-carbon fatty acid onto the appetite-stimulating peptide hormone ghrelin (17, 18).
Hhat, Porcupine, and GOAT are localized in the endoplasmic reticulum (ER) and acylate proteins that travel through the secretory pathway. Because entry into the secretory pathway is necessary for Hhat-mediated palmitoylation of Shh and GOAT-mediated octanoylation of ghrelin (1, 19), the active site of these enzymes is most likely located on the lumenal face of the ER. Knowledge of the topological organization of MBOAT transmembrane helices would considerably enhance our understanding of the mechanism of MBOAT-mediated protein fatty acylation. A recent study of the topology of GOAT identified 11 transmembrane domains (TMDs) and one re-entrant loop, with the invariant His residue in the lumen and the highly conserved Asn residue in the cytoplasm (19). To date, the number and orientation of TMDs within Hhat remains unknown.
To further our understanding of Hhat structure and function, we set out to determine the membrane topology of Hhat. Here, we combine in silico and empirical methods to experimentally determine the topological organization of Hhat across the membrane bilayer. Selective membrane permeabilization coupled with immunofluorescence and an in vitro protease protection assay were used to establish the presence of 10 TMDs and two re-entrant loops within Hhat. The topological organization of Hhat provides a framework for understanding its mechanism of action and may aid in the further design of Hhat inhibitors.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Reagents were purchased from the following vendors: trypsin, digitonin, cycloheximide, chloramphenicol, Triton X-100, and anti-FLAG (Sigma); anti-Shh, anti-Myc, and anti-caveolin antibodies (Santa Cruz Biotechnology, Dallas, TX); anti-HA (Roche Applied Science); anti-PDI (Enzo Life Sciences, Farmingdale, NY); octylglucoside (EMD Millipore, Billerica, MA); [125I]NaI (PerkinElmer Life Sciences).
Mammalian Expression Plasmids
The plasmid encoding HA-tagged Hhat was generated as previously described (1). Hhat constructs with C-terminal FLAG and Myc epitope tags as well as FLAG and HA epitope insertions were generated using site-directed mutagenesis via the QuikChange II XL Site-directed mutagenesis kit (Stratagene, La Jolla, CA). All constructs were confirmed by DNA sequencing.
Cell Culture and Transfections
COS-1 and COS-7 cells were grown in Dulbecco's Modified Eagle's (DMEM) medium supplemented with 10% fetal bovine serum, 1 mm GlutaMAX (Invitrogen), 50 units/ml penicillin, and 50 μg/ml streptomycin. 293FT cells were grown in DMEM medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin, 50 μg/ml streptomycin, 500 μg/ml Geneticin, 1 mm GlutaMAX, 1 mm sodium pyruvate, and 0.1 mm nonessential amino acids. Transfections were carried out using Lipofectamine 2000® (Invitrogen).
Selective Permeabilization and Indirect Immunofluorescence
COS-7 cells were transfected with the indicated Hhat constructs. 24 h post transfection, cells were split onto coverslips in 6-well plates and cultured for an additional 24 h. Cells were fixed and permeabilized as previously described (19) with a few changes. Briefly, to selectively permeabilize the plasma membrane, cells were incubated with 65 μg/ml digitonin in KHM (20 mm HEPES (pH 7.4), 110 mm potassium acetate, 2 mm magnesium acetate) for 10 min on ice and fixed with 3% paraformaldehyde for 10 min at room temperature. To permeabilize all cellular membranes, cells were fixed with 3% paraformaldehyde for 20 min at room temperature and permeabilized with 0.2% Triton X-100 for 5 min at room temperature. Cells were incubated with the indicated primary antibodies and with secondary antibodies (Alexa Flour® 488-conjugated anti-mouse IgG and Alexa Flour® 594-conjugated anti-rabbit IgG) for 45 min each. Slides were mounted with ProLong® Gold Antifade (Invitrogen). Images were collected using a Leica SP5 confocal microscope and analyzed with the Leica Application Suite software.
Protease Protection Assays
P100 membranes were prepared as previously described (1). Briefly, 293FT cells transfected with the indicated Hhat constructs were washed with ice-cold STE (100 mm NaCl, 10 mm Tris, and 1 mm EDTA (pH 7.4)), collected, and centrifuged for 10 min at 1000 × g at 4 °C. Cell pellets were resuspended in hypotonic lysis buffer (10 mm HEPES (pH 7.3) and 0.2 mm MgCl2) and incubated on ice for 10 min followed by Dounce homogenization with 30 strokes. The homogenate was supplemented with 0.25 m sucrose and centrifuged for 45 min at 100,000 × g at 4 °C. The pellets were resuspended in hypotonic lysis buffer supplemented with protease inhibitors and flash-frozen. For each protease protection assay, 50 μg of total membrane protein was incubated at 30 °C for 30 min with 20 μg/ml trypsin in the absence or presence of 1% octylglucoside. The reaction was stopped with the addition of protease inhibitors. After incubation with 2 units of DNase I for 5 min, the samples were solubilized with 2× sample buffer and electrophoresed on 10% SDS-PAGE.
Cell-based Palmitoylation Assay
COS-1 cells expressing Shh and the indicated Hhat constructs were starved for 1 h in DMEM medium containing 2% dialyzed fetal calf serum followed by incubation with 13 μCi/ml [125I]iodopalmitate for 4 h at 37 °C. Cells were washed twice with 2 ml of ice-cold STE buffer and lysed in radioimmune precipitation assay buffer (150 mm NaCl, 50 mm Tris, (pH 7.4), 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 1 mm EDTA). Lysates were clarified by ultracentrifugation at 100,000 × g for 15 min in a Beckman T100.2 rotor. Immunoprecipitations were performed by incubating clarified lysates with 7 μl of anti-Shh and 50 μl of protein A/G+-agarose beads (Santa Cruz Biotechnology) for 16 h at 4 °C. The beads were washed twice with 500 μl of radioimmune precipitation assay buffer, and bead pellets were resuspended in 40 μl of 2 × sample buffer containing 40 mm dithiothreitol. Immunoprecipitated samples were electrophoresed on a 12.5% SDS-PAGE, dried, and exposed by phosphorimaging for 4 days. Screens were analyzed on a TyphoonTM FLA-7000 bioimaging analyzer (GE Healthcare). Labeling experiments were performed in duplicate and repeated three times.
Protein Stability Assay
COS-1 cells were transfected with the indicated Hhat constructs and 48 h post transfection placed in DMEM media supplemented with 10% FBS, 100 μg/ml cycloheximide, and 40 μg/ml chloramphenicol. After incubation for 0 or 24 h, cells were washed twice with 2 ml of STE buffer and scraped in 500 μl of radioimmune precipitation assay buffer supplemented with protease inhibitors. Protein concentrations were determined using DCTM Protein Assay (Bio-Rad), and equal amounts of samples were electrophoresed on SDS-PAGE gels, transferred onto PVDF membranes, and probed with anti-HA, anti-FLAG, or anti-Myc antibodies. Expression levels were quantified from Western blots using Quantity One (Bio-Rad) with a GS-800 Calibrated Densitometer (Bio-Rad). Experiments were performed in duplicate and repeated three times.
RESULTS
Predicted Membrane Topology of Hhat
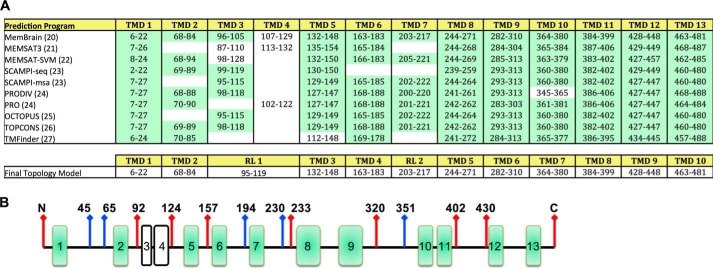
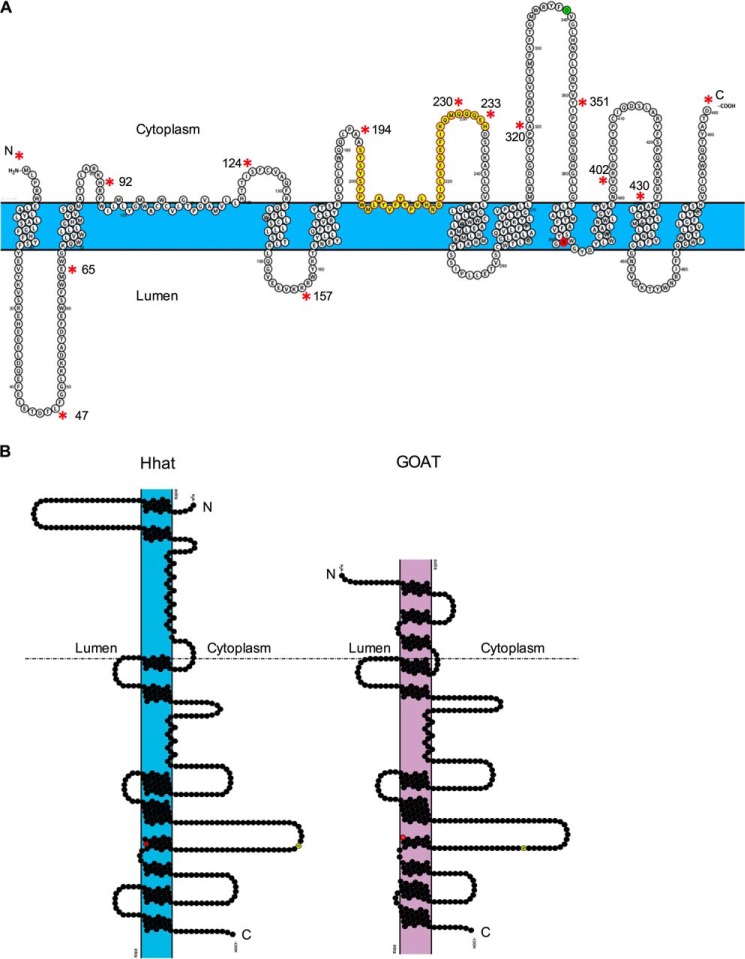
We first performed in silico analysis of the membrane topology of human Hhat (UniProt #Q5VTY9-1) using 10 different prediction programs. Between 10 and 13 TMD helices were predicted by these programs, and no signal peptide was evident (Fig. 1A). Most TMDs, especially helices 5–13, were predicted with remarkable consistency, with the boundaries of potential TMDs marked within a few residues across 7–10 of the algorithms. There was, however, variability among the different prediction programs in the N-terminal region of Hhat. Specifically, the presence of potential TMDs between residues 87–132 was inconsistent. MemBrain (20) predicted one possible re-entrant helix (residues 96–105) and a closely spaced subsequent TMD. MEMSAT3 (21) predicted the presence of two TMDs, and most other programs predicted one TMD (22–27). Among the programs that predicted one TMD in this region, there was considerable variation in the boundaries of the TMD helix. Analysis with ZPRED (28), which predicts the distance of a given residue to the center of the membrane, indicated that residues 95–119 were likely located within the membrane bilayer.
FIGURE 1.
Predicted transmembrane domains for human Hhat. A, TMDs predicted for human Hhat by the indicated programs (top panel). TMDs predicted with high consistency by most programs are highlighted in green. Bottom panel, final topology model of Hhat. RL, re-entrant loop. B, schematic representation of FLAG epitope insert (red arrows) and HA epitope insert (blue arrows) constructs used to map the topology of Hhat.
Identification of the Membrane Topology of Hhat via Differential Membrane Permeabilization
To experimentally determine the topology of Hhat, FLAG and HA epitope tags were introduced within hydrophilic regions between predicted TMDs as well as at either the N or C terminus (Fig. 1B). Each internal epitope insertion construct also contained a C-terminal tag, which allowed us to verify that the epitope insertion does not significantly alter protein topology and “flip” the C terminus across the membrane. FLAG insert constructs were designed with an HA C-terminal tag, and HA insert constructs were designed with either a FLAG or Myc C-terminal tag. The orientation of each epitope tag with respect to the ER membrane was analyzed via differential membrane permeabilization coupled with immunofluorescence microscopy. COS-7 cells expressing individual Hhat constructs were treated with either digitonin, to selectively permeabilize the plasma membrane, or Triton X-100, to permeabilize all cellular membranes. Cells were incubated with antibodies directed against terminal or internal epitope tags. Co-staining for endogenous protein disulfide isomerase (PDI), an ER protein localized to the ER lumen, provided an internal control to verify either complete or selective membrane permeabilization.
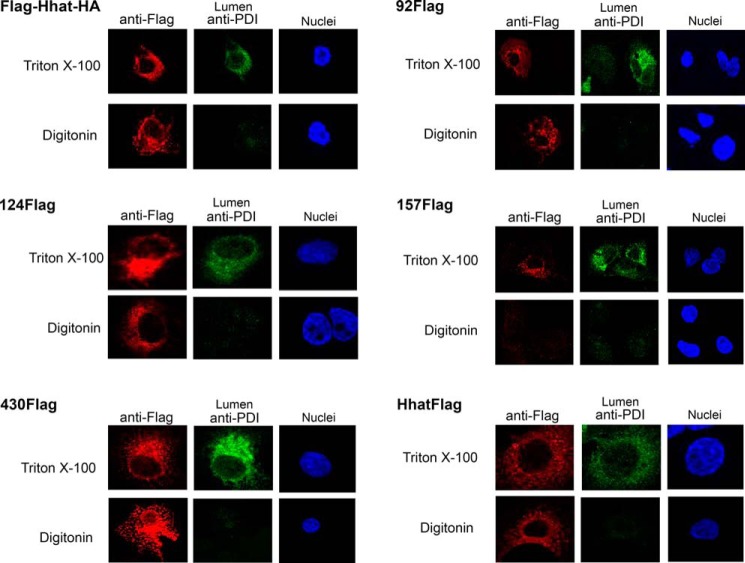
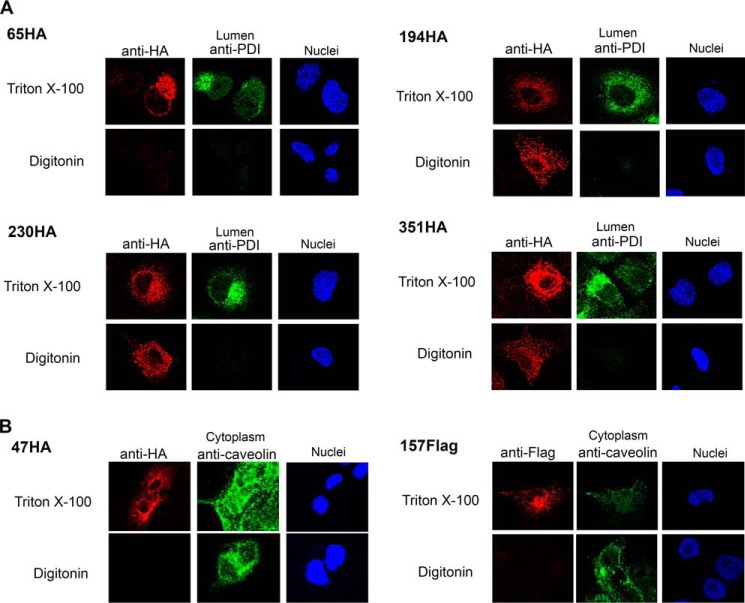
Multiple cytosolic loops were identified using the FLAG and HA-tagged constructs. A FLAG epitope placed at the N terminus of Hhat (FLAG-HhatHA) exhibited a cytosolic localization, as permeabilization with digitonin was sufficient to detect the epitope tag (Fig. 2). FLAG epitopes inserted into six predicted loops, 92FLAG, 124FLAG, 233FLAG, 320FLAG, 402FLAG, and 430FLAG, also displayed cytosolic orientations (Fig. 2, Table 1). The cytosolic orientation of both 92FLAG and 124FLAG epitopes suggests that the hydrophobic segment predicted by most algorithms between residues 87 and 132 does not cross the membrane. An additional cytosolic loop was identified with the 194HA construct (Fig. 3A). Moreover, two HA insert constructs, 230HA and 351HA, contain epitope inserts within the same predicted loops as the 233FLAG and 320FLAG constructs, respectively. Both 230HA and 351HA displayed cytosolic orientations, consistent with the results from the respective FLAG insert constructs (Fig. 3A, Table 1). The cytosolic orientation of epitope tags on either side of the strongly predicted hydrophobic segment from residues 203–217 suggests that this segment does not span the membrane. In addition, these data localize the highly conserved Asp-339 residue to the cytosol. Finally, a FLAG tag at the C terminus of Hhat exhibited a cytosolic orientation (Fig. 2).
FIGURE 2.
Hhat topology mapping using FLAG insert constructs. COS-7 cells were transfected with Hhat cDNA containing the indicated internal FLAG epitope tags. Digitonin was used to selectively permeabilize the plasma membrane, and Triton X-100 was used to permeabilize all cellular membranes. Cells were stained with anti-FLAG antibody to visualize the cytosolic or lumenal orientation of the inserted FLAG tag. Staining of endogenous PDI served as an internal control for a lumenal epitope. Images for each construct were collected using the same conditions on the same day ensuring fair side-by-side comparison.
TABLE 1.
Summary of Hhat membrane topology experiments
| Epitope tag | Location: IF assaya | Location: protease assayb |
|---|---|---|
| N-term FLAG | Cytosol | NDc |
| 47HA | Lumen | ND |
| 65HA | Lumen | ND |
| 92FLAG | Cytosol | Cytosol |
| 124FLAG | Cytosol | Cytosol |
| 157FLAG | Lumen | Lumen |
| 194HA | Cytosol | ND |
| 230HA | Cytosol | ND |
| 233FLAG | Cytosol | ND |
| 320FLAG | Cytosol | ND |
| 351HA | Cytosol | ND |
| 402FLAG | Cytosol | ND |
| 430FLAG | Cytosol | Cytosol |
| C-term FLAG | Cytosol | Cytosol |
| C-term Myc | ND | Cytosol |
FIGURE 3.
Hhat topology mapping using HA insert constructs. A, COS-7 cells were transfected with Hhat cDNA containing the indicated internal HA epitope tags. Cells were treated and visualized as in Fig. 2. B, COS-7 cells were transfected with Hhat cDNA with the indicated internal FLAG or HA epitope tags. Cells were treated as in Fig. 2 with one exception: staining of endogenous caveolin served as an internal control for a cytosolic epitope. Images for each construct were collected using the same conditions on the same day ensuring fair side-by-side comparison.
Lumenal loops were identified when permeabilization with Triton X-100, but not with digitonin, was required to detect the epitope tag insertion. These included 157FLAG as well as 47HA and 65HA, which have HA tags within the same predicted loop (Figs. 2 and 3). Co-staining with an antibody directed against an endogenous caveolin epitope localized in the cytoplasm was also performed. In all cases caveolin fluorescence was detected with digitonin as well as Triton X-100, confirming that selective permeabilization of the plasma membrane was achieved (Fig. 3B, data not shown). Finally, for each construct with an internal epitope insertion, the C-terminal tag was stained in parallel and confirmed to be localized in the cytosol, suggesting that the overall topology of Hhat was not affected by the internal epitope tag insertion (not shown). Taken together, these data suggest that both N and C termini of Hhat are located in the cytosol and establish the presence of six cytosolic and two lumenal loops within Hhat.
Identification of the Membrane Topology of Hhat via Protease Protection Assay
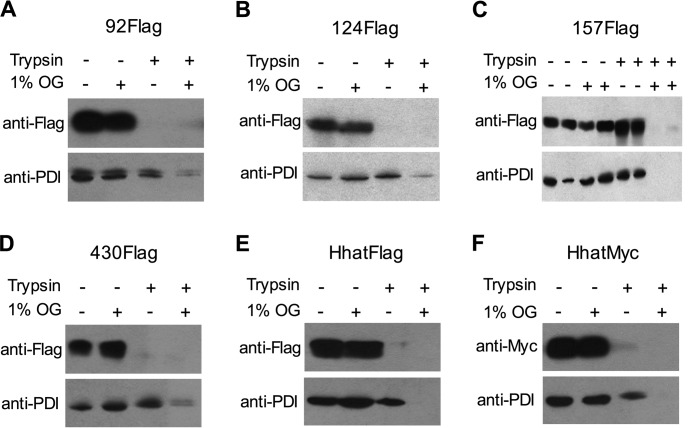
We next used an in vitro protease protection assay as an additional method to analyze the orientation of each epitope tag insertion with respect to the membrane. Lumenal epitopes should be protected from protease digestion in the absence, but not the presence of, detergent. Membranes from 293FT cells expressing individual constructs were incubated with trypsin in the presence or absence of octylglucoside. We chose this detergent as it allows for the purification of an active, functional Hhat (1). PDI served as an internal control for the integrity of membrane preparations and confirmation of membrane permeabilization upon treatment with octylglucoside. FLAG epitope tags within the 92FLAG, 124FLAG, and 430FLAG constructs were cytosolic, as incubation with trypsin alone was sufficient to digest the epitope tag (Fig. 4, A, B, and D). In contrast, the FLAG epitope in the 157FLAG construct was lumenal, as the epitope was only accessible to trypsin-mediated digestion upon membrane permeabilization with octylglucoside (Fig. 4C). Constructs with a C-terminal FLAG or Myc tag displayed a cytosolic orientation (Fig. 4, E and F). These data are consistent with the findings from the immunofluorescence-based assay. Use of the remaining FLAG insert constructs in the protease protection assay generated inconsistent results, which may reflect changes in epitope accessibility or topology resulting from the membrane preparation. Furthermore, HA insert constructs could not be used in this assay, as we were unable to detect the HA tags via immunoblotting despite being able to detect the tags by immunofluorescence. A similar issue with epitope recognition was observed with a subset of internal epitope tags within GOAT (19).
FIGURE 4.
Protease protection assay to determine the topology of Hhat. A–F, membranes were prepared from 293FT cells transfected with 92FLAG (A), 124FLAG (B), 157FLAG (C), 430FLAG (D), HhatFLAG (E), or HhatMyc (F) constructs. Membranes were incubated with trypsin in the absence or presence of 1% octylglucoside (OG). Samples were electrophoresed on 10% SDS-PAGE and probed with the indicated antibodies.
Activity and Stability of Hhat Proteins with Internal Epitope Tags
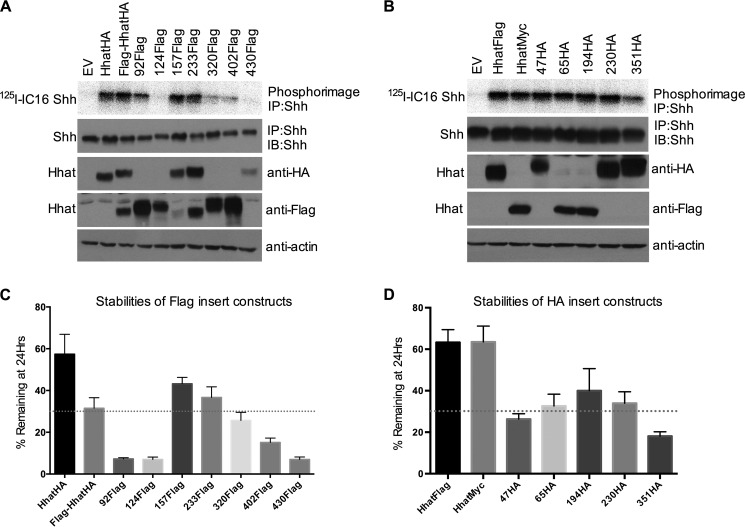
To determine whether epitope tag insertions resulted in gross changes in protein structure or function, we compared the activity and stability of individual constructs to that of Hhat with the corresponding C-terminal tag. We monitored the catalytic activity of each construct using a cell-based palmitoylation assay. COS-1 cells were co-transfected with cDNAs encoding Shh and either empty vector, C-terminally tagged Hhat, or individual epitope insertion Hhat constructs. Cells were then labeled with [125I]iodopalmitate, a radioiodinated palmitate analog. Shh was immunoprecipitated from cell lysates, and the amount of radiolabeled palmitate incorporation into Shh was determined by phosphorimaging analysis after SDS-PAGE.
Hhat constructs with FLAG epitopes at the N terminus or within the N-terminal half of Hhat, with the exception of 124FLAG, retained activity comparable with that of Hhat. In contrast, FLAG insertions within the C-terminal portion of Hhat exhibited significantly decreased activity (Fig. 5A). Four of five HA insert constructs showed activity comparable with that of HhatFLAG or HhatMyc (Fig. 5B). Consistent with the behavior of the FLAG insert constructs, the HA insert closest to the C terminus, 351HA, was also compromised in its catalytic activity (Fig. 5B). Importantly, the subcellular localization of each construct was comparable with that of wild type Hhat (Figs. 2 and 3). The lack of large puncta or aggregates suggests that the reduced catalytic activity of a subset of constructs was not due to gross protein misfolding.
FIGURE 5.
Activity and stability of constructs used to map the topology of Hhat. A and B, COS-1 cells co-expressing Shh and Hhat with the indicated FLAG insert (A) or HA insert (B) were labeled with [125I]iodopalmitate ([125I]IC16) for 4 h. Cell lysates were immunoprecipitated (IP) with anti-Shh antibody and analyzed by SDS-PAGE and phosphorimaging (top panels) or Western blotting (IB) with anti-HA, anti-FLAG, anti-Myc, or anti-actin antibodies. The experiment was performed 3 times in duplicate; a representative image is shown. C and D, COS-1 cells expressing the indicated FLAG insert (C) or HA insert (D) Hhat constructs were treated with 100 μg/ml cycloheximide and 40 μg/ml chloramphenicol for 0 h or 24 h. Cells were lysed and analyzed by Western blotting with an antibody directed against the C-terminal tag (anti-HA, anti-FLAG, or anti-Myc) after SDS-PAGE. The amount of Hhat signal at each time point was determined by densitometry. Each point indicates the mean ± S.D. (n = 3).
We noted variability in the detection of each FLAG insert construct by immunoblotting (Fig. 5A). Two constructs, FLAG-HhatHA and 233FLAG, were detected by both HA and FLAG antibodies, whereas the remaining constructs could only be detected by either the FLAG or HA antibody. With the exception of 430FLAG, constructs that could be detected by the HA antibody exhibited comparable expression to that of HhatHA. Expression of the HA insert constructs was monitored by their C-terminal tags as internal HA epitopes were undetectable by immunoblotting. HA insert constructs showed comparable expression with that of HhatFLAG or HhatMyc (Fig. 5B). Taken together, these data suggest that epitope insertions within the N-terminal portion of Hhat are well tolerated, whereas insertions within the C-terminal MBOAT homology domain led to reduced activity.
We next asked whether decreased stability could account for the reduced catalytic activity exhibited by some constructs. COS-1 cells expressing individual epitope insertion constructs were treated with cycloheximide and chloramphenicol to block new protein synthesis, and levels of protein remaining at 24 h were quantified. Hhat with a C-terminal HA, FLAG, or Myc tag was remarkably stable with about 57–63% of the initial protein remaining after 24 h (Fig. 5, C and D). Three constructs carrying FLAG epitope insertions within the N-terminal half of Hhat (FLAG-HhatHA, 157FLAG, and 233FLAG) were about half as stable as HhatHA. Importantly, the catalytic activity of these constructs was not compromised (Fig. 5A). The 92FLAG and 124FLAG constructs exhibited reduced stability with <10% of initial levels of protein remaining after 24 h (Fig. 5C). Despite low stability, the catalytic activity of 92FLAG was not affected (Fig. 5A). In contrast, the decreased stability of 124FLAG may account for its reduced activity (Fig. 5, A and C). Furthermore, FLAG insertions within the C-terminal portion of Hhat also led to reduced stability, suggesting reduced stability may account for the reduced activity of these constructs (Fig. 5, A and C). HA epitope insertions led to a 2-fold decrease in stability compared with HhatFLAG or HhatMyc (Fig. 5D); however, most of these proteins remained catalytically active (Fig. 5B). 351HA, which exhibited impaired catalytic activity, also displayed a modest reduction in stability compared with the other HA insert constructs. Taken together, these data suggest that although epitope insertions result in reduced stability, they do not necessarily lead to a corresponding decrease in activity.
DISCUSSION
Here we present an experimentally derived model for the membrane topology of Hhat using selective permeabilization coupled with immunofluorescence as well as a protease protection assay. We determined that both N and C termini of Hhat are localized in the cytosol and identified 10 TMDs and two re-entrant loops. These features are incorporated into the topology model depicted in Fig. 6. Most of the Hhat TMDs, especially helices 3–10 (Fig. 6A), were predicted with high consistency by multiple algorithms. This region encompasses the MBOAT homology region and contains the presumed active site. The invariant His-379 residue is located between two adjacent TMD helices, at the boundary of the membrane and the lumen of the ER. The orientation of His-379 within the lumenal side of the membrane is consistent with its role in Hhat catalysis (29). This organization is also observed in the recently published topology model for GOAT (19) and may provide a favorable hydrophobic environment for the catalytic reaction that uses fatty acyl-CoA as a substrate. However, the highly conserved Asp-339 residue is located in the cytoplasm. The tightly packed helices around the invariant His residue and the segregation of the conserved His and Asp residues on opposite sides of the membrane were also observed in the topology model for GOAT.
FIGURE 6.
Model for the membrane topology of Hhat. A, graphical representation of the topology of human Hhat generated with Protter Server (31). The boundaries of TMDs were designated using output from MemBrain (20). The boundaries of the first re-entrant loop (residues 95–119) are determined as discussed in the Discussion. The area of high conservation (residues 196–234) among MBOAT enzymes with protein substrates is highlighted in yellow. The invariant His-379 residue is shown in red, and the highly conserved Asp-339 is shown in green. Positions of FLAG epitope and HA epitope insertions (asterisks) are indicated along with the corresponding residue numbers. B, graphic representation of the topology of human Hhat (left) and human GOAT as previously described (19) (right) generated with Protter Server. The conserved His and Asp residues are shown in red and yellow, respectively. The topological organization of the C-terminal regions of Hhat and GOAT are highly conserved (below the dotted line).
A striking aspect of Hhat topology is the remarkable similarity to the topology of GOAT (Fig. 6B). The overall disposition and location of the TMDs and loops is nearly identical over ⅔ of the Hhat and GOAT proteins despite <20% primary sequence identity. In addition, we note that a large fraction of the protein is located within the cytoplasm (172 cytoplasmic residues versus 87 lumenal residues). Similarly, a to-scale model of GOAT topology reveals that it has long loops in the cytoplasm (161 residues in total) and mostly smaller loops (56 residues in total) in the lumen. The large cytoplasmic loops of these MBOAT enzymes may be required for protein-protein interactions, regulatory modifications, or substrate binding.
Due to the ambiguity in the predictions for TMDs between residues 87–132, we designed two constructs, 92FLAG and 124FLAG, in this region (Fig. 1). Both FLAG epitopes were determined to be cytosolic by two independent assays suggesting that this hydrophobic stretch of residues does not cross the membrane (Figs. 2 and 4). Although the 92FLAG insertion resulted in an unstable protein, the catalytic activity was not affected (Fig. 5). In contrast, the 124FLAG insertion resulted in a significant decrease in both stability and activity (Fig. 5). Although it remains possible that this FLAG tag interrupts a TMD as predicted by MemBrain and MEMSAT-SVM programs, all other algorithms place residue 124 outside potential TMDs (Fig. 1). Furthermore, the subcellular localization of the 124FLAG protein was comparable to that of wild type Hhat, and the C terminus of the construct was localized in the cytosol, suggesting that the membrane insertion and overall topology of this construct is not affected. Based on our data, it is likely that this region forms either a re-entrant loop or folds into a small domain with a hydrophobic interior within the cytosol. We propose that Hhat contains a re-entrant loop between residues 95 and 119 that is consistent with the ZPRED analysis, indicating that these residues are located within the membrane. Three-dimensional structural determination will be required to provide definitive confirmation for this interesting structural feature within Hhat.
We also identified the presence of a second re-entrant loop between residues 203–217 of Hhat. Epitope insertions on either side of this hydrophobic segment (194HA, 230HA, and 233FLAG) were localized in the cytosol, suggesting the hydrophobic segment does not cross the membrane (Fig. 3, Table 1). Importantly, these insertions do not alter the catalytic activity of Hhat (Fig. 5), suggesting that the tertiary structure of the protein is unaltered. It is possible that these residues form an interfacial helix, as the amino acid sequence of this segment (WMLAYVFYYPVLHNG) contains multiple Tyr and Trp residues, known to be enriched in interfacial helices (30). Interestingly, global sequence alignment of Hhat, Porcupine, and GOAT identified a second region of high conservation located between residues 196 and 234 in Hhat (29). This region encompasses the re-entrant loops within Hhat and GOAT, and sequence similarity suggests that Porcupine may also contain a re-entrant loop in this region. In addition, mutation of specific residues (Y207A and G217A) within this region of Hhat reduces catalytic activity (29). Further structure-function studies are needed to determine whether a re-entrant loop in MBOAT enzymes with protein substrates plays a role in substrate recognition, protein stability, and/or catalysis.
Acknowledgments
We thank Raisa Louft-Nisenbaum for expert technical assistance, Dr. Robert Fieldhouse and Dr. Chris Sander for helpful discussions, and the Memorial Sloan Kettering Molecular Cytology Core Facility for providing support with confocal imaging microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grants GM57966 and CA186957. This work was also supported by Cycle for Survival, Memorial Sloan Kettering Cancer Center.
- Hhat
- Hedgehog acyltransferase
- Shh
- Sonic Hedgehog
- MBOAT
- membrane bound O-acyltransferase
- GOAT
- ghrelin O-acyltransferase
- TMD
- transmembrane domain
- ER
- endoplasmic reticulum
- PDI
- protein disulfide isomerase.
REFERENCES
- 1. Buglino J. A., Resh M. D. (2008) Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J. Biol. Chem. 283, 22076–22088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dawber R. J., Hebbes S., Herpers B., Docquier F., van den Heuvel M. (2005) Differential range and activity of various forms of the Hedgehog protein. BMC Dev. Biol. 5, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Micchelli C. A., The I., Selva E., Mogila V., Perrimon N. (2002) Rasp, a putative transmembrane acyltransferase, is required for Hedgehog signaling. Development 129, 843–851 [DOI] [PubMed] [Google Scholar]
- 4. Chen M. H., Li Y. J., Kawakami T., Xu S. M., Chuang P. T. (2004) Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 18, 641–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee J. D., Treisman J. E. (2001) Sightless has homology to transmembrane acyltransferases and is required to generate active Hedgehog protein. Curr. Biol. 11, 1147–1152 [DOI] [PubMed] [Google Scholar]
- 6. Chamoun Z., Mann R. K., Nellen D., von Kessler D. P., Bellotto M., Beachy P. A., Basler K. (2001) Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 293, 2080–2084 [DOI] [PubMed] [Google Scholar]
- 7. Ingham P. W., McMahon A. P. (2001) Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087 [DOI] [PubMed] [Google Scholar]
- 8. Barakat M. T., Humke E. W., Scott M. P. (2010) Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol. Med. 16, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vidal S. J., Rodriguez-Bravo V., Galsky M., Cordon-Cardo C., Domingo-Domenech J. (2014) Targeting cancer stem cells to suppress acquired chemotherapy resistance. Oncogene 33, 4451–4463 [DOI] [PubMed] [Google Scholar]
- 10. Petrova E., Matevossian A., Resh M. D. (2015) Hedgehog acyltransferase as a target in pancreatic ductal adenocarcinoma. Oncogene 34, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Konitsiotis A. D., Chang S. C., Jovanović B., Ciepla P., Masumoto N., Palmer C. P., Tate E. W., Couchman J. R., Magee A. I. (2014) Attenuation of hedgehog acyltransferase-catalyzed sonic Hedgehog palmitoylation causes reduced signaling, proliferation and invasiveness of human carcinoma cells. PLoS ONE 9, e89899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rodriguez-Blanco J., Schilling N. S., Tokhunts R., Giambelli C., Long J., Liang Fei D., Singh S., Black K. E., Wang Z., Galimberti F., Bejarano P. A., Elliot S., Glassberg M. K., Nguyen D. M., Lockwood W. W., Lam W. L., Dmitrovsky E., Capobianco A. J., Robbins D. J. (2013) The hedgehog processing pathway is required for NSCLC growth and survival. Oncogene 32, 2335–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Justilien V., Walsh M. P., Ali S. A., Thompson E. A., Murray N. R., Fields A. P. (2014) The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell 25, 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Petrova E., Rios-Esteves J., Ouerfelli O., Glickman J. F., Resh M. D. (2013) Inhibitors of Hedgehog acyltransferase block Sonic Hedgehog signaling. Nat. Chem. Biol. 9, 247–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hofmann K. (2000) A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem. Sci. 25, 111–112 [DOI] [PubMed] [Google Scholar]
- 16. Rios-Esteves J., Resh M. D. (2013) Stearoyl CoA desaturase is required to produce active, lipid-modified Wnt proteins. Cell Rep. 4, 1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gutierrez J. A., Solenberg P. J., Perkins D. R., Willency J. A., Knierman M. D., Jin Z., Witcher D. R., Luo S., Onyia J. E., Hale J. E. (2008) Ghrelin octanoylation mediated by an orphan lipid transferase. Proc. Natl. Acad. Sci. U.S.A. 105, 6320–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang J., Brown M. S., Liang G., Grishin N. V., Goldstein J. L. (2008) Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell 132, 387–396 [DOI] [PubMed] [Google Scholar]
- 19. Taylor M. S., Ruch T. R., Hsiao P. Y., Hwang Y., Zhang P., Dai L., Huang C. R., Berndsen C. E., Kim M. S., Pandey A., Wolberger C., Marmorstein R., Machamer C., Boeke J. D., Cole P. A. (2013) Architectural organization of the metabolic regulatory enzyme ghrelin O-acyltransferase. J. Biol. Chem. 288, 32211–32228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen H., Chou J. J. (2008) MemBrain: improving the accuracy of predicting transmembrane helices. PLoS ONE 3, e2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones D. T. (2007) Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23, 538–544 [DOI] [PubMed] [Google Scholar]
- 22. Nugent T., Jones D. T. (2012) Detecting pore-lining regions in transmembrane protein sequences. BMC Bioinformatics 13, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bernsel A., Viklund H., Falk J., Lindahl E., von Heijne G., Elofsson A. (2008) Prediction of membrane-protein topology from first principles. Proc. Natl. Acad. Sci. U.S.A. 105, 7177–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Viklund H., Elofsson A. (2004) Best α-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci. 13, 1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Viklund H., Elofsson A. (2008) OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24, 1662–1668 [DOI] [PubMed] [Google Scholar]
- 26. Bernsel A., Viklund H., Hennerdal A., Elofsson A. (2009) TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37, W465–W468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deber C. M., Wang C., Liu L. P., Prior A. S., Agrawal S., Muskat B. L., Cuticchia A. J. (2001) TM Finder: a prediction program for transmembrane protein segments using a combination of hydrophobicity and nonpolar phase helicity scales. Protein Sci. 10, 212–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Granseth E., Viklund H., Elofsson A. (2006) ZPRED: predicting the distance to the membrane center for residues in α-helical membrane proteins. Bioinformatics 22, e191–e196 [DOI] [PubMed] [Google Scholar]
- 29. Buglino J. A., Resh M. D. (2010) Identification of conserved regions and residues within Hedgehog acyltransferase critical for palmitoylation of Sonic Hedgehog. PLoS ONE 5, e11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Viklund H., Granseth E., Elofsson A. (2006) Structural classification and prediction of reentrant regions in α-helical transmembrane proteins: application to complete genomes. J. Mol. Biol. 361, 591–603 [DOI] [PubMed] [Google Scholar]
- 31. Omasits U., Ahrens C. H., Müller S., Wollscheid B. (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 [DOI] [PubMed] [Google Scholar]