Background: The acidic loops of E2 enzymes are important for their Lys-48-ubiquitylation activity.
Results: The presence of Tyr residues modulates the ubiquitin binding property of the acidic loop.
Conclusion: A proper interaction of the acidic loop with the attached donor ubiquitin is important for Lys-48-ubiquitylation activity.
Significance: One of molecular bases to study the mechanism of Lys-48-ubiquitylation is provided.
Keywords: Nuclear Magnetic Resonance (NMR), Structural Biology, Ubiquitin, Ubiquitin-conjugating Enzyme (E2 enzyme), Ubiquitylation (ubiquitination)
Abstract
The ubiquitin E2 enzymes, Ube2g1 and Ube2r1, are able to synthesize Lys-48-linked polyubiquitins without an E3 ligase but how that is accomplished has been unclear. Although both E2s contain essential acidic loops, only Ube2r1 requires an additional C-terminal extension (184–196) for efficient Lys-48-ubiquitylation activity. The presence of Tyr-102 and Tyr-104 in the Ube2g1 acidic loop enhanced both ubiquitin binding and Lys-48-ubiquitylation and distinguished Ube2g1 from the otherwise similar truncated Ube2r11–183 (Ube2r1C). Replacement of Gln-105–Ser-106–Gly-107 in the acidic loop of Ube2r1C (Ube2r1CYGY) by the corresponding residues from Ube2g1 (Tyr-102–Gly-103–Tyr-104) increased Lys-48-ubiquitylation activity and ubiquitin binding. Two E2∼UB thioester mimics (oxyester and disulfide) were prepared to characterize the ubiquitin binding activity of the acidic loop. The oxyester but not the disulfide derivative was found to be a functional equivalent of the E2∼UB thioester. The ubiquitin moiety of the Ube2r1CC93S-[15N]UBK48R oxyester displayed two-state conformational exchange, whereas the Ube2r1CC93S/YGY-[15N]UBK48R oxyester showed predominantly one state. Together with NMR studies that compared UBK48R oxyesters of the wild-type and the acidic loop mutant (Y102G/Y104G) forms of Ube2g1, in vitro ubiquitylation assays with various mutation forms of the E2s revealed how the intramolecular interaction between the acidic loop and the attached donor ubiquitin regulates Lys-48-ubiquitylation activity.
Introduction
The modification of proteins by ubiquitylation is important for the regulation of a large variety of cellular processes that includes cell cycle control, endocytosis, DNA repair, and regulation of transcription (1, 2). E3 ligases mediate the formation of isopeptide bonds between the C-terminal carboxyl group of ubiquitin and a Lys side chain of the target protein (mono-ubiquitylation) or a previously attached ubiquitin (polyubiquitylation). Although many kinds of ubiquitin-ubiquitin linkages are present in vivo (3), Lys-48-linked polyubiquitin chains, which direct ubiquitylated proteins to the 26 S proteasome complex for their degradation, are the most abundant ubiquitin signals. RING-E3 ligases are the dominant E3 species that mediate the direct transfer of ubiquitin (4) from the E2∼UB thioester intermediate to a target protein (2). An interaction between E2∼UB and ubiquitin is likely to be necessary for efficient Lys-48 polyubiquitylation, but such an interaction remains poorly characterized. In contrast, for Lys-63-linked ubiquitylation by the Ubc13 (E2) and Mms2 (ubiquitin-E2 variant) complex, Mms2 recruits an acceptor ubiquitin and positions its Lys-63 side chain in close proximity to the active site Cys of Ubc13 (5, 6). In a recent molecular docking model for Lys-11-ubiquitin linkage formation by Ube2S (7) based on NMR chemical shift perturbation (CSP)4 data and in vitro ubiquitylation assay, Ube2S has binding surfaces for both acceptor and donor ubiquitin to position them and generate Lys-11-linkage polyubiquitin, whereas Ubc13 needs Mms2 to bind to and position an acceptor ubiquitin. In this model the interaction between donor ubiquitin or acceptor ubiquitin and Ube2S was very weak. Analogous mechanisms for the formation of Lys-48 ubiquitin linkages are unknown, possibly because interactions that guide Lys-48-ubiquitylation may be very weak and difficult to identify.
An in vitro E3-independent ubiquitylation assay that used p27-E2 fusion proteins had identified Ube2d2 (UbcH5b) and Ube2b (hHR6b) as the most efficient mediators for the ubiquitylation of p27, but those E2s were found to be less effective for subsequent Lys-48-ubiquitylation in comparison to Ube2r1 (human Cdc34) (8). Recent reports showed that the sequential engagement of two different E2 enzymes may be important for efficient polyubiquitylation of target proteins in an E3-dependent system. UbcH5 was found to be efficient for the first ubiquitylation of IκBα, but then Ube2r1 performed the subsequent Lys-48-ubiquitylation reactions in the presence of Skip/Cullin/F-box E3 complex (9). Ube2r1 is able to synthesize Lys-48-linked polyubiquitin chains efficiently without an E3 ligase. This E3-independent reaction mechanism is likely to be used as well by the E2 during the RING-E3-mediated ubiquitylation.
Both Ube2g1 (human Ubc7) and Ube2r1 possess an acidic loop in the vicinity of the active-site Cys residue and display an exceptional ability to synthesize Lys-48-linked polyubiquitin chains. The acidic loops contain several Asp and Glu residues, and mutations eliminating these negative charges have been shown to decrease the ubiquitylation activity of yeast Cdc34 in the presence of the Skip/Cullin/F-box E3 complex (10). Ube2k, which lacks the acidic loop, has a similar ability for Lys-48-specific polyubiquitylation, whereas most other E2 enzymes cannot synthesize Lys-48 polyubiquitin efficiently in the absence of an E3 ligase (11). Ube2g1 consists of only an E2-core domain, but Ube2r1 has an additional C-terminal extension (residues 184–236). Work from our group previously showed that, unlike full-length Ube2r1, the Ube2r1 core (residues 1–183; Ube2r1C) has a decreased capability to synthesize Lys-48 polyubiquitin chains efficiently in the absence of an E3 (8). Ube2d2, which does not have an acidic loop, also is unable to synthesize Lys-48 polyubiquitin in the absence of an E3 ligase (8) despite having ubiquitin binding affinity almost identical to that of Ube2g1 as assessed by NMR CSP experiments using [15N]UB (12).
In this study we focused on the acidic loops and ubiquitin binding activities of Ube2g1 and Ube2r1C. By comparing ubiquitin binding and polyubiquitin synthesis by Ube2g1, Ube2r1C, and Ube2d2, their different abilities to synthesize Lys-48-linked chains were evaluated with respect to intramolecular interaction between the acidic loop and the attached donor ubiquitin.
EXPERIMENTAL PROCEDURES
Protein Preparations
Genes encoding E2 enzymes (ube2g1, ube2r1, and ube2d2) and ubiquitin variants (His- or non-tagged) were cloned into the pGEX-4T-1 (BamHI/XhoI) and pET15b/pET21a (NdeI/XhoI) vectors, respectively. Point mutations were introduced using the QuikChange site-directed mutagenesis kit (Stratagene). All proteins were expressed in the Escherichia coli strains BL21 DE3 or Rosetta DE3. The proteins were purified using GSTrap and HisTrap chromatography columns (GE Healthcare), respectively. The attached GST or His tags were cleaved by thrombin digestion followed by thrombin inactivation with 1 mm PMSF and benzamidine-HCl. Target proteins were further purified by gel permeation chromatography using Superdex-75 (GE Healthcare); where applicable, subsequent passage through a GSTrap column was used to remove the cleaved GST. Murine E1 protein was purified following a previously reported method (8), and the concentration of the E1 stock solution (∼2.0 mg/ml) was estimated based on band intensity observed after sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and staining with Coomassie Blue.
All of the isotope-enriched proteins for NMR experiments were obtained by growing E. coli in M9 minimal medium supplemented with [15N]NH4Cl (0.1%) and [13C]glucose (0.2%). The (i) random fractional (75∼80%) and (ii) complete (∼97%) deuteration of proteins were done by growing E. coli in M9 minimal medium (99% D2O) supplemented with (i) [13C]glucose (0.2%) and [15N]NH4Cl (0.1%) or (ii) [13C,2H]glucose (0.2%), 13C,2H,15N-labeled Celton powder (0.1%, Cambridge Isotope Laboratory), and [15N]NH4Cl (0.1%), respectively (13, 14).
In Vitro Ubiquitylation Assay
In vitro ubiquitylation was done using 200∼300 μm ubiquitin, 20 μm E2, and ∼0.5 μm E1 in a reaction buffer (pH 7.0, 7.5, or 8.0) composed of 50 mm Tris-HCl, 1 mm DTT, 2 mm ATP, 2 mm MgCl2, 10 mm creatine phosphate, and 1 unit each of pyrophosphatase and creatine phosphokinase at 25 °C. The reaction products were separated by 14∼16% SDS-PAGE. The formation of E2∼UB thioester was evaluated by non-reducing SDS-PAGE in which the reaction mixtures were sampled 20 min after the addition of E1 and then quickly frozen in liquid nitrogen.
To estimate the approximate Km values for E2-mediated Lys-48-ubiquitylation, the in vitro ubiquitylation assays were performed with 1.5 μm E1 and increasing ubiquitin concentrations from 0.2 to 2.4 mm. The amounts of diubiquitin synthesized were estimated by integrating the diubiquitin bands in the Coomassie Blue-stained SDS-PAGE gel using the Multi Gauge program (Fujifilm). The reaction sample containing the highest concentration of the ubiquitin substrate was serially diluted and separated in the same SDS-PAGE as an internal reference to confirm the linearity of the integrated band intensities. Sample loadings were reduced to ensure that the volume integrals of diubiquitin bands correlated linearly to the amounts loaded; two or more experimental replicates were done for each E2 enzyme. The establishment of a stable concentration of E2∼ubiquitin thioester was associated with a time lag during which the product was not synthesized constantly. Therefore, the amount of synthesized di-UB is lower than expected, and this error was high at low concentrations of substrate ubiquitin. Nevertheless, the volume integrals of the diubiquitin bands were fitted with the Michaelis-Menten equation to estimate relative Km values of different E2s.
Preparation of E2-UB Oxyester and E2-UB Disulfide Derivatives
E2-UB oxyesters were synthesized using the active-site Cys-to-Ser mutant E2 protein (Ube2g1C90S or Ube2r1CC93S) in reactions with a UBK48R:E2 molar ratio of 2. Instead of wild-type UB, intact or His6-tagged UBK48R was used here to prevent loss of E2-UB oxyesters caused by Lys-48 diubiquitin formation and to facilitate product purification. The amounts of E1 used were up to 10× larger than was used in the standard ubiquitylation assay; more E1 was required for Ube2g1C90S than for Ube2r1CC93S due to the higher hydrolysis rate of Ube2g1C90S-UBK48R oxyester. The ubiquitin-coupling reactions were done in a buffer (pH 8.0) consisting of 50 mm Tris·HCl, 1 mm DTT, 4 mm ATP, 5 mm MgCl2, 20 mm creatine phosphate, and 2 units of pyrophosphatase and creatine phosphokinase. The reaction temperature and time were 35∼37 °C and 6 h for Ube2g1C90S and 30∼33 °C and 15 h for Ube2r1CC93S.
The synthesized [15N]Ube2g1C90S-UBK48R oxyesters were separated from the remaining Ube2g1C90S by His-tag affinity column chromatography, and the remaining free His-tagged UBK48R was completely removed by gel permeation chromatography using Superdex-75 column in a buffer (pH 7.0) containing 50 mm HEPES and 100 mm NaCl. To minimize hydrolysis during purification, the products (i.e. Ube2g1C90S-[15N]UBK48R or Ube2r1CC93S-[15N]UBK48R oxyester) were purified only by gel permeation chromatography; as a result, those purified E2-UB oxyester proteins contained 20∼30% of the free E2 proteins.
Disulfide-linked complexes of Ube2r1C and [15N]UBK48R/G76C, generated essentially by following the previously reported protocol (15), were made in which N-terminal His-tagged [15N]UBK48R/G76C was attached to Ube2r1 via a disulfide bond. The E2-UB disulfide complex was purified by His-tag affinity column chromatography followed by gel permeation chromatography after thrombin-catalyzed removal of the His tag.
NMR Experiments and Backbone Chemical Shift Assignments
NMR spectra were recorded using the 800- and 900-MHz (cryogenic probe) Bruker spectrometers in the Korea Basic Science Institute at 25 °C. NMR samples were prepared in a buffer (pH 7.0) containing 50 mm HEPES, 100 mm NaCl, and 10% D2O; 1∼2 mm DTT was included, except in the case of E2-UB disulfide complexes. NMR data were processed using the NMRPipe program (16) and analyzed using the SPARKY program (17). Kd values were obtained from titrations by fitting CSP data to a simple binding equation in which the CSP values were measured as [ΔN2 + (6×ΔH)2]1/2 (12).
TROSY triple-resonance experiments were used for the backbone chemical shift assignment of [13C,2H,15N]Ube2g1, and resonance assignments of [13C,15N]Ube2g1 and [13C,15N]Ube2g1Y102G/Y104G were determined by comparing their HSQC and three-dimensional-HNCA spectra with those of [13C,2H,15N]Ube2g1. The HSQC peaks of [13C,15N]Ube2d2 were assigned by using the previously reported chemical shifts (4).
Rosetta Docking Model of Ube2g1C90S-UBK48R Based on the NMR CSP Data
Homology-based model structures of Ube2g1 were generated from the SWISS-MODEL server (18) and CS-Rosetta (19) using the assigned chemical shifts of Ube2g1; the two model structures generated in this way were almost identical (root mean square deviation 0.97 Å). The C90S mutation was generated from the CS-Rosetta model of Ube2g1. We first performed rigid-body docking between the attached donor UBK48R and Ube2g1C90S with Patchdock (20) using the chemical shift perturbation data from both UBK48R and Ube2g1C90S as constraints. The acidic loop (residues Glu-96—Trp-111) was removed during the rigid-body docking step. Starting from the Patchdock rigid models with the incomplete acidic loop, we used the RosettaCM program (21) to build the acidic loop and simultaneously refine the rigid body orientations between UBK48R and Ube2g1C90S. The backbone atoms of both proteins in the complex model were allowed to move with constraint to initial backbone during the flexible refinement, and 2000 refined models are generated. The final ensemble structures were selected by their clustering and interface scores. The overall CSP of Ube2g1C90S-UBK48R oxyester matched well to the previously reported docking models of Ubc1∼UB thioester (22) and Cdc34∼UB thioester (23). All molecular visualizations were done using the Chimera program (24).
RESULTS
Despite Their Similar Domain Structures, the Lys-48 Polyubiquitylation Activity of Ube2r1C Is Much Lower Than That of Ube2g1
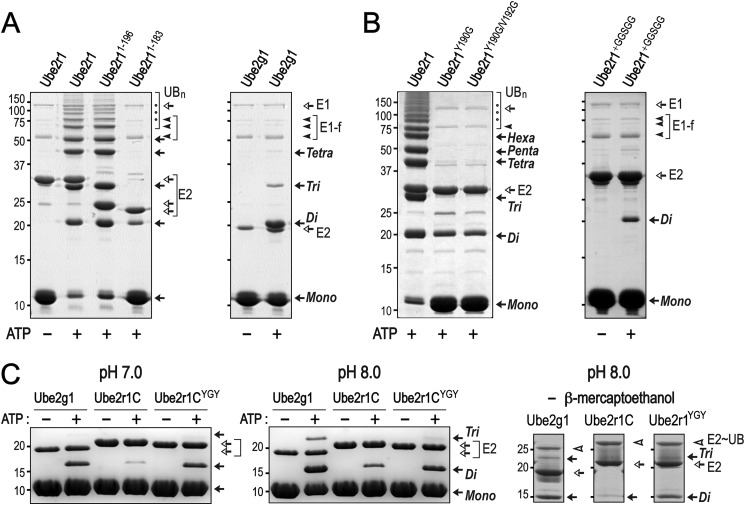
Sequence alignment of several E2 enzymes (Ube2g1, Ube2g2, Ube2r1, yeast Cdc34, Ube2d2, and Ube2k) shows that Ubc7 homologs (Ube2g1 and Ube2g2) and Cdc34 homologs (Ube2r1 and yeast Cdc34) have a distinct 12- or 13-residue insertion sequence in the vicinity of the active-site Cys (Fig. 1); this sequence was designated as an acidic loop due to the presence of many negatively charged Asp and Glu residues in yeast Cdc34 (10) (Fig. 1). Both Ube2g1 and Ube2r1 can synthesize Lys-48-linked polyubiquitin adducts in the absence of E3 ligase. However, despite the close similarity of their sequences (52% identity and 68% similarity), the in vitro ubiquitylation activity of Ube2r1 core enzyme (Ube2r1C, residues 1–183) was much lower than that of Ub2g1; Ube2r1 required a C-terminal extension (at a minimum, residues 184–196) for efficient Lys-48-ubiquitylation (Fig. 2A). Mutations within residues 184–196 of Ube2r1 (i.e. Y190G or Y190G/V192G) significantly impaired Lys-48-ubiquitylation activity, and the insertion of a GGSGG spacer between Val-183 and Pro-184 of Ube2r1 also greatly decreased the ubiquitylation activity (Fig. 2B).
FIGURE 1.

Structure-based sequence alignment of E2 enzymes. The sequence alignment was performed through structural matching of the E2 enzymes using the Chimera program (24). The previously identified non-covalent ubiquitin-binding sites of the Ube2r1 C-terminal tail (26) are marked with a gray highlight, and the acidic loop region is indicated by a yellow highlight. The asterisk represents the active-site Cys residues of the E2 enzymes. Ube2g1 and Ube2r1 residues that when mutated resulted in a decrease in Lys-48-ubiquitylation activity are indicated by open arrows, and those that have no activity change are indicated by closed arrows. The surface of Ube2g1 is roughly divided into four areas for better description of the data; the bottom side (red bar), the acidic loop (green bar), the left side (blue bar), and the right side (gray bar).
FIGURE 2.
E3-independent Lys-48 ubiquitylation activities of Ube2g1, Ube2r1, and Ube2r1 mutants. The in vitro ubiquitylation reactions were performed using E1 (∼0.5 μm), E2 (20 μm), and ubiquitin (0.2 mm) in a buffer containing 50 mm Tris-HCl (pH 7.5). Ubiquitin and the polyubiquitin products are marked Mono, Di, Tri, etc. A, Ube2r1 requires C-terminal residues 184–196 for wild-type activity. The activity of Ube2r11–183 (Ube2r1C) was much lower than that of either Ube2r1 or Ube2g1. B, the Ube2r1 mutant proteins (Y190G and Y190G/V192G) showed significant impairment in their in vitro ubiquitylation activities. The insertion of a GGSGG sequence between Val-183 and Pro-184 of Ube2r1 resulted in a complete loss of Lys-48 ubiquitylation activity. C, Ube2r1CYGY had Gln-105–Ser-106–Gly-107 in the acidic loop replaced by the Tyr-102–Gly-103–Tyr-104 sequence from Ube2g1. Although the Lys-48-ubiquitylation activity of Ube2r1C was lower than that of Ube2g1, Ube2r1CYGY showed activity to a level close to that of Ube2g1 (left panel). The formation of E2∼UB thioester adducts by Ube2g1, Ube2r1C, and Ube2r1CYGY was assessed by non-reducing SDS-PAGE analysis. Samples were prepared in SDS-PAGE loading buffer without β-mercaptoethanol 15 min after the ubiquitylation reaction was initiated with ATP. The three enzymes showed similar amounts of E2∼UB thioester intermediate formation (right panel).
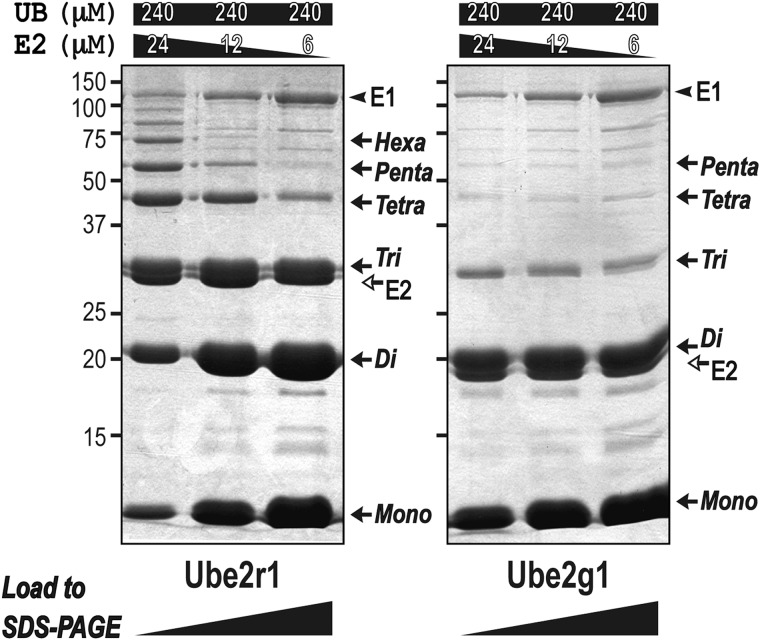
Ubiquitylation assays in which the E2 concentration was varied showed that the length of Lys-48 polyubiquitin products increased with increasing the concentration of Ube2r1 (Fig. 3). This result is consistent with the previous report that the dimerization of Ube2r1 increases its Lys-48-ubiquitylation activity (25). On the other hand, with Ube2g1 the size distribution of Lys-48 polyubiquitin products did not depend on the E2 concentration, and thus catalysis by Ube2g1 likely involves a reaction between the E2∼UB thioester and an acceptor ubiquitin that is not associated with a second E2∼UB molecule. In comparison with Ube2r1, the relatively simple ubiquitylation reaction by Ube2g1 suggested that studies focused on Ube2g1 and comparisons with Ube2d2 and Ube2r1C might reveal a mechanistic relationship between the ubiquitin binding activity and Lys-48-ubiquitylation.
FIGURE 3.
The E2 concentration dependence of in vitro ubiquitylation by Ube2r1 and Ube2g1. The amount of E1 needed to produce maximal Lys-48-ubiquitylation activity was first determined to ensure that the activation of E2 by E1 was not rate-limiting. The synthesis of Lys-48-linked triubiquitin and higher polyubiquitin by Ube2r1 was dependent on the E2 concentration. This result is consistent with a previous report that the dimerization of Ube2r1 increases its Lys-48-ubiquitylation activity. However, the concentration dependence of Ube2g1 indicated that catalysis by Ube2g1 likely involves a reaction between E2-UB thioester and a free acceptor ubiquitin.
Two Aromatic Residues (Tyr-102 and Tyr-104) in the Ube2g1 Acidic Loop Are Important for Non-covalent Ubiquitin Binding
Previous NMR CSP experiments using [15N]UB showed that the ubiquitin binding properties of Ube2d2 are similar to those of Ube2g1 (12), although Ube2d2 lacks an acidic loop and also cannot synthesize Lys-48 polyubiquitin chains efficiently in the absence of an E3 (8). We now have compared the ubiquitin-interacting surfaces of Ube2g1 and Ube2d2 in detail. Interestingly, CSP signals from the acidic loop region of Ube2g1 (Loop) were clearly identified when ubiquitin was added (Fig. 4, A and B, top panel). Two other major non-covalent ubiquitin binding surfaces of Ube2g1 (bottom and L-side) are very similar to those in Ube2d2 (data not shown). The presence of three ubiquitin binding surfaces in Ube2g1 (Kd = 1.25 ± 0.39, 1.32 ± 0.40, and 1.46 ± 0.36 mm for bottom, L-side, and loop regions, respectively) can explain the lower apparent Kd value (0.56 ± 0.17 mm) that was previously determined from the CSP experiments using [15N]UB and Ube2g1 (12). The acidic loop in Ube2g1 is similar to the previously characterized weak ubiquitin-binding site 1 (UBS1) in the C-terminal tail of Ube2r1 (DLFYDDYYED) (26), which also consists of hydrophobic and acidic residues (Fig. 1). The T1, T2, and 1H,15N heteronuclear NOE values measured for Ube2g1 showed that the acidic loop region is more flexible than the bulk of the structured E2 core (data not shown); this finding is consistent with recent NMR 15N relaxation data for Ube2g2 (27).
FIGURE 4.
Comparison of the non-covalent ubiquitin-binding surfaces of Ube2g1 and Ube2g1Y102G/Y104G. A, ubiquitin binding surfaces of Ube2g1 as determined by CSP experiments in the presence of 0.5 mm ubiquitin. The magenta, red, orange, and yellow colors, respectively, indicate high to low magnitude of CSP. The peaks of Phe-25, Ala-27, and Ser-126 disappeared upon the addition of 0.5 mm ubiquitin and are in green. Residues in black represent prolines, which are not visible in the HSQC spectrum due to the absence of an amide proton. The original peak intensity of Leru-112 was too weak to determine the CSP and is, therefore, also in black. The ubiquitin binding surface of Ube2g1 can be separated into three parts, namely the bottom, L-side, and loop. The non-covalent ubiquitin binding affinities (Kd values) were measured by titrations with ubiquitin to be 1.25 ± 0.39, 1.32 ± 0.40, and 1.46 ± 0.36 mm for the bottom, L-side, and loop regions, respectively. B, the CSP data for [15N]Ube2g1 and [15N]Ube2g1Y102G/Y104G in the presence of 0.5 mm ubiquitin are represented as a histogram. The striped bars represent residues for which the resonance peaks disappeared in the presence of 0.5 mm ubiquitin. The negative black and gray bars represent, respectively, Pro residues or residues where peak intensities were too weak to be detected in the CSP experiment. Note that Ube2g1Y102G/Y104G lost ubiquitin interactions only in the acidic loop region.
Sequence alignment of Ube2g1 and Ube2r1C revealed two critical differences in their acidic loop regions: Tyr-102 and Tyr-104 in Ube2g1 correspond to Gln-105 and Gly-107 in Ube2r1 (Fig. 1). To assess the effect of Tyr-102 and Tyr-104 on non-covalent ubiquitin binding by Ube2g1, both were mutated to Gly residues (Ube2g1Y102G/Y104G). After the assignment of the backbone chemical shifts of [13C,15N]Ube2g1Y102G/Y104G, CSPs were measured upon titration with unlabeled ubiquitin. Although the ubiquitin binding properties of the bottom and L-side regions of the mutant of Ube2g1 were unchanged, the CSPs from the loop region completely disappeared (Fig. 4B). Thus, the two Tyr residues are critical for non-covalent ubiquitin binding of the acidic loop in Ube2g1.
Introduction of Two Tyr Residues into the Ube2r1C Acidic Loop Increased Both Ubiquitin Binding Affinity and Lys-48-ubiquitylation Activity
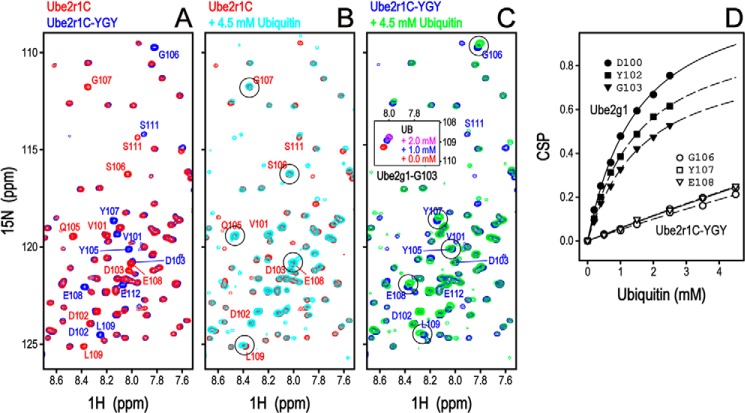
We changed Gln-105–Ser-106–Gly-107 in Ube2r1C to Tyr-105–Gly-106–Tyr-107 (Ube2r1CYGY) to make its acidic loop more similar to that of Ube2g1 and then assessed the in vitro ubiquitylation activity. The introduction of two Tyr residues into the Ube2r1C acidic loop greatly increased Lys-48-ubiquitylation activity to a level close to that of Ube2g1 (Fig. 2C). Non-reducing SDS-PAGE showed that the activity differences between wild-type Ube2r1C and Ube2r1CYGY were not the result of impaired E2∼UB thioester formation (Fig. 2C, right panel).
It has been reported that the C-terminal extension of Ube2r1 includes two weak ubiquitin binding motifs (UBS1, residues 205–214, and UBS2, residues 216–225) and an additional possibility of a third, very weak ubiquitin-binding site (residues 187–196) (26). However, Ube2r1C alone did not detectably bind to [15N]UB (data not shown); the low background with Ube2r1C made identification of ubiquitin binding differences between Ube2r1C and Ube2r1CYGY straightforward. Comparison of the HSQC spectra between Ube2r1C and Ube2r1CYGY showed that the effect of Tyr-105–Gly-106–Tyr-107 substitution was limited to the backbone chemical shifts only of the acidic loop residues (Fig. 5A). The CSP data showed that the acidic loop of Ube2r1CYGY, but not Ube2r1C, had a non-covalent interaction with ubiquitin (Fig. 5). However, the CSP magnitudes of the [15N]Ube2r1CYGY acidic loop were very small even in the presence of 4.5 mm ubiquitin, and thus the exact affinity for ubiquitin could not be determined. This low ubiquitin binding affinity together with the high Lys-48-ubiquitylation activity of Ube2r1CYGY compared with Ube2r1C suggested a possible role for the acidic loop in an intramolecular interaction with E2 thioester-linked donor ubiquitin.
FIGURE 5.
HSQC perturbation experiment of [15N]Ube2r1C and [15N]Ube2r1CYGY in the presence of ubiquitin. A, substitution of the Gln-105–Ser-106–Gly-107 residues in the Ube2r1C acidic loop by the Tyr-102–Gly-103–Tyr-104 sequence from Ube2g1 changed the chemical shifts of several residues. The HSQC peaks that originated from the flexible acidic loop (residues 101–112) and the N- and C-terminal regions (residues 1–6 and 182–183) were assigned by using the 13C,15N-labeled protein. Although the chemical shifts of the region, including the Gln-105–Ser-106–Gly-107 residues of Ube2r1C, were mostly unchanged even in the presence of 4 mm ubiquitin (B), the region changed to Tyr-105–Gly-106–Tyr-107 was clearly perturbed by the addition of ubiquitin (C). The HSQC spectrum of the region representing the Gly-103 residue of [15N]Ube2g1 in the presence of 0.0, 1.0, and 2.0 mm ubiquitin are shown for the reference of CSP amount (inset). D, [15N]Ube2g1 or [15N]Ube2r1CYGY is titrated by ubiquitin, and the titration curves of the selected residues are plotted.
Synthesis of E2-UB Oxyester for NMR Experiments
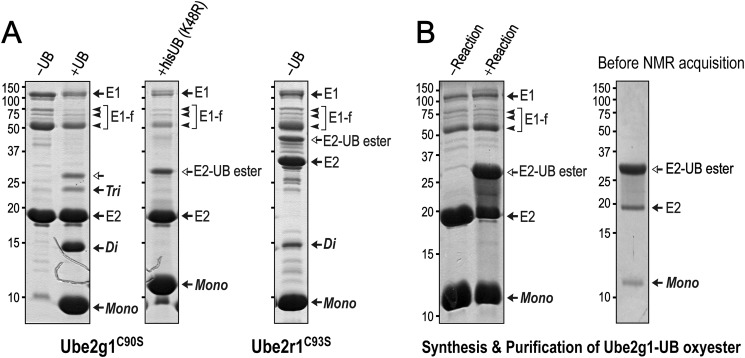
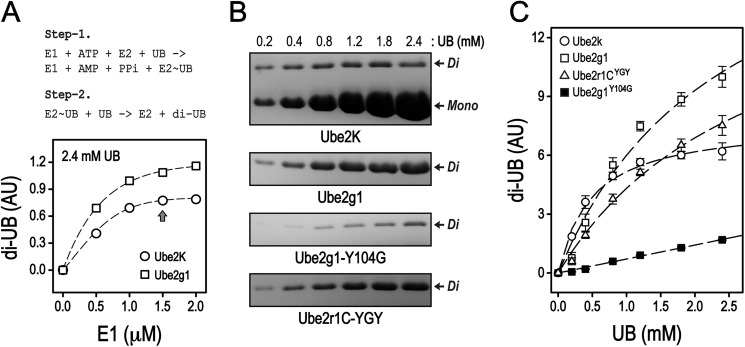
To characterize the intramolecular interaction between the acidic loop and the E2-attached donor ubiquitin used for Lys-48-ubiquitylation, we tried to obtain more stable E2-UB complexes as oxyester adducts. Crystal structures of the Ube2d2-UB oxyester alone (28) and its complex with a HECT-family E3 ligase (29) have been reported, and NMR studies on Ube2d3 (UbcH5c)-UB and Ubc13-UB oxyesters have been described (30). Interestingly, using a high concentration of E1, the ubiquitin-charging reaction with Ube2g1C90S at 35 °C produced not only the Ube2g1C90S-UB oxyester but also appreciable amounts of Lys-48-linked diubiquitin. In comparison, the same reaction using Ube2r1CC93S required less E1 and mostly produced the Ube2r1CC93S-UB oxyester and only a small amount of diubiquitin (Fig. 6A). These results are evidence that the E2-UB oxyester is a functional mimic of the labile E2∼UB thioester. The ubiquitin K48R mutant (UBK48R) was used to prepare the [15N]Ube2g1C90S-UBK48R oxyester on a large scale, but spontaneous hydrolysis of the E2-UB oxyester was detected during the purification even at 4 °C (Fig. 6B). We monitored the spontaneous hydrolysis at 25 °C of the Ube2g1C90S-UBK48R and Ube2r1CC93S-UBK48R oxyesters by SDS-PAGE analysis and found that the Ube2r1CC93S-UBK48R oxyester (t½ ∼ 20 h) was more stable than the Ube2g1C90S-UBK48R oxyester (t½ ∼ 5 h) (Table 1).
FIGURE 6.
Synthesis and purification of the Ube2g1C90S-UBK48R oxyester. A, the in vitro ubiquitylation of Ube2g1C90S with ∼2.0 μm E1 at 35 °C and pH 8.0 produced not only the Ube2g1C90S-UB oxyester but also appreciable amounts of diubiquitin (Di). The Ube2g1C90S-UBK48R oxyester was synthesized with His-tagged UBK48R instead of wild-type UB. The same reaction using Ube2r1C93S mostly produced the Ube2r1C93S-UB oxyester intermediate, and a small amount of diubiquitin product also was detected. B, a longer incubation with concentrations of ∼2.0 μm E1, 200 μm E2, and 400 μm UBK48R produced a large amount of the Ube2g1C90S-UBK48R oxyester (left panel). His-tagged UBK48R was used to facilitate purification of the E2-UB oxyester product. However, the stability of the E2-UB oxyester was relatively low, and its spontaneous hydrolysis was apparent during the purification even at 4 °C (right panel).
TABLE 1.
Spontaneous hydrolysis rates of various E2-UB oxyesters at 25 °C
| E2-UBK48R oxyester | Half-life |
|---|---|
| h | |
| Ube2g1C90S-[15N]UBK48R | 8.8 ± 1.1a |
| Ube2g1C90S/Y102G/Y104G-[15N]UBK48R | 5.8 ± 0.2a |
| Ube2g1C90S-UBK48R | 6.4 ± 1.7b |
| Ube2g1C90S/Y102G/Y104G-UBK48R | 2.8 ± 0.4b |
| Ube2r1C93S-UBK48R | 24.3 ± 5.9b |
| Ube2r1C93S/YGY-UBK48R | 20.0 ± 6.5b |
a The donor ubiquitin was 13C/2H/15N-labeled; the hydrolysis rates (shown here as t½) in buffer (pH 7.0, 50 mm HEPES and 100 mm NaCl) were determined from the several well separated HSQC peaks.
b The hydrolysis rates (i.e., t½ values) at pH 8.0 were determined from two or more separate measurements using non-reducing SDS-PAGE analysis.
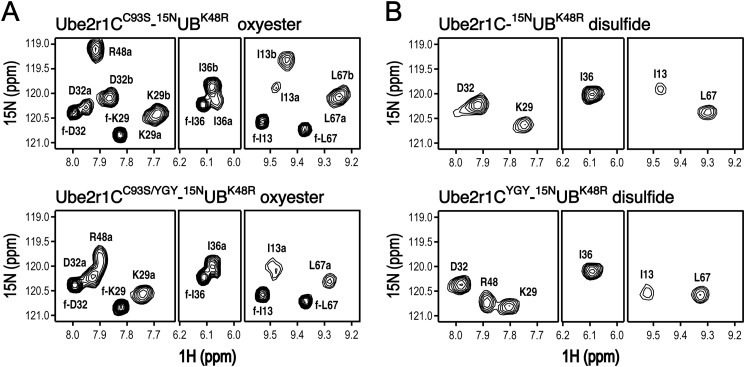
The Donor Ubiquitin of Ube2r1CC93S-[15N]UBK48R Oxyester Exhibited Two-state Conformational Exchange, Whereas the Ubiquitin in the Ube2r1CC93S/YGY-UBK48R Oxyester Preferred One Conformation
Unfortunately, the very complex HSQC spectrum of the [15N]Ube2r1CC93S-UBK48R oxyester made detailed characterization by NMR difficult. The peak intensities of the HSQC spectra overall were very weak and inhomogeneous (data not shown). For more clear analyses by NMR of the oxyester adducts, we reduced the number of HSQC peaks by preparation of the Ube2r1CC93S-[15N]UBK48R and Ube2r1CC93S/YGY-[15N]UBK48R oxyesters. We also prepared chemically stable Ube2r1C-[15N]UBK48R/G76C and Ube2r1CYGY-[15N]UBK48R/G76C disulfide adducts, as Ube2r1C contains only one Cys residue (i.e. Cys-93 at the active site). Interestingly, the HSQC spectra of disulfide-linked Ube2r1C-[15N]UBK48R/G76C were greatly different from those of the Ube2r1CC93S-[15N]UBK48R oxyester (Fig. 7). The HSQC peaks from [15N]UBK48R in the oxyesters were much less intense than those from the disulfide-linked [15N]UBK48R/G76C due to motion (i.e. conformational exchange) in an intermediate-to-slow NMR time scale that caused severe broadening and disappearance of the peaks. The ubiquitin CSPs of the oxyester relative to the chemical shifts of free [15N]UBK48R and also the CSPs of the disulfide adducts relative to free [15N]UBK48R/G76C showed that the [15N]UBK48R moiety of the oxyester had much larger CSPs than with the disulfide-linked [15N]UBK48R/G76C (Fig. 8, A and B). Many peaks of the Ube2r1CC93S-[15N]UBK48R oxyester were split into two sets; these corresponded to the major hydrophobic surface patch that includes residue Ile-44 of ubiquitin. However, with the Ube2r1CC93S/YGY-[15N]UBK48R oxyester, these resonances were not split and remained as one set (Fig. 7A). This effect was much smaller or absent with the disulfide adducts where possible splittings in only a few peaks were barely identified for the Ube2r1C-[15N]UBK48R/G76C disulfide but not for the Ube2r1CYGY-[15N]UBK48R/G76C disulfide (Fig. 7B). Overall, evidence of intramolecular interaction between the donor ubiquitin and Ube2r1C was greatly reduced with the disulfide adduct relative to the oxyester adduct. A possible explanation is that, compared with the E2-UB oxyester linkage, the E2-UB disulfide is a poorer mimic of the physiological thioester (e.g. it is longer by approximately one S atom) and may interfere with the normal intramolecular interaction between the acidic loop and the donor ubiquitin.
FIGURE 7.
HSQC spectra of Ube2r1CC93S- and Ube2r1CC93S/YGY-[15N]UBK48R oxyesters (A) and Ube2r1C- and Ube2r1CYGY-[15N]UBG76C disulfides (B). A, selected regions of the HSQC spectra show that the attached donor ubiquitin of the Ube2r1CC93S-[15N]UBK48R oxyester has two different conformations (top panel) but that of the Ube2r1CC93S/YGY-[15N]UBK48R oxyester has one predominant conformation (bottom panel). Residues labeled with an f- are residues from ubiquitin that had dissociated from the E2. B, the same regions of the HSQC spectra of disulfide-linked Ube2r1C-[15N]UBK48R/G76C and Ube2r1CYGY-[15N]UBK48R/G76C show that in both complexes the attached donor ubiquitin mostly has a single conformation and that the chemical shift differences from free ubiquitin are much smaller than with the oxyester complexes.
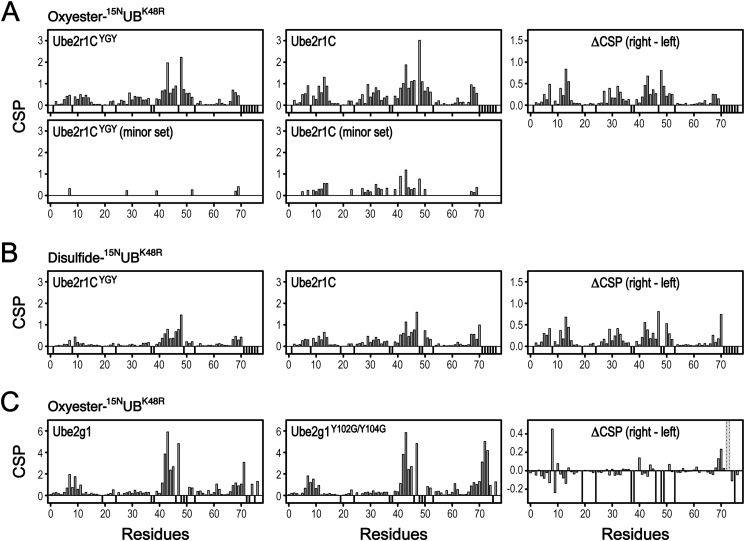
FIGURE 8.
Chemical shift perturbations relative to free [15N]UB of [15N]UB resonances in the Ube2r1CC93S/YGY-[15N]UBK48R and Ube2r1CC93S-[15N]UBK48R oxyesters (A), Ube2r1CYGY-[15N]UBK48R/G76C and Ube2r1C-[15N]UBK48R/G76C disulfides (B), and Ube2g1C90S-[15N]UBK48R and Ube2g1C90S/Y102G/Y104G-[15N]UBK48R oxyesters (C). The chemical shifts of free [15N]UBK48R and [15N]UBK48R/G76C were used to determine the CSPs of E2-[15N]UBK48R oxyesters and disulfides, respectively. The chemical shift differences between the attached [15N]UB molecules of the wild-type and the acidic-loop mutant E2 are shown in the right panels. The negative bars represent the residues including prolines (Pro-19, Pro-37, and Pro-38) for which peaks were not shown in the HSQC spectra. The dotted bars in C (right panel) represent the residues apparent in the HSQC spectra of the Ube2g1C90S-[15N]UBK48R oxyester but not the Ube2g1C90S/Y102G/Y104G-[15N]UBK48R oxyester.
The HSQC spectra indicated that the ubiquitin in the Ube2r1CC93S-[15N]UBK48R oxyester has two different conformations but that one conformation is preferred in the YGY-mutant complex (i.e. the Ube2r1CC93S/YGY-[15N]UBK48R oxyester). The major set of peaks in the HSQC spectrum of the Ube2r1CC93S-[15N]UBK48R oxyester displayed much larger CSPs than did the minor set, whereas the Ube2r1CC93S/YGY-[15N]UBK48R oxyester displayed one set of peaks whose CSP magnitudes were similar to those of the minor set in the HSQC spectrum of the Ube2r1CC93S-[15N]UBK48R oxyester (Fig. 8A). The absence of apparent non-covalent UB binding activity in Ube2r1C can explain why the donor ubiquitin exhibits (at least) two different conformations in its Ube2r1CC93S-[15N]UBK48R oxyester form. Both weak intermolecular interactions can be entropically stabilized in an intramolecular association. The intensified interactions of the donor ubiquitin with both the L-side and the acidic loop of Ube2r1C, by competing with each other, can produce two sets of HSQC peaks. The introduction of the two Tyr residues into the acidic loop of Ube2r1C likely shifts the equilibrium by stabilizing the interaction between the donor UB and the acidic loop. However, we cannot exclude a possibility that the preferred conformation is caused by other mechanisms, such as a conformational change accompanying binding. The chemical shift differences between Ube2r1CC93S-[15N]UBK48R and Ube2r1CC93S/YGY-[15N]UBK48R oxyesters showed that the interaction between the E2 and the hydrophobic patch of the donor ubiquitin was reduced more for Ube2r1CC93S/YGY than for Ube2r1CC93S (Fig. 8A).
Proposed Role of the Ube2g1 Acidic Loop for the Efficient Lys-48-ubiquitylation in the Absence of E3
We prepared Ube2g1-UB oxyesters to obtain more detailed information about the intramolecular interaction between Ube2g1 and the donor ubiquitin. Interestingly, the HSQC spectrum of the [15N]Ube2g1C90S-UBK48R oxyester was well dispersed (Fig. 9A). Previous reports have shown that the [15N]Ube2d3∼UB thioester mostly lost discernible HSQC peaks due to intermolecular interaction mediated by the attached donor ubiquitin and another [15N]Ube2d3∼UB (31). The donor ubiquitin of the Ube2d3C85S/S22R-[15N]UB oxyester has been found in an extended and dynamic state, referred to as an “open conformation,” that is not favorable for its ubiquitylation activity (30). Ube2g1 has UB binding surfaces similar to Ube2d3; in Ube2d3∼UB, these promote the self-association of Ube2d3∼UB. However, because there was no disappearance or significant decrease of HSQC peaks of the UB-oxyester when compared with [15N]Ube2g1, the [15N]Ube2g1C90S-UB oxyester apparently lacks the intermolecular interaction.
FIGURE 9.
Assignment of HSQC spectrum of [15N]Ube2g1C90S-UBK48R oxyester. A, two sets of 1H,15N HSQC peaks were identified resulting from the [15N]Ube2g1-UBK48R oxyester and free [15N]Ube2g1 (labeled with f-), respectively. Nevertheless, the well dispersed spectrum of the purified [15N]Ube2g1C90S-UBK48R oxyester shows the absence of an intermolecular interaction between the attached donor ubiquitin and different Ube2g1 molecules. Inset, interestingly, the peak from the Gly-103 residue of the [15N]Ube2g1-UBK48R oxyester was not strongly perturbed by additional free ubiquitin (0.6 mm) compared with that of free [15N]Ube2g1, suggesting that the Gly-103 residues were affected differently by added (i.e. acceptor) ubiquitin in the presence of the oxyester-attached donor ubiquitin. B, the HSQC-peaks assignment of [15N]Ube2g1C90S-UBK48R oxyester was performed by comparing the HSQC spectra with [15N]Ube2g1C90S-UBK48R oxyester and with [15N]Ube2g1C90S hydrolyzed from its oxyester adduct. The residues displaying very similar peak positions in the HSQC spectrum for the oxyester and free form were assigned unambiguously, and some of additional residues were also assigned based on the HNCO spectrum. C, the CSPs of [15N]Ube2g1C90S between the oxyester and free form are shown as a bar plot. The residues for which chemical shifts were too different between the oxyester and free form to be clearly assigned are indicated using green bars.
Although we could not fully assign the HSQC peaks of the [15N]Ube2g1C90S-UBK48R oxyester, comparisons of the HSQC and three-dimensional-HNCO spectra between [15N]Ube2g1C90S-UBK48R oxyester and free [15N]Ube2g1C90S made it possible to determine the interaction surface on Ube2g1 for the attached donor ubiquitin (Fig. 9). The HSQC peaks of the acidic loop and many other residues, especially those on the L-side, were changed too much to be traced reliably from the spectrum of free [15N]Ube2g1C90S to the [15N]Ube2g1C90S-UBK48R oxyester (Figs. 9C and 10A). This result contrasts with Ube2d3C85S/S22R-[15N]UB oxyester (30) in which the chemical shift differences between the oxyester-bound and free [15N]UB were relatively small. To determine in more detail the binding mode of the donor ubiquitin in the Ube2g1-UB oxyester, we also prepared oxyester complexes of Ube2g1C90S and Ube2g1C90S/Y102G/Y104G using [13C,2H,15N]UBK48R. Due to their low stability in solution and larger chemical shift changes compared with TROSY peaks of free [13C,2H,15N]UBK48R, the 1H,15N TROSY peaks of Ube2g1C90S-[13C,2H,15N]UBK48R were assigned using additional TROSY-based three-dimensional-HNCO spectra. From time-dependent changes in the TROSY spectra, we also determined the half-lives of [15N]Ube2g1C90S- and [15N]Ube2g1C90S/Y102G/Y104G-[2H,15N]UBK48R oxyesters to be 8.8 ± 1.1 and 5.8 ± 0.2 h, respectively (Table 1). The comparison of TROSY peaks from free and attached [13C,2H,15N]UBK48R showed that the hydrophobic patch of the attached donor ubiquitin interacts with the L-side of Ube2g1 (Fig. 8C).
FIGURE 10.
The interaction surfaces between UBK48R and Ube2g1C90S mapped on a model of the oxyester complex. A, intramolecular UBK48R-interaction surfaces on Ube2g1C90S are highlighted on the structure of free Ube2g1. The red and yellow colors, respectively, indicate high and low magnitude CSPs. The residues in which the HSQC peaks shifted too much to be traced are shown in green (see Fig. 9C). B, Ube2g1C90S-interacting surface of UBK48R is highlighted using magenta, red, orange, and yellow colors to indicate high to low magnitude of CSPs, respectively. The residues colored black represent prolines and other residues that were invisible in the HSQC spectrum, and the green-colored residues indicate where resonances disappeared in the oxyester complex. C, the residues of UBK48R showing different chemical shifts between the oxyesters with Ube2g1C90S and Ube2g1C90S/Y102G/Y104G are highlighted as in panel B. D, the residues of Ube2g1 mutated for the ubiquitylation assays (Fig. 12) are shown superimposed onto a ribbon representation of the structure. E, surface presentation shows the donor UBK48R-occupied surface of the E2.
The CSP-based Rosetta docking model of the acidic loop-deleted Ube2g1C90S-UBK48R oxyester together with additional fitting of the loop based on CSPs provided structural insight about the intramolecular interaction between the E2 acidic loop and the attached donor ubiquitin (Fig. 10). Although the exact modeling of the acidic loop structure in the Ube2g1C90S-UBK48R oxyester was difficult, the geometric constraint of the oxyester bond shows that the YGY segment of the acidic loop is likely to be located in the vicinity of Arg-74 of the donor ubiquitin. The active site of the Ube2g1C90S-UBK48R oxyester appears to be accessible only through the R-side of Ube2g1 because the L-side was completely occupied by the attached donor ubiquitin (Fig. 10E). Our data clearly supported the idea that an acceptor ubiquitin approaches the R-side of Ube2g1; this result agrees well with the recently reported docking model of Ube2S∼UB thioester in which L-side positioning of the donor ubiquitin is important for the activity of Lys-11 ubiquitin-linkage formation (7). The chemical shift differences of [15N]UBK48R molecules between Ube2g1C90S-[15N]UBK48R and Ube2g1C90S/Y102G/Y104G-[15N]UBK48R oxyesters were much smaller than those between the free and attached [15N]UBK48R. The acidic loop mutation (Y102G/Y104G) changed the chemical shifts of specific residues of donor [15N]UBK48R that were localized along one edge of the hydrophobic patch extending to the C terminus (Fig. 8C and Fig. 10C). Therefore, the intramolecular interaction between the donor ubiquitin and the L-side of Ube2g1 seemed overall to be identical for both Ube2g1C90S and Ube2g1C90S/Y102G/Y104G. Thus, the complex structure (Fig. 10C) caused by the intramolecular interaction between the donor ubiquitin and the L-side of Ube2g1 or Ube2g1Y102G/Y104G is not the only requirement for Ube2g1 ubiquitylation activity. Our model for the Ube2g1-UB oxyester suggests that the interaction of the acidic loop with the donor ubiquitin helps to stabilize binding of the acceptor ubiquitin. The interaction surface for the acceptor ubiquitin is located on the R-side and is composed of the C-terminal region of the donor ubiquitin, the region of the acidic loop that includes residues Glu-99 and Glu-100, and additional Ube2g1 residues near Cys-90 at the active site.
To estimate the Km values for ubiquitin in E3-independent ubiquitylation reactions by Ube2g1, Ube2g1Y104G, and Ube2r1CYGY, we measured the steady-state rates of diubiquitin formation as a function of ubiquitin concentration (Fig. 11). The Km and kcat values for E3-independent ubiquitylation by Ube2k were reported previously to be 0.58 mm and 0.56 min−1, respectively (11); thus, ubiquitylation by Ube2k was used as a reference to validate our steady-state kinetic analysis. The Km value (∼0.5 mm) we determined for Ube2k agreed well with the reported value. Although we could not determine exact Km values due to a practical upper limit of several millimolar for the ubiquitin substrate in our assays, it was apparent that the Km for Ube2g1Y104G is at least severalfold greater than that of the wild-type Ube2g1 (Fig. 11). The Km data suggest that Tyr-104 in the acidic loop is important for the interaction of Ube2g1∼UB thioester with an acceptor ubiquitin. Importantly, the HSQC peak of Gly-103 in the [15N]Ube2g1C90S-UBK48R oxyester form was shifted much less by the addition of free ubiquitin than that in [15N]Ube2g1 alone (Fig. 9A, inset), which suggests that the intermolecular interaction between the acidic loop and an acceptor ubiquitin in the context of the Ube2g1-UB oxyester is different from that with free Ube2g1.
FIGURE 11.
In vitro ubiquitylation by Ube2k, Ube2g1, Ube2g1Y104G, or Ube2r1CYGY in the presence of various concentrations of ubiquitin. A, the reaction of di-UB synthesis can be divided into two steps (I and Step-2). If the step-1 is faster than the step-2, the concentration of E2∼UB is essentially constant (dt[E2∼UB]/dt ≅ 0). The ubiquitylation activity of Ube2k and Ube2g1 was assessed at pH 7.5 in the presence of increasing concentrations of E1 and with 2.4 mm ubiquitin. The E1 concentration used was ∼1.5 μm for further steady-state kinetic analysis of E3-independent ubiquitylation. AU, absorbance units. B, the in vitro ubiquitylation reactions were performed in the presence of increasing concentration of the substrate ubiquitin with fixed concentrations of E1 and E2. The amount of the diubiquitin product bands increased as the concentration of the substrate ubiquitin increased. C, the Km values of E2-mediated ubiquitylation were estimated for Ube2k, Ube2g1, Ube2g1Y104G, and Ube2r1CYGY from the modified Michaelis-Menten equation (see “Experimental Procedures”). The Km value of Ube2K, Ube2g1, or Ube2r1YGY was 0.5 ± 0.06, 2.5 ± 0.7, or 2.9 ± 0.7 mm, respectively. The Km value of Ube2g1Y104G was too high to be determined by this experiment.
Mutation in the Acidic Loop Region Changed the in Vitro Ubiquitylation Activity of Ube2g1
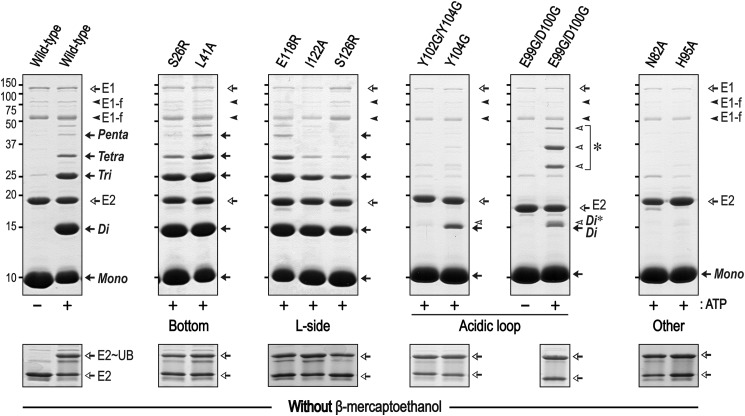
We introduced various point mutations into each ubiquitin-interaction site (bottom, S26R and L41A; L-side, E118R, I122A, and S126R; loop, Y104G, Y102G/Y104G, and E99G/D100G) to explore their effects on the ubiquitylation activity. Our rationale for choosing these mutations is as follows. Mutations in specific Ube2g1 residues (S26R and L41A in the bottom; E118R in the L-side) were determined from the sequence alignment with Ube2r1 (Fig. 1), because Ube2r1C did not display any detectable non-covalent ubiquitin binding. Ser-26 corresponds to Ser-22 of Ube2d3 (UbcH5c), and Ube2d3S22R greatly decreases the ubiquitin binding activity (31). The I122A mutation corresponds to the L125A mutation of Ube2S; it was reported that Leu-125 is important for Lys-11-ubiquitylation activity by positioning the donor ubiquitin into the L-side of Ube2S (7). The results in Fig. 12 show that only the mutations in the acidic loop resulted in severe loss of Lys-48-ubiquitylation activity; none of the mutations in the bottom or L-side of Ube2g1 had significant defects in the ubiquitylation activity. The lower ubiquitylation activity did not result from an impaired formation of E2∼UB thioester by E1 enzyme (Fig. 12, bottom panel). We additionally confirmed that the time-course activations of Ube2g1, Ube2g1S126R, and Ube2g1Y104G by E1 enzyme were almost identical, and their productions of diubiquitin species were gradually increased as increasing the incubation time from 0 to 2 h (data not shown).
FIGURE 12.
In vitro ubiquitylation activities of Ube2g1 mutants in the absence of E3 ligase. The in vitro ubiquitylation reaction was performed using E1 (∼0.5 μm), E2 (20 μm), and ubiquitin (0.3 mm) in a 50 mm Tris-HCl (pH 7.5) buffer. Ubiquitin and ubiquitin oligomers are marked Mono, Di, Tri, etc. Ube2g1 mutants were generated in any of three independent non-covalent ubiquitin binding regions (S26R and L41A for the bottom; E118R, I122A, and S126R for the L-side; Y104G, Y102G/Y104G, E99G/D100G for the acidic loop) or two residues potentially involved directly in catalysis (N82A and H95A). Only the mutations in the ubiquitin binding region of the acidic loop and in the potential catalytic residues displayed a severe defect in E3-independent ubiquitylation. An additional diubiquitin band (Di*) was identified for the Ube2g1 mutants that had decreased activity. Note that, although the apparent molecular size of the Ube2g1E99G/D100G determined by SDS-PAGE analysis was slightly lower than that of other Ube2g1 proteins, the correct molecular mass of 18,526 Da was confirmed by MALDI-TOF mass spectrometry. The reaction with Ube2g1E99G/D100G produced unidentified high molecular sized bands (marked with an asterisk) that probably reflect auto-ubiquitylation of the E2. The formation of Ube2g1∼UB thioesters was assessed by non-reducing SDS-PAGE analysis (bottom). AU, absorbance units.
The activity of the Y104G mutant was lower than that of the wild type, and the Y102G/Y104G mutant was nearly inactive. Other acidic loop mutations in Ube2g1 (i.e. E99G/D100G) also abolished ubiquitylation activity, which is in agreement with a previous report that showed decreased activity in the acidic loop mutants of yeast Cdc34 in the presence of Skip/Cullin/F-box-E3 (10). Residues Glu-99 and Asp-100 are located in the R-side of Ube2g1 (Fig. 10D). Interestingly, the ubiquitylation reaction catalyzed by Ube2g1E99G/D100G produced additional unidentified protein bands (shown by an asterisk in Fig. 12); these most likely reflect autoubiquitylation of the E2 itself.
Recent NMR studies of Ube2g2 proposed that His-94, which is positioned close to Cys-89 in the active site, is likely to have a catalytic role (27). His-94 corresponds to His-95 of Ube2g1 and is also conserved in Ube2r1 (Fig. 1). We confirmed that this His residue, like the Asn-82 residue that is evolutionarily conserved among E2 proteins and is important for the formation of a ubiquitin-protein linkage (32), is important for Lys-48-ubiquitylation activity. Either H95A or N82A mutants of Ube2g1 completely eliminated the ubiquitylation activity (Fig. 12). Other Ube2g1 His-95 mutants (H95R and H95E) similarly failed to show detectable activity (data not shown).
DISCUSSION
We have shown that the intramolecular interaction between the acidic loop of Ube2g1 and the donor ubiquitin plays an important role in the Lys-48-ubiquitylation activity of E2 enzymes. Ubiquitin binding properties of the acidic loop were investigated with two E2∼UB thioester mimics, E2-UB oxyesters, and disulfides, and then the inferences regarding E2 Lys-48-ubiquitylation activity were confirmed by enzymatic assays using various point mutants. The Ube2g1-UB oxyester can serve as a functional analog of the labile E2∼UB thioester intermediate. Ube2g1C90S and Ube2r1CC93S/YGY were able to produce Lys-48 diubiquitin.
The presence of the cognate RING-E3 ligase restricts the dynamics of the donor ubiquitin in the Ube2d3-UB oxyester to a position (closed conformation) that is more favorable for the ubiquitylation reaction (33). Moreover, donor ubiquitins on Ube2S (7), Ubc1 (the yeast homolog of Ube2k) (22), and Ube2r1 (23) have been proposed to adopt closed conformations predicted to facilitate ubiquitylation. We have shown here that the donor ubiquitin in the Ube2g1-UB oxyester interacts with the L-side of Ube2g1 and thus adopts a closed conformation that is similar to the models suggested for Ube2S, Ubc1, and Ube2r1. However, adoption of the closed conformation by the donor ubiquitin through its interaction with the L-side of E2 does not seem to be critical for Lys-48-ubiquitylation by Ube2g1. The donor ubiquitin in Ube2g1C90S/Y102G/Y104G-UBK48R oxyester also can adopt the closed conformation, although no ubiquitylation activity was observed for the Ube2g1Y102G/Y104G mutant. Moreover, the introduction of two Tyr residues into the acidic loop of Ube2r1C stabilized a conformation of the donor ubiquitin that, according to NMR CSP data, reflected a decreased interaction between the hydrophobic patch of the donor ubiquitin and the L-side of Ube2r1CYGY (Fig. 8A). Although both Ube2g1C90S and Ube2r1CC93S/YGY displayed appreciable Lys-48-ubiquitylation activity, the configurations of their donor ubiquitin in the oxyester forms appeared to differ. Smaller CSPs (relative to free [15N]UBK48R) with the Ube2r1CC93S/YGY-[15N]UBK48R oxyester than with the Ube2g1C90S-[15N]UBK48R oxyester indicate reduced intramolecular interaction between the L-side of Ube2r1CC93S/YGY and the hydrophobic patch of the donor ubiquitin. For Ube2g1 or Ube2r1CYGY Lys-48-ubiquitylation activity, it is likely that the local structure formed by the acidic loop and the C-terminal region of the attached donor ubiquitin is more important than the simple positioning of the donor ubiquitin onto the L-side of the E2. These results suggest that Ube2g1, Ube2r1, and Ube2k, which are known to generate Lys-48-linked polyubiquitin efficiently without E3, seem to use a different mechanism to conjugate acceptor and donor ubiquitin. Other evidence supporting this idea is that the UBN60A mutant displayed different Lys-48-ubiquitylation activities as an acceptor for Ube2k or Ube2g1, showing a severe defect with Ube2k, whereas ubiquitylation by Ube2g1 was relatively unaffected by the ubiquitin N60A mutation (12).
In the context of Ube2g1-UB oxyester, the acidic loop mutation (Y102G/Y104G) changes the chemical environment of the C-terminal region and the edge of the hydrophobic patch in the attached ubiquitin (Fig. 8C and Fig. 10C). The estimated Km values of Ube2g1 and Ube2g1Y104G showed that the acidic loop mutation also affected recruitment of the acceptor ubiquitin (Fig. 11). Moreover, production of a tiny amount of diubiquitin with linkage(s) other than to Lys-48 was identified with the E99G/D100G mutation (Fig. 12, see gel band indicated as Di*); this suggests that the acidic loop also may play an important role in proper alignment of an acceptor ubiquitin. The production of non-Lys-48-linked diubiquitin was reported for Ube2d2 and Ube2b in the absence of E3 (8).
The acceptor ubiquitin can approach the active Cys residue via the R-side of Ube2g1∼UB thioester. Recently Ube2g2, a homolog of Ube2g1, was shown to form a functional dimer, and that residue Cys-48 residue is important for the dimerization (34). Because the Cys-48 residue is not conserved in Ube2g1 (Fig. 1) and Ube2g1 ubiquitylation activity is independent of the E2 concentration (Fig. 3), the mechanisms of ubiquitylation between Ube2g1 and Ube2g2 would be different from each other. The cognate E3 ligase(s) of Ube2g1 has not yet been identified; therefore, we have fit our model structure of the Ube2g1-UB oxyester (Fig. 10) to the Ube2g2·gp78-E3 complex structure reported by Das et al. (35). A fit was obtained without any steric clash and in which the R-side of Ube2g1, the candidate surface for an acceptor ubiquitin, is well exposed to solvent (not shown).
Interestingly, the recent docking model of Ube2r1∼UB thioester that was based on the Rosetta force field (and does not contain the acidic loop or the C-terminal extension) shows that the donor ubiquitin is located at the L-side region of Ube2r1, which is similar to our model for the Ube2g1C90S-UBK48R oxyester, where Ile-44 of the donor ubiquitin is close to Ser-129 of Ube2r1, and thus the I44A mutation of the donor ubiquitin can be suppressed by the S129L mutation of Ube2r1 (23). The introduction of two Tyr residues in the acidic loop of Ube2r1C reduced the interaction between the donor ubiquitin and the L-side of Ube2r1CYGY but increased the Lys-48-ubiquitylation activity. The acidic loop of Ube2r1 displayed a lower binding affinity to the donor ubiquitin than did that of Ube2g1, and thus the acidic loop and the C-terminal tail of Ube2r1 might work together during the catalysis. It might be possible that the residues 184–196 of Ube2r1 stabilize the conformation of the donor ubiquitin in a manner similar to this Ube2r1∼UB thioester docking model (23) and thus facilitate the recruitment of an acceptor ubiquitin. Ube2r1 could adapt to its specific function in the presence of SCE-E3 by losing the ubiquitin binding property of its acidic loop.
Acidic loop or tail motifs are known to be important for the ubiquitylation activities of E2 or E3 enzymes; examples are the acidic loops in Ube2g1 and Ube2g2 (36), the acidic tail (residues 888–900) in human Nedd4 (HECT-E3) (37), and both acidic loop and acidic tail in Ube2r1 (10, 26, 38). These motifs possess both acidic and bulky hydrophobic residues and usually are near the active-site Cys residue. Although the acidic tail of Ube2r1 is far from the active site, it is known to interact with a donor ubiquitin of Ube2r1 (26, 39). Although the ubiquitin binding of the acidic tail of Nedd4 has not yet been identified, the others in Ube2g1 and Ube2r1 display a weak ubiquitin binding activity. HECT-E3 ligase could be similar with E2 enzyme, because the active Cys residue forms a labile thioester bond with a donor ubiquitin. We speculate that, as with the acidic loop of Ube2g1, the acidic tail of Nedd4 may interact with donor ubiquitin to enhance its ubiquitylation activity.
This work was supported, in whole or in part, by National Institutes of Health Grant GM097452 (to R. E. C.). This study was also supported by the high-field NMR research program from Korea Basic Science Institute (KBSI; to K.-S. R.) and KBSI Grant T33410 (to H.-K. C.).
- CSP
- chemical shift perturbation
- HSQC
- heteronuclear single quantum coherence
- TROSY
- transverse relaxation optimized spectroscopy
- UB
- ubiquitin
- Ube2r1C
- Ube2r1 (1–183)
- Ube2r1CYGY
- Ube2r1C (Q105Y/S106G/G107Y)
- HSQC
- heteronuclear single quantum correlation.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Pickart C. M. (2004) Back to the future with ubiquitin. Cell 116, 181–190 [DOI] [PubMed] [Google Scholar]
- 3. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 4. Houben K., Dominguez C., van Schaik F. M., Timmers H. T., Bonvin A. M., Boelens R. (2004) Solution structure of the ubiquitin-conjugating enzyme UbcH5B. J. Mol. Biol. 344, 513–526 [DOI] [PubMed] [Google Scholar]
- 5. VanDemark A. P., Hofmann R. M., Tsui C., Pickart C. M., Wolberger C. (2001) Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 105, 711–720 [DOI] [PubMed] [Google Scholar]
- 6. Eddins M. J., Carlile C. M., Gomez K. M., Pickart C. M., Wolberger C. (2006) Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 13, 915–920 [DOI] [PubMed] [Google Scholar]
- 7. Wickliffe K. E., Lorenz S., Wemmer D. E., Kuriyan J., Rape M. (2011) The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ryu K. S., Choi Y. S., Ko J., Kim S. O., Kim H. J., Cheong H. K., Jeon Y. H., Choi B. S., Cheong C. (2008) Direct characterization of E2-dependent target specificity and processivity using an artificial p27-linker-E2 ubiquitination system. BMB Rep. 41, 852–857 [DOI] [PubMed] [Google Scholar]
- 9. Wu K., Kovacev J., Pan Z. Q. (2010) Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell 37, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Petroski M. D., Deshaies R. J. (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123, 1107–1120 [DOI] [PubMed] [Google Scholar]
- 11. Haldeman M. T., Xia G., Kasperek E. M., Pickart C. M. (1997) Structure and function of ubiquitin conjugating enzyme E2–25K: the tail is a core-dependent activity element. Biochemistry 36, 10526–10537 [DOI] [PubMed] [Google Scholar]
- 12. Choi Y. S., Jeon Y. H., Ryu K. S., Cheong C. (2009) 60th residues of ubiquitin and Nedd8 are located out of E2-binding surfaces, but are important for Lys-48 ubiquitin-linkage. FEBS Lett. 583, 3323–3328 [DOI] [PubMed] [Google Scholar]
- 13. Shekhtman A., Ghose R., Goger M., Cowburn D. (2002) NMR structure determination and investigation using a reduced proton (REDPRO) labeling strategy for proteins. FEBS Lett. 524, 177–182 [DOI] [PubMed] [Google Scholar]
- 14. Fiaux J., Bertelsen E. B., Horwich A. L., Wüthrich K. (2004) Uniform and residue-specific 15N-labeling of proteins on a highly deuterated background. J. Biomol. NMR 29, 289–297 [DOI] [PubMed] [Google Scholar]
- 15. Merkley N., Barber K. R., Shaw G. S. (2005) Ubiquitin manipulation by an E2 conjugating enzyme using a novel covalent intermediate. J. Biol. Chem. 280, 31732–31738 [DOI] [PubMed] [Google Scholar]
- 16. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 17. Goddard T. D., Kneller D. G. SPARKY 3. University of California, San Francisco [Google Scholar]
- 18. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 37, D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen Y., Lange O., Delaglio F., Rossi P., Aramini J. M., Liu G., Eletsky A., Wu Y., Singarapu K. K., Lemak A., Ignatchenko A., Arrowsmith C. H., Szyperski T., Montelione G. T., Baker D., Bax A. (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. U.S.A. 105, 4685–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J. (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–W367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song Y., DiMaio F., Wang R. Y., Kim D., Miles C., Brunette T., Thompson J., Baker D. (2013) High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamilton K. S., Ellison M. J., Barber K. R., Williams R. S., Huzil J. T., McKenna S., Ptak C., Glover M., Shaw G. S. (2001) Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure 9, 897–904 [DOI] [PubMed] [Google Scholar]
- 23. Saha A., Lewis S., Kleiger G., Kuhlman B., Deshaies R. J. (2011) Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 25. Gazdoiu S., Yamoah K., Wu K., Escalante C. R., Tappin I., Bermudez V., Aggarwal A. K., Hurwitz J., Pan Z. Q. (2005) Proximity-induced activation of human Cdc34 through heterologous dimerization. Proc. Natl. Acad. Sci. U.S.A. 102, 15053–15058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Choi Y. S., Wu K., Jeong K., Lee D., Jeon Y. H., Choi B. S., Pan Z. Q., Ryu K. S., Cheong C. (2010) The human Cdc34 carboxyl terminus contains a non-covalent ubiquitin binding activity that contributes to SCF-dependent ubiquitination. J. Biol. Chem. 285, 17754–17762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ju T., Bocik W., Majumdar A., Tolman J. R. (2010) Solution structure and dynamics of human ubiquitin conjugating enzyme Ube2g2. Proteins 78, 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sakata E., Satoh T., Yamamoto S., Yamaguchi Y., Yagi-Utsumi M., Kurimoto E., Tanaka K., Wakatsuki S., Kato K. (2010) Crystal structure of UbcH5b approximately ubiquitin intermediate: insight into the formation of the self-assembled E2 approximately Ub conjugates. Structure 18, 138–147 [DOI] [PubMed] [Google Scholar]
- 29. Kamadurai H. B., Souphron J., Scott D. C., Duda D. M., Miller D. J., Stringer D., Piper R. C., Schulman B. A. (2009) Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol. Cell 36, 1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pruneda J. N., Stoll K. E., Bolton L. J., Brzovic P. S., Klevit R. E. (2011) Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme approximately ubiquitin conjugate. Biochemistry 50, 1624–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brzovic P. S., Lissounov A., Christensen D. E., Hoyt D. W., Klevit R. E. (2006) A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 21, 873–880 [DOI] [PubMed] [Google Scholar]
- 32. Wu P. Y., Hanlon M., Eddins M., Tsui C., Rogers R. S., Jensen J. P., Matunis M. J., Weissman A. M., Weisman A. M., Weissman A. M., Wolberger C., Wolberger C. P., Pickart C. M. (2003) A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 22, 5241–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pruneda J. N., Littlefield P. J., Soss S. E., Nordquist K. A., Chazin W. J., Brzovic P. S., Klevit R. E. (2012) Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47, 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu W., Shang Y., Zeng Y., Liu C., Li Y., Zhai L., Wang P., Lou J., Xu P., Ye Y., Li W. (2014) Dimeric Ube2g2 simultaneously engages donor and acceptor ubiquitins to form Lys48-linked ubiquitin chains. EMBO J. 33, 46–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Das R., Liang Y. H., Mariano J., Li J., Huang T., King A., Tarasov S. G., Weissman A. M., Ji X., Byrd R. A. (2013) Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine. EMBO J. 32, 2504–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li W., Tu D., Brunger A. T., Ye Y. (2007) A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 446, 333–337 [DOI] [PubMed] [Google Scholar]
- 37. Maspero E., Valentini E., Mari S., Cecatiello V., Soffientini P., Pasqualato S., Polo S. (2013) Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat. Struct. Mol. Biol. 20, 696–701 [DOI] [PubMed] [Google Scholar]
- 38. Kleiger G., Saha A., Lewis S., Kuhlman B., Deshaies R. J. (2009) Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139, 957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spratt D. E., Shaw G. S. (2011) Association of the disordered C-terminus of CDC34 with a catalytically bound ubiquitin. J. Mol. Biol. 407, 425–438 [DOI] [PubMed] [Google Scholar]