Background: Hereditary Wilms tumors are preceded by WT1(−) clones with an inhibitory chromatin histone pattern established by EZH2.
Results: In amniotic mesenchymal stem cells, WT1 suppresses EZH2, derepresses β-catenin (CTNNB1), and enhances responsiveness to WNT9b.
Conclusion: WT1 regulates transition from the epigenetically silenced chromatin state.
Significance: Developmental blockade in nephrogenic rests may be mediated by loss of the WT1-EZH2-CTNNB1 axis.
Keywords: β-Catenin (B-catenin), Epigenetics, Kidney, Polycomb, Wnt Pathway, Enhancer of Zeste Homolog 2 (EZH2), Epigenetic Silencing, Polycomb Repressor Complex, Wilms Tumor Suppressor 1 (WT1), Nephrogenesis
Abstract
The mammalian kidney is derived from progenitor cells in intermediate mesoderm. During embryogenesis, progenitor cells expressing the Wilms tumor suppressor gene, WT1, are induced to differentiate in response to WNT signals from the ureteric bud. In hereditary Wilms tumors, clonal loss of WT1 precludes the β-catenin pathway response and leads to precancerous nephrogenic rests. We hypothesized that WT1 normally primes progenitor cells for differentiation by suppressing the enhancer of zeste2 gene (EZH2), involved in epigenetic silencing of differentiation genes. In human amniotic fluid-derived mesenchymal stem cells, we show that exogenous WT1B represses EZH2 transcription. This leads to a dramatic decrease in the repressive lysine 27 trimethylation mark on histone H3 that silences β-catenin gene expression. As a result, amniotic fluid mesenchymal stem cells acquire responsiveness to WNT9b and increase expression of genes that mark the onset of nephron differentiation. Our observations suggest that biallelic loss of WT1 sustains the inhibitory histone methylation state that characterizes Wilms tumors.
Introduction
In resting stem cells, genes required for organ-specific differentiation are broadly silenced by repressive Polycomb group protein complexes that methylate the tails of histones associated with target gene promoter regions; this compacts chromatin structure and impedes transcription (1). At the core of the key repressive Polycomb group protein complex, PRC2,2 are three proteins: embryonic ectoderm development (EED), suppressor of zeste 12 homolog (SUZ12), and enhancer of zeste homolog 2 (EZH2), all highly conserved throughout evolution. EZH2, the catalytic subunit of the PRC2, silences gene expression by trimethylating histone H3 lysine 27 (H3K27me3) via its histone methyltransferase Su(var)3–9, Enhancer of zeste, Triothorax (SET) domain (2). Indirectly, EZH2 is also involved in DNA methylation by recruiting DNA methyltransferase1 to the promoter. Both activities contribute to epigenetic silencing of stem cell differentiation genes (3).
To escape the stem cell state, genes regulating the response to inductive signals must be made accessible by erasing the repressive histone H3K27me3 marks catalyzed by EZH2 (1, 4–6). Although EZH2 expression is high in early embryogenesis, levels are sharply decreased as development proceeds, suggesting one plausible mechanism by which epigenetic gene silencing might be relaxed prior to inductive events (7). In ex vivo cultures of rat metanephric mesenchyme, EZH2 levels decline in parallel with progressive epithelial differentiation (8). Interestingly, EZH2 is known to target genes encoding secreted WNT ligands that activate canonical β-catenin signaling (1). However, little is known about how EZH2 is down-regulated in embryonic tissues.
Mammalian kidney arises from progenitor cells of intermediate mesoderm (9). At about embryonic day E8.0, a subset of these cells begins to express the zinc finger transcription factor gene, Wt1. WT1 is essential for responsiveness to the inductive WNT9b signal released by the nearby ureteric bud (10). Mice with homozygous null Wt1 mutations are unable to induce metanephric mesenchyme and are anephric at birth (11). In 2008, Metsuyanim et al. (8) reported that genes encoding all members of the PRC2 complex are up-regulated in Wilms tumor tissue; however, only EZH2 was found to be dynamically regulated during development of the mouse kidney. Interestingly, humans who inherit one mutant WT1 allele develop scattered Wilms tumor precursor lesions (“nephrogenic rests”) that form when the second WT1 allele is lost through sporadic somatic mutations. These mutant WT1 clones lack WNT/β-catenin signaling activity and resemble embryonic progenitor cell clusters that have failed to undergo differentiation (12).
On the basis of these observations, we hypothesized that clonal loss of WT1 causes developmental arrest by permitting sustained EZH2 expression and inappropriate epigenetic silencing of genes required for responsiveness to the inductive WNT9b signal. We used a primary human amniotic fluid cell line to model the relationship between WT1 and EZH2 in embryonic kidney; these cells express the mesodermal marker OSR1 and high levels of EZH2 but have no detectable WT1. We confirmed that WT1 isoforms lacking the KTS (lysine-threonine-serine) motif between zinc fingers 3 and 4 directly suppress EZH2 transcription; previous studies showed that WT1 isoforms lacking the KTS motif have highest affinity for the classical WT1 recognition motif in target gene promoters (13), whereas +KTS isoforms have been implicated in RNA processing (13). We show that WT1−KTS or siRNA inhibition of EZH2 dramatically reduces Lys-27 trimethylation of H3 histone associated with the β-catenin gene (CTNNB1) promoter and derepresses CTNNB1 expression. Finally, we show that suppression of EZH2 heightens responsiveness to an exogenous WNT9b signal and increases expression of genes involved in early stages of nephrogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture
Human amniotic mesenchymal stem cells (amMSC) were isolated from amniotic fluid obtained at the time of diagnostic amniocentesis during the 18th week of gestation in a healthy Caucasian woman. Cells growing in monolayer attached to plastic culture vessels after 6–8 passages in the presence of DMEM culture medium with 15% fetal bovine serum and 1% penicillin/streptomycin were further characterized (14) and used in this study.
Stable Transfections
amMSC were transfected with the four human WT1 isoforms: A (−/−); B (+/−); C (−/+); and D (+/+) cloned into a pcDNA3.1/Zeo (+) vector (kindly provided by Dr. P. Grundy, Calgary, Alberta, Canada) and the empty vector as a control (Invitrogen, Burlington, Ontario, Canada) using FuGENE HD transfection reagent (1:3), according to the manufacturer's instructions (Roche Applied Science, Laval, Quebec, Canada). The transfected cells were selected using ZeocinTM at 500 μg/ml concentration (Invitrogen). Specific WT1 isoforms are denoted by plus or minus symbols, the first indicating the presence or absence of 17-amino acid sequence in exon5 and the second KTS between zinc fingers 3 and 4 status.
Gene Silencing
amMSC were transiently transfected with a negative control or a targeted to EZH2 siRNA (Silencer® Negative Control #1 siRNA and Silencer® Select Pre-designed siRNA s4916 targeting the human EZH2 gene (siEZH2), Invitrogen) using X-treme GENE transfection reagent (Roche Applied Science) according to the manufacturer's instructions.
Luciferase Reporter Transfections and Dual-Luciferase Assay
Transient transfections were done using a negative control plasmid pGL3basic (Promega, San Luis Obispo, CA), prhEZH2-FL, a plasmid containing the 5′-flanking sequence of human EZH2 gene (−1095 to +48) driving the luciferase reporter gene (kindly provided by Dr. K. Helin, Copenhagen, Denmark), and the truncated EZH2 promoters. These constructs were generated by digestion of the prhEZH2FL plasmid using NheI and BlpI, and EagI restriction enzymes (New England Biolabs, Pickering, Ontario, Canada). amMSC stably transfected with WT1 isoforms were seeded in 24-well plates and transfected at 60% confluence with 100 ng of the corresponding EZH2-luciferase reporter plasmids using FuGENE HD transfection reagent according to the manufacturer's instructions (Roche Applied Science). The Renilla luciferase expression vector pRL-SV40 (Promega) was used to normalize for transfection efficiency. Transfections were performed three times in triplicate. Firefly and Renilla luciferase reporter activities were measured after 48 h using the Dual-Luciferase assay system reagents and quantified in a GLOMAX 96 microplate luminometer (Promega). The reporter activity was expressed as a Firefly/Renilla luciferase ratio. To assess canonical WNT signaling activity, cells were transfected with a Super8XTOPFlash or Super8XFOPFlash control reporter vector. They were then co-cultured with L WNT9b (kindly provided by Dr. T. Carroll, Dallas, TX) or L cells (CRL-2648, ATCC, Manassas, VA) for 48 h before assaying for luciferase activity.
Immunoblotting
Protein content was quantified in cellular extracts using the BCA assay (Pierce). Twenty-five μg of protein extract were loaded onto SDS-PAGE gel and subjected to electrophoresis following standard immunoblotting techniques. The following primary antibodies and titers were used: anti-EZH2 (antibody 4095, 1/1000, Cell Signaling, Danvers, MA), anti-WT1 (antibody C19: sc-192, 1/200, Santa Cruz Biotechnology, Santa Cruz, CA), and anti-actin (Ab-1 CP01, 1/10,000, Calbiochem, Laval, Quebec, Canada). Immunoreactive bands were detected using species-specific horseradish peroxidase-conjugated secondary antibodies (1/2000, Cell Signaling) and visualized and analyzed using the GE Healthcare ECL Plus Western blotting detection reagents and the Storm imager scanner and software (GE Healthcare, Mississauga, Ontario, Canada).
RNA Isolation and Real-time PCR Analysis
Total RNA was purified using the Qiagen RNeasy mini according to the manufacturer's instructions (Qiagen, Toronto, Ontario, Canada) and reverse-transcribed with the iScript cDNA synthesis kit (Bio-Rad, Mississauga, Ontario, Canada). Quantitative PCR was performed using the SsoFast EvaGreen supermix with low ROX (Bio-Rad) and specific primer sets in a LightCycler 480 II (Roche Applied Science).
Chromatin Immunoprecipitation Assay-qPCR
Cells were grown, fixed, quenched, and harvested following standard techniques. Sonication was performed on a SonoLab 7.1 (Covaris, Toronto, Ontario, Canada) to obtain 500-bp fragments. The immunoprecipitation was performed using protein A/G-agarose beads. The following antibodies and titers were used: normal rabbit IgG (2729, 2 μg, Cell Signaling), histone H3 (2650, Cell Signaling, 1/100), trimethyl-histone H3 (9733, 1/100 Cell Signaling), and WT1 C19 (sc-192, 2 μg, Santa Cruz Biotechnology). Primers for the qPCR are EpiTect ChIP qPCR assay (200), human CTNNB1 (−) 01 kb, and human EZH2 (−) 01 kb (Qiagen), as well as qPCR primers designed for the detection of a genomic region of the EZH2 gene (gEZH2-L: CACCTCAGTCTTCTCATGGC and gEZH2-R: AGCAAAACCCCGTCTCAAAT).
DNA Methylation Analysis
This analysis was carried out at the Genome Quebec and McGill University Innovation Center. Genomic DNA from the stably transfected amMSC was extracted using the illustraTM tissue and cells genomicPrep Midi Flow Kit (GE Healthcare) following the manufacturer's instructions and eluted in sterile water. A PCR was performed using 30 ng of DNA. The 1× PCR buffer mix contained, MgCl2 (2.45 mm), HotStarTaq (0.1 units/μl) (Kapa Biosystems), dNTPs (Roche Applied Science), and PCR primer pool (0.25 μm). This was followed by a shrimp alkaline phosphatase (SAP) step of 37 °C for 20 min and at 85 °C for 5 min, holding at 4 °C using 0.15 units/μl of shrimp alkaline phosphatase (Sequenom, San Diego, CA), T-cleavage at 37 °C for 3 h using T7 RNA, DNA polymerase (2.86 units/μl), T7 polymerase buffer (0.64×), T-cleavage mix (3.1% volume), DTT (3.14 mm), and RNase A (0.09 mg/ml). The read-out was done on mass spectrometer, and the percentage of methylation was determined by analyzing the ratio of the area under the methylated cytosine peak over the ratio of the area for the unmethylated cytosine peak.
Statistical Analysis
Data are presented as means ± S.E. of three or more independent results. Statistical significance was assessed using t tests or one-way or two-way ANOVA followed by comparison tests.
RESULTS
WT1 Suppresses EZH2 Expression in Human Mesenchymal Stem Cells
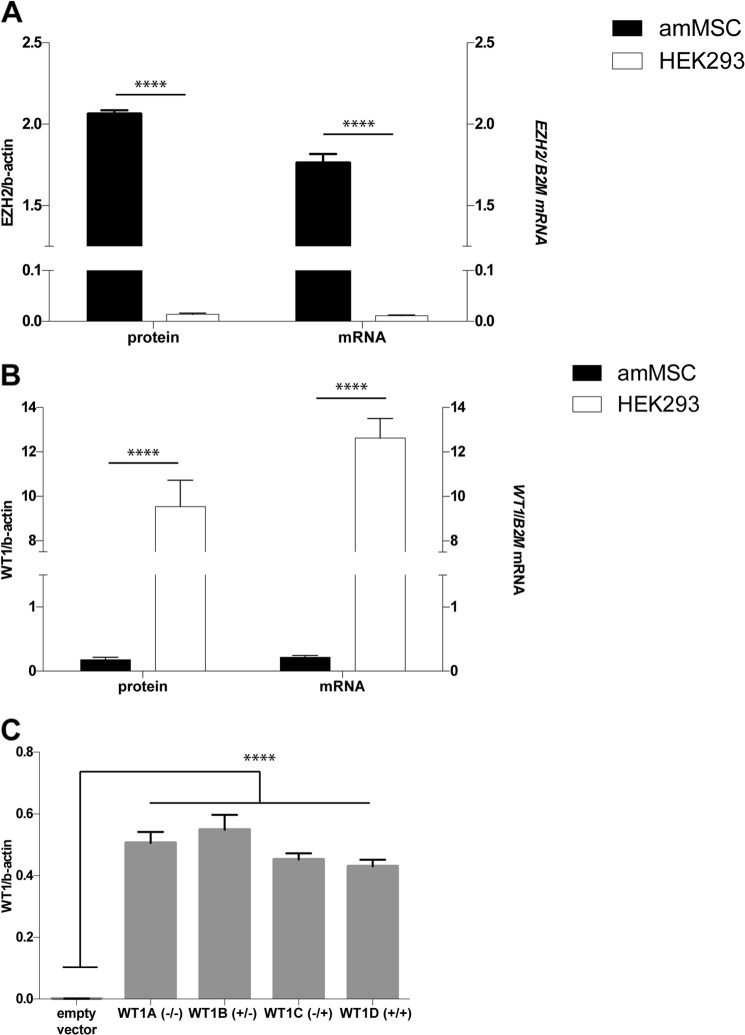
To examine this hypothesis and the molecular mechanisms by which WT1 suppresses EZH2, we identified a primary mesenchymal stem cell line from human amniotic fluid with many characteristics of the intermediate mesoderm lineage prior to onset of WT1 expression (14). These cells (amMSC) offer a convenient model system because they have high levels of EZH2 expression (Fig. 1A) but barely detectable expression of WT1 (Fig. 1B). The amMSC express typical mesenchymal stem cell surface markers (CD90, CD105, CD73, and CD44) and the transcription factor, OSR1, characteristic of lineages derived from the mesoderm (15). The cells lack surface markers for endothelial (CD31) and hematopoietic (CD34, CD45 and HLA-DR) cell lineages (14). To select a WT1 isoform for our studies, we first transfected amMSC with each of the four primary WT1 isoforms and assessed the level of EZH2 expression in each cell line. WT1 isoform levels differed by only 10–30% among the stably transfected lines (Fig. 1C). As seen in Fig. 2, EZH2 transcript (Fig. 2A) and protein (Fig. 2B) levels were substantially reduced in cell lines expressing WT1−KTS isoforms. The strong inhibitory effect of −KTS isoforms could not be attributed to variation in WT1 isoform level among cell lines (Fig. 2C). For convenience, we selected the WT1B (+17aa/−KTS) (where aa indicates amino acids) isoform for further study.
FIGURE 1.
amMSC cells as a model for study of WT1 effects on EZH2. A, levels of EZH2 protein were quantified on Western immunoblots (n = 4) normalized to β-actin (left); levels of EZH2 mRNA were quantified by qRT-PCR (n = 4) and normalized to β2-microglobulin (B2M) (right). Levels in amMSC were compared with those in HEK293 cells. B, levels of WT1 protein and mRNA were compared in amMSC and HEK293 cells as above. C, WT1 protein levels (n = 4) were compared in each stably transfected amMSC cell line; WT1 levels varied by <30% among the four cell lines. ANOVA p < 0.0001 (****).
FIGURE 2.
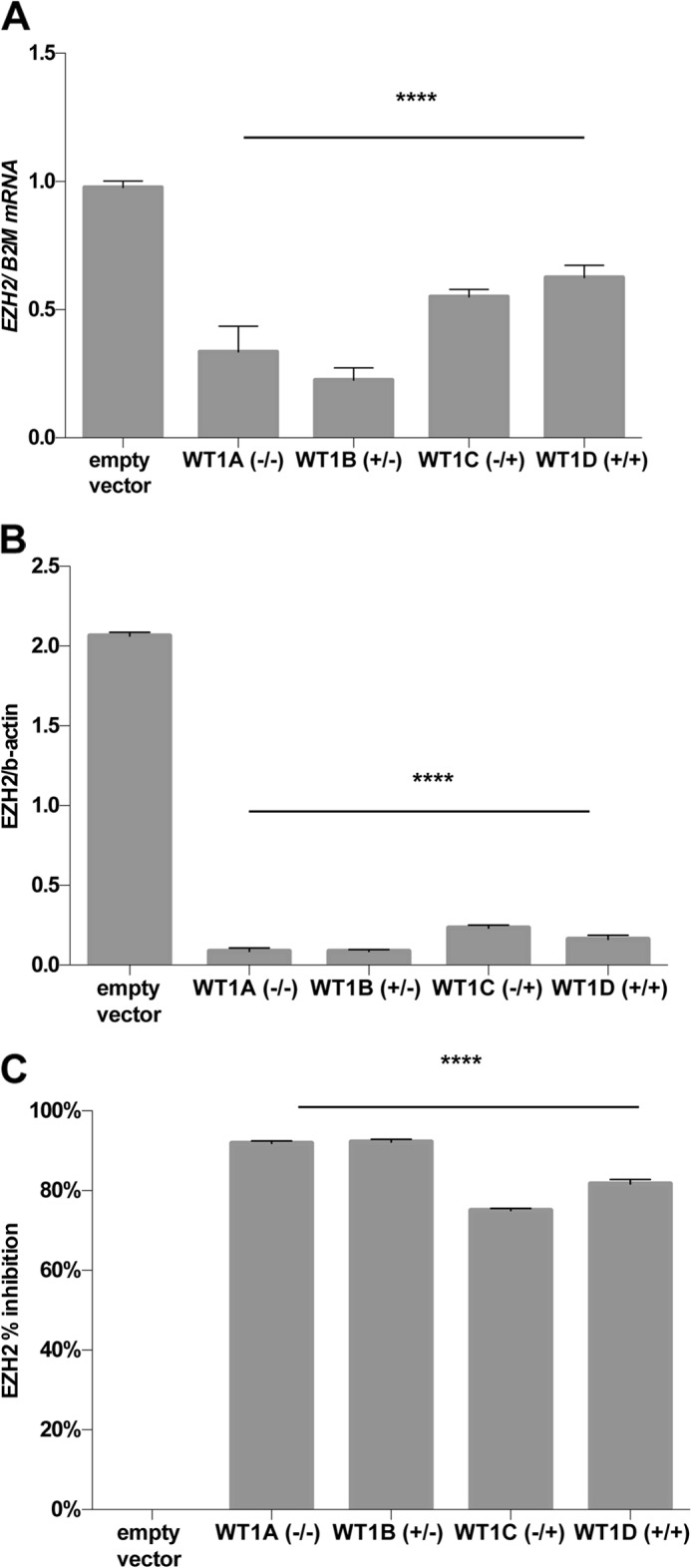
Effect of WT1 on endogenous EZH2 expression. A, EZH2 mRNA was quantified (n = 4) by qRT-PCR with β2-microglobulin (B2M) as the reference transcript in amMSC stably transfected with each of the four major WT1 isoforms. B, EZH2 protein levels were quantified by densitometric analysis of Western immunoblots (n = 4) in each stably transfected amMSC line normalized to β-actin. C, the percentage of inhibition of EZH2 expression was adjusted for relative levels of WT1 protein in each cell line; the two WT1−KTS lines (WT1A and WT1B) gave similar inhibition (92%) of EZH2 as compared with the empty vector control (not significant), whereas WT1+KTS isoforms (WT1C and WT1D) were slightly less effective (75 and 82% inhibition, respectively). ANOVA with post hoc t tests, p < 0.0001 (****).
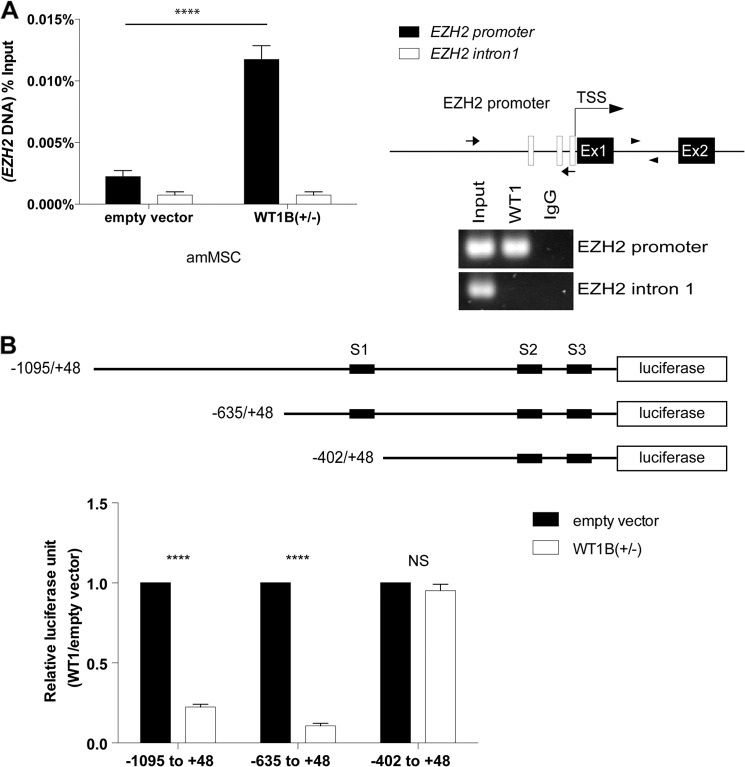
WT1B Directly Represses Transcriptional Activity of the EZH2 Promoter in Vitro
Previous studies have shown that WT1−KTS isoforms exert transcriptional inhibition of target genes by binding to conserved motifs in their promoter regions (16). To ascertain whether WT1B binds the promoter region of EZH2 specifically, we performed ChIP followed by quantitative PCR for the 1-kb 5′-flanking region and an intronic region between exon 1 and exon 2 of EZH2 in amMSCWT1B. As seen in Fig. 3A, the 5′-flanking region of EZH2 is pulled down with an antibody specific for WT1, whereas the intronic DNA sequence is not. This suggests specific binding of WT1B to the EZH2 promoter region. Analysis of the EZH2 5′-UTR promoter sequence using the MatInspector software showed the presence of three classical WT1 binding motifs conserved in mice and humans (29, 42). The sites are located between nucleotides −1095 and +48 in the EZH2 promoter region (Fig. 3B, black boxes). A minimal construct (−635 to +48), containing all three putative WT1 sites (S1–S3), drove luciferase expression in amMSC that was significantly inhibited by WT1B. A truncated construct (−402 to +48) lacking the S1 motif was unresponsive to WT1B (Fig. 3B).
FIGURE 3.
Transcriptional repression of EZH2 by WT1. A, three putative WT1 recognition motifs were identified in the 1143-bp human EZH2 5′-flanking sequence (empty boxes, upper right corner). Different sets of primers were used to quantify the promoter or intron 1 of EZH2 after ChIP using a WT1-specific antibody. Quantification by PCR of immunoprecipitated DNA is shown on the left. TSS, transcription start site. B, amMSC stably expressing WT1B isoforms were transiently transfected with the full-length (−1095 to +48) and the two truncated EZH2 promoter/luciferase constructs; normalized reporter luciferase activity is expressed as the percentage of empty vector control. ANOVA with post hoc t test, p < 0.001(**), not significant (NS).
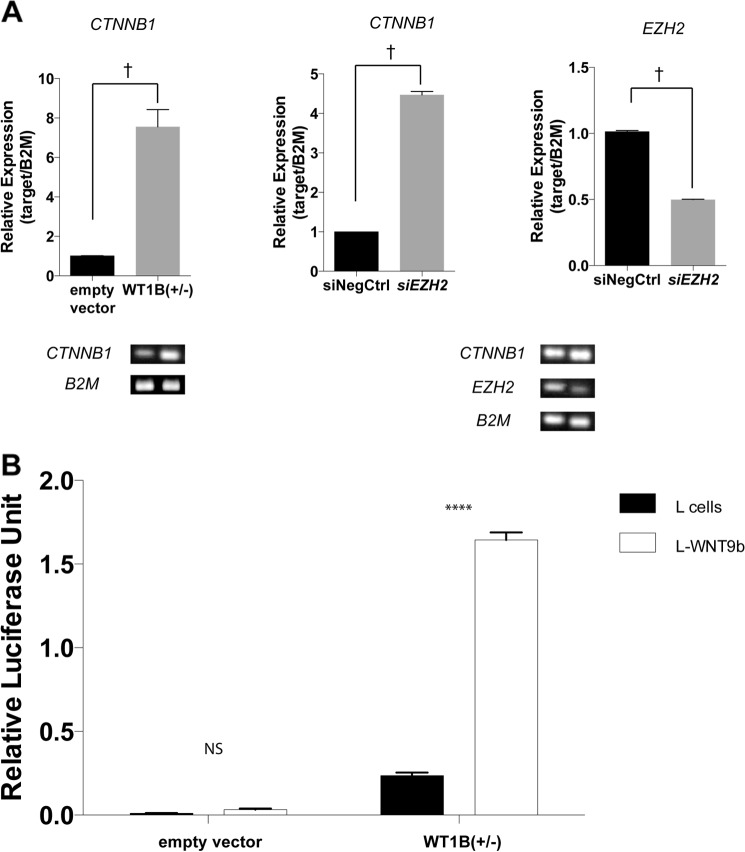
WT1B Enhances amMSC Responsiveness to WNT9b
Several lines of evidence suggest that differentiation of stem cells requires derepression of epigenetically silenced genes. We reasoned that the effects of WT1B on EZH2 might subsequently derepress key genes involved in the nephron progenitor cell response to an inductive WNT9b signal. We measured β-catenin mRNA levels in control and WT1B-expressing amMSC by quantitative RT-PCR and found that CTNNB1 transcription was increased 6–7-fold in the presence of WT1B (Fig. 4A, left panel). To confirm that the effect of WT1B on CTNNB1 expression could be mediated by down-regulation of EZH2, we performed siRNA-targeted knockdown of EZH2 in amMSC. In the presence of siEZH2, EZH2 transcript level was reduced to 50% of control (Fig. 4A, right panel). Paralleling the effects of WT1B, siEZH2 increased CTNNB1 transcript level 4–5-fold (Fig. 4A, middle panel). It follows that, by increasing CTNNB1 expression, WT1 could enhance responsiveness to canonical WNT signals during development. To test this hypothesis, we transfected amMSCWT1B cells with a Super 8X TOPFlash reporter vector. We then co-cultured the transfected cells for 48 h with either control or WNT9b-expressing L-cells. In the absence of WT1B, the β-catenin transcriptional response to WNT9b was negligible (Fig. 4B, left). In the presence of WT1B, baseline reporter activity was modestly increased and was dramatically increased in the presence of WNT9b (Fig. 4B, right).
FIGURE 4.
Effect of WT1 or siEZH2 on β-catenin expression and canonical WNT9b response. A, CTNNB1 mRNA was measured by qRT-PCR in amMSC transfected with WT1B versus empty vector control (left panel) and in amMSC transfected with siEZH2 versus scrambled siRNA negative control (siNegCtrl, middle panel). A representative agarose gel of EZH2 and CTNNB1 qRT-PCR products is shown. EZH2 transcript level normalized to β2-microglobulin (B2M) is shown in the presence or absence of siEZH2 (right panel).B, amMSCWT1B were transiently transfected with the Super 8X TOPFlash reporter construct and exposed to L cells or L cells expressing WNT9b for 48 h. Luciferase activity was normalized to a co-transfected Renilla reporter. t test, p < 0.0001 (†), p < 0.001 (****), not significant (NS).
WT1B Alters the Epigenetic Regulation of CTNNB1
EZH2 can silence genes through epigenetic mechanisms involving either recruitment of DNA methyltransferases (DNMTs) to CpGs in promoter sequences of target genes or direct trimethylation of histone H3 lysine 27 (H3K27me3). We tested these two possibilities in stepwise fashion. We first isolated genomic DNA from amMSC transfected with WT1B or an empty vector and analyzed the methylation pattern of the 5′-flanking sequence (Chr3: 41,239,802–41,241,965) of the CTNNB1 gene (Fig. 5). Bisulfite DNA analysis showed that the CpG methylation pattern in the 5′-flanking sequence and the initial segment of the coding sequence including exon 1 of the CTNNB1 gene were indistinguishable in control versus amMSCWT1B cells. The same was found when amMSC were treated with siEZH2.
FIGURE 5.
DNA methylation analysis of CTNNB1 5′-flanking region. A 2-kb fragment including the 5′ upstream sequence of CTNNB1 and part of the first exon was analyzed for the DNA methylation pattern in parental amMSC and amMSC with empty vector control versus amMSCWT1B or amMSC transiently transfected with siEZH2 versus siRNA negative control (siNegCtrl). Circles denote the approximate location of the CpGs analyzed by bisulfite analysis. Color gradient marks degree of methylation. TSS, transcription start site.
We then assessed WT1B effects on Lys-27 methylation of CTNNB1-associated histone H3 in amMSCWT1B cells. Using ChIP-qPCR assays to analyze the 1-kb CTNNB1 5′-flanking sequence, we found that the H3K27me3 fraction was reduced to about 25% of empty vector control (p < 0.0001) (Fig. 6). To confirm that the effects of WT1B on CTNNB1 histone methylation are mimicked by suppression of EZH2, we repeated this study using specific siEZH2. Again, H3K27me3 was reduced to about 20% of control (Fig. 6).
FIGURE 6.
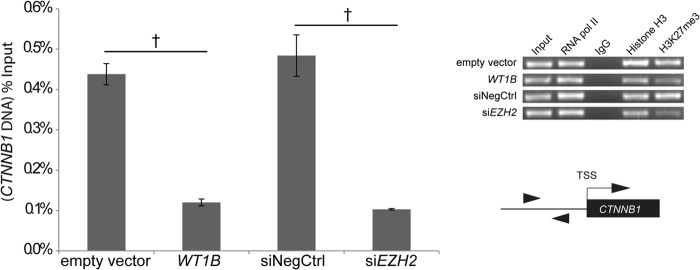
Effect of WT1B or siEZH2 on histone H3 lysine 27 trimethylation (H3K27me3) of the CTNNB1 5′-flanking region. H3K27me3 levels were quantified by ChIP-qPCR in amMSCWT1B versus empty vector control and siEZH2 versus siRNA negative control (siNegCtrl). Results of qPCR, expressed as the percentage of DNA input, are shown in the bar graph. Experimental controls (upper right panel) include: ChIP for RNA polymerase II (RNA pol II,), for total histone H3 (positive), and for rabbit IgG (negative). A representative agarose gel showing the qPCR product within the 1-kb region upstream of the CTNNB1 transcriptional start site is shown for each condition. The diagram shows the approximate location of primers used in the qPCR reaction. t test, p < 0.0001 (†). TSS, transcription start site.
Both WT1B and siEZH2 Suppress Genes Involved in Stem Cell Maintenance and Induce Genes Involved in early Nephrogenesis
In amMSC stably transfected with WT1b, we noted suppression of stem cell maintenance genes NANOG and POU5F1 (also known as OCT4) (Fig. 7A). Genes expressed in uninduced progenitor cells of cap mesenchyme (SIX2, PAX2, and SALL1) were unchanged by WT1B (Fig. 7A). On the other hand, expression of genes associated with the mesenchyme-to-epithelium transition during early stages of nephrogenesis (WNT4, PAX8, RAR-α, FGF8, and LHX1) were uniformly increased 2–8-fold (Fig. 7A). Genes associated with terminal differentiation of the renal proximal tubule (SLC3A1 and AQP1) were unchanged (Fig. 7A). Similar results were noted in amMSC transfected with siEZH2 (Fig. 7B).
FIGURE 7.
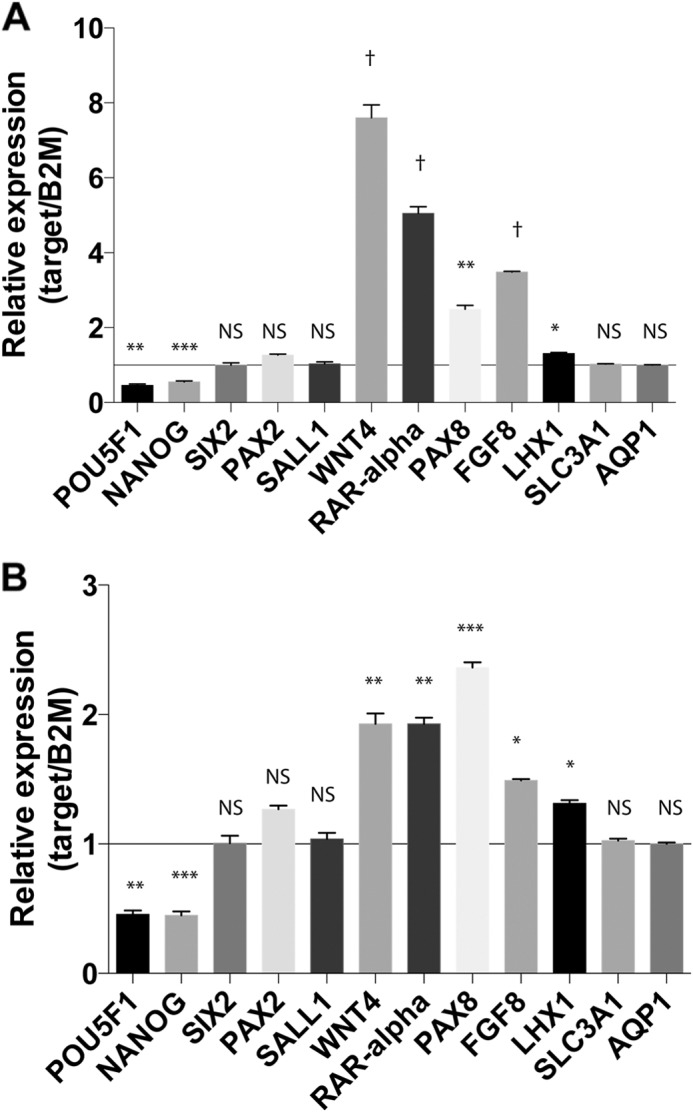
Effect of WT1B or siEZH2 on dynamic expression of genes involved in nephrogenesis. A, in amMSC transfected with WT1b or empty vector control, transcript levels for various markers validated by GUDMAP data were quantified by qRT-PCR normalized for β2-microglobulin (B2M): stem cell maintenance markers (POU5F1 and NANOG); uninduced progenitor cells of cap mesenchyme (SIX2, PAX2, and SALL1); mesenchyme-to-epithelium markers during early stages of nephrogenesis (RAR-α, PAX8, WNT4, FGF8, and LHX1); and terminal differentiation of the proximal tubule (SLC3A1 and AQP1). B, the same genes were quantified in amMSC transfected with siEZH2 versus siNegCtrl. t test, p < 0.0001 (†). NS, not significant.
DISCUSSION
During early embryogenesis, maintenance of the stem cell state requires epigenetic silencing of genes involved in differentiation cascades (6, 17). Recent studies indicate that concerted silencing of these genes is accomplished by Polycomb complexes PRC2 and PRC1 that act in a sequential fashion modifying histones associated with these genes. First, the histone methyltransferase EZH2 directs PRC2 H3K27me3 to generate a docking site for proteins of the PRC1 complex, which then catalyzes mono-ubiquitinylation of lysine 119 on histone H2A. This sequence of events holds the transcriptional activity of key genes in a “bivalent” state with minimal expression, which can be either fully activated or fully repressed by inductive signals (18). These epigenetic mechanisms seem to be crucial for normal development because inactivation of EZH2 results in a severe disturbance of embryogenesis (19, 20). Studies of mouse kidney development by the Genitourinary Development Molecular Anatomy Project (GUDMAP) consortium identify strong EZH2 expression in uninduced metanephric mesenchyme at embryonic day E11.5 and in E15.5 renal progenitor cells clustering at the tips of ureteric buds (21, 22). However, EZH2 is sharply down-regulated in E15.5 S-shaped bodies and terminally differentiated proximal tubules (21, 22).
A clue linking the down-regulation of EZH2 to WT1 during kidney development has come from the study of hereditary Wilms tumors, thought to arise from developmentally arrested clones (nephrogenic rests) of renal progenitor cells that lack evidence of canonical WNT signaling activity (23). Wilms tumors express EZH2 at relatively high levels (8, 24) and exhibit hypermethylation of various genomic regions (25–27). Aiden et al. (28) compared patterns of active (H3K4me3) and inactive (H3K27me3) chromatin in normal adult kidney, embryonic stem cells, and micro-dissected blastemal compartments of Wilms tumors. They found that the chromatin state of Wilms tumors strongly resembled that of embryonic stem cells rather than normal kidney. Tumors showed active chromatin marks for genes that confer pluripotency, expand the renal progenitor cell pool, or maintain the undifferentiated stem cell state. Conversely, there were strong H3K27me3 repressive marks for genes involved in nephron differentiation. Analysis of bivalent histone marks suggested that developmental arrest occurs at an early stage of renal progenitor cell commitment. The authors concluded that Wilms tumors display a characteristic pattern of epigenetic silencing consistent with a “stalled” kidney differentiation program (28).
During kidney development, nephron progenitor cells are propelled out of the resting stem-like state after they come into intimate contact with branches of the arborizing ureteric bud secreting WNT9b (10). In response, nephron progenitor cells surrounding each ureteric bud tip (cap mesenchyme) exhibit canonical WNT/β-catenin signaling activity and then undergo the mesenchyme-to-epithelium transition that accompanies early stages of nephrogenesis (9). It follows that derepression of CTNNB1 must occur if these cells are to activate the β-catenin signaling pathway in response to an inductive WNT9b signal. We used a mesenchymal stem cell (MSC) cell line derived from human amniotic fluid to model the effects of WT1 on EZH2-mediated silencing of CTNNB1. amMSC express the transcription factor, OSR1, a marker of the intermediate mesoderm that gives rise to the kidneys (14, 15). We saw that repression of EZH2 by WT1B or siRNA led to a dramatic increase in both the expression of CTNNB1 and the activity of a β-catenin pathway reporter (TOPFlash) in response to WNT9b.
As a prelude to nephrogenesis, WT1 and EZH2 expression is co-expressed in progenitor cells of the cap mesenchyme, but only WT1 is sustained in the S-shaped body (21, 22, 30). This suggests either that the effect of WT1 on EZH2 transcript level is delayed or that additional factors contribute to the full down-regulation of EZH2 (21, 22). In our amMSC model, WT1B- or siRNA-mediated suppression of endogenous EZH2 erased repressive marks on histone H3 silencing transcriptional activity of the CTNNB1 promoter. This was associated with suppression of stem cell maintenance genes and enhanced expression of genes marking nephrogenesis in pre-tubular aggregates and S-shaped bodies. Interestingly, there was no effect of WT1 on expression of other cap mesenchyme transcription factors. Thus, the effects of WT1 on EZH2 may be related to the mesenchyme-to-epithelium transition but not to events that transiently delay differentiation (e.g. SIX2) in cap mesenchyme. Other groups examined mice with conditional knockdown of Wt1 in embryonic kidney and noted a profound disturbance of mesenchyme-to-epithelium transition; at E18.5 the mutant fetal mice showed overabundance of metanephric mesenchyme versus epithelial structures, reduced formation of progenitor cell caps surrounding ureteric bud branch tips, and reduced nephrogenesis (31). In those studies, Wnt4 expression was absent at E12.5 and Wnt4 chromatin was predominantly in the inactive H3K27me3 state (31). Although WT1 affected genes marking the early differentiation of the nephron, we found no changes in amMSC PAX2 or SIX2 expression. PAX2 exerts an anti-apoptotic effect during kidney development (32), and SIX2 appears to delay differentiation of induced nephron progenitor cells (33). Thus, these genes may not be part of the differentiation cascade controlled by the WT1/EZH2 axis.
In embryonic stem cells, the abundance of most PRC2 target gene transcripts is lower than in differentiated cells; 93% of these target transcripts are up-regulated during ES cell differentiation (6). As a central component of the PRC2 complex, EZH2 participates in histone H3 methylation but may also recruit DNA methyltransferases to direct methylation of cytosines in the promoters of target genes (3, 34). We examined the effects of EZH2 on CTNNB1 in amMSC via each mechanism. Although overexpression of WT1 is reported to increase DNMT3a expression in HEK293 cells (35), we found no effect of WT1B on the methylation pattern of the CTNNB1 promoter. In contrast, ChIP analysis showed a striking decrease in H3K27me3 associated with the CTNNB1 5′-UTR. Our observations are consistent with studies of others, suggesting that PRC2-facilitated DNA methylation is essential for survival of somatic cells, whereas histone methylation may be important for maintenance of the stem cell state (6, 36, 37).
Although we found that WT1 activates CTNNB1 in mesenchymal stem cells, other investigators have noted up-regulation of testicular β-catenin protein in mice with conditional knock-out of Wt1 (38). However, this was attributed to post-transcriptional effects of Wt1 knock-out on β-catenin stabilization that may be specific to the Sertoli cell and involve the WT1+KTS isoforms that modulate tissue-specific mRNAs (39). Kim et al. (40) reported WT1 inhibition of the canonical WNT/β-catenin pathway in mature zebrafish podocytes. The very high levels of WT1 in mature growth-arrested podocytes is thought to have a unique role in inducing the VEGF expression that elicits capillary ingrowth during glomerulogenesis (41). Thus, WT1 can exert a variety of inhibitory effects on the canonical WNT signaling pathway, but these effects are specific to the terminal differentiation of certain tissues and appear to differ from the novel EZH2-dependent mechanism by which WT1 derepresses genes during transition from the renal progenitor cell state.
This work was supported by Operating Grant MOP-119571 from the Canadian Institutes of Health Research (CIHR) and an infrastructure support grant to the McGill University Health Center Research Institute from the Fonds de la Recherche en Santé du Québec (FRSQ).
- PRC2
- Polycomb repressor complex 2
- WT
- Wilms tumor
- amMSC
- amniotic fluid-derived mesenchymal stem cells
- E
- embryonic day
- qPCR
- quantitative PCR
- qRT-PCR
- quantitative RT-PCR
- ANOVA
- analysis of variance.
REFERENCES
- 1. Chen Y. H., Hung M. C., Li L. Y. (2012) EZH2: a pivotal regulator in controlling cell differentiation. Am. J. Transl. Res. 4, 364–375 [PMC free article] [PubMed] [Google Scholar]
- 2. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 3. Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J. M., Bollen M., Esteller M., Di Croce L., de Launoit Y., Fuks F. (2006) The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874 [DOI] [PubMed] [Google Scholar]
- 4. Chou R. H., Yu Y. L., Hung M. C. (2011) The roles of EZH2 in cell lineage commitment. Am. J. Transl. Res. 3, 243–250 [PMC free article] [PubMed] [Google Scholar]
- 5. Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., Koseki H., Fuchikami T., Abe K., Murray H. L., Zucker J. P., Yuan B., Bell G. W., Herbolsheimer E., Hannett N. M., Sun K., Odom D. T., Otte A. P., Volkert T. L., Bartel D. P., Melton D. A., Gifford D. K., Jaenisch R., Young R. A. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 7. Ningxia Z., Shuyan L., Zhongjing S., Ling C., Tianzhong M., Lifeng W., Yan Y., Leili L., Xiancai C., Haibin C. (2011) The expression pattern of Polycomb group protein Ezh2 during mouse embryogenesis. Anat. Rec. (Hoboken) 294, 1150–1157 [DOI] [PubMed] [Google Scholar]
- 8. Metsuyanim S., Pode-Shakked N., Schmidt-Ott K. M., Keshet G., Rechavi G., Blumental D., Dekel B. (2008) Accumulation of malignant renal stem cells is associated with epigenetic changes in normal renal progenitor genes. Stem Cells 26, 1808–1817 [DOI] [PubMed] [Google Scholar]
- 9. Iglesias D. M., Akpa M. M., Goodyer P. (2014) Priming the renal progenitor cell. Pediatr. Nephrol. 29, 705–710 [DOI] [PubMed] [Google Scholar]
- 10. Carroll T. J., Park J. S., Hayashi S., Majumdar A., McMahon A. P. (2005) Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev. Cell 9, 283–292 [DOI] [PubMed] [Google Scholar]
- 11. Kreidberg J. A., Sariola H., Loring J. M., Maeda M., Pelletier J., Housman D., Jaenisch R. (1993) WT-1 is required for early kidney development. Cell 74, 679–691 [DOI] [PubMed] [Google Scholar]
- 12. Royer-Pokora B., Busch M., Beier M., Duhme C., de Torres C., Mora J., Brandt A., Royer H. D. (2010) Wilms tumor cells with WT1 mutations have characteristic features of mesenchymal stem cells and express molecular markers of paraxial mesoderm. Hum. Mol. Genet. 19, 1651–1668 [DOI] [PubMed] [Google Scholar]
- 13. Hohenstein P., Hastie N. D. (2006) The many facets of the Wilms' tumour gene, WT1. Hum. Mol. Genet. 15, Spec. No. 2, R196–R201 [DOI] [PubMed] [Google Scholar]
- 14. Iglesias D. M., El-Kares R., Taranta A., Bellomo F., Emma F., Besouw M., Levtchenko E., Toelen J., van den Heuvel L., Chu L., Zhao J., Young Y. K., Eliopoulos N., Goodyer P. (2012) Stem cell microvesicles transfer cystinosin to human cystinotic cells and reduce cystine accumulation in vitro. PLoS One 7, e42840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Z., Iglesias D., Eliopoulos N., El Kares R., Chu L., Romagnani P., Goodyer P. (2011) A variant OSR1 allele which disturbs OSR1 mRNA expression in renal progenitor cells is associated with reduction of newborn kidney size and function. Hum. Mol. Genet. 20, 4167–4174 [DOI] [PubMed] [Google Scholar]
- 16. Miller-Hodges E., Hohenstein P. (2012) WT1 in disease: shifting the epithelial-mesenchymal balance. J. Pathol. 226, 229–240 [DOI] [PubMed] [Google Scholar]
- 17. Ezhkova E., Pasolli H. A., Parker J. S., Stokes N., Su I. H., Hannon G., Tarakhovsky A., Fuchs E. (2009) Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136, 1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mohn F., Schübeler D. (2009) Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet. 25, 129–136 [DOI] [PubMed] [Google Scholar]
- 19. O'Carroll D., Erhardt S., Pagani M., Barton S. C., Surani M. A., Jenuwein T. (2001) The Polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21, 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen X., Liu Y., Hsu Y. J., Fujiwara Y., Kim J., Mao X., Yuan G. C., Orkin S. H. (2008) EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harding S. D., Armit C., Armstrong J., Brennan J., Cheng Y., Haggarty B., Houghton D., Lloyd-MacGilp S., Pi X., Roochun Y., Sharghi M., Tindal C., McMahon A. P., Gottesman B., Little M. H., Georgas K., Aronow B. J., Potter S. S., Brunskill E. W., Southard-Smith E. M., Mendelsohn C., Baldock R. A., Davies J. A., Davidson D. (2011) The GUDMAP database: an online resource for genitourinary research. Development 138, 2845–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McMahon A. P., Aronow B. J., Davidson D. R., Davies J. A., Gaido K. W., Grimmond S., Lessard J. L., Little M. H., Potter S. S., Wilder E. L., Zhang P., and GUDMAP Project (2008) GUDMAP: the genitourinary developmental molecular anatomy project. J. Am. Soc. Nephrol. 19, 667–671 [DOI] [PubMed] [Google Scholar]
- 23. Fukuzawa R., Heathcott R. W., More H. E., Reeve A. E. (2007) Sequential WT1 and CTNNB1 mutations and alterations of β-Catenin localisation in intralobar nephrogenic rests and associated Wilms tumours: two case studies. J. Clin. Pathol. 60, 1013–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zirn B., Hartmann O., Samans B., Krause M., Wittmann S., Mertens F., Graf N., Eilers M., Gessler M. (2006) Expression profiling of Wilms tumors reveals new candidate genes for different clinical parameters. Int. J. Cancer 118, 1954–1962 [DOI] [PubMed] [Google Scholar]
- 25. Riccio A., Sparago A., Verde G., De Crescenzo A., Citro V., Cubellis M. V., Ferrero G. B., Silengo M. C., Russo S., Larizza L., Cerrato F. (2009) Inherited and sporadic epimutations at the IGF2-H19 locus in Beckwith-Wiedemann syndrome and Wilms' tumor. Endocr. Dev. 14, 1–9 [DOI] [PubMed] [Google Scholar]
- 26. Haruta M., Matsumoto Y., Izumi H., Watanabe N., Fukuzawa M., Matsuura S., Kaneko Y. (2008) Combined BubR1 protein down-regulation and RASSF1A hypermethylation in Wilms tumors with diverse cytogenetic changes. Mol. Carcinog. 47, 660–666 [DOI] [PubMed] [Google Scholar]
- 27. Honda S., Arai Y., Haruta M., Sasaki F., Ohira M., Yamaoka H., Horie H., Nakagawara A., Hiyama E., Todo S., Kaneko Y. (2008) Loss of imprinting of IGF2 correlates with hypermethylation of the H19 differentially methylated region in hepatoblastoma. Br. J. Cancer 99, 1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aiden A. P., Rivera M. N., Rheinbay E., Ku M., Coffman E. J., Truong T. T., Vargas S. O., Lander E. S., Haber D. A., Bernstein B. E. (2010) Wilms tumor chromatin profiles highlight stem cell properties and a renal developmental network. Cell Stem Cell 6, 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakagama H., Heinrich G., Pelletier J., Housman D. E. (1995) Sequence and structural requirements for high-affinity DNA binding by the WT1 gene product. Mol Cell. Biol. 15, 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Georgas K., Rumballe B., Valerius M. T., Chiu H. S., Thiagarajan R. D., Lesieur E., Aronow B. J., Brunskill E. W., Combes A. N., Tang D., Taylor D., Grimmond S. M., Potter S. S., McMahon A. P., Little M. H. (2009) Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev. Biol. 332, 273–286 [DOI] [PubMed] [Google Scholar]
- 31. Essafi A., Webb A., Berry R. L., Slight J., Burn S. F., Spraggon L., Velecela V., Martinez-Estrada O. M., Wiltshire J. H., Roberts S. G., Brownstein D., Davies J. A., Hastie N. D., Hohenstein P. (2011) A Wt1-controlled chromatin switching mechanism underpins tissue-specific Wnt4 activation and repression. Dev. Cell 21, 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dziarmaga A., Eccles M., Goodyer P. (2006) Suppression of ureteric bud apoptosis rescues nephron endowment and adult renal function in Pax2 mutant mice. J. Am. Soc. Nephrol. 17, 1568–1575 [DOI] [PubMed] [Google Scholar]
- 33. Kobayashi A., Valerius M. T., Mugford J. W., Carroll T. J., Self M., Oliver G., McMahon A. P. (2008) Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3, 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bracken A. P., Dietrich N., Pasini D., Hansen K. H., Helin K. (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Szemes M., Dallosso A. R., Melegh Z., Curry T., Li Y., Rivers C., Uney J., Mägdefrau A. S., Schwiderski K., Park J. H., Brown K. W., Shandilya J., Roberts S. G., Malik K. (2013) Control of epigenetic states by WT1 via regulation of de novo DNA methyltransferase 3A. Hum. Mol. Genet. 22, 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McLaughlin N., Wang F., Saifudeen Z., El-Dahr S. S. (2014) In situ histone landscape of nephrogenesis. Epigenetics 9, 222–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McLaughlin N., Yao X., Li Y., Saifudeen Z., El-Dahr S. S. (2013) Histone signature of metanephric mesenchyme cell lines. Epigenetics 8, 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang H., Gao F., Guillou F., Taketo M. M., Huff V., Behringer R. R. (2008) Wt1 negatively regulates β-catenin signaling during testis development. Development 135, 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Larsson S. H., Charlieu J. P., Miyagawa K., Engelkamp D., Rassoulzadegan M., Ross A., Cuzin F., van Heyningen V., Hastie N. D. (1995) Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell 81, 391–401 [DOI] [PubMed] [Google Scholar]
- 40. Kim M. S., Yoon S. K., Bollig F., Kitagaki J., Hur W., Whye N. J., Wu Y. P., Rivera M. N., Park J. Y., Kim H. S., Malik K., Bell D. W., Englert C., Perantoni A. O., Lee S. B. (2010) A novel Wilms tumor 1 (WT1) target gene negatively regulates the WNT signaling pathway. J. Biol. Chem. 285, 14585–14593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eremina V., Sood M., Haigh J., Nagy A., Lajoie G., Ferrara N., Gerber H. P., Kikkawa Y., Miner J. H., Quaggin S. E. (2003) Glomerular-specific alterations of VEGF-A expression lead to distinct congenital and acquired renal diseases. J. Clin. Invest. 111, 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bracken A. P., Pasini D., Capra M., Prosperini E., Colli E., Helin K. (2003) EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22, 5323–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]