Background: The Mycobacterium tuberculosis gene, Rv2745c, leads to induction of downstream genes that is condition-dependent.
Results: Rv2745c is induced during hypoxia and reaeration conditions. An isogenic mutant leads to differential transcriptional profiles.
Conclusion: Rv2745c plays a role during hypoxia and reaeration.
Significance: Rv2745c is important for survival during reaeration, implicating that Rv2745c is important for growth during reactivation in vivo.
Keywords: Hypoxia, Mycobacterium tuberculosis, Post-transcriptional Regulation, Protease, Tuberculosis, clp, Reaeration
Abstract
Mycobacterium tuberculosis (Mtb) is the leading cause of death from an infectious disease worldwide and is the causative agent of tuberculosis (Chao, M. C., and Rubin, E. J. (2010) Annu. Rev. Microbiol. 64, 293–311). Throughout infection, Mtb encounters a variety of host pressures. Thus, responding to these host stresses via the induction of multiple regulatory networks is needed for survival within the host. The Clp protease gene regulator, Rv2745c (clgR), is induced in response to environmental stress conditions, implicating its potential role in Mtb pathogenesis. Transcriptional activation of genes downstream of Rv2745c occurs in a condition-dependent manner. Our isogenic Mtb:ΔRv2745c mutant expresses a significantly different phenotype upon reaeration conditions. Transcriptional analysis revealed differential gene expression profiles relative to wild-type Mtb. Rv2745c is strongly induced in response to hypoxic and reaeration conditions, implicating a role of Rv2745c in vivo during both establishment of infection and reactivation. We found dysregulation of downstream genes within both the σH/σE regulon as well as the dosR regulon in the isogenic mutant, Mtb:ΔRv2745c. Upon hypoxic and reaeration conditions, Clp protease induction occurred within wild-type Mtb, indicating that activation of clgR, which subsequently leads to Clp protease induction, is crucial for degradation of misfolded proteins and ultimately survival of Mtb upon specific stress conditions. Our data indicate the diverse response of Rv2745c, σH and σE in response to a variety of stress conditions. Activation of Rv2745c in response to various stress conditions leads to differential activation of downstream genes, indicating the diverse role of Rv2745c and its importance for Mtb survival in vivo.
Introduction
Mtb2 infects one-third of the world's population (1). In 2012 alone, an estimated 8.6 million people became infected with TB (2). The Mtb virulence lifecycle begins when an actively infected individual spreads Mtb via aerosolized droplets containing bacilli through coughing and sneezing (3). Upon initial infection, Mtb replicates freely within the alveolar macrophage (3). Recruitment of alveolar macrophages to the site of infection is the first stage of granuloma formation and consequently, leads to hypoxic conditions within the central region of the granuloma (4). Granulomas are clearly organized structures that develop extensive fibrotic capsules as they mature (5). As such, late stage granulomas show loss of vascularization that results in necrosis and caseation, which occurs in response to hypoxic conditions (3, 5). Development of a granuloma due to Mtb occurs as a means of bacilli restriction, thus preventing dissemination, but does not lead to eradication (6). The ability of Mtb to survive within the host for extended periods of time, up to decades, is thought to occur in response to adaptations to hypoxic conditions and results in latent TB infections (7, 8). Latent TB occurs in 90% of cases; however, reactivation can occur and does so in response to weakened immune systems, indicating that further elucidation of Mtb transcriptional changes during both latent TB and reactivation are important for better understanding Mtb pathogenesis (3).
The granuloma is a crucial physiological and immunological response that facilitates containment of Mtb, preventing the spread of the bacilli throughout the host lung and ultimately dissemination throughout the body (3). However, a breach in the immune response can lead to reactivation and dissemination of Mtb throughout the lung. Upon reactivation, the Mtb environment shifts from low oxygen tension to an aerated environment. As such, changes within the Mtb transcriptome occur in response to the transition to increased oxygen tension. To study these transcriptional changes, the Wayne model was developed to replicate hypoxic conditions in vitro and facilitates a gradual depletion of oxygen as it is consumed (9, 10). Subsequently, reaeration, in which hypoxic cultures are transferred to aerated cultures, is used to study reactivation in vitro (9).
Mtb has evolved sensing mechanisms to respond to hypoxic conditions via nitric oxide and carbon monoxide levels through the DosR regulon (11, 12). Upon low oxygen tension, the pathogen undergoes metabolic changes that facilitate a non-replicating but viable state (1, 3, 8). Consequently, activation of the DosR regulon leads to dormancy in which there is a shift in the metabolism of Mtb leading to expression of ∼48 genes within the DosR regulon (11). Although the DosR regulon plays a role in response to hypoxic conditions, the enduring hypoxic response (EHR) is an added layer of regulatory mechanisms that facilitate the shift to a reduced metabolic state. The EHR is an important component in that it may play a role during the persistent state of infection upon which Mtb remains in a metabolically inactive but viable state. In this vein, DosR and its sensor kinases are required for the pathogenesis of Mtb in non-rodent animal models like rabbits (13), guinea pigs (13, 14), and macaques.3 Pulmonary granulomas in these model systems are known to caseate and develop hypoxia (15). Furthermore, the expression of DosR regulon members has also been demonstrated to occur in bacilli present in macaque human-like caseous granulomas.3
Clp proteases are crucial to the degradation of intracellular proteins, thus facilitating cellular homeostasis. Clp proteases play a vital role in the virulence of Mtb as well as other bacterial pathogens, such as Listeria monocytogenes (16). In the absence of proteolytic degradation via ClpP1P2, the levels of misfolded proteins can reach toxic levels, leading to cell death. As such, Raju et al. (16) found that functional ClpP1P2 is required for normal growth. In Mtb, clpP1 and clpP2 are encoded on a single operon that is activated by Rv2745c (9). Rv2745c, clgR, is a Clp protease gene regulator that is induced under a variety of conditions, but ultimately induction of Rv2745c leads to differential activation of downstream genes in a condition-dependent manner (9, 17, 18).
The role of Rv2745c during hypoxic conditions has yet to be defined. Rv2745c expression is induced under multiple stress conditions, such as redox stress, SDS treatment, heat shock, and acid stress (19) as well as hypoxia (9, 18) implicating that Rv2745c plays a role in response to various environmental pressures. Rv2745c up-regulation in response to redox stress or envelope damage by thioridazine (20) or vancomycin (21) is dependent upon σH and/or σE levels and does not lead to the induction of clp proteases (9, 17, 19). However, induction of Rv2745c during the late stages of hypoxia as well as during reaeration leads to clp protease induction (18). As to how the induction of Rv2745c by various stressors can lead to different downstream outcomes is, however, not well understood. Thus the role of Rv2745c in responding to changing environmental pressures requires further elucidation.
The vast array of environmental pressures that Mtb encounters during infection has led to the necessity of coordinated regulatory networks to respond to these stressors. However, throughout the course of infection, the types of environmental pressures that Mtb faces change. Thus, there is temporal expression of these differing regulatory networks. Concurrently, there is also partial overlap of some of these regulatory networks that Mtb uses to respond to subtle changes in environmental cues. As such, Mtb has become highly adapted to sensing and responding to shifting environmental cues, thus leading to its success as a human pathogen. In this investigation we studied the growth phenotype and the transcriptomic response of the isogenic Mtb:ΔRv2745c mutant that we described earlier (17) in comparison to the wild-type Mtb strain in response to in vitro hypoxia as well as reaeration. Our results concur with previous reports that describe a strong induction of the Rv2745c gene as well as the downstream clp genes when Mtb is subjected to hypoxia and reaeration. Furthermore, we show that although the Mtb:ΔRv2745c mutant is able to replicate without any defect during hypoxia, it is significantly impaired in its ability to resume its growth during the subsequent reaeration phase. Our results identify potential targets of the Mtb Clp proteolytic machinery during this stress in vitro. These have important implications for Mtb pathogenesis as well as drug development efforts.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
Liquid cultures of Mtb CDC1551 (referred to as Mtb) and Mtb:ΔRv2745c were grown in Middlebrook 7H9 broth (BD Diagnostic Systems) supplemented with 0.1% glycerol, 0.05% Tween 80, and 10% albumin dextrose catalase. Cultures were incubated at 37 °C with shaking at 225 rpm except where noted.
In Vitro Hypoxia and Reaeration Treatment
Cultures were grown to mid-log phase (A260 0.39–0.45) without antibiotics. Hypoxia and reaeration treatment were followed based on Sherrid et al. (18). Briefly, for hypoxia treatment, a modified Wayne model was used in which cultures were transferred to 50-ml conical tubes with a headspace of 0. Cultures were sealed and incubated without shaking at 37 °C. For each hypoxia time point, a 50-ml conical tube was opened for cfu plating and RNA extraction. For hypoxia, samples were taken at t = 0, 1, 5, and 7 days. For the reaeration time points, sealed conical tubes were opened and transferred to disposable Erlenmeyer culture flasks with a 1:5 volume to headspace ratio. Aliquots of 25 ml were taken at t = 1, 4, 6, 12, 24, and 48 h after cultures were reaerated for cfu plating and RNA extraction.
Bacterial RNA Extraction
25 ml of culture was used to extract RNA upon cell lysis via the TRIzol bead beater method and phenol extraction (22). RNA concentrations were quantified using a Nanodrop 2000 (NanoDrop Technologies).
DNA Microarrays and Real-time (RT)-PCR
Mtb-specific DNA microarrays (MYcroarrays, Biodiscovery Llc.) were used to compare transcriptome-wide responses in Mtb and the mutant to stress caused by hypoxia and reaeration. Detailed protocols for array procedures have been described earlier (19, 22, 23). Genes were considered to have a perturbed expression level if they exhibited a 2-fold higher or lower expression in the mycobacterial strain (wild-type or mutant) at a given time point relative to control samples in each of the three biological replicate arrays and in every technical replicate spot on each array. Microarray data have been submitted to the Gene Expression Omnibus. Data can be retrieved using the GEO platform number GPL18320. For RT-PCR, RNA was treated with DNase as previously described (17, 22, 24). RNA was reverse-transcribed following the manufacturers' instructions using the High Capacity RNA-to-cDNA kit (Applied Biosystems) (17, 22, 24). RT PCR was performed as per the manufacturers' instructions using Power SYBR Green PCR Master Mix (Applied Biosystems) and as previously described (17, 22, 24). Expression levels were normalized to sigA rRNA.
Statistical Analysis
Statistical analyses of the growth curves were performed using a Wilcoxon matched pairs signed rank test in GraphPad Prism. Microarray statistical analyses were performed using a t test script in the Spotfire DecisionSite/S+ Array Analyzer.
Regulatory Compliance
The investigators received approval from the Tulane Institutional Biosafety Committee for all procedures involving Mtb.
RESULTS
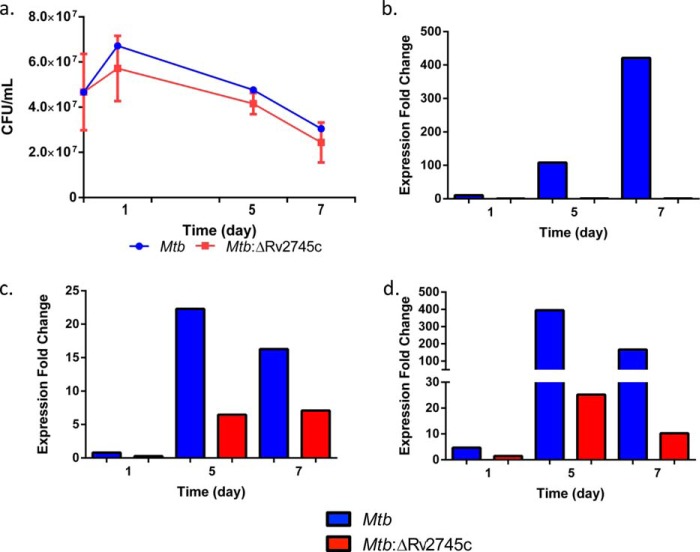
Hypoxia and reaeration result in differential signaling cascades in response to a transition from an anaerobic to oxygen rich state. As such, we employed hypoxic conditions using a static culture model following Sherrid et al. (18) specifically studying cfu as well as transcriptional changes 1, 5, and 7 days after the onset of hypoxia. To achieve reaeration conditions, hypoxic cultures were transferred to cultures flasks and incubated with shaking to allow for reaeration and samples retrieved for experiments 1, 4, 6, 12, 24, and 48 h afterward. Our results showed that the Mtb:ΔRv2745c strain did not exhibit any defect in growth relative to the parental Mtb strain during hypoxia for 1–7 days (Fig. 1a). This was despite the fact that the expression of Rv2745c transcript was strongly induced in Mtb but not in Mtb:ΔRv2745c at this stage (Fig. 1b). As a positive control for the effectiveness of the hypoxia set-up, we also assessed if the expression of hypoxia response genes dosR and acr was induced in this setting. We found that their expression was strongly induced in Mtb at all the time points (Fig. 1, c and d). The expression of dosR was not detected during hypoxia at day 1 in Mtb:ΔRv2745c and detected at lower levels at the two latter time points relative to wild type (Fig. 1c). Similarly, the expression of acr was induced ∼400-fold at the 5-day time point and ∼200-fold at the 7-day time point in Mtb, values that were higher than those observed for the mutant (Fig. 1d).
FIGURE 1.
Hypoxia growth curve and RT-PCR levels of control genes. a, graph representing cfu numbers comparing wild-type Mtb to that of the isogenic mutant, Mtb:ΔRv2745c. During hypoxia, the there is a decline in cfu numbers in both the wild type and isogenic mutant, with a greater decline in the isogenic mutant; however, the differences during hypoxia are not significantly different. Expression levels of control genes Rv2745c (b), dosR (c), and acr (d) are shown.
Despite the fact that Mtb:ΔRv2745c was able to grow at a rate comparable to Mtb in the face of hypoxia, the ability of this mutant strain to reactivate during the reaeration phase was significantly compromised relative to Mtb (Fig. 2). Although a trend in this direction was evident during the initial time points, there was a half-log reduction in Mtb:ΔRv2745c cfu relative to Mtb by 12 h after normoxia was reinstated. The differences increased to 1 log by 24 h and to ∼3 log by 48 h. At this time increased levels of cfu were observed for either strain, but Mtb grew at a significantly higher rate (Fig. 2). The observed differences in cfu during reaeration were found to be statistically significant when comparing wild-type Mtb to the isogenic mutant, Mtb:ΔRv2745c, using a Wilcoxon matched pairs signed rank test (p = 0.0313, n = 3, except for t = 48, in which n = 2 for Mtb) (Fig. 2). The high level of growth in the wild-type strain is likely due to the transition from a dormant to vegetative state. As normoxia was restored, the response of the wild-type strain in terms of growth was significantly better relative to the mutant. Thus, the cfu levels in the wild-type strain were already ∼1 log higher relative to the mutant at the 24-h time point. These differences increased to ∼2 logs by the 48-h time point. Although the doubling time of Mtb is 12–16 h on average, it generally decreases to a much shorter period during mid-log phase. The increase in wild-type cfu is most likely due to this strain reaching the mid-log phase earlier relative to the mutant. Our result suggests that the Mtb:ΔRv2745c mutant fails to adapt to normoxia, likely due to a deficit in the important role that Rv2745c may play during hypoxia.
FIGURE 2.
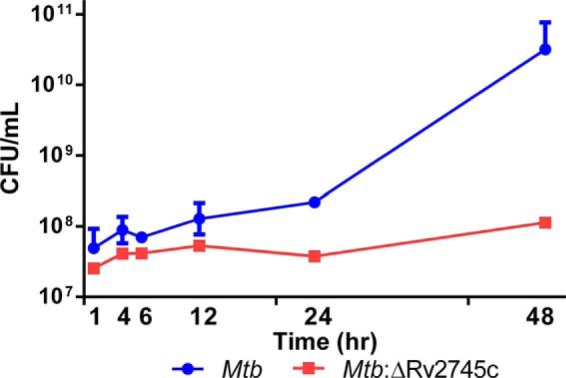
Reaeration growth curve. During reaeration the mutant shows decreased viability up to 48 h post-reaeration, whereas the wild type continues to replicate until 48 h-post reaeration. p = 0.0313. n = 3, except at t = 48 h, n = 2 for wild type.
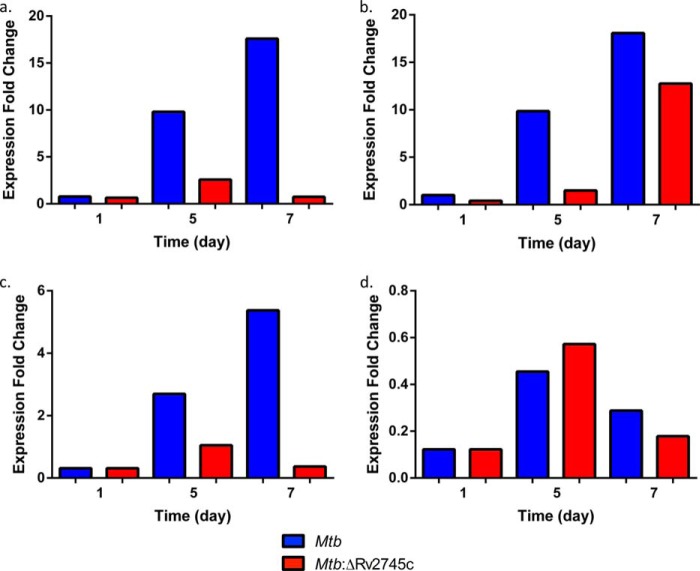
Thus, the survival of the Mtb:ΔRv2745c strain was impaired during the reaeration phase in vitro, which has the potential to model reactivation of the pathogen in vivo from intragranulomatous hypoxia. However, the survival of this mutant was not compromised during hypoxia itself, a condition during which Mtb, but not the mutant itself, exhibited high levels of induction of the Rv2745c transcript. Based on prior publications (9, 18), we hypothesized that the expression of genes involved in Clp proteolysis may be induced during hypoxia, downstream of Rv2745c. We found that the expression of clpP1, clpP2, and clpC was induced to high levels during late stages of hypoxia (days 5 and 7) in Mtb but not in the Mtb:ΔRv2745c mutant (Fig. 3, a–d). Thus, RT-PCR analysis revealed induction of clpP1 as early as 5 days post-hypoxia in Mtb (Fig. 3a). clpP1 levels increased at day 7, in which levels were induced ∼18-fold in the wild type (Fig. 3a). However, clpP1 levels remained low in Mtb:ΔRv2745c, in which levels were induced 2-fold at day 5 post-hypoxia, whereas levels remained unchanged at day 7 within the isogenic mutant (Fig. 3a). A similar pattern was observed with clpP2, with the exception of day 7 post-hypoxia in the isogenic mutant in which levels were induced by ∼15-fold (Fig. 3b). The similar pattern between clpP1 and clpP2 was expected as they are cotranscribed (16). The ATPase adapter clpC also had a similar pattern of induction when compared with clpP1 for both Mtb and Mtb:ΔRv2745c (Fig. 3c). On the contrary, the expression of the additional ATPase adaptor, clpX, was not induced during hypoxia in either strain, indicating that the transcription of this gene may either be unlinked to Rv2745c or be repressed by the Rv2745c-encoded protein or require additional players (Fig. 3d).
FIGURE 3.
RT-PCR of clp genes during hypoxia. RT-PCR of clp proteases during hypoxia. a, clpP1. b, clpP2. c, clpC. d, clpX. Induction of clpP1, clpP2, and clpC occurred by 5 days post-hypoxia in Mtb. clpP2 expression occurred at 7 days post-hypoxia treatment in Mtb:ΔRv2745c, whereas there was no induction of clpP1 and clpC throughout hypoxia treatment. clpX expression -fold change was <1 for all time points and strains throughout hypoxia.
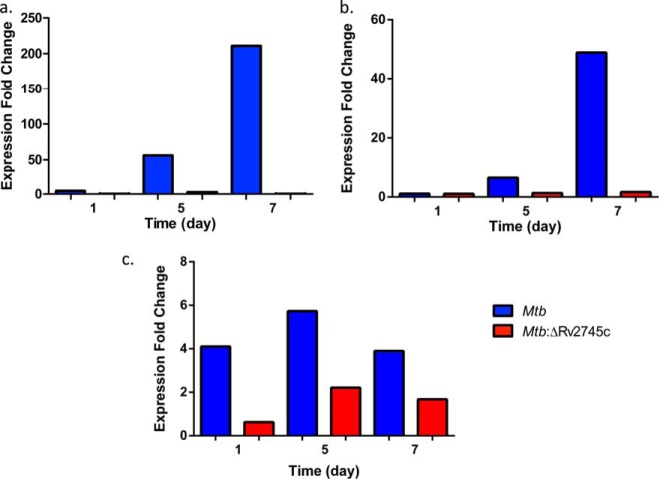
Because Rv2745c was induced in Mtb and not the isogenic mutant and because this gene is transcribed in conjugation with its neighbor Rv2744c, we also assessed the expression of this gene in both strains during hypoxia. As would be expected given the expression profiles of Rv2745c, a similar pattern was seen with Rv2744c induction, although the expression in the wild type when compared with Rv2745c levels were half that of Rv2745c, a result that could be explained by polar effects on a downstream transcript (Fig. 4a). More importantly, the expression of Rv2744c was detected as early as day 5 and by day 7 was >200-fold in hypoxia-stressed Mtb relative to control cultures. Furthermore, levels remained undetected in the mutant strain (Fig. 4a).
FIGURE 4.
RT-PCR of target genes during hypoxia. Expression levels of Rv2744c (a), σH (b), and σE (c) are shown.
Several groups have implicated that the network controlled by σH and σE is transcriptionally interlinked with the expression of Rv2745c during diverse stress conditions (10, 20, 21, 25). We, therefore, also studied the transcription of these two inducible sigma factors during hypoxia in both Mtb and the mutant strain. σH was induced ∼6.5-fold by day 5 hypoxia and ∼49-fold by day 7 post-hypoxia in Mtb, whereas its expression was not induced in Mtb:ΔRv2745c (Fig. 4b). σE was maximally induced at day 5 post-hypoxia in both Mtb and Mtb:ΔRv2745c; however, levels were lower in Mtb:ΔRv2745c (Fig. 4c). Overall, σE levels were induced to a lesser extent upon hypoxia treatment, similar to that of diamide treatment (17).
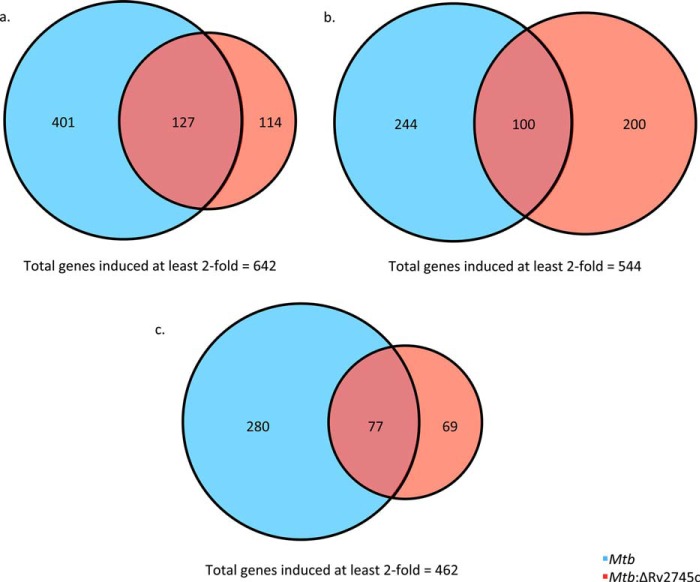
The differential expression pattern found upon RT-PCR analysis led us to investigate the differences at the global level, employing whole genome transcriptomics. There were subtle but key differences in the transcriptional profiles of Mtb and the isogenic mutant, Mtb:ΔRv2745c. After 1 day of hypoxia treatment, a total of 528 genes were induced at least 2-fold in Mtb, whereas only less than half that many (241) genes were induced at least 2-fold in the mutant (Fig. 5a). There was an overlap of 127 genes, or ∼20%, that were induced in both strains, whereas 114 unique genes were induced at least 2-fold in the isogenic mutant by day 1 post-hypoxia (Fig. 5a). By day 5 post-hypoxia, the number of genes induced at least 2-fold in Mtb and the mutant were 344 and 300, respectively, with an overlap of exactly 100 (∼18%) genes (Fig. 5b). By day 7 post-hypoxia, 357 and 146 genes were induced >2-fold in Mtb and the mutant, respectively, of which only 77 (16%) were shared (Fig. 5c). Thus, overall, the number of genes that were induced steadily declined throughout hypoxia treatment in both strains (Fig. 5, a–c). This differential response in the mutant is likely 2-fold in that the hypoxia-induced genes are lower overall for the mutant; however, this is in part due to the deletion of Rv2745c in the isogenic mutant, which consequently results in lower expression of genes in response to stress treatments, such as hypoxia and reaeration conditions.
FIGURE 5.
Venn diagrams of hypoxia-induced genes. Venn diagrams describe the extent of overlap between gene expression upon hypoxia treatment in Mtb (blue circles) and Mtb:ΔRv2745c (red circles). Genes induced at least 2-fold at 1 day (a), 5 days (b), and 7 days (c) post-hypoxia treatment are shown. n = 3.
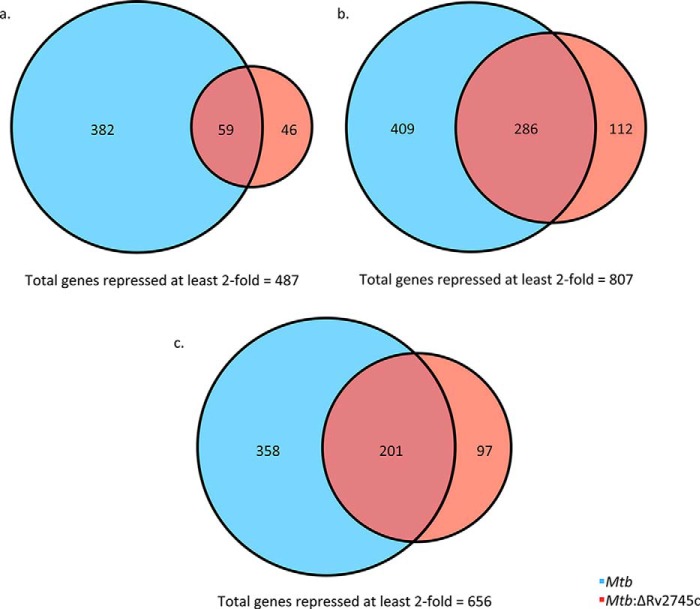
The numbers of genes repressed at least 2-fold in Mtb and the mutant were more divergent after day 1 post-hypoxia treatment (Fig. 6a). 441 and 105 genes were repressed at least 2-fold in Mtb and the mutant, respectively, after 1 day of hypoxia, of which only 59 (∼12%) were commonly repressed (Fig. 6a). By 5 days post-hypoxia, 694 and 398 genes were repressed >2-fold in Mtb and Mtb:ΔRv2745c, respectively, with 286 (∼35%) being similarly repressed in both strains (Fig. 6b). By 7 days post-hypoxia, 559 and 298 were repressed >2-fold in Mtb and the mutant, respectively. Approximately 30% (201) of the total repressed genes were repressed in both strains (Fig. 6c).
FIGURE 6.
Venn diagrams of hypoxia-repressed genes. Venn diagrams describe the extent of overlap between gene expression upon hypoxia treatment in Mtb (blue circles) and Mtb:ΔRv2745c (red circles). Genes repressed at least 2-fold at 1 day (a), 5 days (b), and 7 days (c) post-hypoxia treatment are shown. n = 3.
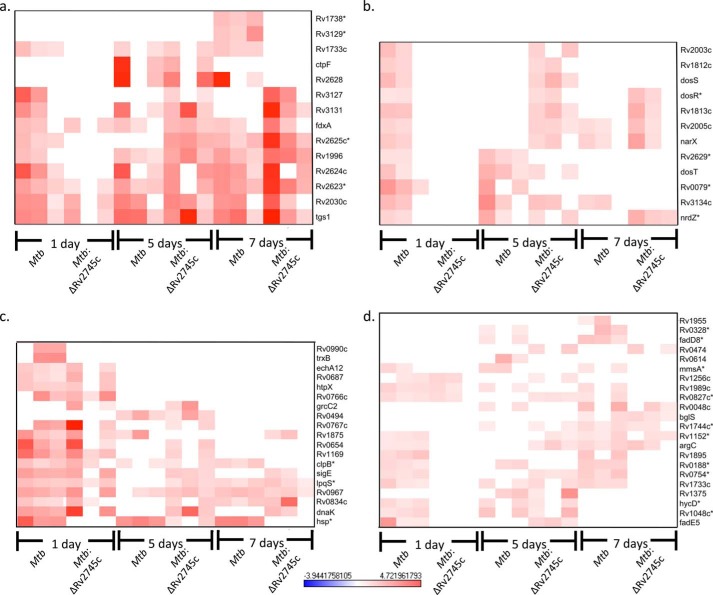
Hypoxic conditions induce many downstream signaling events, such as induction of the DosR regulon in Mtb (26). However, the comparable response within the mutant was delayed and reduced in magnitude. Thus, within the DosR regulon, genes that were induced maximally in Mtb were also induced within the Mtb:ΔRv2745c but to a lesser degree in the mutant (Fig. 7a). However, genes with a small magnitude of initial expression in Mtb (day 1 post-hypoxia) were induced at day 5 post-hypoxia in Mtb:ΔRv2745c (Fig. 7b). Additionally, these genes remained induced by day 7 post-hypoxia in the isogenic mutant, whereas expression levels remained unchanged in the wild type (Fig. 7b). Within the DosR regulon, the delayed response of genes that are minimally induced within the isogenic mutant may contribute to the greater decrease in growth of Mtb:ΔRv2745c throughout the hypoxia treatment. This disruption in DosR regulon signaling also implicates a role of Rv2745c in response to low oxygen levels. However, of note, fdxA, a ferredoxin that acts as an alternative electron acceptor upon low oxygen levels, was equally induced in both the wild type and the isogenic mutant (Fig. 7a) (12). Furthermore, tgs1, which is maximally induced upon dosR expression, encodes for a triacylglycerol synthase, and is used to synthesize triacylglycerol as a proposed energy source in Mtb during dormancy, was induced in both the wild type and the isogenic mutant (Fig. 7a) (11). Another important gene that is induced upon dosR activation is nrdZ, a ribonucleoside diphosphate reductase, that was also induced in Mtb and the isogenic mutant (Fig. 7b). However, nrdZ induction occurred at days 1 and 5 post-hypoxia in Mtb and at days 5 and 7 post-hypoxia in Mtb:ΔRv2745c, indicating that Rv2745c may perhaps play a role via clp protease activation (Fig. 7b).
FIGURE 7.
DosR regulon and EHR during hypoxia. Heat maps show the results of supervised hierarchical clustering. a, genes maximally induced in the DosR regulon. b, genes minimally induced in the DosR regulon. c, genes maximally induced in the EHR. d, genes minimally induced in the EHR. Genes induced in both the DosR regulon and the EHR in Mtb are induced as early as day 1 hypoxia, whereas there is a delayed response in Mtb:ΔRv2745c of genes that induced minimally in these pathways indicating that Rv2745c plays a role in these regulatory networks via direct or indirect regulation. n = 3. Red indicates induction, whereas blue indicates repression relative to the control channel. The intensity of each color corresponds to the magnitude. *, p < 0.05 for at least 1 time point.
As hypoxia also involves additional regulation through the EHR, we assessed the change in the EHR upon hypoxic conditions and found a similar pattern. Thus the response to the EHR was both reduced in magnitude as well as breadth and time. Of the genes that were induced to the highest level in Mtb, these genes were also induced in Mtb:ΔRv2745c (Fig. 7c). However, genes with a minimal level of induction in the wild type were not induced in the isogenic mutant until 5 days post-hypoxia, whereas these genes were induced by day 1 in the wild type (Fig. 7d). However, by days 5 and 7 post-hypoxia, there was a subset of genes that was no longer induced in the wild type that were induced at day 1 (Fig. 7c). This subset of genes is not induced within the isogenic mutant at any stage, also implying that Rv2745c is indeed facilitating a regulatory change that ultimately alters the expression levels of genes within the EHR.
The disruption of both the DosR regulon and EHR also occurs within the σH regulon. However, hsp, which is maximally induced in the wild type at all time points, was not induced in the isogenic mutant until day 5 post-hypoxia and was induced to a lesser extent than that of Mtb (Fig. 8). Again, there was a disruption of induction of specific genes within the σH regulon, such as trxB, Rv2745c, and σB within Mtb:ΔRv2745c (Fig. 8). These genes were induced at day 1 post-hypoxia in Mtb but not the mutant (Fig. 8). However, the expression levels of these genes dropped back to baseline by 5 days post-hypoxia in the wild-type strain (Fig. 8). Additionally, we observed a repression in the expression of Rv2743c and Rv2744c, two neighboring genes that the Rv2745c-encoded protein may also regulate, in the Mtb:ΔRv2745c mutant. This result implied that these genes are indeed cotranscribed upon Rv2745c activation (Fig. 8). Upon microarray analysis of σH expression levels, these levels are similar between that of the wild-type and isogenic mutant, which also supports a differential role of Rv2745c in a condition-dependent manner (Fig. 8).
FIGURE 8.
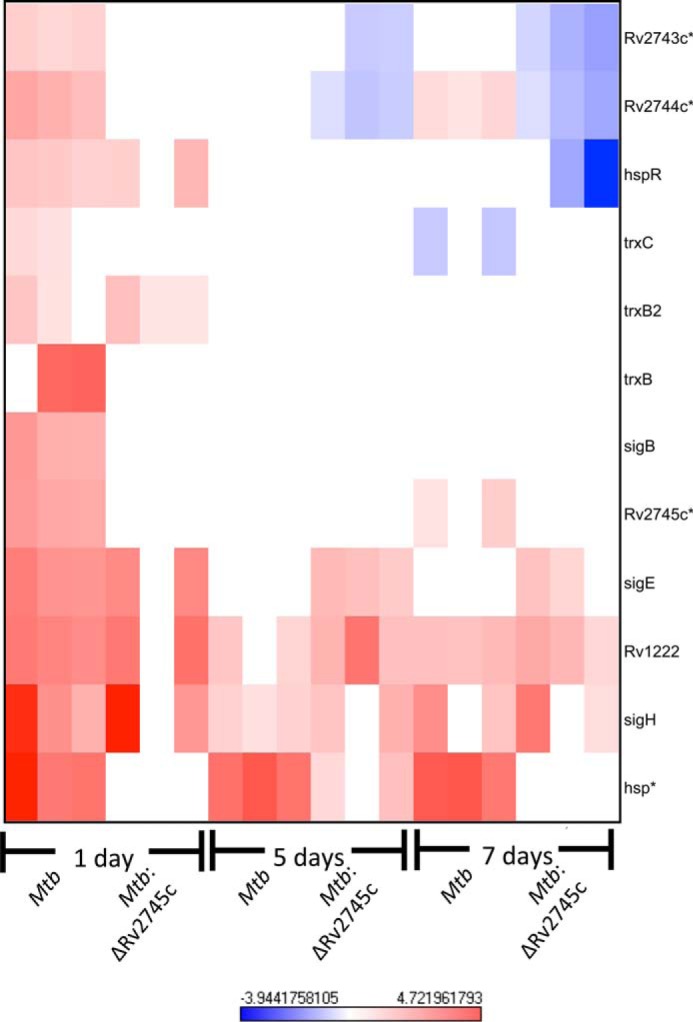
σH regulon during hypoxia. The heat map shows results of supervised hierarchical clustering focusing on the genes with the highest magnitude of change. Genes within the σH regulon are induced in Mtb during the initial stages of hypoxia, whereas only a subset of this regulon is induced in Mtb:ΔRv2745c, indicating that Rv2745c plays a differential regulatory role in a condition-dependent manner via direct or indirect regulation. n = 3. Red indicates induction, whereas blue indicates repression relative to the control channel. The intensity of each color corresponds to the magnitude. *, p < 0.05 for at least 1 time point.
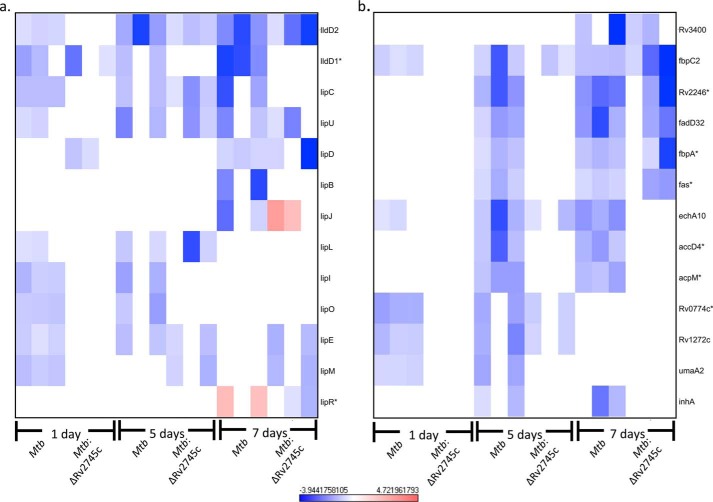
Upon hypoxic conditions, Mtb down-regulates multiple genes involved in metabolism, thus facilitating a transition from a metabolically active to dormant state. As such, we assessed the transcriptional profile of genes involved in both fatty acid metabolism and mycolic acid synthesis (Fig. 9, a and b). By day 1 post-hypoxia, several genes, such as lipL, lipO, and lipE, were down-regulated within the wild type (Fig. 9a). However, lipL and lipO levels remained at baseline within the isogenic mutant (Fig. 9a), whereas down-regulation of lipE occurred by day 5 post-hypoxia in the isogenic mutant (Fig. 9a). This further implicates that the transition from a metabolically active state to a dormant state is dysregulated in the isogenic mutant, which also supports the decreased cfu numbers that occur throughout the hypoxia treatment within the isogenic mutant relative to wild type.
FIGURE 9.
Fatty acid metabolism and mycolic acid synthesis repression during hypoxia. Heat maps show the results of supervised hierarchical clustering focusing on the genes with the highest magnitude of change. Shown are fatty acid metabolism (a) and mycolic acid synthesis genes (b). Genes involved in fatty acid metabolism and mycolic acid synthesis were repressed in Mtb during the initial stages of hypoxia, whereas only a subset of these genes were repressed in Mtb:ΔRv2745c at later stages of hypoxia indicating that Rv2745c plays a differential regulatory role in a condition-dependent manner via direct or indirect regulation. n = 3. Red indicates induction, whereas blue indicates repression relative to the control channel. The intensity of each color corresponds to the magnitude. *, p < 0.05 for at least 1 time point.
Our analysis revealed disruption of mycolic acid synthesis as well. As such, repression of several genes, umaA2, Rv1272c, and Rv0774c, was observed after 1 day of hypoxia treatment in Mtb (Fig. 9b). However, repression of Rv0774c and Rv1272c did not occur until day 5 post-hypoxia treatment in Mtb:ΔRv2745c (Fig. 9b). Hierarchical clustering revealed repression of acpM, accD4, fas, fbpA, fadD32, and Rv2246 by 5 days post-hypoxia in Mtb but not the isogenic mutant (Fig. 9b). However, by day 7 of hypoxia treatment, repression of fas, fbpA, fadD32, and Rv2246 occurred within the isogenic mutant (Fig. 9b). Expression levels remained unchanged in the isogenic mutant for acpM and accD4 at day 7 post-hypoxia (Fig. 9b). Again, the transition from a metabolically active state to a dormant state is dysregulated within Mtb:ΔRv2745c. Of particular note, fas and accD4, which are involved in lipid metabolism, are shown to be down-regulated upon hypoxic conditions, which we also found within our study (7).
As only a subset of Clp protease targets have yet to be identified, we scanned our expression profiles of Mtb and Mtb:ΔRv2745c to identify additional potential targets. Interestingly, σB, rpsL, dnaK, and Rv3334 have been identified as Clp protease substrates (27). As such, upon day 1 of hypoxia, these genes were induced in Mtb but were not induced at later time points (Fig. 10). However, rpsL and dnaK were induced by 1 day post-hypoxia in Mtb:ΔRv2745c. Induction of rpsL remained elevated at 5 days post-hypoxia in the isogenic mutant but returned to baseline by day 7 post-hypoxia, whereas dnaK remained induced throughout the entire hypoxia time course in Mtb:ΔRv2745c (Figs. 10 and 7, respectively). Not surprisingly, the return to baseline expression levels of these proposed Clp protease substrates in Mtb occurred concordantly with the induction of Rv2745c, clpP1, and clpP2 (Figs. 1b and 3, a and b). A similar pattern of induction followed by a return to baseline levels in Mtb were also seen with htpX, rpoB, rpsI, Rv1072, and Rv1073. Although these have yet to be identified as Clp protease substrates, they are possible targets.
FIGURE 10.
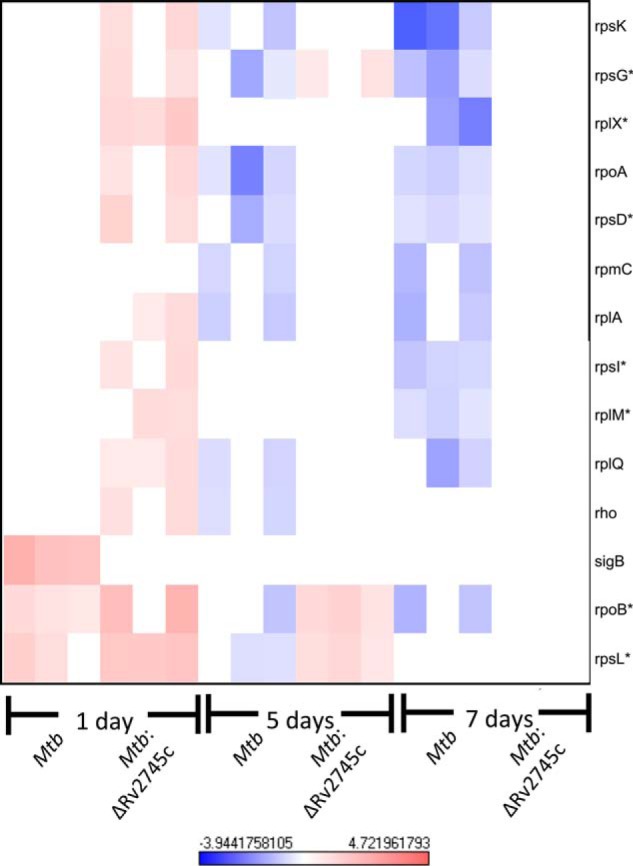
Known and potential Clp protease targets during hypoxia. The heat map shows the results of supervised hierarchical clustering focusing on the genes with the highest magnitude of change. Known and potential protease targets are displayed. Genes with reduced expression levels by days 5 and 7 post-hypoxia but expressed in the mutant at either 1 day or 5 days post-hypoxia were identified as potential Clp protease targets. n = 3. Red indicates induction, whereas blue indicates repression relative to the control channel. The intensity of each color corresponds to the magnitude. *, p < 0.05 for at least 1 time point.
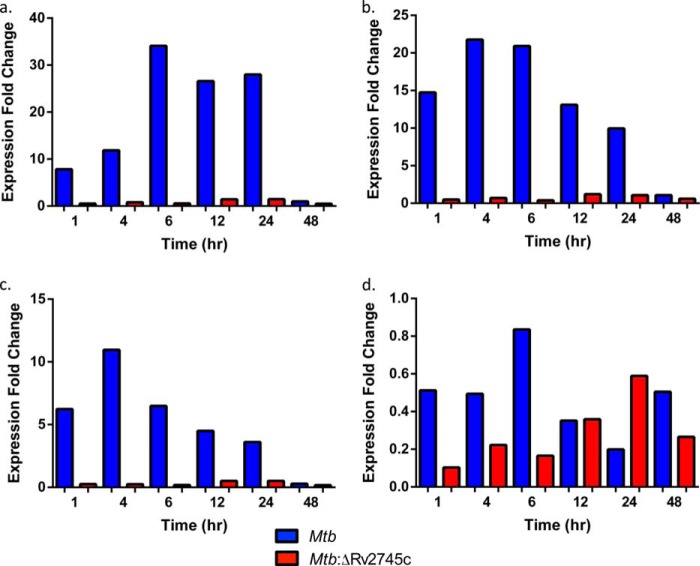
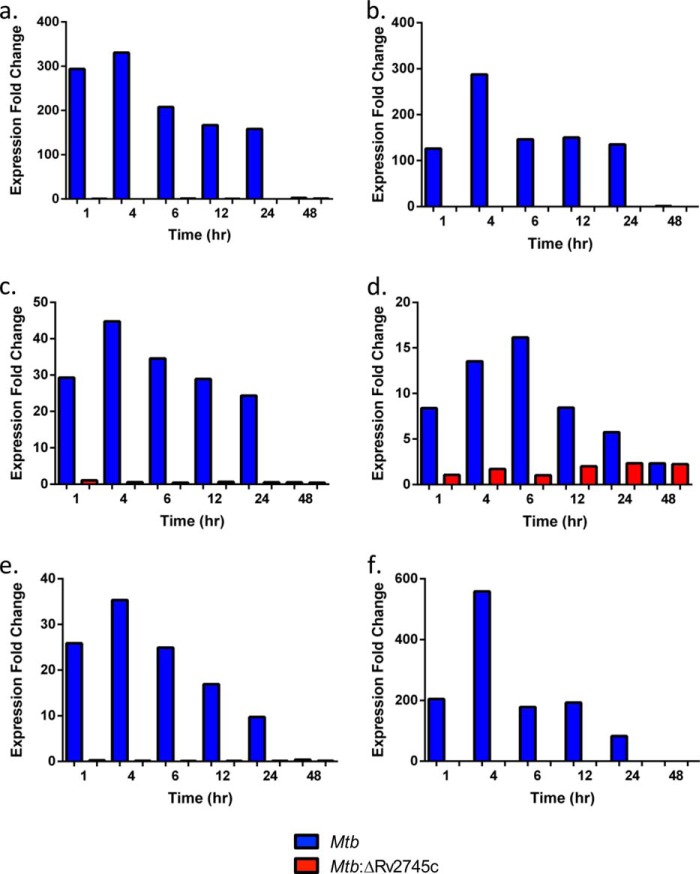
We found maximal clpP1 induction by 4 h upon RT-PCR analysis of Mtb, with our reported levels being higher than previously reported with the H37Rv strain (Fig. 11a). However, there was no clpP1 induction in the isogenic mutant, as expected, as Rv2745c encodes clgR, the Clp protease gene regulator (Fig. 11a). We also found higher levels of clpP2 induction in Mtb in which clpP2 was maximally induced by 4 h post-reaeration in our study (Fig. 11b). Again, there is no induction of clpP2 in the isogenic mutant, as expected (Fig. 11b). clpC induction follows a similar pattern as previously reported, with the exception that we again found higher levels of induction in Mtb (Fig. 11c) (18). However, we found that clpX was not induced in both Mtb and Mtb:ΔRv2745c (Fig. 11d). By 48 h post-reaeration, all clp gene expression levels returned to baseline (Fig. 11, a–d).
FIGURE 11.
RT-PCR of clp genes during reaeration. RT-PCR of clp proteases during reaeration clpP1 (a), clpP2 (b), clpC (c), and clpX (d) is shown. Induction of clpP1, clpP2, and clpC occurred by 1 h post-reaeration Mtb. However, there was no induction of clpP1, clpP2, and clpC throughout reaeration treatment within the isogenic mutant, Mtb:ΔRv2745c. clpX expression -fold change was <1 for all time points and strains throughout reaeration.
Upon RT-PCR analysis of Rv2745c by Sherrid et al. (18), they found maximal induction at 6 h post-reaeration followed by a drop in expression levels at 12 h post-reaeration, at which point Rv2745c expression stabilized. We found maximal induction by 4 h post-reaeration in Mtb and stabilization in expression by 12 h post-reaeration (Fig. 12a). As expected, the isogenic mutant did not express Rv2745c (Fig. 12a). Upon analysis of the neighboring gene, Rv2744c, we found a similar pattern of induction as compared with Rv2745c, albeit at lower expression levels, in Mtb (Fig. 12b). However, the isogenic mutant showed no expression of Rv2744c (Fig. 12b). The lack of Rv2744c expression, which also occurred upon hypoxia treatment, again implicates that Rv2745c and Rv2744c are cotranscribed. As we have previously shown that Rv2745c plays a role in the positive feedback loop of the σH regulon, we also assessed σH and σE expression levels upon reaeration (17). We found that upon reaeration, there is disruption of σH expression in Mtb:ΔRv2745c upon RT-PCR analysis (Fig. 12c). However, σH was induced in Mtb and followed a similar pattern of induction with respect to Rv2745c, except with lower levels of expression (Fig. 12c). σE showed low levels of expression, in which the highest levels were reached at 12 h post-reaeration (2.3-fold change) in Mtb:ΔRv2745c (Fig. 12d). Within Mtb, σE was maximally induced by 6 h post-reaeration (Fig. 12d). As dosR and acr are expressed throughout hypoxia, we analyzed these expression levels to show a transition from dormancy to a replicating state. As such, dosR levels decreased to ∼10-fold induction by 24 h post-reaeration, indicating that Mtb sensed increasing oxygen levels (Fig. 12e). By 48 h, both dosR and acr levels returned to baseline (Fig. 12, e and f). In the isogenic mutant, dosR induction did not occur, whereas at 7 days post-hypoxia levels were ∼10-fold, indicating that a transition to a vegetative state does occur with Mtb:ΔRv2745c but does not occur optimally, as dosR expression is needed to transition from a dormant to vegetative state (Fig. 12e) (8). Similar patterns with respect to dosR expression were also found with acr for both Mtb and Mtb:ΔRv2745c (Fig. 12f).
FIGURE 12.
RT-PCR of target genes during reaeration. Expression levels of Rv2745c (a), Rv2744c (b), σH (c), σE (d), dosR (e), and acr (f) are shown.
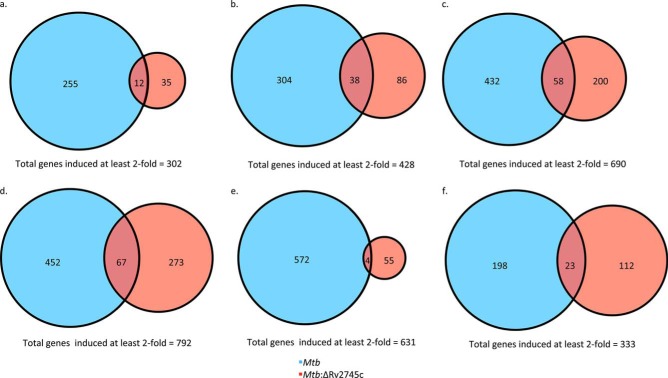
The disruption in clp protease expression within the isogenic mutant led us to perform additional analyses of the transcriptome. Upon 1 h post-reaeration, a total of 302 genes were induced >2-fold; however, only 35 were uniquely induced in the Mtb:ΔRv2745c and only 12 were induced in both Mtb and Mtb:ΔRv2745c (Fig. 13a). Within Mtb, a total of 255 genes were uniquely induced >2-fold (Fig. 13a). 88% of all genes induced >2-fold were induced within Mtb (Fig. 13a). This indicates there is dysregulation within Mtb:ΔRv2745c upon the transition out of a dormant state, which perhaps occurs because the isogenic mutant has a delayed response to increased oxygen levels. By 4 h, there was an increase in the number of genes that were induced at least 2-fold, in which a total of 446 genes were induced (Fig. 13b). However, of these 446 genes, a total of 297 were uniquely induced in Mtb, and 104 were uniquely induced in Mtb:ΔRv2745c (Fig. 13b). However, only 45 genes were induced in both strains, accounting for only 10% of genes induced at least 2-fold by 4 h post-reaeration (Fig. 13b). By 6 h post-reaeration, the number of genes induced >2-fold rose to 690 (Fig. 13c). Again, a majority (70%) of these genes were induced in Mtb, with a total of 490 (including uniquely induced and genes induced in both strains), whereas only 258 were induced in the mutant (Fig. 13c). By 12 h post-reaeration, the highest number of genes were induced in which 792 were expressed >2-fold (Fig. 13d). Again, a large number of genes induced >2-fold were expressed by Mtb, with 452 being uniquely expressed, whereas only 273 were uniquely expressed by Mtb:ΔRv2745c (Fig. 13d). However, by 24 h post-reaeration, there was a large disruption in signaling events within the isogenic mutant. As such, only 59 genes were induced >2-fold within Mtb:ΔRv2745c, with only 4 genes also induced in Mtb as well (Fig. 13e). An even greater number of genes were induced >2-fold in Mtb in which 576 genes were induced (Fig. 13e). By 48 h post-reaeration, a total of 333 genes were induced in both Mtb and Mtb:ΔRv2745c, of which only 23 (∼7%) genes were commonly induced >2-fold in both strains, which may account for the large differences in cfu numbers at this time point (Fig. 13f). By 48 h post-reaeration a total of 59% of genes were induced at least 2-fold in the parental strain (Fig. 13f).
FIGURE 13.
Venn diagrams of reaeration-induced genes. Venn diagrams describe the extent of overlap between gene expression upon reaeration treatment in Mtb (blue circles) and Mtb:ΔRv2745c (red circles). Genes induced at least 2-fold at 1 h (a), 4 h (b), 6 h (c), 12 h (d), 24 h (e), and 48 h (f) post-reaeration treatment are shown. n = 3.
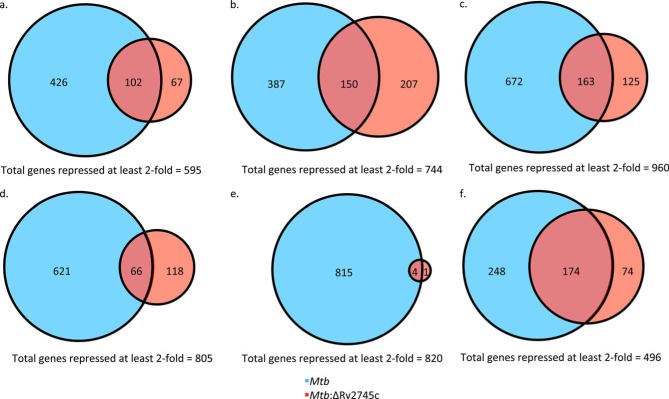
The disruption in expression within Mtb:ΔRv2745c was also reflected in the repressed genes profiles. By 1 h post-reaeration, a total of 595 genes were repressed >2-fold (Fig. 14a). Of these repressed genes, a total of 426 were uniquely repressed in Mtb, 102 were repressed in both strains, and 67 were uniquely repressed in Mtb:ΔRv2745c (Fig. 14a). By 4 h post-reaeration, a total of 744 genes were repressed, with ∼52% of genes (387) being uniquely repressed within Mtb, 20% of genes (150) being repressed in both strains, and ∼28% being repressed within Mtb:ΔRv2745c (Fig. 14b). However, by 6 h post-reaeration the number of genes repressed >2-fold jumps to a total of 960 genes, in which a majority (70%) of these genes were uniquely repressed within Mtb (Fig. 14c). However, 163 genes were repressed in both strains, and only 13% (125 genes) of genes were uniquely repressed within the isogenic mutant (Fig. 14c). However, by 12 h post-reaeration, the number of repressed genes dropped to 805, in which 621 genes were uniquely repressed in Mtb, and only 118 were repressed within the mutant (Fig. 14d). By 24 h post-reaeration, there was a drastic disruption in signaling in Mtb:ΔRv2745c, in which only 1 gene was uniquely repressed >2-fold, whereas 815 were uniquely repressed in Mtb and 4 genes were commonly repressed >2-fold (Fig. 14e). This lack of coordinated repression was more than likely due to the buildup of proteins to toxic levels, thus skewing feedback loops in the isogenic mutant and leading to cell death, as shown by the decreased cell viability at the 24 h post-reaeration time point (Fig. 2). However, by 48 h cell viability was restored, which was also reflected in the number of genes that were repressed >2-fold in the isogenic mutant, which is 248 genes in total, with 174 being commonly repressed by at least 2-fold in Mtb (Fig. 14f). A total of 248 genes were uniquely repressed in Mtb, with a total of 322 being repressed in the wild type at 48 h post-reaeration (Fig. 14f).
FIGURE 14.
Venn diagrams of reaeration-repressed genes. Venn diagrams describe the extent of overlap between gene expression upon reaeration treatment in Mtb (blue circles) and Mtb:ΔRv2745c (red circles). Genes induced at least 2-fold at 1 h (a), 4 h (b), 6 h (c), 12 h (d), 24 h (e), and 48 h (f) post-reaeration treatment is shown. n = 3.
Upon hierarchical clustering, the expression of DosR regulon was found to steadily decrease upon reaeration within Mtb (Fig. 15). However, the regulatory response of the DosR regulon of Mtb:ΔRv2745c was ablated in which expression levels return to baseline within 1 h post-reaeration (Fig. 15). This indicates that there is a differential response within the isogenic mutant that is perhaps in response to the lower levels of dosR expression throughout the hypoxia treatment. Consequently, this implied that Mtb:ΔRv2745c did not enter a complete dormant state upon hypoxia treatment. Thus, upon increasing oxygen conditions, the transition to a vegetative state was disrupted. Furthermore, it has been shown that expression of dosR is crucial for the optimal shift from a dormant, nonreplicating state to a vegetative state (8). As such, loss of proper dosR signaling and activation of the dosR regulon may contribute to decreased viability of Mtb:ΔRv2745c. Again as was found with hypoxia, the dosR regulon antigen Rv1733c was expressed within Mtb at each of the time points up to 48 h post-reaertion, pointing to the idea that this antigen facilitates a role in the immune response and perhaps contributing to the survival of Mtb within the host (Fig. 15). However, Rv1733c was not expressed within the isogenic mutant, further pointing to an altered antigenic profile with respect to the wild type at earlier stages of reaeration (Fig. 15). By 48 h, the expression levels of a majority of genes within the dosR regulon went back to baseline in both the wild type and the isogenic mutant (Fig. 15).
FIGURE 15.
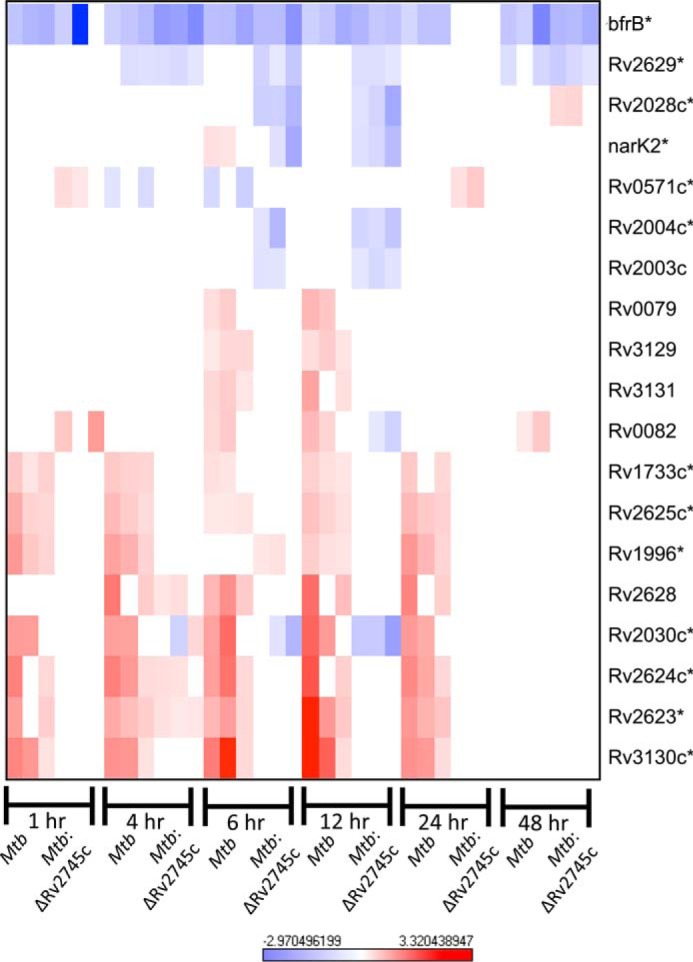
DosR regulon during reaeration. The heat map shows the results of supervised hierarchical clustering focusing on the genes with the highest magnitude of change. The DosR regulon is induced in Mtb, whereas the response in Mtb:ΔRv2745c is ablated, indicating that Rv2745c plays a role in this regulatory network. n = 3. Red indicates induction, whereas blue indicates repression relative to the control channel. The intensity of each color corresponds to the magnitude. *, p < 0.05 for at least 1 time point.
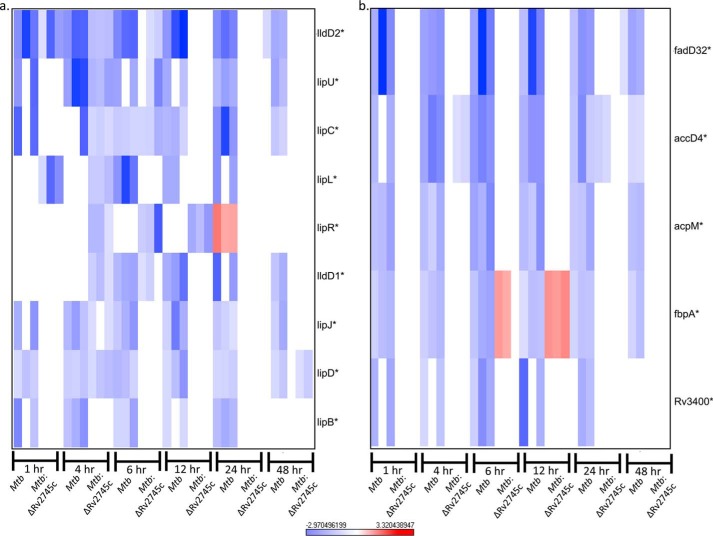
Upon reaeration, there was still down-regulation of fatty acid genes within Mtb (Fig. 16a). This may be in response to a slower return to a metabolically active state of Mtb. Again, genes involved in fatty acid metabolism within the isogenic mutant were at basal levels, also implying a defect in the transition to a replicating state (Fig. 16a). By 48 h post-reaertion this down-regulation within the wild-type was ablated. Concurrently, a similar pattern was seen with mycolic acid synthesis genes in that there was down-regulation within the wild type and no change in expression levels within the isogenic mutant, with ablation of expression levels by 48 h in Mtb (Fig. 16b). Repression of fatty acid metabolism genes and mycolic acid synthesis genes may be beneficial in the transition to a vegetative state in that the pathogen maintains lower levels of energy requirements as mycolic acid synthesis is costly in terms of energy requirements. However, of note, fbpA expression levels were induced in Mtb:ΔRv2745c, whereas they were down-regulated in Mtb at both 6 and 12 h post-reaeration (Fig. 16b). As such, this altered fbpA expression may play a role in pathogenesis. A ΔfbpA mutant in the H37Rv strain is attenuated in macrophages as the cell wall is deficient in lipids and is more susceptible to oxidative stress employed by macrophages (28). Thus, fbpA contributes to the cell wall profile of Mtb. As such, this gene more than likely plays a role in the transition from a dormant to replicating state upon the introduction of oxygen levels; thus this differential profile of Mtb:ΔRv2745c may be a compensatory change that the mutant undergoes, as its cell wall profile may be different from that of the wild type upon hypoxic and reaeration treatments.
FIGURE 16.
Fatty acid metabolism and mycolic acid synthesis repression during reaeration. Heat maps show the results of supervised hierarchical clustering focusing on the genes with the highest magnitude of change. Shown are fatty acid metabolism (a) and mycolic acid synthesis genes (b). Genes involved in fatty acid metabolism and mycolic acid synthesis were repressed in Mtb during reaeration, whereas these genes are not repressed in Mtb:ΔRv2745c during reaeration. n = 3. Red indicates induction, whereas blue indicates repression relative to the control channel. The intensity of each color corresponds to the magnitude. *, p < 0.05 for at least 1 time point.
Thus far, RpsL has been identified as a Clp protease target under normal culturing conditions (27). Upon expression profile analysis, we found no expression of rpsL within Mtb at early stages or reaeration, whereas there was increased expression of rpsL at later stages of reaeration within the mutant (Fig. 17). By 24 h reaeration, there was down-regulation of rpsL within Mtb (Fig. 17). By comparing genes that are differentially regulated in Mtb and the mutant (i.e. down-regulation in wild type and up-regulation in the mutant), we identified additional genes that may be potential Clp protease targets (Fig. 17). Similar to what we found upon hypoxia treatment, we found that rpoB follows this pattern. Both rpsL and rpoB genes encode for ribosomal proteins, which are fairly stable proteins, and thus regulation at the transcriptional level would require longer periods of time to lead to decreased levels at the proteomic level. As such, it is more likely that these proteins would be targeted for proteolytic degradation, as Mtb requires more rapid changes in protein levels due to the nature of its virulence lifecycle and the necessity to respond to rapidly changing environmental stressors it encounters upon infection.
FIGURE 17.
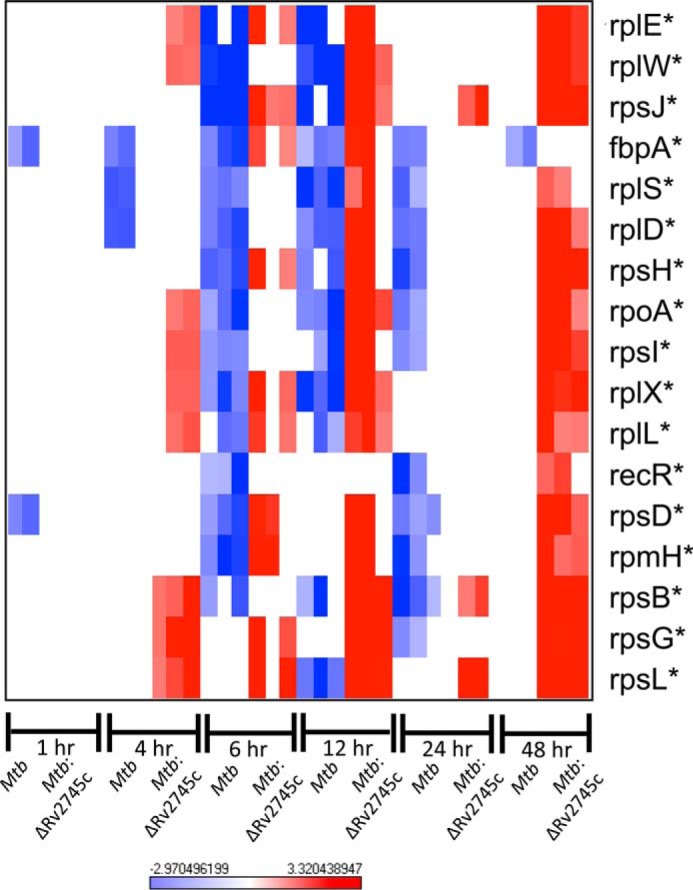
Known and potential Clp protease targets during reaeration. The heat map shows results of supervised hierarchical clustering focusing on the genes with the highest magnitude of change. Known and potential protease targets are displayed. Genes with reduced expression levels by 6 and 12 h post-reaeration but expressed in the mutant at either 4, 6, or 12 h post-reaeration were identified as potential Clp protease targets. n = 3. Red indicates induction, whereas blue indicates repression relative to the control channel. The intensity of each color corresponds to the magnitude. *, p < 0.05 for at least 1 time point.
DISCUSSION
Mtb has evolved into a very successful pathogen of human lung tissues (29), in part due to its ability to modulate its gene expression in response to changes in its environment, which undoubtedly occur during the different parts of its lifecycle. Mtb achieves this by invoking the expression of multiple stress induced sigma factor-anti-sigma factor networks, eukaryotic-like protein kinases, two-component regulatory systems, and transcriptional activators and repressors (22, 30–34). In addition to transcriptional regulation, it is increasingly being recognized that post-translational regulation of gene expression is a key component of Mtb phase variation (27). Thus, several dozen toxins encoded by the Mtb genome contain the potential to target and degrade specific transcripts and are regulated by their cognate antitoxin pairs yet exhibit promiscuous binding affinities (35, 36). Numerous small RNA molecules with the ability to disrupt gene expression have been identified (37).
In this context the role of ATP-dependent Clp proteolysis, which also has the potential to regulate Mtb gene expression at the post-transcriptional level, is noteworthy. It has been suggested that Clp proteolysis is essential for the survival of Mtb, though this system is currently understudied. Furthermore, the Clp proteolysis complex is increasingly being recognized as a key druggable target (38), especially in Gram-positive bacteria (39, 40), including Mtb (41–43). There are key implications of Rv2745c and the ATP-dependent Clp proteolysis genes being induced during stress. The interplay between this regulon and stress-induced sigma factor modules further indicates that clgR and clp genes are induced in and play an important role during Mtb infection. Therefore, elucidating the mechanisms of regulation of these networks is likely to significantly extend our knowledge about how Mtb is able to survive in vivo in the face of host immune mechanisms. However, there are specific shortcomings in our understanding of this process. We do not completely understand how the protein encoded by the clgR or Rv2745c gene results in the induction of the downstream proteolytic enzymes under some but not all of the conditions in which its own levels are highly induced (17).
Our key results show that although the isogenic mutant did not exhibit a significant defect in growth during 7-day hypoxia, it failed to recover in the subsequent reaeration phase to such that when compared with Mtb, its growth was significantly slower. Taken in the context of latency and reactivation, it is likely that the isogenic mutant would be defective during the initial stages of hypoxia and reactivation, implicating that Rv2745c induction, which leads to clp expression upon reaeration, is important for later stages of survival upon transitioning from a dormant to replicating state that occurs within the lung during reactivation. It is also possible that the mutant would exhibit a phenotype during long term hypoxia. Or a lack of growth-restriction phenotype in the mutant during hypoxia may simply reflect the generally slow replication rates of mycobacteria in that condition. Thus, hypoxia affects the replication rate of Mtb in a profound manner, impacting the expression of a large percentage of its genome. It is, therefore, conceivable that accumulation of misfolded proteins is a lesser impediment to the pathogen during hypoxia. However, during reaeration, rapid influx of oxygen allows Mtb to replicate at significantly higher rates and results in the transcription and translation of a majority of its genome. It is under these conditions that the Mtb:ΔRv2745c mutant exhibits sensitivity to its inability to induce the Clp proteolytic regulon. Our results are mirrored by those from the Lewis laboratory (43) that has shown that chemical disruption in Clp proteolysis kills Mtb.
Interestingly, we detected higher expression levels of the ATPase adaptor, clpC1, when compared with the ATPase adaptor protein, clpX, during both hypoxia and reaeration. It has been proposed by others that perhaps ClpX interacts with ClpP1 and ClpP2 to degrade Ssr-A tagged proteins, whereas ClpC1 interacts with ClpP2 to facilitate degradation of untagged proteins (9). As such, higher levels of ClpC1 during hypoxia and reaeration may occur in response to protein turnover as opposed to Ssr-A-tagged proteins that would occur in response to transcriptional errors, such as ribosomal stalling. However, another group did find increased expression of clpX during reaeration, although expression levels remained low, with maximal induction 2-fold higher relative to the control (18). These differences in results may be in response to different parental strains for the studies in which our studies used CDC1551, whereas the other group used the laboratory adapted strain, H37Rv.
Targeted degradation of Clp protease substrates can lead to a positive feedback loop in which mRNA levels become depleted. Although a majority of Clp protease substrates are currently unknown in Mtb, a small subset of substrates have been identified using a conditional knock down under normal culturing conditions (27). Interestingly, σB, rpsL, dnaK, and Rv3334 have been identified as Clp protease substrates (27). As such, upon day 1 of hypoxia, these genes were induced in Mtb but were not induced at later time points (Fig. 10). However, rpsL and dnaK were induced by 1 day post-hypoxia in Mtb:ΔRv2745c. Induction of rpsL remained elevated at 5 days post-hypoxia in the isogenic mutant but returned to baseline by day 7 post-hypoxia, whereas dnaK remained induced throughout the entire hypoxia time course in Mtb:ΔRv2745c (Figs. 10 and 7, respectively). Not surprisingly, the return to baseline expression levels of these proposed Clp protease substrates in Mtb occurred concordantly with the induction of Rv2745c, clpP1, and clpP2 (Figs. 1b and 3, a and b). A similar pattern of induction followed by a return to baseline levels in Mtb was also seen with htpX, rpoB, rpi, Rv1072, and Rv1073. Although the proteins encoded by these genes have yet to be identified as Clp protease substrates, they are possible targets.
rpsL encodes for the ribosomal protein S12 subunit, and mutations within rpsL are associated with streptomycin resistance (44). rpoB encodes for DNA-dependent RNA polymerase, and mutations within rpoB can result in rifampicin resistance (44, 45). dnaK encodes heat shock protein that is the most abundant protein within Mtb and plays a crucial role in preventing partially misfolded proteins from aggregating by binding to hydrophobic residues, which then allows these proteins to refold (46). As such, coordinated transcriptional and proteomic regulation of these known and proposed Clp protease substrates is crucial to the transition from a metabolically active to dormant state. Additionally, these proteins are relatively stable; thus transcriptional regulation would not lead to lower protein levels in a timely manner in response to environmental stressors. As such, targeting these proteins at the proteolytic level would lead to depletion of these proteins at a rate that is necessary for responding to changes in environmental cues.
Several known antigens within the DosR regulon were induced earlier and to higher levels in Mtb when compared with Mtb:ΔRv2745c. For instance, Rv1733c, Rv1738, and Rv2628 are known antigens that have been studied in murine models (8). Specifically, mice infected with Mtb recognize the Rv1733c gene product, which is one of the top ten antigens that is recognized by individuals exposed to Mtb (8). Interestingly, Rv1733c is induced at earlier stages of hypoxia within Mtb and is expressed at higher levels when compared with Mtb:ΔRv2745c (Fig. 7a). Furthermore, DosR regulon antigens are preferentially recognized by T cells of latently infected TB patients and not active TB individuals, implicating that the DosR regulon antigenic profile contributes to host immune pressures that facilitate containment of the bacilli to the granuloma (8). As such, the isogenic mutant has slower activation of the DosR regulon as well as less expression of this regulon. This altered transcriptional profile of Mtb:ΔRv2745c indicates that Clp proteases may perhaps play a role in facilitating the antigenic profile of Mtb during the hypoxic conditions that are found during the latent stages of infection.
Although there was disruption of genes involved within several important regulatory networks that play a role in response to hypoxia, the transcriptional profiles of Mtb and Mtb:ΔRv2745c are not as drastically different relative to diamide treatment. This is more than likely is due to the regulatory role of Rv2745c induction upon hypoxia treatment, which leads to Clp protease activity; thus the transition from a metabolically active state to a dormant state also relies more heavily upon changes at the proteomic level rather than at the transcriptional level, as was observed with diamide treatment (17).
As such, the disruption of signaling seen with the isogenic mutant in conjunction with the lack of protease expression and decreased viability throughout the course of reaeration indicate that Rv2745c facilitates viability through induction of Clp proteases. The absence of Clp proteases within the isogenic mutant leads to dysregulation at the transcriptional level, as the feedback loop regulating protein levels is skewed. Additionally, the inability of targeted proteolytic degradation of proteins more than likely leads to the build up of proteins that ultimately reached toxic levels in Mtb:ΔRv2745c. Consequently, this leads to cell death upon reaeration conditions. The ablated response within the DosR regulon of the isogenic mutant as well as decreased viability indicates that Rv2745c induces Clp proteases as a means to facilitate the transition from a dormant state to a vegetative state.
The role of Rv2745c under varying stress conditions and its implication in Mtb survival in vivo needs further elucidation. Further study that links proteomic to transcriptional changes in response to stress conditions that lead to differential induction of clp proteases is also of particular interest. As such, these studies are currently under way in our laboratory. The condition-dependent induction of Clp proteases upon Rv2745c activation and their role during infection also needs further study. Mtb Clp proteases have become an ideal drug target recently, as disruption of Clp protease machinery leads to cell death as protein concentrations increase, leading to toxic levels. In this investigation we clearly show a role for both Rv2745c and Clp proteases during reaeration in vitro, a representative condition of reactivation in vivo. As one-third of the world's population is infected with Mtb, reactivation of latent TB infections poses a serious threat. The current state of therapeutics is far from ideal, and as such development of more effective and less toxic treatments that a target-specific gene(s) are becoming increasingly appealing.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AI089323.
S. Mehra, T. W. Foreman, P. J. Didier, M. H. Ahsan, T. A. Hudock, R. Kissee, N. Golden, U. S. Gautam, X. Alvarez, K. E. Russell-Lodrigue, L. A. Doyle, C. J. Roy, J. L. Blanchard, S. A. Khader, A. A. Lackner, D. R. Sherman, and D. Kaushal, submitted for publication.
- Mtb
- M. tuberculosis
- TB
- tuberculosis
- EHR
- enduring hypoxic response
- RT
- real-time.
REFERENCES
- 1. Chao M. C., Rubin E. J. (2010) Letting sleeping dos lie: does dormancy play a role in tuberculosis? Annu. Rev. Microbiol. 64, 293–311 [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization. (2013) Global Tuberculosis Report 2013, p. 1, World Health Organization, Geneva [Google Scholar]
- 3. Singh S., Saraav I., Sharma S. (2014) Immunogenic potential of latency associated antigens against Mycobacterium tuberculosis. Vaccine 32, 712–716 [DOI] [PubMed] [Google Scholar]
- 4. Philips J. A., Ernst J. D. (2012) Tuberculosis pathogenesis and immunity. Annu. Rev. Pathol. 7, 353–384 [DOI] [PubMed] [Google Scholar]
- 5. Russell D. G. (2007) Who puts the tubercle in tuberculosis? Nat Rev. Microbiol. 5, 39–47 [DOI] [PubMed] [Google Scholar]
- 6. Cilfone N. A., Perry C. R., Kirschner D. E., Linderman J. J. (2013) Multi-scale modeling predicts a balance of tumor necrosis factor-α and interleukin-10 controls the granuloma environment during Mycobacterium tuberculosis infection. PloS ONE 10.1371/journal.pone.0068680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Galagan J. E., Minch K., Peterson M., Lyubetskaya A., Azizi E., Sweet L., Gomes A., Rustad T., Dolganov G., Glotova I., Abeel T., Mahwinney C., Kennedy A. D., Allard R., Brabant W., Krueger A., Jaini S., Honda B., Yu W. H., Hickey M. J., Zucker J., Garay C., Weiner B., Sisk P., Stolte C., Winkler J. K., Van de Peer Y., Iazzetti P., Camacho D., Dreyfuss J., Liu Y., Dorhoi A., Mollenkopf H. J., Drogaris P., Lamontagne J., Zhou Y., Piquenot J., Park S. T., Raman S., Kaufmann S. H., Mohney R. P., Chelsky D., Moody D. B., Sherman D. R., Schoolnik G. K. (2013) The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499, 178–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen T., He L., Deng W., Xie J. (2013) The Mycobacterium DosR regulon structure and diversity revealed by comparative genomic analysis. J. Cell. Biochem. 114, 1–6 [DOI] [PubMed] [Google Scholar]
- 9. Personne Y., Brown A. C., Schuessler D. L., Parish T. (2013) Mycobacterium tuberculosis ClpP proteases are co-transcribed but exhibit different substrate specificities. PloS ONE 10.1371/journal.pone.0060228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rustad T. R., Harrell M. I., Liao R., Sherman D. R. (2008) The enduring hypoxic response of Mycobacterium tuberculosis. PloS ONE 10.1371/journal.pone.0001502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chauhan S., Tyagi J. S. (2009) Powerful induction of divergent tgs1-Rv3131 genes in Mycobacterium tuberculosis is mediated by DevR interaction with a high-affinity site and an adjacent cryptic low-affinity site. J. Bacteriol. 191, 6075–6081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhat S. A., Singh N., Trivedi A., Kansal P., Gupta P., Kumar A. (2012) The mechanism of redox sensing in Mycobacterium tuberculosis. Free Radic. Biol. Med. 53, 1625–1641 [DOI] [PubMed] [Google Scholar]
- 13. Converse P. J., Karakousis P. C., Klinkenberg L. G., Kesavan A. K., Ly L. H., Allen S. S., Grosset J. H., Jain S. K., Lamichhane G., Manabe Y. C., McMurray D. N., Nuermberger E. L., Bishai W. R. (2009) Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect. Immun. 77, 1230–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malhotra V., Sharma D., Ramanathan V. D., Shakila H., Saini D. K., Chakravorty S., Das T. K., Li Q., Silver R. F., Narayanan P. R., Tyagi J. S. (2004) Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 231, 237–245 [DOI] [PubMed] [Google Scholar]
- 15. Via L. E., Lin P. L., Ray S. M., Carrillo J., Allen S. S., Eum S. Y., Taylor K., Klein E., Manjunatha U., Gonzales J., Lee E. G., Park S. K., Raleigh J. A., Cho S. N., McMurray D. N., Flynn J. L., Barry C. E., 3rd (2008) Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76, 2333–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Raju R. M., Unnikrishnan M., Rubin D. H., Krishnamoorthy V., Kandror O., Akopian T. N., Goldberg A. L., Rubin E. J. (2012) Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog. 10.1371/journal.ppat.1002511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McGillivray A., Golden N. A., Gautam U. S., Mehra S., Kaushal D. (2014) The Mycobacterium tuberculosis Rv2745c plays an important role in responding to redox stress. PloS ONE 10.1371/journal.pone.0093604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sherrid A. M., Rustad T. R., Cangelosi G. A., Sherman D. R. (2010) Characterization of a Clp protease gene regulator and the reaeration response in Mycobacterium tuberculosis. PloS ONE 10.1371/journal.pone.0011622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mehra S., Dutta N. K., Mollenkopf H. J., Kaushal D. (2010) Mycobacterium tuberculosis MT2816 encodes a key stress-response regulator. J. Infect. Dis. 202, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dutta N. K., Mehra S., Kaushal D. (2010) A Mycobacterium tuberculosis sigma factor network responds to cell-envelope damage by the promising anti-mycobacterial thioridazine. PloS ONE 10.1371/journal.pone.0010069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Provvedi R., Boldrin F., Falciani F., Palù G., Manganelli R. (2009) Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology 155, 1093–1102 [DOI] [PubMed] [Google Scholar]
- 22. Mehra S., Kaushal D. (2009) Functional genomics reveals extended roles of the Mycobacterium tuberculosis stress response factor σH. J. Bacteriol. 191, 3965–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manganelli R., Voskuil M. I., Schoolnik G. K., Dubnau E., Gomez M., Smith I. (2002) Role of the extracytoplasmic-function σ factor σH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45, 365–374 [DOI] [PubMed] [Google Scholar]
- 24. Gautam U. S., Mehra S., Ahsan M. H., Alvarez X., Niu T., Kaushal D. (2014) Role of TNF in the altered interaction of dormant Mycobacterium tuberculosis with host macrophages. PloS ONE 10.1371/journal.pone.0095220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barik S., Sureka K., Mukherjee P., Basu J., Kundu M. (2010) RseA, the SigE specific anti-sigma factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Mol. Microbiol. 75, 592–606 [DOI] [PubMed] [Google Scholar]
- 26. Rustad T. R., Sherrid A. M., Minch K. J., Sherman D. R. (2009) Hypoxia: a window into Mycobacterium tuberculosis latency. Cell Microbiol. 11, 1151–1159 [DOI] [PubMed] [Google Scholar]
- 27. Raju R. M., Jedrychowski M. P., Wei J. R., Pinkham J. T., Park A. S., O'Brien K., Rehren G., Schnappinger D., Gygi S. P., Rubin E. J. (2014) Post-translational regulation via Clp protease is critical for survival of Mycobacterium tuberculosis. PLoS Pathog. 10.1371/journal.ppat.1003994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katti M. K., Dai G., Armitige L. Y., Rivera Marrero C., Daniel S., Singh C. R., Lindsey D. R., Dhandayuthapani S., Hunter R. L., Jagannath C. (2008) The Delta fbpA mutant derived from Mycobacterium tuberculosis H37Rv has an enhanced susceptibility to intracellular antimicrobial oxidative mechanisms, undergoes limited phagosome maturation, and activates macrophages and dendritic cells. Cell. Microbiol. 10, 1286–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Russell D. G. (2013) The evolutionary pressures that have molded Mycobacterium tuberculosis into an infectious adjuvant. Curr. Opin. Microbiol. 16, 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaushal D., Schroeder B. G., Tyagi S., Yoshimatsu T., Scott C., Ko C., Carpenter L., Mehrotra J., Manabe Y. C., Fleischmann R. D., Bishai W. R. (2002) Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. U.S.A. 99, 8330–8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manganelli R., Provvedi R., Rodrigue S., Beaucher J., Gaudreau L., Smith I. (2004) Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186, 895–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sachdeva P., Misra R., Tyagi A. K., Singh Y. (2010) The sigma factors of Mycobacterium tuberculosis: regulation of the regulators. FEBS J. 277, 605–626 [DOI] [PubMed] [Google Scholar]
- 33. Av-Gay Y., Everett M. (2000) The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8, 238–244 [DOI] [PubMed] [Google Scholar]
- 34. Bretl D. J., Demetriadou C., Zahrt T. C. (2011) Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis. Microbiol. Mol. Biol. Rev. 75, 566–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sala A., Bordes P., Genevaux P. (2014) Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins 6, 1002–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sala A., Calderon V., Bordes P., Genevaux P. (2013) TAC from Mycobacterium tuberculosis: a paradigm for stress-responsive toxin-antitoxin systems controlled by SecB-like chaperones. Cell Stress Chaperones 18, 129–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arnvig K. B., Comas I., Thomson N. R., Houghton J., Boshoff H. I., Croucher N. J., Rose G., Perkins T. T., Parkhill J., Dougan G., Young D. B. (2011) Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 10.1371/journal.ppat.1002342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raju R. M., Goldberg A. L., Rubin E. J. (2012) Bacterial proteolytic complexes as therapeutic targets. Nat. Rev. Drug Discov. 11, 777–789 [DOI] [PubMed] [Google Scholar]
- 39. McGillivray S. M., Tran D. N., Ramadoss N. S., Alumasa J. N., Okumura C. Y., Sakoulas G., Vaughn M. M., Zhang D. X., Keiler K. C., Nizet V. (2012) Pharmacological inhibition of the ClpXP protease increases bacterial susceptibility to host cathelicidin antimicrobial peptides and cell envelope-active antibiotics. Antimicrob. Agents Chemother. 56, 1854–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Conlon B. P., Nakayasu E. S., Fleck L. E., LaFleur M. D., Isabella V. M., Coleman K., Leonard S. N., Smith R. D., Adkins J. N., Lewis K. (2013) Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ollinger J., O'Malley T., Kesicki E. A., Odingo J., Parish T. (2012) Validation of the essential ClpP protease in Mycobacterium tuberculosis as a novel drug target. J. Bacteriol. 194, 663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Parish T. (2014) Targeting mycobacterial proteolytic complexes with natural products. Chem. Biol. 21, 437–438 [DOI] [PubMed] [Google Scholar]
- 43. Gavrish E., Sit C. S., Cao S., Kandror O., Spoering A., Peoples A., Ling L., Fetterman A., Hughes D., Bissell A., Torrey H., Akopian T., Mueller A., Epstein S., Goldberg A., Clardy J., Lewis K. (2014) Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 21, 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee A. S., Ong D. C., Wong J. C., Siu G. K., Yam W. C. (2012) High-resolution melting analysis for the rapid detection of fluoroquinolone and streptomycin resistance in Mycobacterium tuberculosis. PloS ONE 10.1371/journal.pone.0031934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ocheretina O., Escuyer V. E., Mabou M. M., Royal-Mardi G., Collins S., Vilbrun S. C., Pape J. W., Fitzgerald D. W. (2014) Correlation between genotypic and phenotypic testing for resistance to rifampin in Mycobacterium tuberculosis clinical isolates in Haiti: investigation of cases with discrepant susceptibility results. PloS ONE 10.1371/journal.pone.0090569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Choudhary E., Bishai W., Agarwal N. (2014) Expression of a subset of heat stress induced genes of Mycobacterium tuberculosis is regulated by 3′,5′-cyclic AMP. PloS ONE 10.1371/journal.pone.0089759 [DOI] [PMC free article] [PubMed] [Google Scholar]