Background: LGR receptors play important roles in many developmental processes.
Results: The structure of human LGR4-Rspo1 complex was solved.
Conclusion: Diverse mechanisms are utilized by LGRs in ligand recognition.
Significance: Our structures are important for potential drug design.
Keywords: Hormone, Protein Structure, Receptor, Wnt Signaling, X-ray Crystallography
Abstract
Leucine-rich repeat G-protein-coupled receptors (LGRs) are a unique class of G-protein-coupled receptors characterized by a large extracellular domain to recognize ligands and regulate many important developmental processes. Among the three groups of LGRs, group B members (LGR4–6) recognize R-spondin family proteins (Rspo1–4) to stimulate Wnt signaling. In this study, we successfully utilized the “hybrid leucine-rich repeat technique,” which fused LGR4 with the hagfish VLR protein, to obtain two recombinant human LGR4 proteins, LGR415 and LGR49. We determined the crystal structures of ligand-free LGR415 and the LGR49-Rspo1 complex. LGR4 exhibits a twisted horseshoe-like structure. Rspo1 adopts a flat and β-fold architecture and is bound in the concave surface of LGR4 in the complex through electrostatic and hydrophobic interactions. All the Rspo1-binding residues are conserved in LGR4–6, suggesting that LGR4–6 bind R-spondins through an identical surface. Structural analysis of our LGR4-Rspo1 complex with the previously determined LGR4 and LGR5 structures revealed that the concave surface of LGR4 is the sole binding site for R-spondins, suggesting a one-site binding model of LGR4–6 in ligand recognition. The molecular mechanism of LGR4–6 is distinct from the two-step mechanism of group A receptors LGR1–3 and the multiple-interface binding model of group C receptors LGR7–8, suggesting LGRs utilize the divergent mechanisms for ligand recognition. Our structures, together with previous reports, provide a comprehensive understanding of the ligand recognition by LGRs.
Introduction
G-protein-coupled receptors (GPCRs),4 a large group of cell surface receptors, play key roles in many cellular processes. GPCRs generally contain a seven-transmembrane (7TM) domain responsible for ligand binding and downstream G-protein activation. Leucine-rich repeat G-protein-coupled receptors (LGRs) are a unique class of GPCRs characterized by a large extracellular domain (ectodomain) that harbors multiple copies of leucine-rich repeat (LRR) (1). The ectodomain mediates ligand binding to modulate downstream intracellular signaling pathways. LGRs are subdivided into three subgroups (group A, B, and C), according to their sequence similarities (1, 2). Group A LGRs have seven to nine LRRs in their ectodomain and long hinge regions connecting the LRR domain to the 7TM domain. Group A receptors include LGR1, LGR2, and LGR3, which recognize follicle-stimulating hormone (FSH), luteinizing hormone, and thyroid-stimulating hormone, respectively (3, 4). Group C LGRs have similar numbers of LRRs but contain a low density lipoprotein receptor class A domain motif at the N terminus and a short hinge region between the LRR domain and the 7TM domain. Group C LGRs include the relaxin receptor LGR7 and INSL3 (insulin-like peptide 3) receptor LGR8 (1, 3, 4). The molecular mechanisms of ligand recognition by group A and C receptors have been extensively studied and are relatively well understood.
The group B receptors include LGR4, LGR5, and LGR6, which are characterized by a long ectodomain containing 17 LRR repeats (2, 4). LGR4–6 share ∼50% sequence identity and play key roles in stem cell development. The 17 LRR repeats of the LGR4–6 receptors are flanked by the N-terminal cysteine-rich LRRNT region and the C-terminal cysteine-rich LRRCT region (4). The ligands of LGR4–6 remained unidentified for a long time. Recently, the secreted R-spondin proteins (Rspo1–4) were identified as the endogenous ligands for the group receptors to stimulate the Wnt signaling pathways, regulating cell proliferation, differentiation, and adult stem cell maintenance (5, 6).
The R-spondin (Rspo) family of proteins include four members (Rspo1–4) and are conserved in vertebrates. R-spondins are involved in numerous developmental processes (7–9), including sex (10, 11), lung, limb, hair follicle (9, 12), and nail development (13–15). The four R-spondin proteins share ∼40–60% pairwise sequence homology and adopt a common domain architecture that consists of an N-terminal secretory signal peptide sequence, two tandem furin-like cysteine-rich (Fu-CRD) domains, a thrombospondin type I repeat (TSP) domain, and a C-terminal basic amino acid-rich (BR) domain. Among the four subdomains, the two central tandem Fu-CRD domains have been demonstrated to be essential and sufficient for R-spondin stimulation of Wnt signaling (16–18). Previous studies had revealed that the ectodomains of LGR4–6 specifically recognize the two Fu-CRD domains of R-spondins (16, 17).
Although LGR4–6 contain a 7TM domain as the group A and group C receptors, the stimulation of Wnt signaling by LGR4–6 was found to be independent of downstream G-protein activation, suggesting the functional difference of LGR4–6 from LGR1–3 and LGR7–8. Here, we successfully utilized the “hybrid LRR technique” to obtain human LGR4 proteins from insect cells. We determined the crystal structure of human LGR4 alone at a resolution of 2.2 Å, as well as its complex structure with Rspo1 at a resolution of 2.25 Å. Our studies, together with the previously determined structures (19–22), suggest that LGR4–6 recognize R-spondins via a distinct mechanism from those of LGR1–3 and LGR7–8.
EXPERIMENTAL PROCEDURES
Plasmids, Reagents, and Mutagenesis
To express human LGR4 and Rspo1 in insect cells, the LGR415 and LGR49 hybrids (Fig. 1) were cloned into the pFastBac1 vector (Invitrogen) with a gp67 secretory signal peptide at the N terminus and a Fc-9×His tag with a tobacco etch virus protease cleavage site at the C terminus. The two furin-like cysteine-rich regions of Rspo1 (residues 34–135) were cloned into the pFastBac1 vector with an N-terminal gp67 signal peptide. For TOP/FOPFlash assays, full-length wild-type and mutant LGR4 were cloned into the pCDNA4 vector with a CD8 secretory signal peptide and a FLAG tag at the N terminus. Full-length Rspo1 and its mutants were cloned into the pCDNA4 vector with a C-terminal HA tag. All site-directed mutageneses were performed with the standard overlap-extension PCR method, and all plasmids were verified by DNA sequencing.
FIGURE 1.
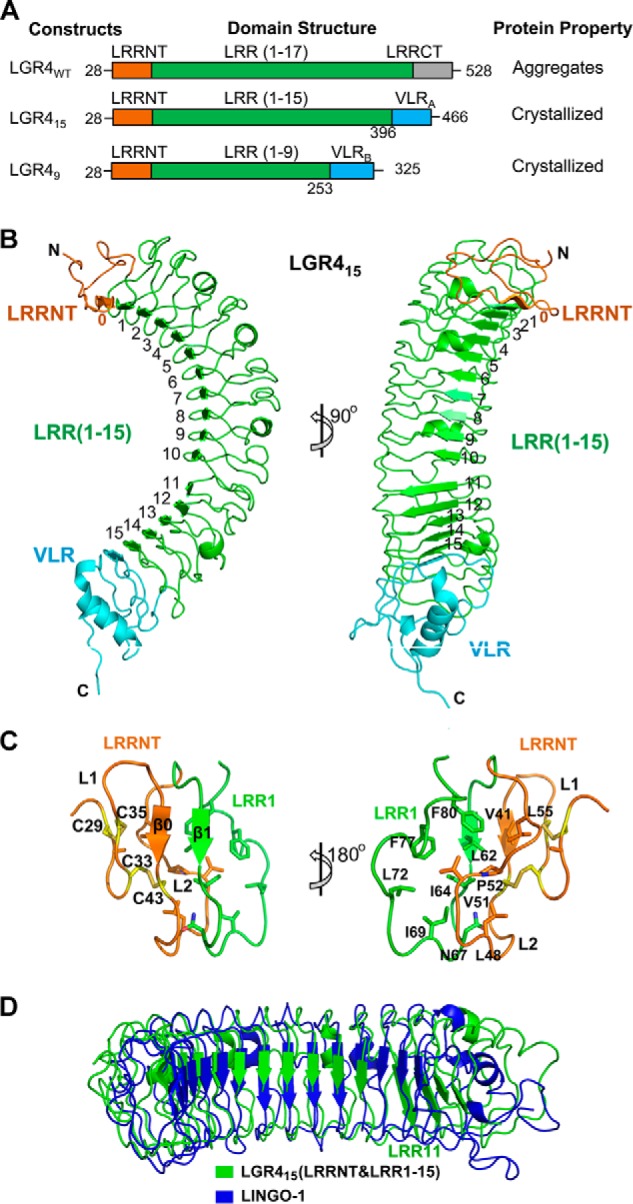
Crystal structure of LGR415. A, LGR4 constructs designed by using the hybrid LRR technique to enhance protein stability. The ectodomain of LGR4 contains LRRNT (orange), LRR (green), and LRRCT (gray). LGR415 was designed by fusing the N-terminal LRRNT and 15 LRR repeats with a hagfish VLRA module (cyan, residues 131–200, Asp-131 and Thr-132 were mutated to alanine and serine). LGR49 was designed by fusing the N-terminal LRRNT and nine LRR repeats with a VLRB module (cyan, residue 129–200). B, overall structure of LGR415. The central 15 LRR repeats of LGR415 are colored in green and labeled as indicated. The LRRNT and the VLR module are colored in orange and cyan, respectively. C, structure of LRRNT and its interaction with LRR1. LRRNT and LRR1 are colored in orange and green, respectively. The four cysteine residues are shown as yellow sticks and form two disulfide bonds (Cys-29–Cys-35, Cys-33–Cys-43) in LRRNT. D, structural comparison of LINGO-1 and LGR415 (LRRNT and LRR(1–15)). LGR415 and LINGO-1 are colored in blue and yellow, respectively. The LRR11 repeat of LGR415 is labeled.
Protein Expression and Purification
Recombinant proteins were expressed in Sf9 insect cells using the Bac-to-Bac baculovirus expression system (Invitrogen). LGR415 and LGR49 recombinant proteins were purified from cell medium using nickel-nitrilotriacetic acid resins. After tobacco etch virus protease digestion to remove the C-terminal Fc-9×His tag, the proteins were further purified by size exclusion chromatography using Superdex 200 10/300 GL column (GE Healthcare). Both purified proteins were concentrated to 10 mg/ml in the Tris-HCl buffer containing 50 mm Tris-HCl, pH 8.0, and 150 mm NaCl for crystallization. To obtain the recombinant LGR4-Rspo1 complex, Sf9 cells were co-infected with baculoviruses encoding LGR49 and human Rspo1 (residues 34–135). After 60 h post-infection, the LGR49-Rspo1 complex was purified from cell medium using nickel-nitrilotriacetic acid column and further purified using a Mono Q column (GE Healthcare). The purified LGR49-Rspo1 complex was concentrated to 40 mg/ml in the buffer containing 50 mm Tris-HCl, pH 8.0, and 150 mm NaCl for crystallization.
Crystallization and Data Collection
All crystallization experiments were carried out with the hanging-drop vapor diffusion method at 16 °C. In initial crystal screening, crystals of LGR415 and LGR49 appeared after 3 days. Crystals of LGR415 were selected for structural determination. After extensive optimization, diffraction-qualified crystals of LGR415 were obtained in the well condition containing 0.1 m HEPES, pH 7.5, and 2.0 m ammonium sulfate. Crystals of LGR415 were transferred into the well solution supplemented with 15% glycerol as the cryoprotectant and then flash-cooled into liquid nitrogen for data collection. The LGR49-Rspo1 complex was crystallized after 7 days in the well solution containing 0.1 m MES, pH 6.5, 0.2 m sodium thiocyanate, and 22% PEG 3350. The crystals were cryoprotected with the well solution supplemented with 20% glycerol in liquid nitrogen. All diffraction data were collected on the BL17U1 beamline of Shanghai Synchrotron Radiation Facility and processed with the HKL-2000 package (23).
Structural Determination
The LGR415 structure was solved with the molecular replacement method using the partial model of LRR repeats from TLR5 (PDB code 3V44) (24) as the search mode in the Phaser program (25) in CCP4 (26). The final structure was refined to 2.2 Å with Rwork/Rfree of 20.7%/23.4% in Refmac5 (27).There is one LGR415 molecule in an asymmetric unit. The LGR49-Rspo1 complex structure was solved with the molecular replacement method using LGR415 and VLR (residues 129–200) as the search modes. The final structure of LGR49-Rspo1 complex was refined to 2.25 Å with Rwork/Rfree of 21.3%/24.2% in Refmac5. There are two LGR49-Rspo1 complex molecules in an asymmetric unit. Because of the lack of electron density, the N-terminal 18 residues and C-terminal 5 residues of Rspo1 were missed in the final complex model. All model building was performed in Coot (28). All structures were checked with the program Procheck (29). All structural pictures were prepared in PyMOL. Statistics of data collection and refinement are listed in Table 1.
TABLE 1.
Data collection and refinement statistics
| LGR415 | LGR49-Rspo1 | |
|---|---|---|
| Data collection | ||
| Space group | I213 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 173.76, 173.76, 173.76 | 44.87, 137.91, 82.56 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 100.13, 90.00 |
| Wavelength | 1.0000 | 0.97925 |
| Resolution (Å)a | 50-2.20 (2.28-2.20) | 50-2.20 (2.28-2.25) |
| Rsym or Rmerge | 0.116 (0.610) | 0.090 (0.650) |
| I/σI | 56.9 (13.1) | 37.7 (11.3) |
| Completeness (%) | 100 (100) | 98.8 (98.3) |
| Redundancy | 28.1 (27.4) | 5.0 (5.2) |
| Refinement | ||
| Resolution (Å) | 50-2.2 | 50-2.25 |
| No. of reflections | 41,816 | 43,609 |
| Rwork/Rfree(%)b | 20.7/23.4 | 21.3/24.2 |
| No. of atoms | ||
| Protein | 3,477 | 5,859 |
| Water/ligand | 267 | 291 |
| B-factors (Å2) | ||
| Protein | 30.34 | 28.49 |
| Water/ligand | 35.04 | 17.40 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.007 | 0.009 |
| Bond angles (°) | 1.171 | 1.267 |
| Ramachandran plot | ||
| Preferred regions(%) | 92.6 | 92.9 |
| Allowed regions(%) | 7.4 | 7.1 |
a The data for the highest resolution shell are shown in parentheses.
b Rfree is calculated using 10% of the total number of reflections.
TOP/FOPFlash Assays
TOP/FOPFlash assays were carried out similarly as described (30). To test the effect of Rspo1 mutations, HEK293T cells cultured in 12-well plates were transfected with 500 ng of Super 8× TOPFlash or FOPFlash firefly luciferase reporter, 10 ng of Renilla luciferase reporter, and 750 ng of wild-type LGR4 plasmids. Wild type and mutant Rspo1 conditional medium (CM) were prepared by culturing HEK293T cells in 6-well plates, which were transiently transfected with 2 μg of Rspo1 variant plasmids in each well. Wnt3a CM was obtained from mouse L cells stably expressing Wnt3a. After a 24-h co-stimulation by Wnt3a and wild-type or mutant Rspo1 CM, the transfected HEK293T cells were harvested for Dual-Luciferase® reporter assays by using the Dual-Glo luciferase assay kit (Promega). To test the effect of LGR4 mutations on the Rspo1 stimulation of Wnt signaling, HEK293T cells cultured in 12-well plates were transfected with 500 ng of Super 8× TOPFlash or FOPFlash firefly luciferase reporter, 10 ng of Renilla luciferase reporter, and 1 μg of wild-type or mutant LGR4 plasmids. After stimulation for 24 h with Wnt3a and wild-type Rspo1 CM, the cells were harvested for Dual-Luciferase reporter assays. The relative TOP/FOPFlash luciferase activities presented were normalized against the levels of vector control.
RESULTS AND DISCUSSION
Production of Recombinant Human LGR4 and LGR4-Rspo1 Complex Proteins
The ectodomain of human LGR4 includes residues 28–528. The two central tandem Fu-CRD domains of human Rspo1, which are required for binding the LGRs, include residues 34–135. When expressed in insect cells, the human LGR4 ectodomain severely aggregated during purification. To enhance the solubility and stability of the LGR4 ectodomain, we designed LGR4 chimeras that contain the LRR repeats of the LGR4 ectodomain and the hagfish VLR protein using the hybrid LRR technique, which was previously used to facilitate the soluble expression and crystallization of TLR4 (Fig. 1A) (31). Previous studies have suggested that the N-terminal residues of LGRs are important for interaction with R-spondins (16). Therefore, we attempted to fuse the N-terminal LRR repeats of LGR4 with the C-terminal regions of the hagfish VLR protein according to the “LXXLXLXXNXLXXL” consensus sequence of LRR modules.
After many attempts, we successfully expressed and crystallized two LGR4 ectodomain fusion proteins, LGR415 and LGR49 (Fig. 1, A and B). In LGR415, the N-terminal 15 LRRs of LGR4 (residues 28–396) were fused with the VLR residues 131–200 (VLRA). In LGR49, the N-terminal nine LRRs of LGR4 (residues 28–253) were fused with the C-terminal 72 residues of VLR (residues 129–200, VLRB). Both chimeric proteins formed stable protein complexes with Rspo1 during purification, suggesting that our designed LGR4 proteins contain the entire Rspo1-binding region. LGR415 crystals were selected for structural determination because the construct contains more residues of LGR4. The LGR415 structure was finally refined to 2.2 Å with Rwork/Rfree values of 20.7%/23.4% (Table 1). Because LGR415 precipitated in polyethylene glycols, which are extensively used in the crystallization of protein complexes, we co-expressed LGR49 with the two central tandem Fu-CRD domains of Rspo1 (residues 34–135) in insect cells to assemble the receptor-ligand complex for crystallization. We successfully crystallized the LGR49-Rspo1 complex and determined its structure to 2.25 Å with high geometry quality (Table 1). Details of the data collection and structural refinement statistics are summarized in Table 1.
Human LGR4 Ectodomain Structure
The final model of the ligand-free LGR415 structure includes all residues of the designed hybrid construct. The overall architecture of LGR415 adopts a typical twisted solenoid-like structure (Fig. 1B). As expected, the central 15 LRR repeats of LGR415 are flanked by the N-terminal LRRNT (residues 28–57) and the C-terminal VLR module (Fig. 1B). In total, 17 β-strands are present in the LGR415 structure, of which 15 originate from the LRR repeats, one from LRRNT, and one from VLR. The 17 β-strands form the inner concave surface of LGR415, which interacts with Rspo1 in the LGR4-Rspo1 complex structure (details below). The outer convex surface of LGR415 is composed of 10 linking loops and four short linking α-helices from the LRR repeats (Fig. 1B).
At the LGR415 N terminus, LRRNT consists of two loops (L1 and L2) and one β-strand (β0). It forms a capping structure to protect the first LRR repeat (LRR1) from solvent exposure (Fig. 1C). LRRNT binds to LRR1 through extensive interactions. The β0 strand of LRRNT forms a parallel β-sheet with β1 of LRR1. The hydrophobic residues Val-41, Leu-48, Pro-52, and Leu-55 from β0 and L2 in LRRNT strongly stack with Leu-62, Ile-64, Asn-67, Ile-69, Leu-72, Phe-77, and Phe-80 of LRR1 (Fig. 1C). LRRNT contains four cysteine residues, which form two disulfide bonds (Cys-29–Cys-35 and Cys-33–Cys-43) to stabilize the L1 loop. These cysteine residues are conserved in LGR5 and LGR6, suggesting that the cysteine residues play important roles in maintaining the LRRNT cap-like structure. At the C terminus of LGR415, the VLR protein contains one α-helix, one β-strand, and three loops. It adopts the identical conformation as reported previously (Fig. 1B) (31). VLR stabilizes the LGR4 C terminus through a parallel β-sheet and extensive hydrophobic interactions with LRR15.
A structural homology search using the LGR415 structure (LRRNT and LRR(1–15)) as the bait in the Dali server (32) revealed that LGR415 shares significant structural similarities with several LRR receptors, including LINGO-1, TLR2, TLR4, and TLR5 (24, 31, 33, 34). LGR415 exhibits the highest structural similarity to the extracellular LRR subdomain of the human neuron co-receptor LINGO-1 (PDB code 2ID5) with a Z-score value of 32.4 and a root mean square deviation value of 4.4 Å for 347 aligned residues (Fig. 1D) (34). LINGO-1 forms a ternary complex with NgR1 and p75 to constitute the Nogo receptor for myelin and mediates the inhibition of axonal growth (34). Although the overall structure of LGR415 is highly similar to the extracellular LRR subdomain of LINGO-1, the C terminus of LGR415 (LRR11–15) is largely different from that of LINGO-1 (Fig. 1D).
Structure of the Human LGR4-Rspo1 Complex
The recombinant LGR4-Rspo1 complex for crystallization was prepared from the co-expression of LGR49 with the two central Fu-CRD domains of Rspo1 in insect cells (Fig. 2A). LGR49 formed a stable 1:1 complex with Rspo1 during size exclusion chromatography. We successfully crystallized the LGR49-Rspo1 complex and determined its structure by the molecular replacement method using LGR415 and VLR as the search models. In the final structural model of the LGR49-Rspo1 complex, there are two LGR49-Rspo1 complex copies related by the noncrystallography symmetry in an asymmetric unit. Because the two complex molecules adopt identical conformation, hereafter, we describe one of them in our structural analysis.
FIGURE 2.
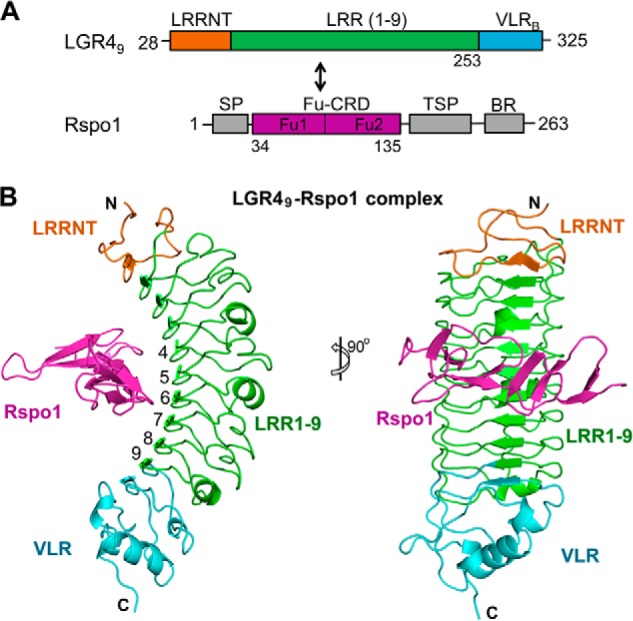
Structure of the LGR49-Rspo1 complex. A, domain structures of LGR49 and Rspo1. LGR49 and the two central furin-like domains (Fu1 and Fu2, colored in purple) of Rspo1 were co-expressed for the LGR4-Rspo1 complex. B, structure of the LGR49-Rspo1 complex. Rspo1 is colored in purple. The central nine LRR repeats of LGR49 are colored in green. The LRRNT and VLR subdomains of LGR49 are colored in orange and cyan, respectively.
Like the ligand-free LGR415 structure, Rspo1-bound LGR49 adopts a short twisted solenoid-like structure and contains the N-terminal LRRNT, the central nine leucine-rich repeats (LRR1–9), and the C-terminal hagfish VLR region (Fig. 2B). All of the residues in the designed LGR49 chimera are included in the final LGR49 model. In the complex, Rspo1 horizontally crosses the inner concave surface of LGR49 from one side to the other side and binds to the concave surface through extensive interactions with the central six LRR repeats (LRR4–9) of LGR49 (Fig. 2B).
Rspo1 Structure in the LGR4-Rspo1 Complex
Rspo1 in complex with LGR4 is composed of eight β-strands that form an elongated and flat overall architecture (Fig. 3A). The final Rspo1 model only includes the central 78 residues, which constitute eight β-strands (β1–β8) in the structure (Fig. 3B). The N-terminal 18 residues (Ala-34–Asn-51) and the C-terminal five residues (Glu-131–Ala-135) of Rspo1 are missing in the structure due to lack of electron density. These residues are not conserved in the four R-spondin proteins (Fig. 3B) and do not participate in any crystal contact.
FIGURE 3.
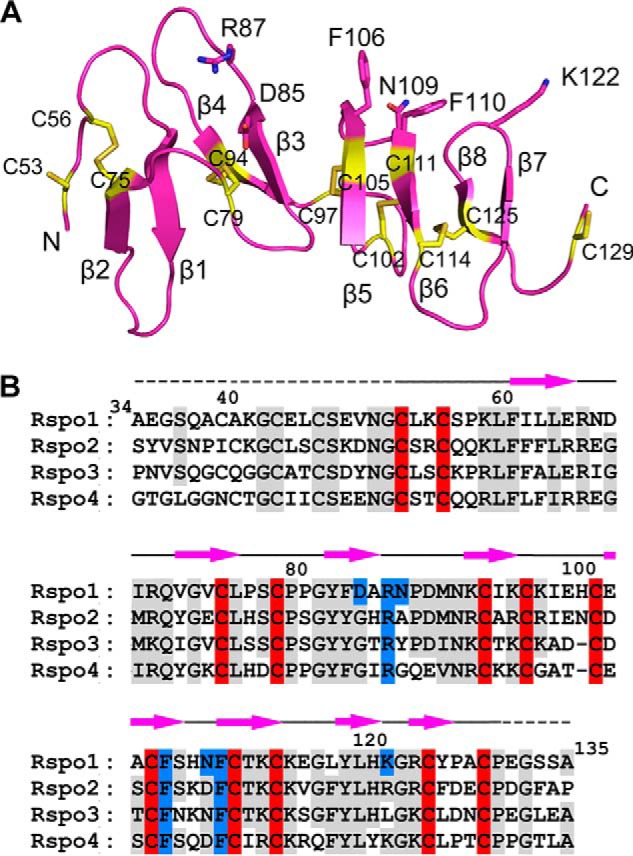
Rspo1 structure in the LGR49-Rspo1 complex. A, structure of Rspo1. The LGR4-binding residues are shown as sticks. The Fu1 subdomain includes the first four β-strands (β1–β4). The Fu2 subdomain includes the C-terminal four β-strands (β5–β8). All cysteine residues are shown as yellow sticks and are labeled as indicated. B, sequence alignment of human Rspo1–4. The residues conserved in Rspo1–4 are colored in gray. The conserved cysteine residues are specifically highlighted in red. The LGR4-binding residues of Rspo1 are highlighted in blue. The missed residues in the Rspo1 final model are marked with dashed lines above the sequences.
The eight Rspo1 β-strands constitute four tandem β-hairpins in the order of β2-β1, β4-β3, β5-β6, and β8-β7. Each hairpin contains a two-stranded antiparallel β-sheet and a linking loop. The N-terminal two hairpins (β2-β1 and β4-β3) are longer than the C-terminal hairpins (β5-β6 and β8-β7) (Fig. 3A). The linking loops of the β4-β3, β5-β6, and β8-β7 hairpins, which are located on the same side of Rspo1, are responsible for binding LGR4 in the complex structure. In total, 12 cysteine residues are present in the Rspo1 structure, 10 of which form five disulfide bonds (Cys-56–Cys-75, Cys-79–Cys-94, Cys-97–Cys-105, Cys-102–Cys-111, and Cys-114–Cys-125) (Fig. 3B). These disulfide bonds bridge the adjacent hairpins or linking loops to maintain the elongated Rspo1 structure.
Rspo2–4 share high sequence homology with Rspo1 in their furin-like domains (Fig. 3B) (7, 18), suggesting that the furin-like domains of Rspo2–4 also adopt the elongated flat β-fold structures similar to Rspo1. The 10 cysteine residues of Rspo1, which form five disulfide bonds within the hairpins, are highly conserved in all R-spondin proteins (Fig. 3B). Previous clinical studies have revealed that the mutations of the cysteine residues completely disrupt Rspo4 function and cause human autosomal-recessive anonychia (14, 15), suggesting that the five disulfide bonds are critical for maintaining the flat structures of R-spondins. Furin-like cysteine-rich regions also exist in a variety of proteins that are involved in signal transduction via receptor tyrosine kinases (35). Our structural homology search revealed that Rspo1 just exhibits low structural similarity to subdomain IV, the furin-like region of the extracellular region of the EGF receptors HER2, -3, and -4 (36–38). Subdomain IV of EGF receptors has been reported to mediate the inter-receptor dimerization of HER2, -3, and -4 to regulate cellular growth and differentiation (36–38). Rspo1 was superposed with subdomain IV only at its middle hairpin β3-β4 in structural superimposition.
Interface between LGR4 and Rspo1
In the complex structure, Rspo1 binds to the concave surface of LGR4 through the residues Asp-85, Arg-87, Phe-106, Asn-109, Phe-110, and Lys-122, which are located in the linking loops of the β4-β3, β5-β6, and β8-β7 hairpins, respectively (Fig. 3A). The interface between Rspo1 and LGR4 buries a surface area of ∼765 Å2 and involves two differently charged regions on the LGR4 surface, a hydrophilic binding region (region 1) and a hydrophobic binding region (region 2) (Figs. 2B and 4A).
FIGURE 4.
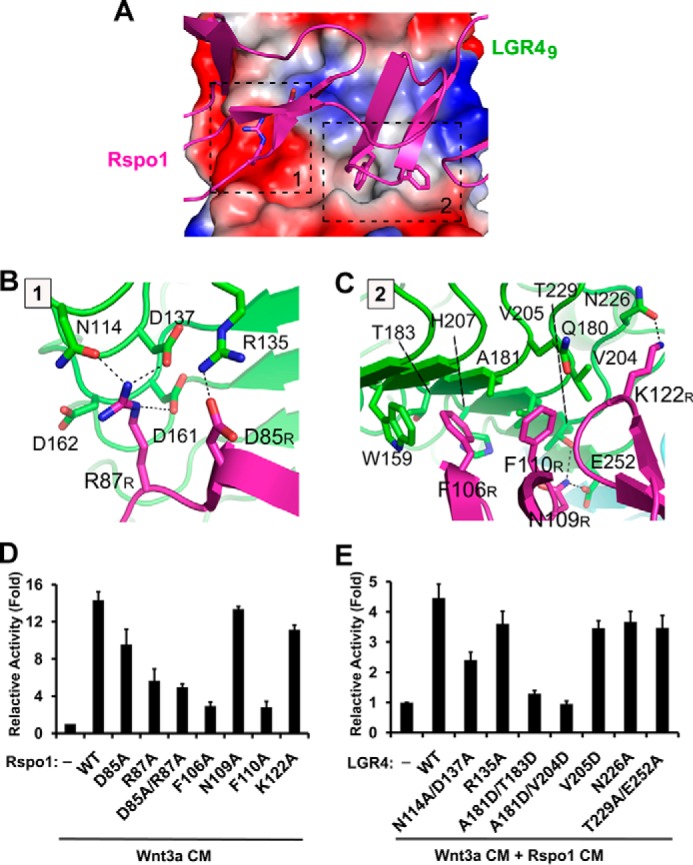
Interface between LGR4 and Rspo1. A, interface between LGR4 and Rspo1. The negatively and positively charged surface and hydrophobic surface of LGR4 are colored in red, blue, and gray, respectively. The LGR4-interacting residues of Rspo1 are represented as sticks. The two interaction regions (regions 1 and 2) in LGR4 are indicated by black dashed lines. B and C, detailed interactions in regions 1 and 2. LGR4 and Rspo1 are colored in green and purple, respectively. The LGR4-interacting residues in Rspo1 are labeled with the R subscripts. D, effects of Rspo1 mutations on the stimulation of Wnt signaling. The potentiation of Wnt signaling by Rspo1 mutants was examined using β-catenin-driven Super 8× TOP/FOPFlash luciferase reporter assays. HEK293T cells were transiently transfected with full-length LGR4, TOPFlash, or FOPFlash firefly luciferase, and Renilla luciferase plasmids and were then stimulated with the CM of Rspo1 mutants in the presence of Wnt3a CM. E, effects of LGR4 mutations on the stimulation of Wnt signaling. HEK293T cells were transfected with the LGR4 mutants, TOPFlash or FOPFlash firefly luciferase, and Renilla luciferase plasmids and were then stimulated with the CM of wild-type Rspo1 and Wnt3a.
In the hydrophilic binding region, the positively charged Arg-87 of Rspo1 interacts with an open acidic surface in LGR4. The side chain of Arg-87 in Rspo1 forms three hydrogen bonds with Asn-114, Asp-137, and Asp-161 of LGR4 (Fig. 4B). Additionally, Asp-85 of Rspo1 interacts with Arg-135 of LGR4 through a salt bridge. In the hydrophobic binding region, Phe-106 and Phe-110 of Rspo1 form strong interactions with the concave surface of LGR4 (Fig. 4C). Phe-106 of Rspo1 stacks with the side chain of Trp-159 of LGR4 and is inserted into a hydrophobic pocket in the concave surface of LGR4, which is formed by Trp-159, Ala-181, Thr-183, and His-207 of LGR4. Phe-110 of Rspo1 interacts with Gln-180, Ala-181, Val-204, and Val-205 of LGR4. In addition to the central hydrophobic interactions, Asn-109 and Lys-122 of Rspo1 interact with Thr-229, Glu-252, and Asn-226 of LGR4 via three hydrogen bonds (Fig. 4C).
Mutagenesis Analysis
To examine the interactions observed in the complex structure, we mutated the interacting residues of Rspo1 and LGR4 and carried out the β-catenin-driven luciferase reporter assays (Fig. 4, D and E). A single mutation of the nonconserved Asp-85 of Rpso1 to alanine just had a minor effect on the stimulation of Wnt signaling. But the R87A mutant and the D85A/R87A mutant of Rspo1 could not sufficiently stimulate Wnt signaling (Fig. 4D), which indicates that the conserved Arg-87 plays a key role in the LGR4-Rspo1 interaction, and the electrostatic interactions in region 1 are required for Rspo1 recognition by LGR4. The Rspo1 mutations (F106A and F110A) of Phe-106 and Phe-110 at the hydrophobic binding region completely abolished the Rspo1-mediated stimulation of Wnt signaling (Fig. 4D), which suggests that the hydrophobic interactions between Rspo1 and LGR4 are also essential for the LGR4-Rspo1 interaction, and the conserved residues Phe-106 and Phe-110 of Rspo1 are critical for the interactions. Different from the F106A and F110A mutations, the N119A and K122A mutants of Rspo1 slightly affected the stimulation of Wnt signaling, suggesting that the nonconserved Asn-119 and Lys-122 of Rspo1 just play supplementary roles in the interactions between Rspo1 and LGR4. The critical residues Arg-87, Phe-106, and Phe-110 of Rspo1 are located in the first furin-like domain (β1–β4) and the second furin-like domain (β5–β8), respectively, which well explains why both furin-like domains of R-spondins are required for the recognition by LGRs.
We also mutated the interacting residues of LGR4 to evaluate their effects on the Rspo1 stimulation of Wnt signaling (Fig. 4E). Consistent with the mutational analysis of Rspo1, the mutations of Arg-135, Asn-226, and Thr-229/Glu-252 in LGR4, which interact with the nonconserved residues of Rspo1, had a minor effect on the Rspo1 stimulation of Wnt signaling. The double mutation (N114A/D137A) of Asn-114 and Asp-137 of LGR4, which interact with Arg-87 of Rspo1 in the hydrophilic binding region, severely affected the Rspo1 stimulation of Wnt signaling. The A181D/T183D and A181D/V204D mutations, which disrupt the hydrophobic binding region of LGR4, also completely abolished the Rspo1 stimulation of Wnt signaling. Therefore, both the electrostatic and hydrophobic interactions between Rspo1 and LGR4 are required for Rspo1 recognition by LGR4.
Comparison of LGR4 with LGR5 and LGR6
We further examined the conformational changes of LGR4 upon Rspo1 binding. The structure of the LGR49-Rspo1 complex was superimposed with ligand-free LGR415. As shown in Fig. 5, LRRNT and the central LRR1–9 repeats of LGR49 adopt identical conformations as those of LGR415 (Fig. 5A). The main chain and secondary structures of LGR4 do not undergo conformational changes upon Rspo1 binding, suggesting that LGR4 utilizes a rigid structural fold to recognize R-spondins to stimulate Wnt signaling. Our LGR49 and Rspo1 structures were also well superimposed with the previously determined LGR4-Rspo1 complex structure (PDB code 4KT1) (Fig. 5B) (21). All results of our luciferase assays for the LGR4 and Rspo1 mutations are consistent with the previously published data (20, 21), which further confirm our structures and mutagenesis analysis.
FIGURE 5.
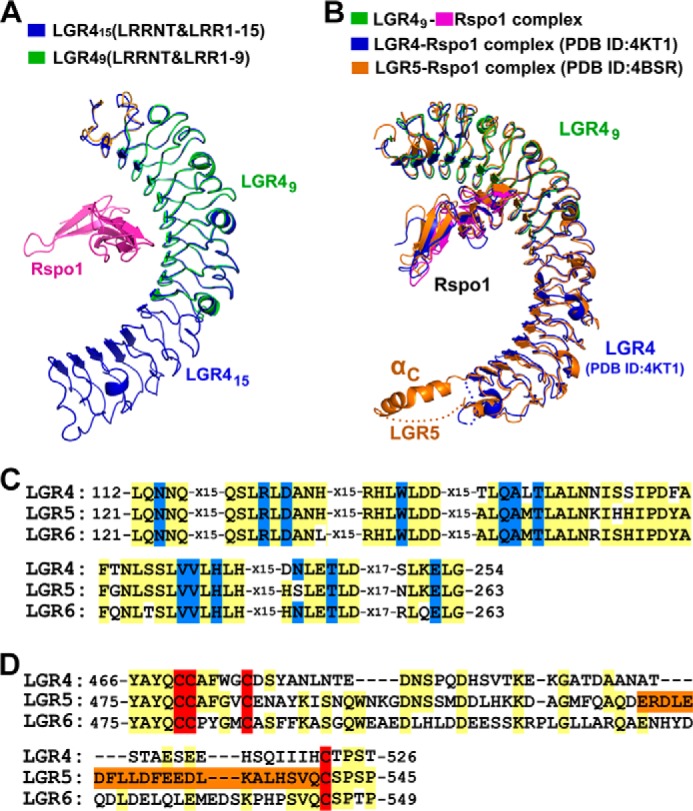
Comparing LGR4 with LGR5 and LGR6. A, structural comparison of Rspo1-bound LGR49 and ligand-free LGR415. LGR49 (LRRNT and LRR(1–9)) was superimposed with LGR415 (LRRNT and LRR(1–15)). The VLR modules in LGR415 and LGR49 were omitted in structural superimposition. LGR49 and Rspo1 are colored as in Fig. 2B. LGR415 is colored in blue. B, structural comparison of the LGR49-Rspo1 complex with the previously determined LGR4-Rspo1 and LGR5-Rspo1 structures. The LGR49-Rspo1 complex is colored in green (LGR49) and purple (Rspo1). The previously determined LGR4-Rspo1 and LGR5-Rspo1 structures are colored in blue and orange, respectively. C, sequence alignment of the Rspo1-binding residues. The Rspo1-binding residues are colored in blue. All the Rspo1-binding residues of LGR4 are conserved in LGR5 and LGR6. D, sequence alignment of the residues around αC in the LRRCT regions. The cysteine residues of disulfide bonds in the LRRCT regions are colored in red. The sequence of the αC helix of LGR5 is colored in orange.
LGR5 and LGR6 serve as markers of specific stem cells and play important roles in stem cell maintenance. The ectodomains of LGR5 and LGR6 have significant sequence similarities to that of LGR4. Previous studies had reported the LGR5-Rpso1 complex structures (20). Structural comparison between LGR4 and LGR5 revealed that the ectodomain of LGR5 adopts a similar horseshoe-like structure as that of LGR4 (Fig. 5B). But the C-terminal LRRCT region of LGR5 contains an additional α-helix (αC), which mediated the dimerization of the LGR5-Rspo1 complex in crystals (details below) (20). Sequence alignment showed that the residues near the αC region are not conserved in LGR4–6 (Fig. 5D).
Because all the Rspo1-interacting residues of LGR4 and LGR5 are conserved in LGR6 (Fig. 5C), LGR6 recognizes R-spondins through an identical surface as LGR4 and LGR5, which includes the central LRR4–9 repeats. All R-spondin proteins bind to the LGRs, but the binding affinity varies slightly among the members (30). Rspo1 exhibits ∼40% sequence similarity with the other three members (7, 18). The critical LGR4-binding residues Arg-87, Phe-106, and Phe-110 of Rspo1 are conserved in the four R-spondin proteins (Fig. 3B). But the interacting residues Asp-85, Asn-109, and Lys-122 of Rspo1 are highly divergent in Rspo2–4 (Fig. 3B). Therefore, the limited difference in the binding affinities between Rspo1–4 and LGRs is likely determined by these variable interacting residues in the four R-spondins.
Divergence of Ligand Recognition by LGRs
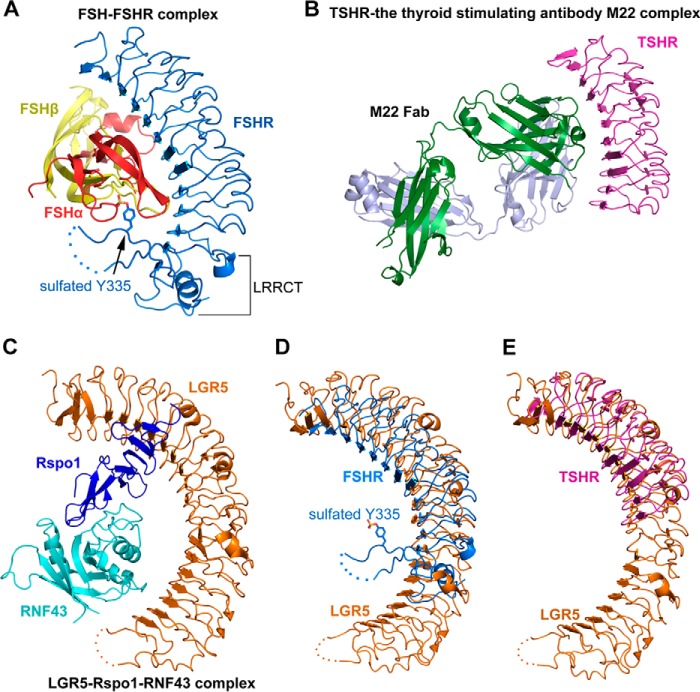
To uncover the differences in ligand recognition, we structurally analyzed the molecular mechanisms of all LGRs. The molecular mechanisms of group A (LGR1–3) and group C receptors (LGR7–8) have been extensively studied. The complex structures of LGR1 (FSH receptor, FSHR) with FSH revealed that the heterodimeric FSH contains similarly β-folded α- and β-chains and is bound into the concave surface of LGR1 in a hand-clasp fashion (Fig. 6A) (39, 40). In addition, LGR1 provides a sulfotyrosine (sulfated Tyr-335) in the LRRCT/hinge region as a second interaction site with FSH (Fig. 6A). The concave surface in the LRR region of LGR1 represents a high affinity FSH-binding site. Interestingly, the monoclonal TSHR-stimulating antibody M22 also binds to the corresponding concave surface of LGR3 (TSHR) in the LGR3-M22 complex (Fig. 6B) (41), suggesting that the concave surface of LGR1 is the major binding site for FSH. The high affinity binding site of LGR1 was suggested to recruit FSH and mediate its conformational changes to form a sulfotyrosine-binding pocket for the sulfated Tyr-335 insertion. Tyr-335 of LGR1 is conserved in all group A receptors, and the tyrosine sulfation is also essential for the activation of LGR2 and LGR3 by luteinizing hormone and thyroid-stimulating hormone, respectively (42), suggesting that group A LGR universally utilizes the two-step mechanism of LGR1 for ligand recognition.
FIGURE 6.
Structural comparison of the receptor-ligand complexes of LGR1, LGR3, and LGR5. A, crystal structure of FSH-FSHR (LGR1) complex (PDB code 4AY9). The α- and β-chain of FSH are colored in red and yellow, respectively. FSHR (LGR1) are colored in blue. The sulfated Tyr-335 is shown as sticks. The disordered region in the LRRCT/hinge region of FSHR is shown as dashed lines. B, crystal structure of TSHR (LGR3) in complex with the thyroid-stimulating antibody M22 (PDB code 3G04). The two chains of the M22 Fab are colored in green and gray, respectively. TSHR is colored in purple. C, crystal structure of the LGR5-Rspo1-RNF43 ternary complex (PDB code 4KNG). LGR5, Rspo1, and RNF43 are colored in orange, blue, and cyan, respectively. D, structural comparison of LGR5 with FSHR (LGR1). LGR5 and FSHR are colored in orange and blue, respectively. E, structural comparison of LGR5 with the M22-bound TSHR (LGR3). LGR5 and TSHR are colored in orange and purple, respectively.
Relaxin and INSL3 are two-chain peptide hormones and play important roles in the control of adult fertility (43). Relaxin and INSL3 are specifically recognized by LGR7 and LGR8, respectively (44). Distinct from R-spondins and FSH, Relaxin and INSL3 are α-folded heterodimers of A- and B-chain linked by disulfide bonds, which are structurally similar to insulin. Previous biochemical studies had revealed that the recognition of relaxin and INSL3 by LGR7 and LGR8 requires multiple interactions between the ligands and various receptor domains. The primary binding occurs via the interactions between B-chain of relaxin/INSL3 with the inner β-sheets of the LRRs in the ectodomains of LGR7 and LGR8 (45, 46). The primary binding site provides a high affinity interface for the ligands. A secondary low affinity binding site is located at the extracellular loops in the 7TM domains of LGR7 and LGR8, which binds the A chains of relaxin and INSL3 (47, 48). Therefore, both the LRR regions and the 7TM extracellular loops are involved in the relaxin/INSL3 recognition by LGR7 and LGR8, suggesting a unique multiple-interface mechanism of group C receptors.
Our LGR4-Rspo1 complex structure and previously determined LGR4–5 structures revealed that the LRR region of LGR4 is the sole binding site for Rspo1, suggesting that LGR4–6 recognize R-spondins via a one-site binding mechanism (Fig. 2) (20, 21). Although the transmembrane E3 ubiquitin ligase ZNRF3 and its homologue RNF43 were reported to interact with R-spondins and form complexes with LGR4–6 (49, 50), the LGR5-Rspo1-RNF43 ternary complex structure demonstrated that LGR5 does not interact with RNF43 to affect Rspo1 binding (Fig. 6C) (19). In addition, LGR5 contains more redundancy of LRR repeats than LGR1 and LGR3 (Fig. 6, D and E). The distinct one-site binding mechanism of R-spondin recognition by LGR4–6, together with the structural differences between LGR5 and LGR1, suggests the mechanism divergence in ligand recognition of LGRs.
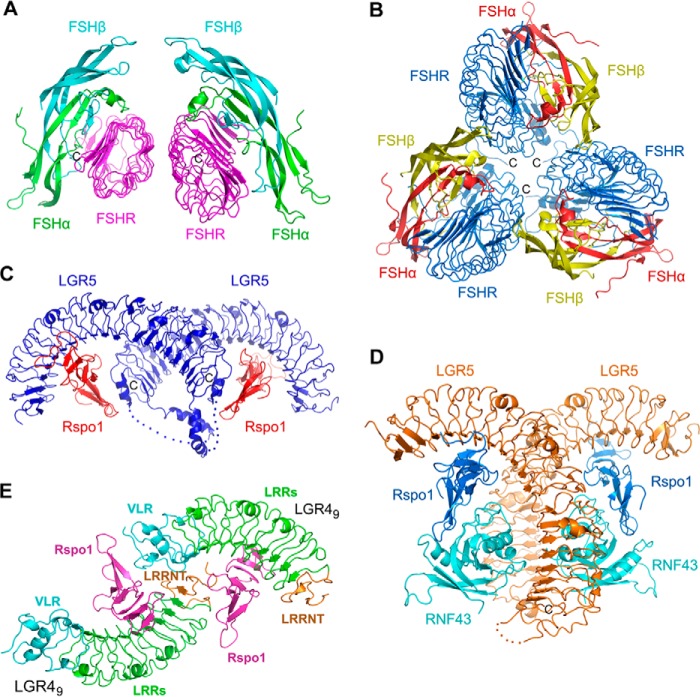
Potential Oligomerization of LGRs in Ligand Recognition
It was suggested that glycoprotein hormone receptors undergo dimerization in living cells (51, 52). The complex of FSH and LGR1 without the LRRCT/hinge region was demonstrated to form a dimer at high concentration in solution (39). The crystal structure of FSH with the hinge-truncated LGR1 revealed that the dimer was formed by two FSH-LGR1 complex molecules via three β-strands on the convex surface of LGR1 in an asymmetric unit (Fig. 7A). The small dimerization interface indicated the weak interactions between the two protomers (Fig. 7A), consistent with that the dimerization was just detected by the chemical cross-linking assays (39). In addition, recent studies suggested that LGR1 underwent trimerization upon FSH binding when the conserved Tyr-335 residue in the LRRCT/hinge domain was sulfated (Fig. 7B) (40, 53). The trimerization of LGR1 provided an additional binding site for FSH, which involves the interaction of the convex surface of LGR1 with a neighboring FSH (40). The trimerization was also observed in the crystals of LGR1 with FSH that was deglycosylated at Asn-52 in the α-chain (53). As the trimerization buried a large surface area of each protomer, the FSH-binding capacity of LGR1 was increased to 3-fold for the deglycosylated FSH (53). Although the trimer model of LGR1 in FSH recognition could well explain some observation in biochemical and functional studies (53), the in vivo relevance of the LGR1-FSH trimerization and the actual oligomerization form in living cells still need to be determined.
FIGURE 7.
Oligomizeration of LGRs in crystals. A, dimerization of FSH-FSHR complex in an asymmetric unit (PDB code 1XWD). The LRRCT/hinge-truncated ectodomain of FSHR is colored in purple. The α- and β-chain of FSH are colored in green and cyan, respectively. B, trimerization of FSH-FSHR complex in an asymmetric unit (PDB code 4AY9). The entire ectodomain of FSHR is colored in blue. The α- and β-chains of FSH are colored in red and yellow, respectively. C, dimerization of the LGR5-Rspo1 binary complex in an asymmetric unit (PDB code 4BSR). LGR5 and Rspo1 are colored in blue and red, respectively. The disordered loops in the LRRCT region of LGR5 are shown as dashed lines. D, dimerization of the LGR5-Rspo1-RNF43 ternary complex in an asymmetric unit (PDB code 4KNG). LGR5, Rspo1, and RNF43 are colored in orange, blue, and cyan, respectively. E, two LGR49-Rspo1 copies in an asymmetric unit in our complex structure. The two LGR4-Rspo1 copies interact with each other via the engineered VLR module and the LRRNT region.
Dimerization of the LGR5-Rspo1 complex was also observed in crystals, although LGR5 formed a 1:1 complex with Rspo1 in solution (Fig. 7C) (20). In the LGR5-Rspo1 binary complex structure (PDB code 4BSR and space group P22121) (20), the dimerization of the LGR5-Rspo1 complex was mediated by the LGR5 αC-helices and the Rspo1-LGR5 LRRCT interactions, forming a cross-dimer of LGR5-Rspo1 complex in an asymmetric unit (Fig. 7C). But in the LGR5-Rspo1-RNF43 ternary complex structure (19) (PDB code 4KNG and space group P212121), a back-to-back dimer of the LGR5-Rspo1-RNF43 complex was formed by the LGR5-LGR5 and LGR5-RNF43 trans-interactions (Fig. 7D). It is unknown which form of dimerization functions in cells and whether the two dimerization forms represent different states of LGR5 during ligand recognition. Different from the LGR5-Rspo1 complex, the LGR4-Rspo1 complex did not dimerize in the previously determined structure (21) and our structures. The previously determined LGR4-Rspo1 structure (PDB code 4KT1 and space group P32) only contained one LGR4-Rspo1 molecule in an asymmetric unit. Although there are two LGR4-Rspo1 molecules in our structure, the two copies do not form a real dimer because the two copies interact with each other via the engineered VLR module and the LRRNT region (Fig. 7E).
Interestingly, Xenopus LGR4 was found to dimerize both in solution and in crystals (22). The dimerization of Xenopus LGR4 was mediated by the side-by-side interactions between LRRNT and the first four LRRs of the two LGR4 copies in an asymmetric unit, forming a tail-to-tail dimer in an asymmetric unit (22). But our human LGR415, which contains the intact LRRNT and the first four LRR repeats, did not dimerize either in solution or in crystals. Given that human LGR4 has significant sequence similarities to that of Xenopus, it is possible that the C-terminal engineered VLR module of LGR415 has negative effects on the human LGR4 dimerization. LGR7 and LGR8 were also reported to homodimerize and heterodimerize in cells, which regulated the negative cooperativity in relaxin/INSL3 binding (54). Dimerization and negative cooperativity in ligand binding have been generally demonstrated for the insulin receptor (55), β2-adrenergic receptor (56), and chemokine receptors (57). It will be interesting to investigate the negative cooperativity in R-spondin binding and its correlation with the dimerization of LGR4–6. Taken together, LGRs likely undergo different types of oligomerization during ligand recognition.
Closing Remarks
LGR4–6 receptors are important for stem cell maintenance by stimulating Wnt signaling. It is important to uncover the molecular mechanism of R-spondin recognition by LGR4–6. In this study, we utilized the hybrid LRR technique to obtain functional human LGR4 proteins, and we determined the crystal structure of the LGR4-Rspo1 complex. Our studies, together with previous reports, suggest that LGR4–6 recognize R-spondins via a one-site binding mechanism, which involves the electrostatic and hydrophobic interactions between R-spondins and the concave surface in the LRR region of LGR4–6. Our analysis reveals the structural differences of the three groups of LGRs in ligand recognition, highlighting the divergence in the mechanisms of ligand recognition. Because of the important roles of LGR4–6 receptors and R-spondin proteins in embryonic development and in many human diseases, our structures are important for potential drug design.
Acknowledgments
We thank the staff at the Shanghai Synchrotron Radiation Facility (Shanghai, China) for assistance with data collection. We also thank members of the Zhu laboratory for helpful discussions and technical assistance.
This work was supported by China's Fundamental Research Funds for the Central Universities, Research Grants 31370722 and 81322024 from the NSFC, Grant LR13C05001 from Natural Science Foundation of Zhejiang Province, and Grant J20130639 from Zhejiang University K. P. Chao's High Technology Development Foundation (to Y. Zhu).
The atomic coordinates and structure factors (codes 4QXE and 4QXF) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GPCR
- G-protein-coupled receptor
- LGR
- leucine-rich repeat G-protein-coupled receptor
- LRR
- leucine-rich repeat
- FSH
- follicle-stimulating hormone
- FSHR
- FSH receptor
- TSHR
- thyroid-stimulating hormone receptor
- PDB
- Protein Data Bank
- 7TM
- seven-transmembrane
- CM
- conditional medium
- Rspo
- R-spondin
- Fu-CRD
- furin-like cysteine-rich.
REFERENCES
- 1. Luo C. W., Hsueh A. J. (2006) Genomic analyses of the evolution of LGR genes. Chang Gung Med. J. 29, 2–8 [PubMed] [Google Scholar]
- 2. Barker N., Tan S., Clevers H. (2013) Lgr proteins in epithelial stem cell biology. Development 140, 2484–2494 [DOI] [PubMed] [Google Scholar]
- 3. Van Loy T., Vandersmissen H. P., Van Hiel M. B., Poels J., Verlinden H., Badisco L., Vassart G., Vanden Broeck J. (2008) Comparative genomics of leucine-rich repeats containing G protein-coupled receptors and their ligands. Gen. Comp. Endocrinol. 155, 14–21 [DOI] [PubMed] [Google Scholar]
- 4. Hsu S. Y., Kudo M., Chen T., Nakabayashi K., Bhalla A., van der Spek P. J., van Duin M., Hsueh A. J. (2000) The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol. Endocrinol. 14, 1257–1271 [DOI] [PubMed] [Google Scholar]
- 5. Clevers H., Nusse R. (2012) Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 [DOI] [PubMed] [Google Scholar]
- 6. Niehrs C. (2012) The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol. 13, 767–779 [DOI] [PubMed] [Google Scholar]
- 7. Kim K. A., Zhao J., Andarmani S., Kakitani M., Oshima T., Binnerts M. E., Abo A., Tomizuka K., Funk W. D. (2006) R-Spondin proteins: a novel link to β-catenin activation. Cell Cycle 5, 23–26 [DOI] [PubMed] [Google Scholar]
- 8. Jin Y. R., Yoon J. K. (2012) The R-spondin family of proteins: emerging regulators of WNT signaling. Int. J. Biochem. Cell Biol. 44, 2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kazanskaya O., Glinka A., del Barco Barrantes I., Stannek P., Niehrs C., Wu W. (2004) R-Spondin2 is a secreted activator of Wnt/β-catenin signaling and is required for Xenopus myogenesis. Dev. Cell 7, 525–534 [DOI] [PubMed] [Google Scholar]
- 10. Schuijers J., Clevers H. (2012) Adult mammalian stem cells: the role of Wnt, Lgr5, and R-spondins. EMBO J. 31, 2685–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parma P., Radi O., Vidal V., Chaboissier M. C., Dellambra E., Valentini S., Guerra L., Schedl A., Camerino G. (2006) R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat. Genet. 38, 1304–1309 [DOI] [PubMed] [Google Scholar]
- 12. Bell S. M., Schreiner C. M., Wert S. E., Mucenski M. L., Scott W. J., Whitsett J. A. (2008) R-spondin 2 is required for normal laryngeal-tracheal, lung, and limb morphogenesis. Development 135, 1049–1058 [DOI] [PubMed] [Google Scholar]
- 13. Aoki M., Mieda M., Ikeda T., Hamada Y., Nakamura H., Okamoto H. (2007) R-spondin3 is required for mouse placental development. Dev. Biol. 301, 218–226 [DOI] [PubMed] [Google Scholar]
- 14. Blaydon D. C., Ishii Y., O'Toole E. A., Unsworth H. C., Teh M. T., Rüschendorf F., Sinclair C., Hopsu-Havu V. K., Tidman N., Moss C., Watson R., de Berker D., Wajid M., Christiano A. M., Kelsell D. P. (2006) The gene encoding R-spondin 4 (RSPO4), a secreted protein implicated in Wnt signaling, is mutated in inherited anonychia. Nat. Genet. 38, 1245–1247 [DOI] [PubMed] [Google Scholar]
- 15. Bergmann C., Senderek J., Anhuf D., Thiel C. T., Ekici A. B., Poblete-Gutierrez P., van Steensel M., Seelow D., Nürnberg G., Schild H. H., Nürnberg P., Reis A., Frank J., Zerres K. (2006) Mutations in the gene encoding the Wnt-signaling component R-spondin 4 (RSPO4) cause autosomal recessive anonychia. Am. J. Hum. Genet. 79, 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Lau W., Barker N., Low T. Y., Koo B. K., Li V. S., Teunissen H., Kujala P., Haegebarth A., Peters P. J., van de Wetering M., Stange D. E., van Es J. E., Guardavaccaro D., Schasfoort R. B., Mohri Y., Nishimori K., Mohammed S., Heck A. J., Clevers H. (2011) Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 [DOI] [PubMed] [Google Scholar]
- 17. Glinka A., Dolde C., Kirsch N., Huang Y. L., Kazanskaya O., Ingelfinger D., Boutros M., Cruciat C. M., Niehrs C. (2011) LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 12, 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim K. A., Wagle M., Tran K., Zhan X., Dixon M. A., Liu S., Gros D., Korver W., Yonkovich S., Tomasevic N., Binnerts M., Abo A. (2008) R-Spondin family members regulate the Wnt pathway by a common mechanism. Mol. Biol. Cell 19, 2588–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen P. H., Chen X., Lin Z., Fang D., He X. (2013) The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 27, 1345–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peng W. C., de Lau W., Forneris F., Granneman J. C., Huch M., Clevers H., Gros P. (2013) Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell Rep. 3, 1885–1892 [DOI] [PubMed] [Google Scholar]
- 21. Wang D., Huang B., Zhang S., Yu X., Wu W., Wang X. (2013) Structural basis for R-spondin recognition by LGR4/5/6 receptors. Genes Dev. 27, 1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu K., Xu Y., Rajashankar K. R., Robev D., Nikolov D. B. (2013) Crystal structures of Lgr4 and its complex with R-spondin1. Structure 21, 1683–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. Yoon S. I., Kurnasov O., Natarajan V., Hong M., Gudkov A. V., Osterman A. L., Wilson I. A. (2012) Structural basis of TLR5-flagellin recognition and signaling. Science 335, 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collaborative Computation Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 27. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Laskowski A. R., MacArthur W. M., Moss S. D., Thornton M. J. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 30. Carmon K. S., Gong X., Lin Q., Thomas A., Liu Q. (2011) R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 11452–11457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim H. M., Park B. S., Kim J. I., Kim S. E., Lee J., Oh S. C., Enkhbayar P., Matsushima N., Lee H., Yoo O. J., Lee J. O. (2007) Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell 130, 906–917 [DOI] [PubMed] [Google Scholar]
- 32. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jin M. S., Kim S. E., Heo J. Y., Lee M. E., Kim H. M., Paik S. G., Lee H., Lee J. O. (2007) Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 34. Mosyak L., Wood A., Dwyer B., Buddha M., Johnson M., Aulabaugh A., Zhong X., Presman E., Benard S., Kelleher K., Wilhelm J., Stahl M. L., Kriz R., Gao Y., Cao Z., Ling H. P., Pangalos M. N., Walsh F. S., Somers W. S. (2006) The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition. J. Biol. Chem. 281, 36378–36390 [DOI] [PubMed] [Google Scholar]
- 35. Raz E., Schejter E. D., Shilo B. Z. (1991) Interallelic complementation among DER/flb alleles: implications for the mechanism of signal transduction by receptor-tyrosine kinases. Genetics 129, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eigenbrot C., Ultsch M., Dubnovitsky A., Abrahmsén L., Härd T. (2010) Structural basis for high affinity HER2 receptor binding by an engineered protein. Proc. Natl. Acad. Sci. U.S.A. 107, 15039–15044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cho H. S., Leahy D. J. (2002) Structure of the extracellular region of HER3 reveals an interdomain tether. Science 297, 1330–1333 [DOI] [PubMed] [Google Scholar]
- 38. Bouyain S., Longo P. A., Li S., Ferguson K. M., Leahy D. J. (2005) The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. Proc. Natl. Acad. Sci. U.S.A. 102, 15024–15029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fan Q. R., Hendrickson W. A. (2005) Structure of human follicle-stimulating hormone in complex with its receptor. Nature 433, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiang X., Liu H., Chen X., Chen P. H., Fischer D., Sriraman V., Yu H. N., Arkinstall S., He X. (2012) Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor. Proc. Natl. Acad. Sci. U.S.A. 109, 12491–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanders J., Chirgadze D. Y., Sanders P., Baker S., Sullivan A., Bhardwaja A., Bolton J., Reeve M., Nakatake N., Evans M., Richards T., Powell M., Miguel R. N., Blundell T. L., Furmaniak J., Smith B. R. (2007) Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody. Thyroid 17, 395–410 [DOI] [PubMed] [Google Scholar]
- 42. Costagliola S., Panneels V., Bonomi M., Koch J., Many M. C., Smits G., Vassart G. (2002) Tyrosine sulfation is required for agonist recognition by glycoprotein hormone receptors. EMBO J. 21, 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hsu S. Y., Nakabayashi K., Nishi S., Kumagai J., Kudo M., Bathgate R. A., Sherwood O. D., Hsueh A. J. (2003) Relaxin signaling in reproductive tissues. Mol. Cell. Endocrinol. 202, 165–170 [DOI] [PubMed] [Google Scholar]
- 44. Hsu S. Y., Nakabayashi K., Nishi S., Kumagai J., Kudo M., Sherwood O. D., Hsueh A. J. (2002) Activation of orphan receptors by the hormone relaxin. Science 295, 671–674 [DOI] [PubMed] [Google Scholar]
- 45. Scott D. J., Wilkinson T. N., Zhang S., Ferraro T., Wade J. D., Tregear G. W., Bathgate R. A. (2007) Defining the LGR8 residues involved in binding insulin-like peptide 3. Mol. Endocrinol. 21, 1699–1712 [DOI] [PubMed] [Google Scholar]
- 46. Büllesbach E. E., Schwabe C. (2005) The trap-like relaxin-binding site of the leucine-rich G-protein-coupled receptor 7. J. Biol. Chem. 280, 14051–14056 [DOI] [PubMed] [Google Scholar]
- 47. Sudo S., Kumagai J., Nishi S., Layfield S., Ferraro T., Bathgate R. A., Hsueh A. J. (2003) H3 relaxin is a specific ligand for LGR7 and activates the receptor by interacting with both the ectodomain and the exoloop 2. J. Biol. Chem. 278, 7855–7862 [DOI] [PubMed] [Google Scholar]
- 48. Diepenhorst N. A., Petrie E. J., Chen C. Z., Wang A., Hossain M. A., Bathgate R. A., Gooley P. R. (2014) Investigation of interactions at the extracellular loops of the relaxin family peptide receptor 1 (RXFP1). J. Biol. Chem. 289, 34938–34952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hao H. X., Xie Y., Zhang Y., Charlat O., Oster E., Avello M., Lei H., Mickanin C., Liu D., Ruffner H., Mao X., Ma Q., Zamponi R., Bouwmeester T., Finan P. M., Kirschner M. W., Porter J. A., Serluca F. C., Cong F. (2012) ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200 [DOI] [PubMed] [Google Scholar]
- 50. Koo B. K., Spit M., Jordens I., Low T. Y., Stange D. E., van de Wetering M., van Es J. H., Mohammed S., Heck A. J., Maurice M. M., Clevers H. (2012) Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669 [DOI] [PubMed] [Google Scholar]
- 51. Roess D. A., Horvat R. D., Munnelly H., Barisas B. G. (2000) Luteinizing hormone receptors are self-associated in the plasma membrane. Endocrinology 141, 4518–4523 [DOI] [PubMed] [Google Scholar]
- 52. Horvat R. D., Barisas B. G., Roess D. A. (2001) Luteinizing hormone receptors are self-associated in slowly diffusing complexes during receptor desensitization. Mol. Endocrinol. 15, 534–542 [DOI] [PubMed] [Google Scholar]
- 53. Jiang X., Fischer D., Chen X., McKenna S. D., Liu H., Sriraman V., Yu H. N., Goutopoulos A., Arkinstall S., He X. (2014) Evidence for follicle-stimulating hormone receptor as a functional trimer. J. Biol. Chem. 289, 14273–14282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Svendsen A. M., Vrecl M., Knudsen L., Heding A., Wade J. D., Bathgate R. A., De Meyts P., Nøhr J. (2009) Dimerization and negative cooperativity in the relaxin family peptide receptors. Ann. N.Y. Acad. Sci. 1160, 54–59 [DOI] [PubMed] [Google Scholar]
- 55. de Meyts P., Roth J., Neville D. M., Jr., Gavin J. R., 3rd, Lesniak M. A. (1973) Insulin interactions with its receptors: experimental evidence for negative cooperativity. Biochem. Biophys. Res. Commun. 55, 154–161 [DOI] [PubMed] [Google Scholar]
- 56. Limbird L. E., Meyts P. D., Lefkowitz R. J. (1975) β-Adrenergic receptors: evidence for negative cooperativity. Biochem. Biophys. Res. Commun. 64, 1160–1168 [DOI] [PubMed] [Google Scholar]
- 57. Springael J. Y., Urizar E., Parmentier M. (2005) Dimerization of chemokine receptors and its functional consequences. Cytokine Growth Factor Rev. 16, 611–623 [DOI] [PubMed] [Google Scholar]