Background: A distinct mechanism linking fasting-associated mitochondrial protein acetylation and increased fat oxidation is unknown.
Results: Acetylation of heat shock protein 10 enhances medium-chain acyl-CoA dehydrogenase folding, enzyme activity, and fat oxidation.
Conclusion: A novel acetylation-dependent mechanism modulates fat oxidation via regulating mitochondrial metabolic enzyme folding.
Significance: A Sirtuin 3 deacetylase-linked mechanism to control fat oxidation via modulation of enzyme folding has been identified.
Keywords: Fatty Acid Oxidation, Heat Shock Protein (HSP), Mitochondria, Protein Folding, Sirtuin, SIRT3
Abstract
Although Sirtuin 3 (SIRT3), a mitochondrially enriched deacetylase and activator of fat oxidation, is down-regulated in response to high fat feeding, the rate of fatty acid oxidation and mitochondrial protein acetylation are invariably enhanced in this dietary milieu. These paradoxical data implicate that additional acetylation modification-dependent levels of regulation may be operational under nutrient excess conditions. Because the heat shock protein (Hsp) Hsp10-Hsp60 chaperone complex mediates folding of the fatty acid oxidation enzyme medium-chain acyl-CoA dehydrogenase, we tested whether acetylation-dependent mitochondrial protein folding contributes to this regulatory discrepancy. We demonstrate that Hsp10 is a functional SIRT3 substrate and that, in response to prolonged fasting, SIRT3 levels modulate mitochondrial protein folding. Acetyl mutagenesis of Hsp10 lysine 56 alters Hsp10-Hsp60 binding, conformation, and protein folding. Consistent with Hsp10-Hsp60 regulation of fatty acid oxidation enzyme integrity, medium-chain acyl-CoA dehydrogenase activity and fat oxidation are elevated by Hsp10 acetylation. These data identify acetyl modification of Hsp10 as a nutrient-sensing regulatory node controlling mitochondrial protein folding and metabolic function.
Introduction
Mitochondrial fuel metabolism is tightly regulated by nutrient availability coupled to organelle energy demand. This concept is evident where the rate of fatty acid oxidation correlates with the level of circulating free fatty acids and where fat oxidation increases in response to impaired glucose uptake associated with insulin resistance (1). These changes in fat catabolism can be acute, where circulating free fatty acids are elevated in response to fasting associated lipolysis, or chronic, where nutrient overload promotes extra-adipocyte lipid deposition, which can exacerbate insulin resistance with increased reliance on fat oxidation.
Regulatory nodes controlling enhanced β-oxidation are evident at the transcriptional and posttranslational level. The regulation of genes encoding enzymes controlling fat oxidation include the modulation of transcription factors such as peroxisome proliferator-activated receptors and peroxisome proliferator-activated receptor γ coactivator regulatory protein activation, which themselves may be modulated by fatty acid ligands (2). In parallel, posttranslational regulatory pathways, including nutrient-sensing components such as signatures of the energetic status regulation through the AMP kinase pathway and the modification of metabolic pathway enzymes by the NAD-dependent sirtuin deacetylases, are implicated in substrate metabolism (3, 4).
Recent studies found conflicting data pertaining to mitochondrial protein acetylation and the regulatory control of mitochondrial fat oxidation. On one hand, the mitochondrial sirtuin SIRT3 targets numerous enzymes involved in fat oxidation. Here SIRT3 deacetylates and activates long-chain acyl-CoA dehydrogenase and medium-chain acyl-CoA dehydrogenase (MCAD)3 (5, 6). In parallel, under dietary restricted conditions, multiple fatty acid oxidation enzymes show increased lysine residue deacetylation in control versus SIRT3 knockout mice, and SIRT3 null mice have increased accumulation of acylcarnitines, a finding consistent with reduced fat oxidation (7). On the other hand, in response to fat feeding, the acetylation of β-hydroxyacyl CoA dehydrogenase results in activation of enzyme activity in muscle cells (8). Furthermore, muscle mitochondrial proteins extracted from fasted mice showed increased acetylation in parallel with higher in vivo rates of fatty acid oxidation (9), and, in response to high fat feeding, mice exhibit increased mitochondrial protein acetylation and fat oxidation (10). The complexity of these counterregulatory findings is further evident where increased levels of mitochondrial protein acetylation and increased fatty acid oxidation rates are evident in SIRT3 KO mice (9, 10).
Because chronic high fat feeding is linked to the down-regulation of SIRT3 (11–13) and to increased mitochondrial protein acetylation, we questioned whether an additional level of regulation linking SIRT3, elevated fatty acid levels, mitochondrial protein acetylation, and fat oxidation may be operational to account for some of these inconsistencies.
In proteomic screening, we and others have identified Hsp10 as a candidate for SIRT3-dependent deacetylation (14–16). At the same time, the Hsp10-Hsp60 chaperoning protein folding complex is instrumental in the appropriate folding of the fatty acid oxidation enzyme MCAD (17). Taking these findings into consideration, we explored whether the regulation of Hsp10-Hsp60 modulation of mitochondrial protein folding could play a SIRT3-dependent role in controlling fat oxidation in response to the known mobilization of fatty acids in response to fasting.
To test this hypothesis, we first explored whether Hsp10 was a functional target of SIRT3. In the SIRT3 null background, Hsp10 shows enhanced acetylation. Moreover, the modulation of SIRT3 activity concordantly modulated the extent of Hsp10 acetylation. Because the nutrient-sensing role of SIRT3 is most evident during fasting (5, 14), we explored mitochondrial protein folding by comparing fed and fasted wild-type and SIRT3 knockout mice. Although mitochondrial protein folding was similar in the fed state, in response to fasting, SIRT3-deficient mice elicited fewer misfolded mitochondrial proteins. The genetic modulation of SIRT3 levels altered mitochondrial protein folding and enzyme activity. We found that acetylation of the Hsp10 Lys-56 residue is critical in mitochondrial protein folding and that acetylation of Hsp10 Lys-56 modifies the interaction between Hsp10-Hsp60 and the rate of fat oxidation. Together, these data support the hypothesis that the acetylation of Hsp10 functions in nutrient-sensing SIRT3-dependent regulation of mitochondrial fat oxidation, in part, via the control of the dynamic interaction between the Hsp10-Hsp60 chaperonins in mediating mitochondrial protein folding.
EXPERIMENTAL PROCEDURES
Animal Studies
Wild-type and SIRT3 KO mice from a heterozygous breeding scheme were fasted for 48 h, as employed previously, to evaluate sirtuin deacetylase substrates (5, 14, 18). Liver mitochondria were extracted from fed and fasted mice for analysis. All animal experiments were approved by the NHLBI/National Institutes of Health Animal Care and Use Committee.
Materials
Stable control and SIRT3 knockdown cells were generated using puromycin-selective lentiviral shRNA constructs. We previously raised an anti-SIRT3 murine antibody (19). Additional antibodies included Hsp60 and acetylated lysine residues (Cell Signaling Technology), Hsp10 (Sigma), and MCAD (Cayman Chemicals). The lentiviral shRNA constructs were from Sigma, and pcDNA-SIRT3 was from Addgene.
Two-dimensional DIGE
Fifty micrograms of WT and SIRT3 KO mice liver mitochondria were labeled with Cy3 and Cy5, respectively. Samples were pooled and separated by two-dimensional gel analysis. Images of the differential gel electrophoresis (DIGE) gels were quantitatively analyzed using Progenesis Same Spots software v3.3 (Nonlinear Dynamics USA, Durham, NC). A cutoff ratio of <0.5 or >1.5 was employed to identify differentially expressed proteins. Proteins of interest were extracted from pick gels and identified by LC-MS/MS. The molecular weight and isoelectric point of the identified protein were cross-checked with the gel position of excised spots.
In Vitro Deacetylation Assay
SIRT3 and Hsp10 proteins were immunoprecipitated from wild-type liver mitochondria or Hepa1c1c7 cell lysates using appropriate antibodies. Immunoprecipitated SIRT3 and Hsp10 were incubated in SDAC (sirtuin deacetylation buffer (50 m Tris-HCl, pH 9.0, 4 m MgCl2, 50 m NaCl, 0.5 m DTT)) with either NAD+ (1 mm) or nicotinamide (10 mm) for 3 h at 32 °C, as described previously (14). Reactions were stopped by adding lithium dodecyl sulfate sample buffer prior to Western blot analysis.
Mitochondrial Isolation and Misfolded Protein Analysis
Mitochondria were isolated as described previously (14), and then incubated in lysis buffer with or without Nonidet P-40 detergent (2% or 0.2%) (20). The Nonidet P-40-insoluble aggregates were pelleted, resuspended, and visualized by silver staining or immunoblot analysis.
Flow Cytometry
Flow cytometry studies were performed to assess native mtGFP expression as a measure of appropriate mitochondrial GFP folding. Cytosolic GFP was used as a control. mtGFP or GFP were transfected into Hepa1c1c7 cells with different Hsp10 constructs, as shown in Figs. 2, E and F, and 4, E and F. Forty-eight hours after transfection, cells were harvested, and the GFP signal was analyzed using BD FACSCanto (BD Biosciences).
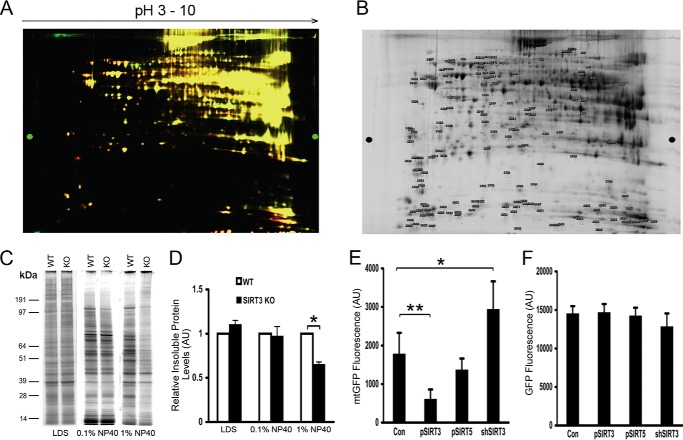
FIGURE 2.
SIRT3 deficiency reduces mitochondrial protein misfolding. A, two-dimensional DIGE showing liver mitochondrial protein levels of WT and SIRT3 KO mice following a 48-h fast. A cutoff ratio of <0.5 or >1.5 was employed to identify differentially expressed proteins. B, proteins of interest were extracted from pick gels and identified by LC-MS/MS. C and D, liver mitochondria were isolated, and insoluble aggregates compared following treatment with different concentrations of Nonidet P-40 were assessed using SDS-PAGE gel and silver stain from WT and SIRT3 KO mice. AU indicates arbitrary units with WT normalized to = 1, showing relative change in KO proteins. LDS, lithium dodecyl sulfate sample buffer. E and F, flow cytometric quantification of the fluorescence of mtGFP and cytoplasmic localized GFP in Hepa1c1c7 cells representing appropriate folding in response to modulation of SIRT3 and SIRT5 levels (n = 3 experiments). Con, control. *, p < 0.05 and **, p <0.01.
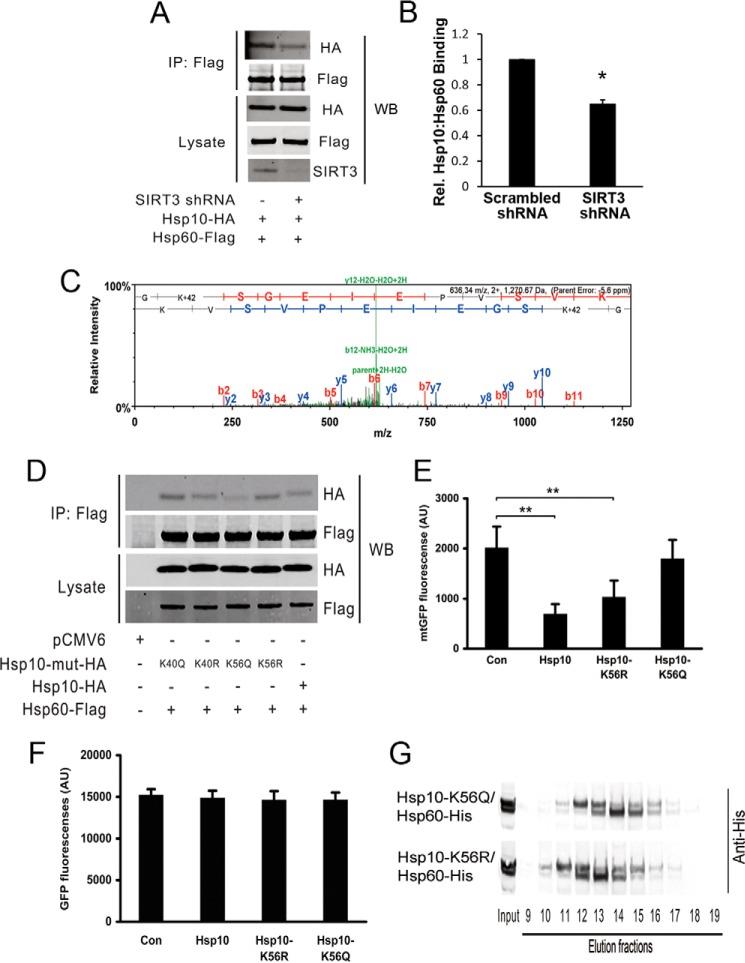
FIGURE 4.
Acetylation of Hsp10 Lys-56 modulates Hsp10-Hsp60 binding affinity and chaperone complex function. A and B, Hepa1c1c7 cells were cotransfected with either SIRT3 shRNA or scrambled shRNA together with Hsp10 and Hsp60. Hsp10-Hsp60 complexes were immunoprecipitated (IP) employing an anti-FLAG antibody. Hsp10 binding affinity was quantified by Western blot (WB) analysis (n = 3 experiments). Rel., relative. *, p <0.05. C, Hsp10 Lys-56 was acetylated in SIRT KO mouse mitochondria as assessed by LC-MS/MS. Shifts in fragment masses confirmed that Lys-56 was the acetylation site within this peptide. The spectrum of a relative peptide containing Lys-56 is shown (n = 4 experiments). D, Hepa1c1c7 cells were cotransfected with either the lysine site mutant or wild-type Hsp10 together with Hsp60. Hsp10-Hsp60 complexes were immunoprecipitated, and mutant Hsp10 binding affinity was assayed by Western blot analysis (n = 3 experiments). E and F, Hepa1c1c7 cells were cotransfected with either mtGFP or GFP together with wild-type or mutant Hsp10, respectively, and fluorescence was measured by FACS (n = 3 experiments). Con, control. **, p <0.01. AU, arbitrary units from flow cytometer. G, Hepa1c1c7 cells were cotransfected with mutant Hsp10 together with Hsp60. Purification and HPLC gel filtration of the Hsp10-Hsp60 complex were performed, and the temporal evaluation of eluates was detected by Western blot analysis (n = 3 experiments).
Blue Native Gel Electrophoresis
Mitochondrial pellets were dissolved in sample buffer (1% η-dodecyl-β-d-maltoside and 750 mm ϵ-amino-N-caproic acid). Lysates were kept on ice for 1 h with occasional vortexing, and then the residual insoluble content was extracted by centrifugation at 20,000 × g for 30 min at 4 °C. The supernatant was quantified, and the sample was mixed with 1/10 5% G-250 sample blue additive and run on NativePAGE 4–16% BisTris gels (Invitrogen) for subsequent Western blot analysis.
Hsp10 Mutagenesis Studies
The QuikChange II site-directed mutagenesis kit (Agilent Technologies) was employed to substitute amino acids according to the instructions of the kit. The template DNA employed was pCMV6-Hsp10-HA, and the following amino acid substitutions were performed: K40Q (AAA-CAA), 5′-ccagaaaagtctc aaggacaagtgttgcaagcaacgg-3′; K40R (AAA-AGA), 5′-ccagaaaagtctcaaggaagagtgttgca agcaacg-3′; K56Q (AAG-CAG), 5′-gggtcaggaggg aaaggacagagtggagagattg-3′; and K56R (AAG-AGG), 5′-ggtcaggagggaaaggaaggagtggagagattga-3′.
Gel Filtration
Mutant Hsp10 constructs were cotransfected with pCMV6-Hsp60-His into Hepa1c1c7 cells. Forty-eight hours after transfection, cells were harvested, and the Hsp10-Hsp60 complex was purified with a nickel-nitrilotriacetic acid column (Qiagen) according to the instructions of the manufacturer (except for the reduction of NaCl to 100 mm). Purified proteins were loaded onto HPLC gel filtration columns (TSKgel G3000SW, 7.5 inner diameter × 300, TOSOH Bioscience) equilibrated with 50 mm sodium phosphate (pH 7.43), 100 mm NaCl, and 0.1% Tween 20. Eluates were collected at 1-min intervals, concentrated with trichloroacetic acid, and then visualized by Western blot analysis employing an anti-His antibody (Sigma-Aldrich). HPLC gel filtration columns were calibrated using six standard proteins obtained from GE Life Sciences and Sigma-Aldrich.
Fatty Acid Oxidation Analysis
Measurements of mitochondrial fatty acid oxidation was performed using a Seahorse XF24 bioanalyzer following the protocol of the manufacturer. Briefly, Hepa1c1c7 cells were seeded in XF24 cell culture plates at 40,000 cells/well and cultured overnight in growth medium. The following day, fatty acid oxidation was measured in KHB buffer (111 m NaCl, 4.7 m KCl, 2 m MgSO4, 1.2 m Na2HPO4, 2.5 m glucose, 0.5 m carnitine) with 200 μm palmitate-BSA substrate, followed by mitochondrial stress compounds (2.5 μg/ml oligomycin, 1 μm carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone, and 2 μm rotenone/4 μm antimycin A). After completion, cells were harvested, and the rates were adjusted relative to protein content. Cells transfected with mutant Hsp10 were seeded 1 day after transfections.
The rate of fatty acid oxidation was concurrently assayed by quantifying 3H2O production from [9,10-3H(N)]palmitic acid (American Radiolabeled Chemicals) as described previously (21). Briefly, plated hepa1c1c7 cells were transfected with wild-type or mutant Hsp10 constructs. Two days after transfection, cells were incubated at 37 °C in PBS containing 125 μm [3H]palmitic acid, 38 μm BSA. and 1 mm carnitine. Two hours later, medium was collected and precipitated with trichloroacetic acid. Supernatants were neutralized with NaOH and applied to ion exchange columns packed with Dowex 1 × 2-400 resin (Sigma). The radioactive product was eluted with water and quantitated by liquid scintillation (PerkinElmer analyzer). Oxidation rates were normalized to protein content and expressed as nanomoles of fatty acid oxidized per hour per milligram of cell protein.
MCAD Enzyme Activity
MCAD activity was measured on the basis of the method published by Spiekerkoetter and co-workers (22). Hepa1c1c7 cells were resuspended in medium (200 μm ferrocenium hexafluorophosphate in 100 mm Tris-HCl (pH 8.0)). The enzyme reaction was initiated by adding octanoyl-CoA (C8:0-CoA, Sigma) as substrate, and the reaction was maintained at 37 °C for 10 min. MCAD assay products were measured by HPLC using a Phenomenex C18(2) Luna column (100 × 2.0 mm × 3 μm). The acyl-CoA esters were detected at 254 nm and quantified on the basis of peak area ratios.
Detection of Reactive Oxygen Species
Hepa1c1c7 cells were seeded on 96-well plates and transfected with wild-type or mutant Hsp10 constructs. Two days after transfection, cells were incubated in PBS containing 500 μm palmitic acid coupled with BSA for 6 h at 37 °C. After treatment with fatty acid, cells were washed twice with PBS and then incubated with 10 μm 2′, 7′-dichlorodihydrofluorescein diacetate (Invitrogen) for 1 h at 37 °C. Oxidation of the dye by intracellular reactive oxygen species generates a fluorescent 2,7-dicholorofluorescein signal. Fluorescence was measured using a Tecan M200 microplate reader at wavelengths of 485/530 nm and normalized to protein content.
Statistical Analysis
Differences between data groups were evaluated for significance using two-tailed Student's t test. Multiple comparison analysis was performed using analysis of variance. Statistical analysis was performed using GraphPad Prism, and data are expressed as mean ± S.E. p = 0.05 was considered statistically significant.
RESULTS
Hsp10 Is a Direct SIRT3 Deacetylase Target
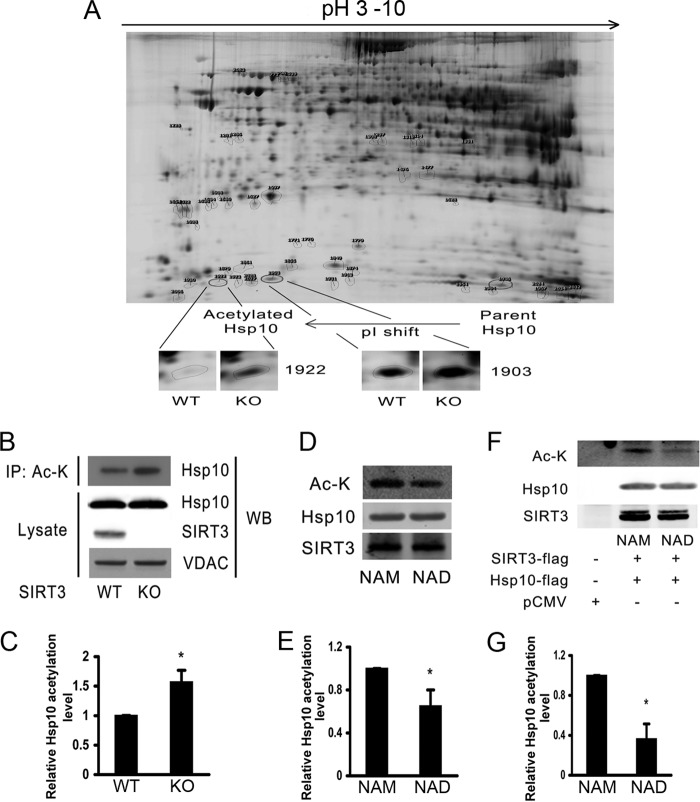
Although Hsp10 has been identified as a SIRT3 target in numerous acetylome screens (14–16), the direct interaction and effect of SIRT3 on Hsp10 has not been investigated previously. We initially confirmed by two-dimensional DIGE that Hsp10 acetylation was increased in SIRT3 KO mice and found a wide pH-dependent electromobility protein shift in parallel with altered Hsp10 acetylation when comparing WT and SIRT3 KO liver mitochondrial proteins (Fig. 1A). In parallel, acetyl-lysine immunoprecipitation of liver mitochondrial lysates confirmed significantly higher levels of endogenous Hsp10 acetylation in SIRT3 KO mice (Fig. 1, B and C). We then performed in vitro SIRT3 deacetylation assays to determine whether SIRT3 directly deacetylated Hsp10. Both endogenous Hsp10 and overexpressed Hsp10 were more highly acetylated in the presence of the SIRT3 antagonist nicotinamide and less acetylated in the presence of the SIRT3 agonist NAD (Fig. 1, D–G).
FIGURE 1.
Hsp10 is a SIRT3 substrate. A, two-dimensional DIGE showing liver mitochondrial Hsp10 levels of WT and SIRT3 KO mice following a 48-h fast. B and C, immunoprecipitation (IP) of the acetylated proteins with anti-acetylated lysine (Ac-K) antibody and quantification of acetylated Hsp10 levels by Western blot (WB) analysis with the corresponding histogram showing relative Hsp10 acetylation levels from four separate experiments. VDAC, voltage-dependent anion channel. D and E, in vitro deacetylation assay employing SIRT3 and Hsp10 immunoprecipitated from liver mitochondrial lysates using SIRT3 and Hsp10 antibodies, respectively. The corresponding histogram shows relative Hsp10 acetylation levels from four separate experiments. F and G, immunoblot showing an in vitro deacetylation assay where SIRT3 and Hsp10 were overexpressed in Hepa1c1c7 cells and immunoprecipitated with anti-FLAG antibody in the presence of a sirtuin inhibitor (nicotinamide (NAM)) or activator (NAD) with a corresponding histogram showing relative Hsp10 acetylation levels from four separate experiments. *, p < 0.05 versus respective controls.
SIRT3 Levels Modulate Mitochondrial Protein Folding
Because Hsp10, a component of the Hsp10-Hsp60 chaperonin complex, regulates mitochondrial protein folding, we investigated whether mitochondrial protein levels were different between WT and SIRT3 KO mouse livers. Proteins were differentially fluorescence-labeled and combined for two-dimensional gel electrophoresis and mass spectrometry analysis. The majority of proteins were found to have similar levels in the wild-type and KO mice. However, of the proteins that were differentially expressed, ≈80% of these showed higher levels in the SIRT3 KO mice (Fig. 2A and B, and data not shown).
Because protein folding is critical to mitochondrial integrity, we investigated whether WT and SIRT3 KO mice exhibited different levels of misfolded mitochondrial proteins. At baseline, and in response to a mild detergent (0.1% Nonidet P-40), no genotype differences were appreciated in analysis of detergent-insoluble misfolded protein aggregates. However, when the mitochondria were interrogated using the more stringent conditions of 1% Nonidet P-40, an ≈35% reduction in misfolded proteins was evident in the SIRT3 KO compared with WT controls (Fig. 2C).
To directly assay the SIRT3 role in mitochondrial folding, we measured the fluorescence levels of mtGFP. The appropriate folding of mtGFP within the mitochondrial matrix is necessary for robust emission of this fluorophore (23). Here mtGFP fluorescence was higher in Hepa1c1c7 cells where SIRT3 had been genetically depleted and diminished by ≈66% when SIRT3 was overexpressed (Fig. 2D). The levels of mtGFP fluorescence were not significantly changed following the overexpression of SIRT5, and the fluorescence intensity of cytosolic GFP was not altered by modulation of SIRT3 or SIRT5 levels (Fig. 2, E and F).
The Folding of Fatty Acid Metabolic Proteins Are Modulated by SIRT3
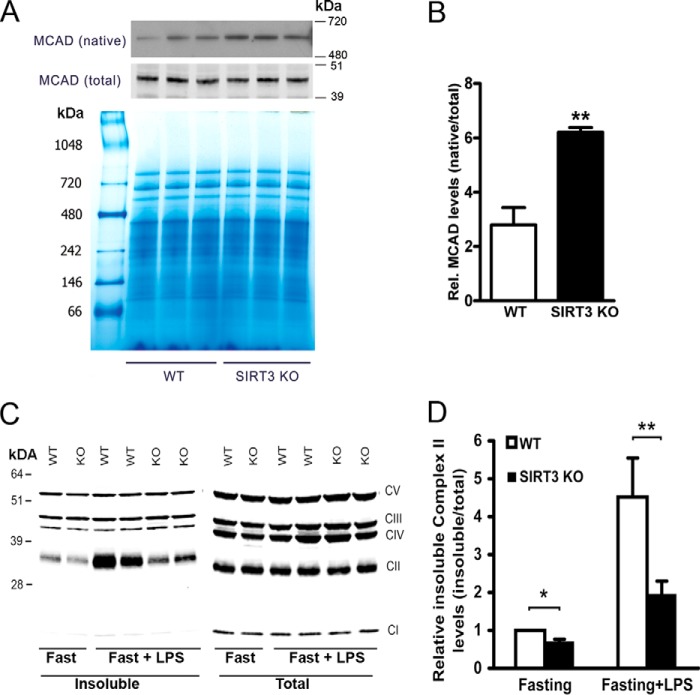
As described previously, MCAD is a substrate of the Hsp10-Hsp60 protein folding machinery (17, 24). We therefore explored the levels of tetrameric MCAD proteins in liver mitochondria by blue native PAGE. Interestingly, SIRT3 KO mice showed higher levels of native tetrameric MCAD compared with the wild-type controls despite no difference in total denatured MCAD levels (Fig. 3, A and B). To assess whether this difference extended to other mitochondrial proteins, we evaluated the folding of mitochondrial protein complexes in the electron transfer chain. It was interesting to note that only the fat oxidation intermediate-dependent complex, namely complex II, showed less misfolding in SIRT3 KO mice (≈30%) compared with control electron transfer chain protein complexes (Fig. 3C). To evaluate whether this difference could be exacerbated further, the mice were exposed to lipopolysaccharide, a known complex II activity inhibitor (25). Here the difference in complex II folding was accentuated, and the levels of unfolded complex II levels was ≈56% lower in the SIRT3 KO mice (Fig. 3D).
FIGURE 3.
SIRT3 regulates folding of fatty acid metabolic proteins. A, Western blots showing native tetrameric MCAD levels in liver mitochondria from WT and SIRT3 KO mice using blue native gel with total MCAD as the control. MW, molecular weight. B, quantification of relative (Rel.) native MCAD levels representing three experiments. **, p <0.01. C, liver mitochondria were isolated from mice following a 24-h fast with exposure to LPS (0.5 mg/kg) or saline administered intraperitoneally 18 h into fasting. Insoluble (after 1% Nonidet P-40 treatment) and total mitochondrial complex levels were measured by Western blot analysis. CV, mitochondrial respiratory chain complex V; CIII, complex III; CIV, complex IC; CII, complex II; CI, complex I. D, histograms showing quantification of relative insoluble complex II levels from four separate experiments. **, p <0.01.
Acetylation of Hsp10 Lys-56 Plays a Regulatory Role in the Hsp10-Hsp60 Chaperone Complex
The mechanisms and regulatory control of mitochondrial protein folding within the Hsp60-Hsp10 machinery have predominantly been investigated in Escherichia coli, where the GroEL-GroES protein complex has been exploited to model this biology (26). GroES, the Hsp10 ortholog, functions as the “lid” covering the GroEL (Hsp60 ortholog) chamber. Mitochondrial protein folding occurs within the chamber, where the combination of ATP hydrolysis (27), the dynamic interaction (28), and conformational changes of the GroEL-GroES (29) supercomplex function in concert to facilitate this processing.
To begin to interrogate this system in response to acetylation, we explored whether the steady-state interaction between these chaperonins was modified by SIRT3. Hsp10 and Hsp60 were cotransfected into a scrambled control or SIRT3 knockdown cells, followed by Hsp10-Hsp60 complex immunoprecipitation. Interestingly, the interaction between these chaperones was diminished in response to SIRT3 depletion (Fig. 4, A and B).
To explore whether the acetylation of Hsp10 is a necessary component of this SIRT3-dependent protein folding regulation, we began by identifying lysine residues that were altered in a SIRT3-dependent manner, as described previously (14). Using LC-MS/MS analysis comparing liver mitochondrial proteins from wild-type and SIRT3 KO mice, we identified that Lys-28, Lys-40, Lys-54, and Lys-56 exhibited differentially acetylated sites (Fig. 4C).
Repeated LC-MS/MS analyses showed that the Lys-40 and Lys-56 residues were most uniformly acetylated in SIRT3 KO liver mitochondria. These two sites were targeted for mutational analyses to mimic deacetylation (K-R) and acetylation (K-Q), respectively. The site-specific modified Hsp10 constructs were then cotransfected with Hsp60 to assay relative binding affinities. Immunoprecipitation analysis showed that the Hsp10-Hsp60 interaction was the weakest when Lys-56 was acetylated (Fig. 4D), suggesting that Hsp10 Lys-56 is a pivotal lysine residue in modifying SIRT3-dependent alteration in the interaction between Hsp10-Hsp60. To functionally confirm the role of Hsp10-K56 acetylation the mtGFP folding assay was repeated in the presence of the various Hsp10 constructs. Here the wild-type or mutant Hsp10 constructs and mtGFP were coexpressed, and the level of GFP fluorescence was assayed. Interestingly, the Hsp10-K56Q acetylated construct exhibited a similar level of mtGFP florescence as the empty vector transfections, whereas both wild-type and Hsp10-K56R construct overexpression resulted in dramatic reductions in mtGFP folding (Fig. 4E). The latter findings are consistent with the prior observation that enhanced Hsp10-Hsp60 interaction inhibits protein folding (30). As with the modulation of SIRT3 levels, the different Hsp10 constructs had no effect on cytosolic GFP fluorescence levels (Fig. 4F). Because lysine deacetylation modification confers a positive charge to lysine residues, we hypothesized that the acetyl modification of Hsp10 may evoke a conformational change to the interaction of the Hsp10-Hsp60 supercomplex. We employed gel filtration analysis to ascertain whether the change in Hsp10 Lys-56 acetylation modifies the Hsp10-Hsp60 complex elution rate (31). These data revealed that Hsp10-K56R·Hsp60 was eluted in earlier fractions compared with Hsp10-K56Q·Hsp60 (Fig. 4G), a finding compatible with an acetylation status-dependent conformational effect on this chaperonin supercomplex (31, 32).
The Acetylation of Hsp10 Lys-56 Increases Fatty Acid Oxidation
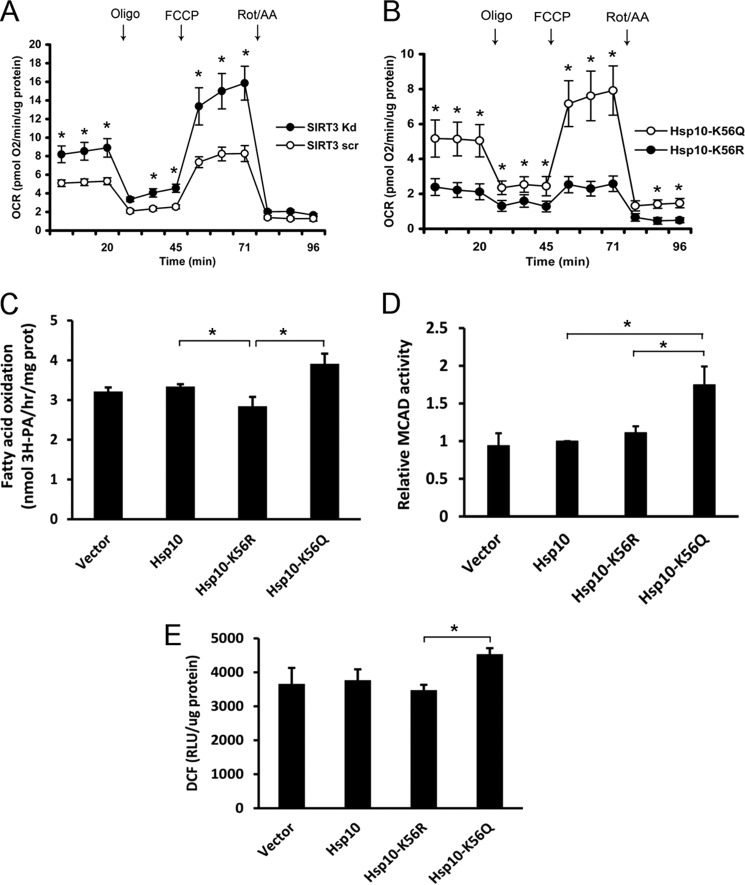
Because a fatty acid oxidation enzyme is a folding substrate of Hsp10-Hsp60 (24) and because the administration of fatty acids as substrates result in mitochondrial protein acetylation and the induction of fatty acid enzyme activity (8), we investigated whether SIRT3 and the acetylation status of Hsp10 Lys-56 were operational under these conditions. We show that cellular oxygen consumption using BSA-conjugated palmitate as a substrate is increased in SIRT3 knockdown cells (Fig. 5A) and that the overexpression of Hsp10-K56Q evokes a higher rate of oxygen consumption compared with Hsp10-K56R (Fig. 5B). In parallel, the direct measurement of radiolabeled intermediates showed a modest blunting of palmitate oxidation in Hsp10-K56R-transfected cells and a significant increase in oxidation in Hsp10-K56Q-overexpressing cells (Fig. 5C). To directly assess the effect of Hsp10 Lys-56 acetylation on MCAD activity, the wild-type and modified Hsp10 constructs were expressed in Hepa1c1c7 cells. Under these conditions, we found that MCAD activity was increased significantly when Hsp10 Lys-56 was constitutively acetylated compared with the activity in the presence of the other Hsp10 plasmids (Fig. 5D). Because increased fat oxidation is usually associated with increased mitochondrial reactive oxygen species production (33), we measured reactive oxygen species levels by H2DCFDA fluorescence. These data correlated with the rate of fat oxidation with higher fluorescence in Hsp10-K56Q-overexpressing cells (Fig. 5E).
FIGURE 5.
Hsp10 Lys-56 acetylation increases fatty acid oxidation and MCAD activity. A and B, fatty acid oxidation was measured using a Seahorse analyzer in Hepa1c1c7 cells with either scrambled or SIRT3 knockdown or mutant Hsp10 plasmid overexpression, respectively (n = 3 experiments). FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone. OCR, oxygen consumption rate; Rot/AA, rotenone/antimycin A. *, p < 0.05. C, measurement of tritiated water release to measure palmitate oxidation in Hsp10 wild-type and mutant-overexpressing cells (n = 3 experiments). prot, protein. 3H-PA, [9,10-3H(N)] palmitic acid. *, p <0.05. D, Hepa1c1c7 cells were transfected with vector, wild-type, or mutant Hsp10. MCAD enzyme activity was measured using HPLC with octanoyl-CoA as the substrate (n = 5 experiments). *, p <0.05. E, measurement of relative 2,7-dicholorofluorescein (DCF) fluorescence to assay the relative reactive oxygen species levels in Hepa1c1c7 cells following overexpression of control, wild-type, or mutant Hsp10 constructs (n = 3 experiments). RLU, relative light units. *, p <0.05.
DISCUSSION
Accumulating evidence supports the hypothesis that the nutrient- and stress-dependent modification of mitochondrial proteins by acetylation plays an instrumental role in mitochondrial function (34), stress susceptibility, and intrinsic organelle quality control (35). Although a component of mitochondrial quality control involves mitochondrial protein folding, whether this homeostatic program is directly modified and/or regulated by the acetyl posttranslational modification appears to not have been well characterized (36). In this study, we show that mitochondrial protein folding is modified by acetylation and that Hsp10 is a direct SIRT3 deacetylase substrate. Furthermore, our data identify a pivotal lysine residue on Hsp10 that changes the dynamic interaction between Hsp10 and Hsp60 as a probable component in the role of this chaperonin in controlling mitochondrial protein folding. Furthermore, the increased acetylation of Hsp10 promotes fat oxidation and suggests that this regulatory program may contribute to increased fatty acid oxidation when SIRT3 is down-regulated in response to high fat feeding (10). Together, these data identify a novel nutrient level-dependent control node in mitochondrial homeostasis.
The acetylation-deacetylation system is emerging as an important protein modifier controlling cellular proteostasis biology, autophagy, and apoptosis. Recent studies have found that changes in acetylation of protein lysine residues can result in the degradation of cytosolic proteins via ubiquitination and activation of the proteasome (37, 38) or via chaperon-mediated autophagy (39). Acetyl modification also modulates the endoplasmic reticulum unfolded protein response (UPRER) and the cytosolic UPR (38, 40, 41).
How and whether acetyl modification regulates mitochondrial proteostasis is less well characterized. Although a recent study showed that NAD+-dependent SIR2.1 signaling augments the mitochondrial UPR (UPRmt) via activation of the FOXO (forkhead box) transcription factor DAF-16 (42). In our study, we provided evidence that the acetyl modification of the mitochondrial chaperonin Hsp10, as a posttranslational event, can modulate mitochondrial protein folding. We show that the acetyl modification of Hsp10 Lys-56 prevented accumulation of misfolded proteins in parallel with the enhanced efficiency of the Hsp10-Hsp60 machinery. To what extent this regulation coordinates with the transcriptional components of the UPRmt and whether additional mitochondrial mediators of this regulation are modified by acetylation remains to be clarified.
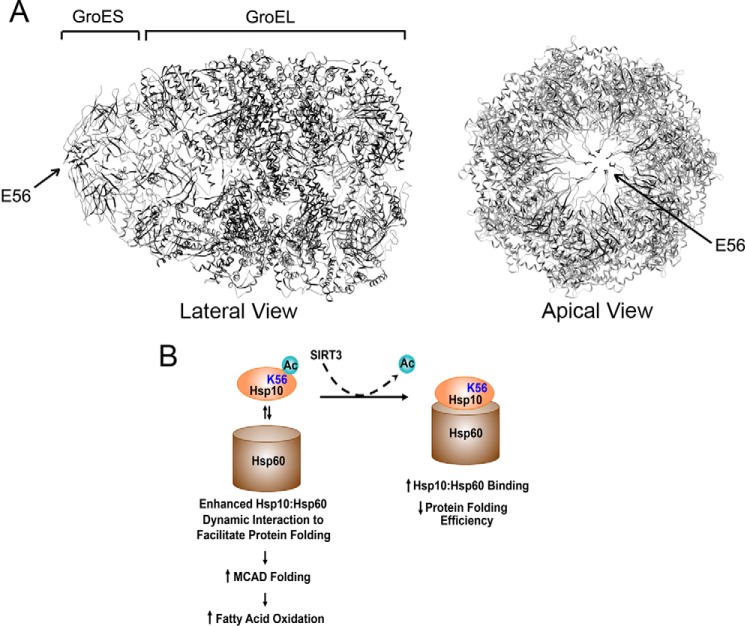
As discussed previously, the regulatory control of mitochondrial protein folding within the Hsp60-Hsp10 machinery has been investigated predominantly in E. coli, where the GroEL-GroES protein complex has been explored to model this biology (26). In light of the regulatory importance of Lys-56 in this study, we compared the sequence alignment at amino acid position 56 between the E. coli GroES and the mammalian Hsp10. In E. coli, this residue is occupied by a glutamic acid rather than a lysine, representing a substitution of an acid with a basic residue from bacteria to mammalia. The E. coli chaperonin complex with the position of the Glu-56 residue highlighted is shown in Fig. 6A. The acetylation of Lys-56 would neutralize the positive charge of this residue, and whether or how this affects the aperture of the orifice within the lid and/or the rate of association and dissociation between Hsp10 and Hsp60 is not understood at this time. Nevertheless, our data show that the retention of acetylation of residue 56 of Hsp10 (K56Q construct) results in greater efficiency of mitochondrial protein folding compared with the deacetylation of this residue (K56R). In parallel, the Hsp10-Hsp60 binding affinity was diminished when Lys-56 was acetylated versus deacetylated. This modification in Hsp10-Hsp60 binding is also evident in the in vivo cellular environment with the genetic knockdown of SIRT3. Gel filtration analysis furthermore supports that the acetyl modification of Lys-56 elicits a conformational change in the Hsp10-Hsp60 supercomplex structure. More refined structure-function studies will be required to integrate the observations that the acetylation of Lys-56 modifies protein folding and the conformation of the Hsp10-Hsp60 supercomplex and changes the binding affinity between these chaperonins. Additionally, why the restricted overexpression of wild-type Hsp10 disrupts mitochondrial protein folding is currently unexplained, although this same interrogation has been shown previously to inhibit ATP hydrolysis of Hsp60 (30).
FIGURE 6.
Model of the role of Hsp10 Lys-56 acetylation in the modulation of protein folding. A, lateral and apical views of the GroES-GroEL complex with Glu-56 labeled within the rim of the aperture of GroES. The mammalian ortholog residue in Hsp10 is represented by Lys-56. B, schematic of acetyl (Ac) modification regulating the dynamic interaction of Hsp10-Hsp60 with the subsequent consequences on protein folding and fat oxidation.
An intriguing concept that has emerged from this study is that a canonical Hsp10-Hsp60 substrate, i.e. MCAD, shows enhanced folding and increased enzyme activity in the absence of the mitochondrial deacetylase SIRT3. These data support the evidence showing increased fatty acid oxidation in the presence of excess fat and mitochondrial protein acetylation (8, 9). Contrasting studies also show that the direct deacetylation of lysine residues on the fatty acid oxidation enzymes themselves can activate metabolic activity (5, 6). Whether this reflects differential regulation of the same metabolic pathway at the levels of protein folding and enzyme activity is an intriguing concept that requires clarification.
Finally, it should be noted the modification of Hsp10 Lys-56 is unlikely to be an exclusive acetyl modification of this complex in governing protein folding. Additionally, recent work has also shown that, in the context of redox or direct proteotoxic stressors, SIRT3 in its capacity to enhance excess reactive oxygen species biology has an inverse, albeit indirect, and ameliorative effect on the mitochondrial protein folding machinery (43). Our study nevertheless advances the understanding of a mechanism that may explain the paradoxical findings, where nutrient excess and caloric restriction have profound effects on mitochondrial homeostasis and quality control (44, 45). Further investigations into the mechanisms underpinning acetylation on the mitochondrial folding and unfolded protein responses should advance our insight into why caloric load can have direct effects on mitochondrial biology (44).
On the basis of our findings, we propose the inclusion of an additional regulatory node into the current model governing the dynamic interaction between Hsp10-Hsp60 in the control of mitochondrial protein folding (Fig. 6B). In brief, we find that the modification of the acetylation of the Lys-56 residue of Hsp10 alters the probable conformation and the binding affinities between Hsp10 and Hsp60. This, in turn, modifies the capacity to appropriately fold MCAD, which alters both enzyme activity and the overall rate of fat oxidation. Because this protein folding is a dynamic process, we propose that the modulation of the acetylation of Hsp10 may play a “fine-tuning” role in modulating MCAD folding with cognate downstream metabolic effects. At the same time, to demonstrate this acetylation-dependent component in the control of mitochondrial protein folding, we employed constructs of Hsp10 with fixed modifications of Lys-56 acetylation. Ultimately, an in vivo system with the capacity to dynamically modify Hsp10 acetylation would be required to quantify the absolute contribution of this modification to the appropriate folding of MCAD and to the regulation of fatty acid oxidation.
In conclusion, we find that the SIRT3-dependent deacetylation of Hsp10 plays a functional role in controlling mitochondrial protein folding via the modulation of the interaction of Hsp10 within the Hsp10-Hsp60 chaperonin complex. This mechanism identifies a novel nutrient-sensing regulatory program that could contribute toward the overall control of the rate of mitochondrial fat oxidation.
Acknowledgments
We thank Rodney L. Levine (NHLBI/National Institutes of Health) for advice regarding protein structural analysis and Duck-Yeon Lee from the NHLBI/National Institutes of Health Biochemistry Core for assistance with gel filtration assays. We also thank Dr. Fred Alt (Harvard Medical School) for SIRT3+/− mice and Dr. Chunxin Wang (NINDS/National Institutes of Health for mtGFP.
This work was supported, in whole or in part, by NHLBI/National Institutes of Health Intramural Research Program HL006047-01 and by NHLBI/National Institutes of Health K22 Award K22HL109236.
- MCAD
- medium-chain acyl-CoA dehydrogenase
- mtGFP
- mitochondrial GFP
- UPR
- unfolded protein response.
REFERENCES
- 1. Sack M. N. (2009) Type 2 diabetes, mitochondrial biology and the heart. J. Mol. Cell Cardiol. 46, 842–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leone T. C., Kelly D. P. (2011) Transcriptional control of cardiac fuel metabolism and mitochondrial function. Cold Spring Harbor Symp. Quant. Biol. 76, 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cantó C., Auwerx J. (2009) PGC-1α, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 20, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sack M. N., Finkel T. (2012) Mitochondrial metabolism, sirtuins, and aging. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a013102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., Newgard C. B., Farese R. V., Jr., Alt F. W., Kahn C. R., Verdin E. (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bharathi S. S., Zhang Y., Mohsen A. W., Uppala R., Balasubramani M., Schreiber E., Uechi G., Beck M. E., Rardin M. J., Vockley J., Verdin E., Gibson B. W., Hirschey M. D., Goetzman E. S. (2013) Sirtuin 3 (SIRT3) Protein regulates long-chain acyl-CoA dehydrogenase by deacetylating conserved lysines near the active site. J. Biol. Chem. 288, 33837–33847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hallows W. C., Yu W., Smith B. C., Devries M. K., Devires M. K., Ellinger J. J., Someya S., Shortreed M. R., Prolla T., Markley J. L., Smith L. M., Zhao S., Guan K. L., Denu J. M. (2011) Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol. Cell 41, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jing E., O'Neill B. T., Rardin M. J., Kleinridders A., Ilkeyeva O. R., Ussar S., Bain J. R., Lee K. Y., Verdin E. M., Newgard C. B., Gibson B. W., Kahn C. R. (2013) Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes 62, 3404–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alrob O. A., Sankaralingam S., Ma C., Wagg C. S., Fillmore N., Jaswal J. S., Sack M. N., Lehner R., Gupta M. P., Michelakis E. D., Padwal R. S., Johnstone D. E., Sharma A. M., Lopaschuk G. D. (2014) Obesity-induced lysine acetylation increases cardiac fatty acid oxidation and impairs insulin signalling. Cardiovasc. Res. 103, 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bao J., Scott I., Lu Z., Pang L., Dimond C. C., Gius D., Sack M. N. (2010) SIRT3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radic. Biol. Med. 49, 1230–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Borengasser S. J., Lau F., Kang P., Blackburn M. L., Ronis M. J., Badger T. M., Shankar K. (2011) Maternal obesity during gestation impairs fatty acid oxidation and mitochondrial SIRT3 expression in rat offspring at weaning. PLoS ONE 10.1371/journal.pone.0024068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kendrick A. A., Choudhury M., Rahman S. M., McCurdy C. E., Friederich M., Van Hove J. L., Watson P. A., Birdsey N., Bao J., Gius D., Sack M. N., Jing E., Kahn C. R., Friedman J. E., Jonscher K. R. (2011) Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem. J. 433, 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu Z., Bourdi M., Li J. H., Aponte A. M., Chen Y., Lombard D. B., Gucek M., Pohl L. R., Sack M. N. (2011) SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep. 12, 840–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fritz K. S., Galligan J. J., Hirschey M. D., Verdin E., Petersen D. R. (2012) Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice. J. Proteome Res. 11, 1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rardin M. J., Newman J. C., Held J. M., Cusack M. P., Sorensen D. J., Li B., Schilling B., Mooney S. D., Kahn C. R., Verdin E., Gibson B. W. (2013) Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc. Natl. Acad. Sci., U.S.A. 110, 6601–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saijo T., Welch W. J., Tanaka K. (1994) Intramitochondrial folding and assembly of medium-chain acyl-CoA dehydrogenase (MCAD): demonstration of impaired transfer of K304E-variant MCAD from its complex with hsp60 to the native tetramer. J. Biol. Chem. 269, 4401–4408 [PubMed] [Google Scholar]
- 18. Nakagawa T., Lomb D. J., Haigis M. C., Guarente L. (2009) SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137, 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bao J., Lu Z., Joseph J. J., Carabenciov D., Dimond C. C., Pang L., Samsel L., McCoy J. P., Jr., Leclerc J., Nguyen P., Gius D., Sack M. N. (2010) Characterization of the murine SIRT3 mitochondrial localization sequence and comparison of mitochondrial enrichment and deacetylase activity of long and short SIRT3 isoforms. J. Cell Biochem. 110, 238–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moisoi N., Klupsch K., Fedele V., East P., Sharma S., Renton A., Plun-Favreau H., Edwards R. E., Teismann P., Esposti M. D., Morrison A. D., Wood N. W., Downward J., Martins L. M. (2009) Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 16, 449–464 [DOI] [PubMed] [Google Scholar]
- 21. Djouadi F., Bonnefont J. P., Munnich A., Bastin J. (2003) Characterization of fatty acid oxidation in human muscle mitochondria and myoblasts. Mol. Genet. Metab. 78, 112–118 [DOI] [PubMed] [Google Scholar]
- 22. ter Veld F., Mueller M., Kramer S., Haussmann U., Herebian D., Mayatepek E., Laryea M. D., Primassin S., Spiekerkoetter U. (2009) A novel tandem mass spectrometry method for rapid confirmation of medium- and very long-chain acyl-CoA dehydrogenase deficiency in newborns. PLoS ONE 10.1371/journal.pone.0006449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bie A. S., Palmfeldt J., Hansen J., Christensen R., Gregersen N., Corydon T. J., Bross P. (2011) A cell model to study different degrees of Hsp60 deficiency in HEK293 cells. Cell Stress Chaperones 16, 633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Corydon T. J., Hansen J., Bross P., Jensen T. G. (2005) Down-regulation of Hsp60 expression by RNAi impairs folding of medium-chain acyl-CoA dehydrogenase wild-type and disease-associated proteins. Mol. Genet. Metab. 85, 260–270 [DOI] [PubMed] [Google Scholar]
- 25. Song Y., Pinniger G. J., Bakker A. J., Moss T. J., Noble P. B., Berry C. A., Pillow J. J. (2013) Lipopolysaccharide-induced weakness in the preterm diaphragm is associated with mitochondrial electron transport chain dysfunction and oxidative stress. PLoS ONE 10.1371/journal.pone.0073457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Horwich A. L., Fenton W. A. (2009) Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q. Rev. Biophys. 42, 83–116 [DOI] [PubMed] [Google Scholar]
- 27. Lin Z., Rye H. S. (2006) GroEL-mediated protein folding: making the impossible, possible. Crit. Rev. Biochem. Mol. Biol. 41, 211–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang D., Ye X., Lorimer G. H. (2013) Symmetric GroEL:GroES2 complexes are the protein-folding functional form of the chaperonin nanomachine. Proc. Natl. Acad. Sci., U.S.A. 110, E4298–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen D. H., Madan D., Weaver J., Lin Z., Schröder G. F., Chiu W., Rye H. S. (2013) Visualizing GroEL/ES in the act of encapsulating a folding protein. Cell 153, 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dubaquié Y., Looser R., Rospert S. (1997) Significance of chaperonin 10-mediated inhibition of ATP hydrolysis by chaperonin 60. Proc. Natl. Acad. Sci., U.S.A. 94, 9011–9016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghosh P., Kornfeld S. (2003) Phosphorylation-induced conformational changes regulate GGAs 1 and 3 function at the trans-Golgi network. J. Biol. Chem. 278, 14543–14549 [DOI] [PubMed] [Google Scholar]
- 32. Feramisco J. D., Radhakrishnan A., Ikeda Y., Reitz J., Brown M. S., Goldstein J. L. (2005) Intramembrane aspartic acid in SCAP protein governs cholesterol-induced conformational change. Proc. Natl. Acad. Sci., U.S.A. 102, 3242–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Du X., Edelstein D., Obici S., Higham N., Zou M. H., Brownlee M. (2006) Insulin resistance reduces arterial prostacyclin synthase and eNOS activities by increasing endothelial fatty acid oxidation. J. Clin. Invest. 116, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anderson K. A., Hirschey M. D. (2012) Mitochondrial protein acetylation regulates metabolism. Essays Biochem. 52, 23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Webster B. R., Scott I., Traba J., Han K., Sack M. N. (2014) Regulation of autophagy and mitophagy by nutrient availability and acetylation. Biochim. Biophys. Acta 1841, 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jensen M. B., Jasper H. (2014) Mitochondrial proteostasis in the control of aging and longevity. Cell Metabolism 20, 214–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang W., Wang S., Xiao M., Lin Y., Zhou L., Lei Q., Xiong Y., Guan K. L., Zhao S. (2011) Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell 43, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang F., Chan C. H., Chen K., Guan X., Lin H. K., Tong Q. (2012) Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene 31, 1546–1557 [DOI] [PubMed] [Google Scholar]
- 39. Lv L., Li D., Zhao D., Lin R., Chu Y., Zhang H., Zha Z., Liu Y., Li Z., Xu Y., Wang G., Huang Y., Xiong Y., Guan K. L., Lei Q. Y. (2011) Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol. Cell 42, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boyault C., Zhang Y., Fritah S., Caron C., Gilquin B., Kwon S. H., Garrido C., Yao T. P., Vourc'h C., Matthias P., Khochbin S. (2007) HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 21, 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Westerheide S. D., Anckar J., Stevens S. M., Jr., Sistonen L., Morimoto R. I. (2009) Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323, 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mouchiroud L., Houtkooper R. H., Moullan N., Katsyuba E., Ryu D., Cantó C., Mottis A., Jo Y. S., Viswanathan M., Schoonjans K., Guarente L., Auwerx J. (2013) The NAD+/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154, 430–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Papa L., Germain D. (2014) SirT3 regulates the mitochondrial unfolded protein response. Mol. Cell Biol. 34, 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pagel-Langenickel I., Bao J., Pang L., Sack M. N. (2010) The role of mitochondria in the pathophysiology of skeletal muscle insulin resistance. Endocr. Rev. 31, 25–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koves T. R., Ussher J. R., Noland R. C., Slentz D., Mosedale M., Ilkayeva O., Bain J., Stevens R., Dyck J. R., Newgard C. B., Lopaschuk G. D., Muoio D. M. (2008) Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 7, 45–56 [DOI] [PubMed] [Google Scholar]