Background: Channel gating is uncoupled from agonist binding when nicotinic receptors lack activating lipids.
Results: Gating is also uncoupled with ELIC, but engineered aromatic interactions at the M4-M1/M3 interface restore coupling.
Conclusion: The strength of M4-M1/M3 interactions impacts both coupling and lipid sensitivity.
Significance: Variable levels of intramembrane aromatics in eukaryotic homologs likely lead to variable lipid sensitivities.
Keywords: Electrophysiology, Infrared Spectroscopy (IR Spectroscopy), Ion Channel, Lipid Bilayer, Membrane Biophysics, Membrane Lipid, Membrane Protein, Membrane Reconstitution, Nicotinic Acetylcholine Receptor (nAChR)
Abstract
Although the Torpedo nicotinic acetylcholine receptor (nAChR) reconstituted into phosphatidylcholine (PC) membranes lacking cholesterol and anionic lipids adopts a conformation where agonist binding is uncoupled from channel gating, the underlying mechanism remains to be defined. Here, we examine the mechanism behind lipid-dependent uncoupling by comparing the propensities of two prokaryotic homologs, Gloebacter and Erwinia ligand-gated ion channel (GLIC and ELIC, respectively), to adopt a similar uncoupled conformation. Membrane-reconstituted GLIC and ELIC both exhibit folded structures in the minimal PC membranes that stabilize an uncoupled nAChR. GLIC, with a large number of aromatic interactions at the interface between the outermost transmembrane α-helix, M4, and the adjacent transmembrane α-helices, M1 and M3, retains the ability to flux cations in this uncoupling PC membrane environment. In contrast, ELIC, with a level of aromatic interactions intermediate between that of the nAChR and GLIC, does not undergo agonist-induced channel gating, although it does not exhibit the expected biophysical characteristics of the uncoupled state. Engineering new aromatic interactions at the M4-M1/M3 interface to promote effective M4 interactions with M1/M3, however, increases the stability of the transmembrane domain to restore channel function. Our data provide direct evidence that M4 interactions with M1/M3 are modulated during lipid sensing. Aromatic residues strengthen M4 interactions with M1/M3 to reduce the sensitivities of pentameric ligand-gated ion channels to their surrounding membrane environment.
Introduction
The ability of the Torpedo nicotinic acetylcholine receptor (nAChR)2 to undergo agonist-induced channel gating is sensitive to lipids (1–6). Increasing levels of both cholesterol and anionic lipids in 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC) membranes stabilize increasing proportions of agonist-responsive nAChRs (4, 7–10). In the absence of activating lipids, the nAChR adopts an uncoupled conformation that binds agonist but does not usually undergo agonist-induced conformational transitions (11). Defining the mechanisms underlying lipid-dependent uncoupling of agonist-binding and channel-gating is central to understanding nAChR-lipid interactions. Such studies may have broader implications because neuronal nAChRs that are functionally uncoupled have been observed in heterologous expression systems and may play a role in the response to nicotine (12, 13). Lipid-dependent mechanisms for “awakening” uncoupled nAChRs have been identified and could play a role modulating synaptic communication (14). The lipid-dependent uncoupled conformation may also be germane to the interpretation of crystal structures of detergent-solubilized pLGICs (15).
Lipids may exert their effects on nAChR function by interacting with interstitial sites between α-helices in the transmembrane domain (16, 17) or by interacting with the lipid-protein interface (11, 16, 18, 19). The lipid-exposed transmembrane α-helix M4 from each subunit probably plays a role in both lipid sensing and the formation of an uncoupled state (11, 14, 20–24). Mutations along the lipid-facing surface of M4 alter gating, showing that altered M4-lipid interactions influence the allosteric pathway leading from the agonist site to the channel gate (25–29). One model of uncoupling proposes that lipids modulate interactions between M4 and the adjacent transmembrane α-helices, M1 and M3, with enhanced M4-M1/M3 interactions promoting effective coupling between the agonist binding domain and the transmembrane domain (TMD) to enhance channel gating (11, 14).
Definitive insight into the mechanisms of lipid-dependent uncoupling in pentameric ligand-gated ion channels (pLGICs) will require the ability to characterize the effects of changing lipid environments on channel mutants designed to test mechanistic hypotheses, such as the M4 lipid sensor model described above. In contrast to the nAChR (30), the homologous prokaryotic pLGICs, GLIC and ELIC (Fig. 1A), are both expressed in relatively large quantities in Escherichia coli and are thus amenable to such biophysical studies. Membrane-reconstituted GLIC has a higher denaturation temperature and a reduced structural/functional sensitivity to lipids compared with the nAChR. In membranes that lock the nAChR in an uncoupled conformation, GLIC retains the ability to undergo agonist-induced conformational change (31).
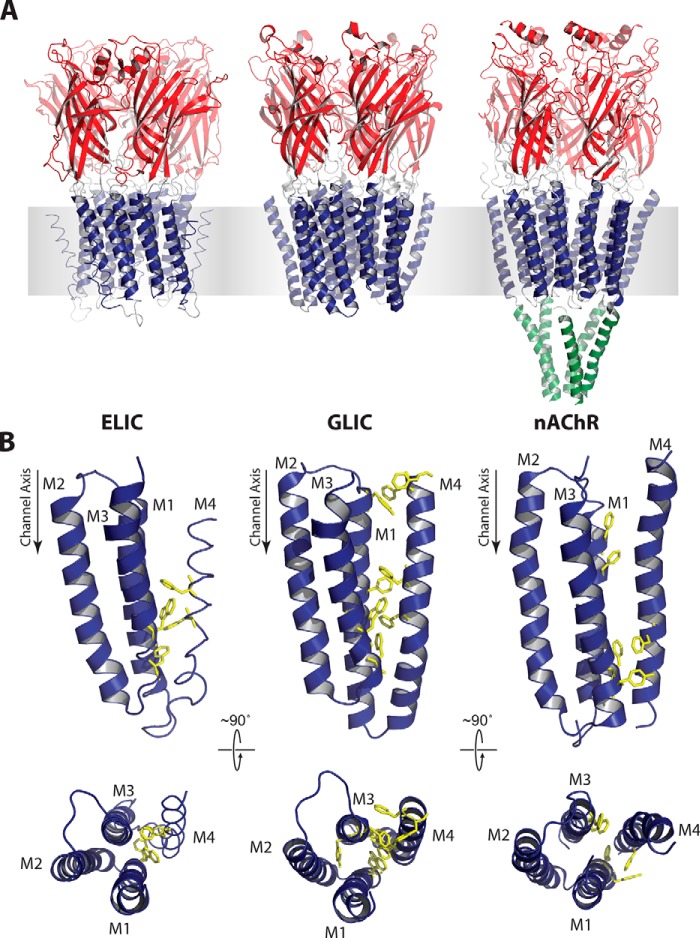
FIGURE 1.
Three-dimensional structure and TMD aromatics in ELIC, GLIC, and the Torpedo nAChR. A, three-dimensional structures of ELIC (2VLO; left), GLIC (3EAM; center), and the Torpedo nAChR (2BG9; right) colored to highlight the functional agonist binding (red), transmembrane gating (blue), and cytoplasmic (green) domains. B, side and top views of a single TMD from each pLGIC (α-subunit for the nAChR) highlighting aromatic residues at the M4-M1/M3 interface as yellow sticks.
Crystal structures of GLIC reveal an extensive network of aromatic interactions between M4 and M1/M3 relative to that observed in the nAChR (Fig. 1B). Given that aromatic interactions are the key interactions that energetically drive M4 binding to M1/M3 during folding (32), we hypothesized that the multiple aromatic interactions in GLIC strengthen M4-M1/M3 interactions, leading to a more stable TMD with a reduced propensity to adopt an uncoupled conformation. Interestingly, the number of aromatic interactions at the M4-M1/M3 interface in ELIC is intermediate between that of the nAChR and GLIC. If aromatic residues at this interface enhance M4-M1/M3 interactions, then the reduced number of aromatic residues in ELIC relative to GLIC should lead to weaker M4-M1/M3 interactions and thus a greater propensity to adopt an uncoupled conformation.
Here, we test whether aromatic interactions at the M4-M1/M3 interface influence the propensity of a pLGIC to adopt a lipid-dependent uncoupled conformation. We find that in contrast to GLIC, ELIC does not undergo agonist-induced channel gating in the minimal PC membranes that stabilize an uncoupled nAChR. Engineering new aromatic residues to enhance the intrinsic strength of M4-M1/M3 interactions, however, restores the ability to gate open in these same PC membranes. Our data highlight the important role played by M4 in pLGIC lipid sensing and suggest that intramembrane aromatic interactions govern the sensitivity of a pLGIC to its membrane environment.
EXPERIMENTAL PROCEDURES
Materials
Soybean asolectin (l-α-phosphatidylcholine, type II-S) and cysteamine were from Sigma. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine was from Avanti (Alabaster, AL). Dodecylmaltoside was from Affymetrix (Santa Clara, CA). A pET-26b expression vector containing the pelB signal sequence followed by a His10 tag and then the maltose-binding protein fused to the N terminus of ELIC through a herpes simplex 3C protease site was kindly provided by Dr. Raimund Dutzler. A C-terminal alanine, a cloning artifact not present in the GenBankTM sequence (accession number POC7B7), was removed. The mutant ELIC was created using QuikChangeTM site-directed mutagenesis kits (Agilent) and verified by sequencing.
Preparation of ELIC Proteoliposomes
ELIC was expressed, purified, and reconstituted into proteoliposomes as described previously for GLIC (31) but with several modifications. ELIC was expressed in the C41 or BL21 strain of E. coli. Cultures of transformed cells were grown in Terrific Broth containing 50 μg/ml kanamycin at 37 °C to an A600 of ∼1.2 absorbance units and then induced overnight at 26 °C with 200 μm isopropyl 1-thio-β-d-galactopyranoside. Cells were harvested, resuspended in Buffer A (150 mm NaCl, 50 mm NaH2PO4, pH 8.0) in the presence of either Roche CompleteTM antiprotease tablets (Branford, CT) or an analogous mix of locally prepared antiproteases and lysed with an Avestin Emulsiflex-C3 homogenizer (Ottawa, Canada). Membrane fractions were solubilized in 1% dodecylmaltoside in Buffer A, and the ELIC fusion protein was bound to an amylose affinity resin. After treatment with herpes simplex 3C protease (Calbiochem), ELIC was eluted in 0.02% dodecylmaltoside and further purified in Buffer B (150 mm NaCl, 10 mm NaH2PO4, pH 8.0) on a Superose 6 10/300 gel filtration column (GE Healthcare).
The purified ELIC in 0.02% dodecylmaltoside in Buffer B was slowly diluted to a ratio of at least 1:4 (v/v) with a solution of lipids solubilized in 0.625% cholate in Buffer B to give a 2:1 (w/w) lipid/protein ratio (33). After gently mixing for ∼30 min, the protein/detergent/lipid mixture was dialyzed five times at 4 °C against 2 liters of Buffer B, leading to turbid solutions of proteoliposomes, which were harvested by ultracentrifugation. The incorporation of ELIC into the liposomes was assessed by discontinuous sucrose density gradient centrifugation (11).
Fourier Transform Infrared Spectroscopy (FTIR)
Infrared spectra were recorded on either a Digilab (now Agilent Technologies; Santa Clara, CA) FTS40 or FTS575 spectrometer. The hydrogen/deuterium infrared spectra (Fig. 3A) were recorded using a Golden-GateTM attenuated total internal reflection accessory (SpecAc, Oprington (Kent, UK)). 5–10-μl aliquots were dried gently under N2 gas. 128 scan spectra were recorded at 4 cm−1 resolution both before and after rehydration with 2H2O (31).
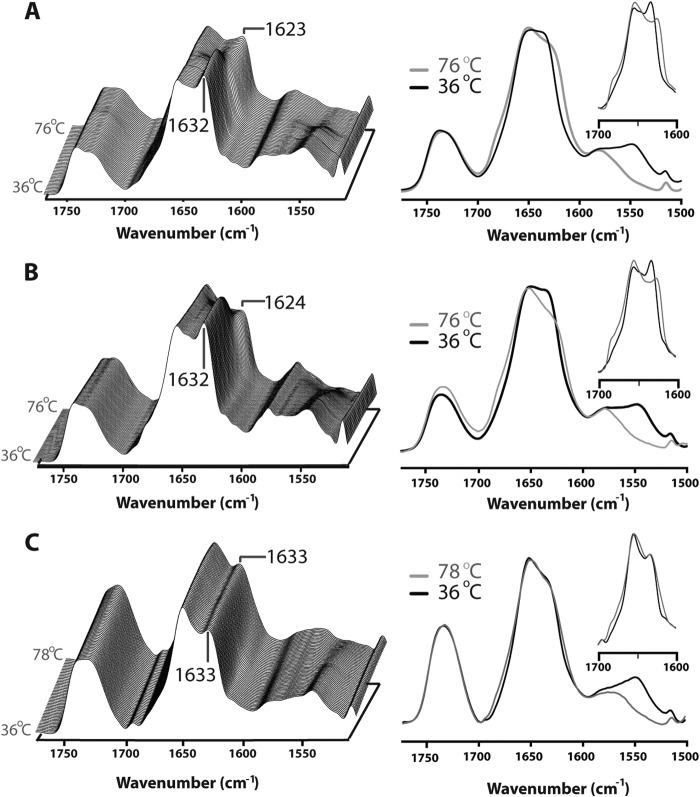
FIGURE 3.
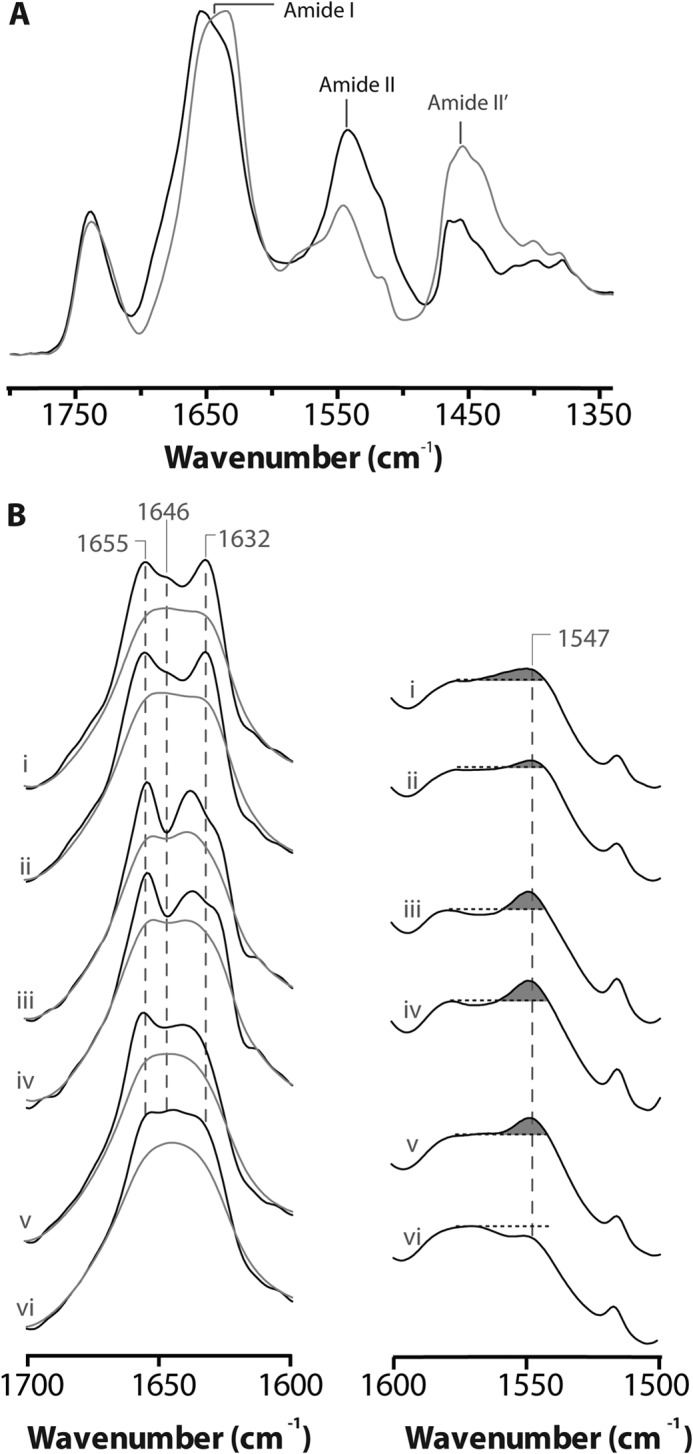
Structural comparisons of membrane reconstituted ELIC, GLIC and the nAChR as probed by infrared spectroscopy. A, infrared spectra of aso-ELIC recorded after gentle drying from 1H2O buffer (black) and immediately after the addition of 2H2O (gray). Note the immediate changes in amide I band shape (from 1700 to 1600 cm−1) and the immediate decrease in amide II band intensity (1547 cm−1), both indicative of the rapid peptide N-1H/N-2H exchange of solvent-exposed peptide hydrogens. B, the structure-sensitive amide I (left) and amide II (right) bands in infrared spectra recorded after 24 h of equilibration in 2H2O at 4 °C from aso-ELIC (i) and PC-ELIC (ii) and after 72 h equilibration in 2H2O at 4 °C for aso-GLIC (iii), PC-GLIC (iv), aso-nAChR (v), and PC-nAChR (vi) (11, 31). The amide I bands are shown both before (gray) and after resolution enhancement (black). Spectra are the averages of at least three spectra recorded from two different purifications/reconstitutions. See Fig. 2 for details.
For more detailed amide I band analyses, samples were exchanged into 2H2O Buffer A for precisely 24 h at 4 °C and then stored at −80 °C until use (spectra are recorded in 2H2O to eliminate the strong overlap between the absorptions of 1H2O and the protein vibrations of interest). Approximately 200 μg of ELIC was then deposited on a CaF2 window with a gentle stream of N2 gas, followed by rehydration with 8 μl of 2H2O. 4000 scan spectra were recorded at 2 cm−1 resolution and then processed with GRAMS/AI software (Thermo Scientific, Waltham, MA) (11). Resolution enhancement was performed between 1900 and 1300 cm−1 using a γ = 7 and a Bessel smoothing function set to 70%. Intensity changes in the amide I band at either 1680 or 1620 cm−1 were plotted as a function of temperature to monitor the thermal denaturation (Td). The Td was calculated by fitting the data (GraphPad Prism, GraphPad Software, Inc., La Jolla, CA) with a Boltzmann sigmoidal, where the fraction of denatured Fd = yinitial + ((yfinal − yinitial)/(1 + exp((Td − x)/mb))).
Electrophysiology
Electrophysiology was performed using a two-electrode voltage clamp apparatus (OC-725C oocyte clamp, Holliston, MA). Oocytes were injected with either 0.2 ng of ELIC cRNA or 100–200 ng of membrane-reconstituted ELIC and allowed to incubate overnight at 16 °C in ND96+ buffer (5 mm HEPES, 96 mm NaCl, 2 mm KCl, 1 mm MgCl2, 1 mm CaCl2, and 2 mm pyruvate). Injected oocytes were placed in a RC-1Z oocyte chamber (Harvard Apparatus; Hamden, CT) containing HEPES buffer (150 mm NaCl, 0.5 mm BaCl2, and 10 mm HEPES, pH 7.0). Currents through the plasma membrane in response to cysteamine concentration jumps were measured with the transmembrane voltage clamped at either −40 or −60 mV. The oocyte chamber was perfused with HEPES buffer (10 mm HEPES, 150 mm NaCl, 0.5 mm BaCl2, pH 7.0) at a rate of ∼5 ml/min.
Homology Model
The homology model of ELIC was constructed with Swiss-Model (34) using GLIC (Protein Data Bank code 3EHZ) as a template. The majority (95%) of the φ and ψ angles of the model (the entire ELIC structure) are found in allowable regions of the Ramachandran plot (35). The ELIC3 mutant (V260Y/G318F/I319F) was generated in silico using PyMOL.
RESULTS
Our goal was to test the proposed role of M4 in lipid-dependent uncoupling by examining whether the variable levels of aromatic interactions at the M4-M1/M3 interfaces of ELIC, GLIC, and the nAChR influence the propensity of each pLGIC to adopt an uncoupled conformation. Although specific probes for this electrically silent conformation are not available, careful biophysical studies have identified features that distinguish uncoupled from coupled resting and desensitized conformations. The uncoupled nAChR exhibits more extensive peptide N-1H/N-2H exchange after exposure to 2H2O than do coupled nAChRs (see Fig. 3B), and displays both a lower denaturation temperature and a reduced cooperativity of unfolding (11, 14). Although the specific structural changes in the nAChR associated with uncoupling have yet to be defined, it has been proposed that an increased physical separation leading to weakened interactions between the agonist binding domain and TMD could account for the lack of functional “coupling” between the agonist site and channel gate. Increased separation between the agonist binding domain and TMD leading to greater solvent penetration would also account for the increased levels of peptide N-1H/N-2H exchange (11).
To probe whether ELIC adopts an uncoupled conformation, we first needed to establish the biophysical properties of ELIC in a membrane that stabilizes an activatable conformation. For the latter, we chose to reconstitute ELIC into membranes composed of soybean asolectin, which are effective at stabilizing functional conformations of both GLIC and the nAChR. Also, the aggregation of membrane-reconstituted GLIC is minimized in asolectin membranes (36). As demonstrated below, ELIC reconstituted into asolectin membranes (aso-ELIC) retains the ability to undergo agonist-induced channel gating.
Membrane Reconstitution of ELIC
We initially used a membrane reconstitution protocol that consistently leads to functional membrane-incorporated GLIC (31). Although this protocol was efficient at membrane incorporation (Fig. 2B, inset), it was difficult to detect the thermal denaturation of the reconstituted ELIC. Using an infrared microsampling accessory (37), we established that ELIC maintains its secondary structure and peptide hydrogen exchange characteristics throughout both purification and membrane reconstitution (data not shown). Specifically, ELIC retained an amide I band shape indicative of a mixed α-helix/β-sheet structure, with no signs of unfolding. ELIC also maintained the peptide N-1H/N-2H exchange-resistant core that is characteristic of a folded membrane-incorporated pLGIC.
FIGURE 2.
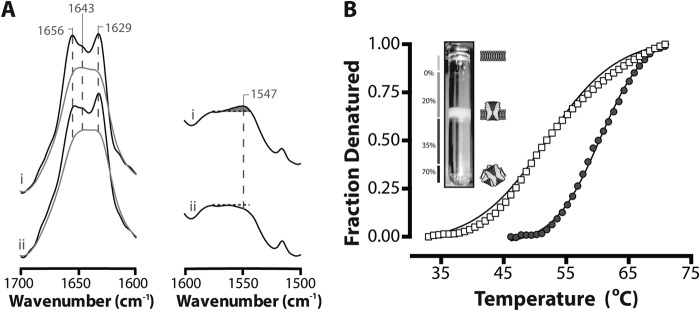
Secondary structure and biophysical properties of ELIC exposed to low ionic strength buffer. A, infrared spectra showing aso-ELIC exchanged into 2H2O Buffer A (i) or 2H2O phosphate buffer (no added salt) (ii). The amide I bands both before (gray) and after deconvolution (black) are presented on the left. Spectra were deconvolved using GRAMS/AI version 7.01 software (Thermo Galactic) with γ = 7 and a Bessel smoothing function set at 70%. The non-deconvolved amide II bands are presented on the right. The relative intensity of the amide II vibration (proportional to unexchanged peptide N-1H) is best assessed relative to the intensity of the adjacent broad peak between 1560 and 1600 cm−1 due to aspartic and glutamic acid. B, thermal denaturation of aso-ELIC exchanged into 2H2O Buffer A (●) and 2H2O phosphate buffer (□). Inset, a sucrose density gradient showing that ELIC incorporates efficiently into proteoliposomes. Protein-free liposomes run at the interface between 0 and 20% sucrose. Low lipid/protein ratio unincorporated protein aggregates run at the interface between 35 and 70% sucrose (11).
We found that a sucrose density gradient step, typically used to purify proteoliposomes from unincorporated and aggregated protein (11), contributed to a lack of reproducibility in the thermal denaturation data. Also, brief exposure of the ELIC proteoliposomes to low ionic strength 2H2O buffer (a technical step in the preparation of samples for infrared spectroscopy) led to marked effects on ELIC structure, including a decrease in the cooperativity of unfolding, a 5–10 °C drop in the thermal denaturation temperature, and a slight decrease in the proportion of exchange-resistant peptide N-1H groups (Fig. 2). These changes are strikingly similar to the biophysical changes that occur in the nAChR upon formation of the uncoupled state (11, 14). Moreover, they highlight the heightened structural sensitivity of ELIC to its surrounding environment compared with GLIC, consistent with the hypothesis that intramembrane aromatic interactions enhance TMD stability. Previous studies have shown that TMD stability impacts the overall stability of a pLGIC (38).
Based on the noted structural sensitivity of ELIC to its surrounding environment, we modified our purification, reconstitution, and sample preparation protocols to minimize potentially detrimental effects on ELIC structure. Because ELIC incorporates efficiently into proteoliposomes, the sucrose density gradient purification step was eliminated. The ELIC proteoliposomes were exchanged into 2H2O Buffer A, instead of low ionic strength 2H2O phosphate buffer (i.e. buffer lacking added salt). As a precautionary measure, the sample exchange time in 2H2O was reduced to 24 h. 24 h in 2H2O Buffer A is sufficient to reach consistent and relatively stable levels of peptide N-1H/N-2H exchange. These minor adjustments led to folded and functional membrane-incorporated ELIC (Figs. 2–5) with clearly defined thermal denaturation profiles.
FIGURE 4.
Thermal denaturation spectra of aso-ELIC (A), PC-ELIC (B), and PC-ELIC3 (C). The left panel depicts a stacked plot of spectra collected for thermal stability characterization of the reconstituted ELIC. Representative traces are shown from 36 to 76 °C, front to back, for aso-ELIC and PC-ELIC, and from 36 to 78 °C for PC-ELIC3. Spectra were collected at 1 °C increments and deconvolved using GRAMS/AI version 7.01 software (Thermo Galactic) with γ = 7 and a Bessel smoothing function set at 70%. The right panel depicts spectra collected at 36 °C (black line) and 76 or 78 °C (gray line) showing full exchange of the N-1H to N-2H upon denaturation. Inset, deconvolved amide I bands (from 1700 to 1600 cm−1) at 36 °C (black) and 76 or 78 °C (gray).
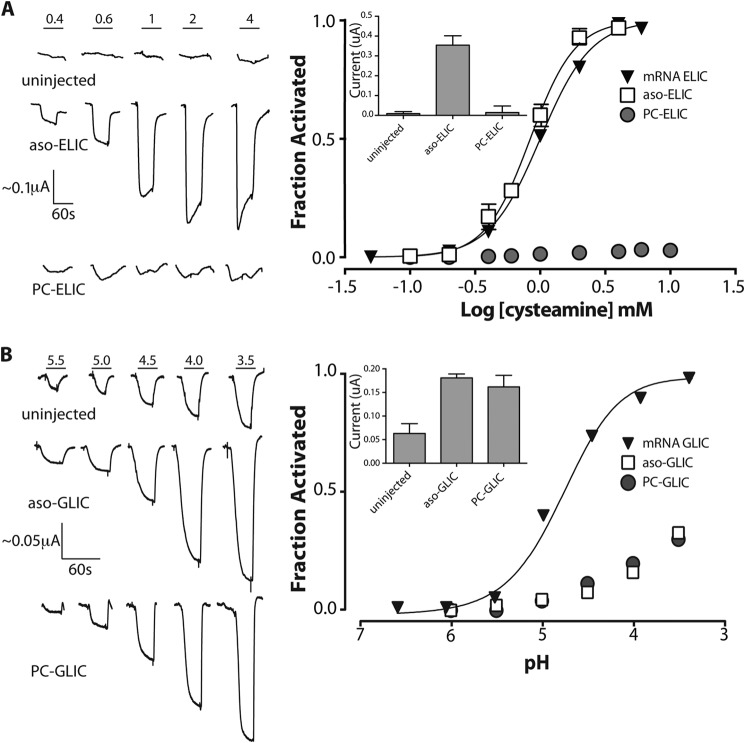
FIGURE 5.
Cysteamine-induced response of membrane-reconstituted ELIC. A, representative whole cell electrophysiology recordings measured at a −60 mV holding potential in response to cysteamine jumps from uninjected oocytes and oocytes injected with aso-ELIC and PC-ELIC (left). Shown are averaged dose-response curves to cysteamine (aso-ELIC, n = 10; PC-ELIC, n = 24) (right). Error bars reflecting the S.E. are shown where they are larger than the symbols. The inset represents the peak current achieved at a cysteamine concentration of 4 mm. The reported values are the averages ± S.E. from at least six different recordings performed on at least six different oocytes. B, representative whole cell electrophysiology recordings measured at −20 mV in response to pH jumps from uninjected oocytes and oocytes injected with aso-GLIC and PC-GLIC (left) (data from Ref. 31). Shown is averaged dose-response to pH (right). Inset, peak current achieved upon exposure of oocytes injected with the indicated reconstituted membranes at pH 3.5. The dose-response curves from oocytes injected with membrane-reconstituted GLIC were normalized assuming a pH50 of 2.90. These proton-induced currents are sensitive to the inhibitor amantadine, whereas the endogenous acid-sensitive channels are not (see Ref. 31).
In light of the sensitivity of ELIC structure to low ionic strength, we also re-evaluated the secondary structure and thermal stability of both membrane-reconstituted GLIC and the nAChR but observed no effects of low ionic strength with either protein (Table 1). Previous studies have shown that the nAChR concentrates endogenous divalent cations throughout purification and membrane reconstitution, even in the presence of divalent cation chelators (39, 40). The ability to concentrate cations may render the nAChR structure relatively insensitive to low ionic strength.
TABLE 1.
Thermal stability of ELIC, GLIC, and the nAChR
| Reconstitution | Tda | n |
|---|---|---|
| °C | °C | |
| Aso-ELIC | 59.7 ± 3.5 (51.6 ± 1.3) | 4 (4) |
| PC-ELIC | 65.1 ± 2.2 | 4 |
| Aso-GLIC | 68.5 (68.8 ± 1.5)b | 1 (5) |
| PC-GLIC | (70.4 ± 0.9)b | 4 |
| Aso-nAChR | 53.0 ± 2.1 (55.4 ± 1.0)b | 2 (2) |
| PC-nAChRb | (52.4 ± 0.1)c | 6 |
a Thermal denaturation temperatures of membrane-incorporated pLGICs after 24-h equilibration in 2H2O Buffer A at 4 °C. Values in parentheses refer to the thermal denaturation temperatures after 72-h equilibration in 2H2O phosphate buffer at 4 °C. See “Results.”
b From Labriola et al. (31).
c From daCosta and Baenziger (11).
Secondary Structure and Thermal Stability of Aso-ELIC
We characterized the secondary structure and peptide N-1H/N-2H exchange characteristics of aso-ELIC using infrared spectroscopy. The amide I band (from 1700 to 1600 cm−1; primarily C=O stretching) in spectra recorded from proteoliposomes gently dried from 1H2O exhibits a peak maximum near 1655 cm−1, due to the vibrations of α-helix and loop/random secondary structures, and a prominent broad shoulder between 1640 and 1625 cm−1, due primarily to the vibrations of β-sheet (Fig. 3A). Immediately after exposure to 2H2O buffer, the amide I band shape becomes more symmetric because the rapid exchange of solvent-exposed α-helical/loop (probably the latter) peptide N-1H to N-2H shifts their frequency from 1655 cm−1 down to near 1645 cm−1. This rapid peptide N-1H/N-2H exchange also leads to an immediate loss of amide II band intensity (centered near 1547 cm−1; primarily N-1H bending) as this vibration shifts down in frequency to near 1450 cm−1 (primarily N-2H bending). Similar rapid spectral changes upon exposure to 2H2O are also observed with GLIC and both the Torpedo nAChR and the human α4β2 nAChR (11, 30, 31). These changes are probably characteristic of the large solvent-exposed extracellular domain.
A more detailed assessment of secondary structure was performed after recording higher fidelity spectra from ELIC proteoliposomes exchanged into 2H2O for 24 h at 4 °C (Fig. 2B). Curve fitting suggests roughly 30% α-helix and 35% β-sheet, estimates that compare well with the crystal structure (roughly 35% α-helix and 30% β-sheet) (Table 2). Note that the strong intensity remaining near 1655 cm−1 due to protiated, and thus solvent-shielded, α-helical/loop structures (probably the former) suggests the existence of a large population of exchange-resistant and probably transmembrane α-helices, as has also been characterized in both GLIC and the nAChR (11, 30, 31, 41, 42).
TABLE 2.
Secondary structure of aso-ELIC, PC-ELIC, and PC-ELIC3
| Band frequency | Band assignment | Aso-ELIC | PC-ELIC | PC-ELIC3 |
|---|---|---|---|---|
| cm−1 | ||||
| 1697 | T | 2 | 1 | 2 |
| 1682 | T | 2 | 1 | 3 |
| 1674 | T + β | 12 | 12 | 11 |
| 1656 | α | 28 | 29 | 36 |
| 1643 | Loop/random | 22 | 22 | 9 |
| 1636 | β | 9 | 9 | 11 |
| 1629 | β | 26 | 25 | 28 |
To assess the thermal stability of aso-ELIC, infrared spectra were recorded as a function of increasing temperature. These spectra reveal changes in the amide I band shape, in particular an increase in intensity near 1620 cm−1, which are characteristic of unfolded proteins (Fig. 4A). Unfolding also leads to the complete exchange of the previously “exchange-resistant” peptide hydrogens. Monitoring amide I changes with increased temperature led to a measured thermal denaturation temperature for aso-ELIC of 59.7 ± 3.5 °C, which is intermediate between that of aso-GLIC (∼69 °C) and aso-nAChR (∼54 °C) (Table 1).
Note that resolution enhancement and curve fitting of the amide I bands show that the spectra of both GLIC and ELIC exhibit two components in the β-sheet region near 1635 and 1630 cm−1, but in ELIC, the latter vibration is relatively more intense. Because denaturation is characterized by an increase in intensity of the overlapping vibration centered near 1620 cm−1, the greater intensity near 1630 cm−1 in spectra of ELIC makes it inherently more difficult to accurately monitor the thermal unfolding process. Thermal denaturation curves yielded consistent thermal denaturation temperatures but greater variability in the cooperativity of unfolding (defined by the Boltzmann slope) (see “Experimental Procedures”). Given this variability, we do not report Boltzmann slopes for the denaturation curves.
Gating of Aso-ELIC
The relatively low lipid/protein ratio ELIC proteoliposomes are ideal for spectroscopy but not useful for direct measurements of channel activity using electrophysiology approaches. We indirectly probed the activity of membrane-reconstituted ELIC by first injecting the aso-ELIC into Xenopus laevis oocytes and then monitoring the appearance of cysteamine-induced currents across the oocyte plasma membranes (43, 44). A similar indirect approach has been used previously to characterize agonist-induced gating of membrane-reconstituted GLIC and the nAChR (31).
Cysteamine did not elicit measurable dose responses in uninjected oocytes (Fig. 5). In contrast, oocytes injected with aso-ELIC gave reproducible currents that respond to cysteamine in a dose-dependent manner, yielding an EC50 of 1.12 ± 0.4 mm (n = 10), a value similar to that observed for oocytes expressing ELIC on the membrane surface after cRNA injection (0.94 ± 0.16 mm, n = 23). The membrane-incorporated aso-ELIC thus retains the ability to gate open in response to cysteamine binding.
Effects of an Uncoupling Lipid Environment on Wild-type ELIC Structure
We next characterized the biophysical properties of ELIC reconstituted into PC membranes (PC-ELIC) to test whether this minimal membrane environment leads to an uncoupled conformation similar to the uncoupled PC-nAChR. The structure and biophysical properties of the homolog GLIC are essentially indistinguishable in asolectin and PC membranes, suggesting that GLIC does not adopt an uncoupled conformation in the minimal PC membranes (Fig. 3B) (31), an interpretation confirmed by the functional measurements discussed below.
Similarly, we detect little if any spectral difference between aso-ELIC and PC-ELIC (Fig. 4). The amide I and amide II bands of aso-ELIC and PC-ELIC are virtually superimposable, with similar levels of peptide N-1H/N-2H exchange and essentially no variability in amide I component band intensity near 1655 and 1645 cm−1. Furthermore, the thermal denaturation temperature of PC-ELIC (65.1 ± 2.2 °C) is actually higher than that of aso-ELIC (59.7 ± 3.5 °C) (Fig. 4 and Table 1), a trend also observed with aso- versus PC-GLIC. PC-ELIC thus does not exhibit any of the biophysical characteristics expected for the uncoupled state. The infrared data suggest that PC-ELIC does not adopt a conformation equivalent to the uncoupled nAChR.
Effects of an Uncoupling Lipid Environment on ELIC Function
The ability of PC-ELIC to undergo cysteamine-induced channel gating was next examined using our electrophysiological assay. Previous studies have shown that the injection of both aso-nAChR and PC-nAChR into X. laevis oocytes leads to the appearance of nAChR binding sites on the plasma membrane, but only aso-nAChR gives rise to acetylcholine-induced currents (31). The lack of channel activity is consistent with PC-nAChR adopting a conformation where agonist binding is “uncoupled” from channel gating, a finding confirmed by other biophysical assays (11). In contrast to the nAChR, the injection of either aso-GLIC or PC-GLIC into oocytes led to proton-activated currents across the plasma membrane, suggesting that GLIC is functional in both asolectin and PC membranes (Fig. 5B). This conclusion is consistent with the infrared data, which suggest a resting-like conformation for GLIC in both asolectin and PC membranes (31).
Given that our infrared data suggest a resting-like conformation for ELIC in PC membranes, we expected that the injection of PC-ELIC into oocytes would lead to the appearance of cysteamine-induced currents across the oocyte plasma membrane, as observed with aso-ELIC. Despite numerous attempts under varying conditions (varying amounts of injected PC-ELIC and varying incubation times after injection), however, we were unable to detect a dose response to cysteamine (Fig. 5A). Given that the injection of PC-nAChR, PC-GLIC, and PC-ELIC3 (an ELIC mutant; see below) into oocytes each led to the appearance of the nAChR, GLIC, and ELIC3, respectively, in the oocyte plasma membrane, the lack of current with PC-ELIC-injected oocytes suggests that agonist binding and channel gating in wild type ELIC are, at best, weakly coupled in PC membranes.
One interpretation of this finding is that PC-ELIC remains locked in a non-functional conformation even when subsequently surrounded by the oocyte plasma membrane lipids. Another is that the inhospitable PC environment remains tightly associated with ELIC, even after many hours within the oocyte membrane environment. Consistent with the latter interpretation, aso-nAChR remains indefinitely in clusters that probably retain the reconstituted lipid environment after fusion with the oocyte plasma membrane (44). Furthermore, the nAChR binding sites that are observed on the oocyte plasma membrane surface do not recover the ability to gate open in response to agonist (31). Regardless of the interpretation of our data, it appears that although ELIC does not adopt an uncoupled conformation identical to that of the nAChR, ELIC function is more sensitive to its surrounding membrane environment than that of GLIC. A simple PC bilayer is not sufficient to promote agonist-induced channel gating in ELIC.
Effects of Enhanced M4-M1/M3 Interactions on the Gating and Lipid Sensitivity of ELIC
Our working model proposes that lipids influence the coupling of agonist binding and channel gating by modulating interactions between the lipid-exposed transmembrane α-helix M4 and the adjacent transmembrane α-helices, M1 and M3. In light of the biophysical data showing that PC-ELIC adopts a resting state-like conformation, but does not undergo cysteamine-induced channel gating, we speculated that the aromatic interactions that are located on the cytoplasmic side of the M4-M1/M3 interface in ELIC may be sufficient to stabilize a conformation during purification and reconstitution where M4 is effectively bound to M1/M3, thus leading to what appears to be a coupled conformation. The absence of aromatic interactions near the extracellular side of the M4-M1/M3 interface of ELIC may, however, weaken interactions between post-M4 and M1/M3 so that in PC membranes lacking activating lipids, there is ineffective coupling between the agonist site and channel gate.
To test this hypothesis, we engineered aromatic interactions into the M4-M1/M3 interface of ELIC to strengthen interactions between post-M4 and M3. To gain insight into the residues on post-M4 that probably interact with adjacent residues on M3, we generated an ELIC homology model based on the GLIC structure (the final five residues in M4 are not observed in the ELIC crystal structure) (Fig. 6C). Note that the homology model reproduces the other “natural” aromatic interactions between M4 and M1/M3 that are defined in the crystal structure. We then generated three mutations in silico (V260Y (on M3) and G318F and I319F (on M4); referred to as ELIC3) to mimic the aromatic interactions that occur between post-M4 and M3 in GLIC. The in silico mutant exhibits relatively tight packing of the engineered aromatic residues and thus close π-π contacts, as expected given the observed interactions of these residues in GLIC (Fig. 1B).
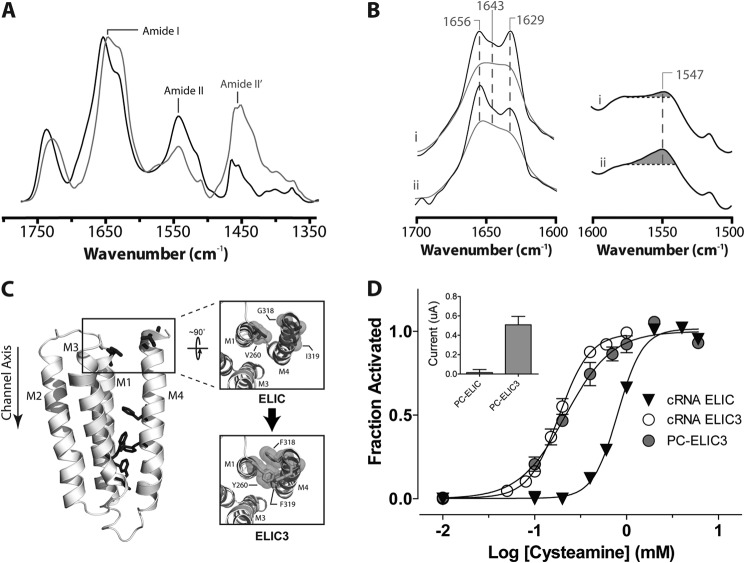
FIGURE 6.
Structural and functional characterization of PC-ELIC3. A, infrared spectra of PC-ELIC3 recorded after gentle drying from 1H2O buffer (black) and immediately after the addition of 2H2O (gray) as in Fig. 3. B, the structure-sensitive amide I (left) and amide II (right) bands in infrared spectra (recorded after 24-h equilibration in 2H2O at 4 °C) from PC-ELIC (i) and PC-ELIC3 (ii). The amide I bands are shown both before (gray) and after resolution enhancement (black). Spectra are the averages of at least three spectra recorded from two different purifications/reconstitutions. C, a homology model of a single subunit of ELIC (based on the GLIC structure) showing aromatic interactions at the M4-M1/M3 interface (dark gray sticks) as well as residues near the C terminus of M4 that are mutated in ELIC3 to aromatic residues as described under “Experimental Procedures.” D, averaged dose-response curves to cysteamine for oocytes injected with the cRNA for wild type ELIC and ELIC3 (n = 9 for both) as well as for oocytes injected with reconstituted PC-ELIC3 (n = 7). Error bars, S.E. Inset, peak current achieved upon exposure of oocytes with either PC-ELIC or PC-ELIC3 (see Fig. 5).
We first examined whether the aromatic additions influence the gating of ELIC. Injection of ELIC3 cRNA into oocytes led to robust cysteamine-activated currents across the oocyte membrane. Significantly, the ELIC3 dose response to cysteamine was left-shifted, with the resulting EC50 (0.18 ± 0.02 mm) indicating that ELIC3 requires only 10–20% of the concentration of cysteamine that is required to elicit half-maximal channel gating in wild type ELIC. Because the aromatic substitutions are distant from the cysteamine binding site, the left shift in EC50 can be attributed to enhanced coupling between the agonist site and channel gate. The left-shifted EC50 shows that enhanced interactions between post-M4 and M1/M3 promote channel function, consistent with the M4 lipid sensor model.
ELIC3 was next expressed in E. coli and reconstituted into PC membranes (PC-ELIC3). Infrared spectra of PC-ELIC3 exhibit amide I bands suggestive of an enhanced α-helical character relative to wild type PC-ELIC, with a slightly larger population of exchange-resistant peptide hydrogens (Fig. 6, A and B). Infrared spectra recorded as a function of temperature to assess the thermal stability of PC-ELIC3 detected the start of a thermal denaturation event. In contrast to wild type PC-ELIC, however, our accessible temperature range (maximum 78 °C) did not lead to complete unfolding of PC-ELIC3 (Fig. 4C). Noting that the M4 α-helix in the ELIC crystal structure is partially unwound with the final five residues unstructured, the enhanced α-helical character and reduced levels of peptide N-1H/N-2H exchange of PC-ELIC3 could reflect a more structured M4 α-helix that interacts tightly with M1/M3, leading to a more stable TMD.
The effects of the engineered aromatic interactions on the gating of ELIC3 in the reconstituted membranes were also assessed. Consistent with our hypothesis and in contrast to wild type PC-ELIC, injection of PC-ELIC3 into oocytes led to the appearance of cysteamine-induced currents on the oocyte membrane surface. Furthermore, the dose response of the cysteamine-induced currents was left-shifted relative to wild type, yielding an EC50 value (0.18 ± 0.04 mm; n = 7) that is similar to that found for cRNA-injected oocytes expressing ELIC3 (Fig. 6D). The engineered aromatic interactions at the M4-M1/M3 interface thus render ELIC3 active in the minimal PC membranes that stabilize both an uncoupled nAChR and an inactive form of wild type ELIC. Intramembrane aromatic interactions both reduce the sensitivity of ELIC to its membrane environment and enhance coupling between the agonist site and channel gate.
DISCUSSION
Cholesterol and anionic lipids are required in a reconstituted membrane to obtain optimal nAChR function (1–9, 45, 46), although membrane physical properties are also important (14). In the absence of these activating lipids, the nAChR adopts an uncoupled conformation that binds agonist but does not usually undergo agonist-induced conformational change (11). Our working model highlights a role for the M4 transmembrane α-helix in uncoupling. M4 is located on the periphery of the TMD, where it makes extensive contact with the lipid bilayer (Fig. 1). The C terminus of M4 (post-M4) extends beyond the lipid bilayer to interact directly with the Cys-loop, one of the key structures implicated in the allosteric pathway leading from the agonist site to the channel gate (47, 48).
The M4 lipid sensor model proposes that lipids influence coupling by altering the association of M4 with its adjacent transmembrane α-helices, M1 and M3, thus ultimately influencing post-M4/Cys-loop interactions. The model suggests that strong M4-M1/M3 interactions promote channel gating, whereas ineffective M4-M1/M3 interactions lead to formation of the uncoupled state. The model also predicts that the intrinsic strength of M4-M1/M3 interactions will influence the sensitivity of a pLGIC to changes in its surrounding lipid environment, which influence M4-M1/M3 interactions. Intrinsically strong M4-M1/M3 interactions should render a pLGIC less sensitive to its membrane environment.
The nAChR, GLIC, and ELIC exhibit strikingly different levels of aromatic interactions at the M4-M1/M3 interface. Given that aromatic interactions are key energetic drivers for M4 binding to M1/M3 (32), the M4 lipid sensor model predicts that the three pLGICs should exhibit distinct propensities to adopt an uncoupled conformation. Consistent with this proposal, we find that all three retain a native-like secondary structure that undergoes agonist-induced channel gating in membranes composed of complex soybean asolectin lipids. When reconstituted into minimal PC membranes lacking activating lipids, however, distinct lipid sensitivities emerge: PC-nAChR adopts an uncoupled conformation; PC-ELIC does not gate open in response to agonist, although it does not adopt a conformation equivalent to the uncoupled PC-nAChR; and PC-GLIC retains an activatable resting state-like conformation that fluxes cations, despite the absence of activating lipids.
Significantly, engineering aromatic residues into the M4-M1/M3 interface to promote strong M4-M1/M3 interactions restores the ability of ELIC to gate open in these minimal PC membranes (Fig. 6). Furthermore, the EC50 for agonist-induced activation of the mutant, referred to as ELIC3, is left-shifted relative to the EC50 of wild type ELIC, suggesting that enhanced M4-M1/M3 interactions promote intrinsic coupling between the agonist site and channel gate.
These observations suggest that modulatable associations of M4 with M1/M3 are central to lipid sensing and that aromatic residues govern the intrinsic strength of M4-M1/M3 associations to alter the propensity of a pLGIC to adopt an uncoupled state. Specifically, the extensive aromatic network at the M4-M1/M3 interface in GLIC leads to relatively strong M4-M1/M3 interactions along the entire length of M4 (Fig. 1B), which probably contribute to the stabilization of a coupled conformation even in membranes lacking activating lipids. ELIC exhibits fewer aromatic interactions at the M4-M1/M3 interface than GLIC, and these are located exclusively toward the cytoplasmic surface of the bilayer. The available aromatic interactions appear to be sufficient to stabilize a conformation where M4 interacts with M1/M3 in the cytoplasmic half of the bilayer so that PC-ELIC does not exhibit features indicative of the uncoupled state. The absence of aromatic interactions near the extracellular surface, however, appears to lead to post-M4 interactions that are too weak to promote effective channel gating. Finally, the nAChR exhibits only one potential aromatic interaction between M4 and M1, which is close to the cytoplasmic surface of the bilayer. As a result of the relative paucity of aromatic residues at the M4-M1/M3 interface, effective M4-M1/M3 interactions may require the presence of activating lipids. Weakened interactions between M4 and M1/M3 along the entire length of M4 with PC-nAChR may lead to formation of the uncoupled state.
The proposed role for intramembrane aromatic interactions in governing pLGIC sensitivity to its surrounding membrane environment is supported further by reconstitution and crystallization studies performed here and elsewhere. Previous work has shown that the nAChR must be solubilized in the presence of lipids to retain an ability to undergo agonist-induced conformational transitions (49). In contrast, both GLIC and ELIC can be solubilized and purified in the absence of stabilizing lipids and yet retain the ability to gate open when reconstituted into asolectin membranes.
Both ELIC and GLIC are more thermally stable than the nAChR, with the denaturation temperature of each pLGIC correlating with the relative abundance of aromatic interactions at the M4-M1/M3 interface (Table 2). Furthermore, the ELIC3 mutant with enhanced M4-M1/M3 interactions exhibits a slightly greater α-helical character (Fig. 6, A and B, and Table 2), less extensive peptide N-1H/N-2H exchange when exposed to 2H2O buffer (Fig. 6B), and enhanced thermal stability relative to wild type ELIC (Fig. 4), all consistent with a structurally more stable TMD. Although additional features are likely to be important, the greater abundance of aromatic interactions at the M4-M1/M3 interface in GLIC, ELIC, and the ELIC3 mutant (Figs. 1B and 6C) probably contributes to the increased stabilities of the respective TMDs relative to that of the nAChR, thus reducing the sensitivities of GLIC, ELIC, and ELIC3 to the perturbing effects of detergent.
The enhanced functional properties of the ELIC3 mutant highlight the importance of post-M4 in both channel gating and lipid sensing. The importance of post-M4 is further suggested by the crystal structures of GLIC and ELIC. As noted, M4 interacts tightly with M1/M3 along the entire length of M4 in the GLIC crystal structures. A widening of the GLIC channel pore is observed upon the binding of agonist (in this case protons), suggesting that crystallized GLIC undergoes agonist-induced conformational transitions (50–52). In contrast, although M4 associates with M1/M3 in the ELIC crystal structure, the C-terminal half of M4 is tilted away from the remainder of the TMD (Fig. 1B) (53). Significantly, agonist binding does not lead to a widening of the channel pore in ELIC, even with mutants that show no propensity to undergo desensitization (54, 55). Although wild type ELIC appears less sensitive to detergent than the nAChR, the absence of aromatic residues in the post-M4 region may weaken post-M4-M1/M3 interactions so that detergent solubilization leads to a non-functional conformation (15), as does reconstitution into the minimal PC membranes.
In this context, it is important to note that the nAChR structure presented in Fig. 1 was solved from electron microscopy images recorded from the nAChR in its native membrane environment (i.e. in the absence of detergent) (21). Crystallization attempts using detergent-solubilized nAChR have not been successful. Furthermore, detergent solubilization of the nAChR in the absence of activating lipids leads to the formation of an uncoupled conformation that does not undergo agonist-induced conformational transitions (49).
Finally, it is intriguing that whereas ELIC exhibits a greater propensity than GLIC to adopt a non-activatable conformation in PC membranes, the EC50 for cysteamine-induced channel gating is the same with oocytes that are injected with either aso-ELIC or ELIC cRNA. The same EC50 values were also observed with oocytes injected with either PC-ELIC3 or ELIC3 cRNA. In contrast, the EC50 for proton-induced gating of GLIC is different with oocytes injected with aso-GLIC or PC-GLIC versus GLIC cRNA, as well as with GLIC reconstituted into asolectin proteoliposomes (56). Proton-induced gating of GLIC is thus sensitive to its membrane environment. One speculative interpretation is that the difference in EC50 between GLIC in asolectin versus Xenopus membranes reflects the involvement of an intramembrane protonation site in gating (57); such a site could be sensitive to surrounding lipids. A role for an intramembrane proton binding site in channel gating would also account for the varying EC50 values observed when GLIC is expressed in HEK cells versus frog oocytes (57, 58).
In summary, we have refined a protocol for the membrane reconstitution of ELIC and have used this protocol to compare the propensities of both GLIC and ELIC to adopt an uncoupled conformation. In asolectin membranes, both ELIC and GLIC retain the ability to undergo agonist-induced channel gating. In minimal PC membranes, however, ELIC does not gate open, whereas GLIC retains the ability to undergo channel gating. Enhanced aromatic interactions between M4 and M1/M3 in ELIC restore the ability to gate open in minimal PC membranes. Our data show that the M4 transmembrane α-helix plays a direct role in lipid sensing. We also show that aromatic interactions at the M4-M1/M3 interface govern the functional sensitivity of a pLGIC to its surrounding lipid environment.
This work was supported by Canadian Institutes of Health Research (CIHR) Grant 111243 and Natural Sciences and Engineering Research Council of Canada Grant 113312, as well as CIHR Training Program in Neurodegenerative Lipidomics Grant TGF-96121.
- nAChR
- nicotinic acetylcholine receptor
- TMD
- transmembrane domain
- ELIC
- Erwinia ligand-gated ion channel
- GLIC
- Gloebacter ligand-gated ion channel
- aso-ELIC
- ELIC in asolectin membranes
- aso-GLIC
- GLIC in asolectin membranes
- aso-nAChR
- nAChR in asolectin membranes
- ELIC3
- ELIC with engineered aromatic residues (V260Y/G318F/I319F) as described under “Experimental Procedures”
- pLGIC
- pentameric ligand-gated ion channel
- PC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- PC-ELIC
- ELIC in PC membranes
- PC-ELIC3
- ELIC3 in PC membranes
- PC-GLIC
- GLIC in PC membranes
- PC-nAChR
- nAChR in PC membranes
- Td
- thermal denaturation temperature.
REFERENCES
- 1. Criado M., Eibl H., Barrantes F. J. (1982) Effects of lipids on acetylcholine receptor. Essential need of cholesterol for maintenance of agonist-induced state transitions in lipid vesicles. Biochemistry 21, 3622–3629 [DOI] [PubMed] [Google Scholar]
- 2. Fong T. M., McNamee M. G. (1986) Correlation between acetylcholine receptor function and structural properties of membranes. Biochemistry 25, 830–840 [DOI] [PubMed] [Google Scholar]
- 3. Sunshine C., McNamee M. G. (1992) Lipid modulation of nicotinic acetylcholine receptor function: the role of neutral and negatively charged lipids. Biochim. Biophys. Acta 1108, 240–246 [DOI] [PubMed] [Google Scholar]
- 4. Rankin S. E., Addona G. H., Kloczewiak M. A., Bugge B., Miller K. W. (1997) The cholesterol dependence of activation and fast desensitization of the nicotinic acetylcholine receptor. Biophys. J. 73, 2446–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barrantes F. J. (2002) Lipid matters: nicotinic acetylcholine receptor-lipid interactions. Mol. Membr. Biol. 19, 277–284 [DOI] [PubMed] [Google Scholar]
- 6. Baenziger J. E., Corringer P. J. (2011) 3D structure and allosteric modulation of the transmembrane domain of pentameric ligand-gated ion channels. Neuropharmacology 60, 116–125 [DOI] [PubMed] [Google Scholar]
- 7. Baenziger J. E., Morris M. L., Darsaut T. E., Ryan S. E. (2000) Effect of membrane lipid composition on the conformational equilibria of the nicotinic acetylcholine receptor. J. Biol. Chem. 275, 777–784 [DOI] [PubMed] [Google Scholar]
- 8. daCosta C. J. B., Ogrel A. A., McCardy E. A., Blanton M. P., Baenziger J. E. (2002) Lipid-protein interactions at the nicotinic acetylcholine receptor. A functional coupling between nicotinic receptors and phosphatidic acid-containing lipid bilayers. J. Biol. Chem. 277, 201–208 [DOI] [PubMed] [Google Scholar]
- 9. Hamouda A. K., Sanghvi M., Sauls D., Machu T. K., Blanton M. P. (2006) Assessing the lipid requirements of the Torpedo californica nicotinic acetylcholine receptor. Biochemistry 45, 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. daCosta C. J. B., Medaglia S. A., Lavigne N., Wang S., Carswell C. L., Baenziger J. E. (2009) Anionic lipids allosterically modulate multiple nicotinic acetylcholine receptor conformational equilibria. J. Biol. Chem. 284, 33841–33849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. daCosta C. J. B., Baenziger J. E. (2009) A lipid-dependent uncoupled conformation of the acetylcholine receptor. J. Biol. Chem. 284, 17819–17825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li P., Steinbach J. H. (2010) The neuronal nicotinic α4β2 receptor has a high maximal probability of being open. Br. J. Pharmacol. 160, 1906–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vallejo Y. F., Buisson B., Bertrand D., Green W. N. (2005) Chronic nicotine exposure upregulates nicotinic receptors by a novel mechanism. J. Neurosci. 25, 5563–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. daCosta C. J. B., Dey L., Therien J. P., Baenziger J. E. (2013) A distinct mechanism for activating uncoupled nicotinic acetylcholine receptors. Nat. Chem. Biol. 9, 701–707 [DOI] [PubMed] [Google Scholar]
- 15. daCosta C. J. B., Baenziger J. E. (2013) Gating of pentameric ligand-gated ion channels: structural insights and ambiguities. Structure 21, 1271–1283 [DOI] [PubMed] [Google Scholar]
- 16. Jones O. T., McNamee M. G. (1988) Annular and nonannular binding sites for cholesterol associated with the nicotinic acetylcholine receptor. Biochemistry 27, 2364–2374 [DOI] [PubMed] [Google Scholar]
- 17. Brannigan G., Hénin J., Law R., Eckenhoff R., Klein M. L. (2008) Embedded cholesterol in the nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. U.S.A. 105, 14418–14423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marsh D., Watts A., Barrantes F. J. (1981) Phospholipid chain immobilization and steroid rotational immobilization in acetylcholine receptor-rich membranes from Torpedo marmorata. Biochim. Biophys. Acta 645, 97–101 [DOI] [PubMed] [Google Scholar]
- 19. Ellena J. F., Blazing M. A., McNamee M. G. (1983) Lipid-protein interactions in reconstituted membranes containing acetylcholine receptor. Biochemistry 22, 5523–5535 [DOI] [PubMed] [Google Scholar]
- 20. Blanton M. P., Cohen J. B. (1992) Mapping the lipid-exposed regions in the Torpedo californica nicotinic acetylcholine receptor [published erratum appears in Biochemistry 1992 Jun 30;31(25):5951]. Biochemistry 31, 3738–3750 [DOI] [PubMed] [Google Scholar]
- 21. Unwin N. (2005) Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J. Mol. Biol. 346, 967–989 [DOI] [PubMed] [Google Scholar]
- 22. Xu Y., Barrantes F. J., Luo X., Chen K., Shen J., Jiang H. (2005) Conformational dynamics of the nicotinic acetylcholine receptor channel: a 35-ns molecular dynamics simulation study. J. Am. Chem. Soc. 127, 1291–1299 [DOI] [PubMed] [Google Scholar]
- 23. Williamson P. T., Zandomeneghi G., Barrantes F. J., Watts A., Meier B. H. (2005) Structural and dynamic studies of the g-M4 trans-membrane domain of the nicotinic acetylcholine receptor. Mol. Membr. Biol. 22, 485–496 [DOI] [PubMed] [Google Scholar]
- 24. Antollini S. S., Xu Y., Jiang H., Barrantes F. J. (2005) Fluorescence and molecular dynamics studies of the acetylcholine receptor gM4 transmembrane peptide in reconstituted systems. Mol. Membr. Biol. 22, 471–483 [DOI] [PubMed] [Google Scholar]
- 25. Li L., Schuchard M., Palma A., Pradier L., McNamee M. G. (1990) Functional role of the cysteine 451 thiol group in the M4 helix of the γ subunit of Torpedo californica acetylcholine receptor. Biochemistry 29, 5428–5436 [DOI] [PubMed] [Google Scholar]
- 26. Lee Y. H., Li L., Lasalde J., Rojas L., McNamee M., Ortiz-Miranda S. I., Pappone P. (1994) Mutations in the M4 domain of Torpedo californica acetylcholine receptor dramatically alter ion channel function. Biophys J. 66, 646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lasalde J. A., Tamamizu S., Butler D. H., Vibat C. R., Hung B., McNamee M. G. (1996) Tryptophan substitutions at the lipid-exposed transmembrane segment M4 of Torpedo californica acetylcholine receptor govern channel gating. Biochemistry 35, 14139–14148 [DOI] [PubMed] [Google Scholar]
- 28. Bouzat C., Roccamo A. M., Garbus I., Barrantes F. J. (1998) Mutations at lipid-exposed residues of the acetylcholine receptor affect its gating kinetics. Mol. Pharmacol. 54, 146–153 [DOI] [PubMed] [Google Scholar]
- 29. Bouzat C., Barrantes F., Sine S. (2000) Nicotinic receptor fourth transmembrane domain: hydrogen bonding by conserved threonine contributes to channel gating kinetics. J. Gen. Physiol. 115, 663–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. daCosta C. J. B., Michel Sturgeon R., Hamouda A. K., Blanton M. P., Baenziger J. E. (2011) Structural characterization and agonist binding to human a4b2 nicotinic receptors. Biochem. Biophys. Res. Commun. 407, 456–460 [DOI] [PubMed] [Google Scholar]
- 31. Labriola J. M., Pandhare A., Jansen M., Blanton M. P., Corringer P. J., Baenziger J. E. (2013) Structural sensitivity of a prokaryotic pentameric ligand-gated ion channel to its membrane environment. J. Biol. Chem. 288, 11294–11303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haeger S., Kuzmin D., Detro-Dassen S., Lang N., Kilb M., Tsetlin V., Betz H., Laube B., Schmalzing G. (2010) An intramembrane aromatic network determines pentameric assembly of Cys-loop receptors. Nat. Struct. Mol. Biol. 17, 90–98 [DOI] [PubMed] [Google Scholar]
- 33. Carswell C. L., Rigden M. D., Baenziger J. E. (2008) Expression, purification, and structural characterization of CfrA, a putative iron transporter from Campylobacter jejuni. J. Bacteriol. 190, 5650–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Ca geometry: f,y and Cb deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 36. Velisetty P., Chakrapani S. (2012) Desensitization Mechanism in Prokaryotic Ligand-gated Ion Channel. J. Biol. Chem. 287, 18467–18477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. daCosta C. J. B., Baenziger J. E. (2003) A rapid method for assessing lipid:protein and detergent:protein ratios in membrane-protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 59, 77–83 [DOI] [PubMed] [Google Scholar]
- 38. Tol M. B., Deluz C., Hassaine G., Graff A., Stahlberg H., Vogel H. (2013) Thermal unfolding of a mammalian pentameric ligand-gated ion channel proceeds at consecutive, distinct steps. J. Biol. Chem. 288, 5756–5769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sturgeon R. M., Baenziger J. E. (2010) Cations mediate interactions between the nicotinic acetylcholine receptor and anionic lipids. Biophys. J. 98, 989–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. daCosta C. J. B., Wagg I. D., McKay M. E., Baenziger J. E. (2004) Phosphatidic acid and phosphatidylserine have distinct structural and functional interactions with the nicotinic acetylcholine receptor. J. Biol. Chem. 279, 14967–14974 [DOI] [PubMed] [Google Scholar]
- 41. Méthot N., Ritchie B. D., Blanton M. P., Baenziger J. E. (2001) Structure of the pore-forming transmembrane domain of a ligand-gated ion channel. J. Biol. Chem. 276, 23726–23732 [DOI] [PubMed] [Google Scholar]
- 42. Baenziger J. E., Méthot N. (1995) Fourier transform infrared and hydrogen/deuterium exchange reveal an exchange-resistant core of α-helical peptide hydrogens in the nicotinic acetylcholine receptor. J. Biol. Chem. 270, 29129–29137 [DOI] [PubMed] [Google Scholar]
- 43. Pandhare A., Hamouda A. K., Staggs B., Aggarwal S., Duddempudi P. K., Lever J. R., Lapinsky D. J., Jansen M., Cohen J. B., Blanton M. P. (2012) Bupropion binds to two sites in the Torpedo nicotinic acetylcholine receptor transmembrane domain: a photoaffinity labeling study with the bupropion analogue [125I]-SADU-3-72. Biochemistry 51, 2425–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morales A., Aleu J., Ivorra I., Ferragut J. A., Gonzalez-Ros J. M., Miledi R. (1995) Incorporation of reconstituted acetylcholine receptors from Torpedo into the Xenopus oocyte membrane. Proc. Natl. Acad. Sci. U.S.A. 92, 8468–8472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Criado M., Eibl H., Barrantes F. J. (1984) Functional properties of the acetylcholine receptor incorporated in model lipid membranes: differential effects of chain length and head group of phospholipids on receptor affinity states and receptor-mediated ion translocation. J. Biol. Chem. 259, 9188–9198 [PubMed] [Google Scholar]
- 46. Ryan S. E., Demers C. N., Chew J. P., Baenziger J. E. (1996) Structural effects of neutral and anionic lipids on the nicotinic acetylcholine receptor: an infrared difference spectroscopy study. J. Biol. Chem. 271, 24590–24597 [DOI] [PubMed] [Google Scholar]
- 47. Jha A., Cadugan D. J., Purohit P., Auerbach A. (2007) Acetylcholine receptor gating at extracellular transmembrane domain interface: the Cys-loop and M2-M3 linker. J. Gen. Physiol. 130, 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee W. Y., Free C. R., Sine S. M. (2009) Binding to gating transduction in nicotinic receptors: Cys-loop energetically couples to pre-M1 and M2-M3 regions. J. Neurosci. 29, 3189–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heidmann T., Sobel A., Popot J. L., Changeux J. P. (1980) Reconstitution of a functional acetylcholine receptor: conservation of the conformational and allosteric transitions and recovery of the permeability response; role of lipids. Eur. J. Biochem. 110, 35–55 [DOI] [PubMed] [Google Scholar]
- 50. Sauguet L., Shahsavar A., Poitevin F., Huon C., Menny A., Nemecz À., Haouz A., Changeux J. P., Corringer P. J., Delarue M. (2014) Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc. Natl. Acad. Sci. U.S.A. 111, 966–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bocquet N., Nury H., Baaden M., Le Poupon C., Changeux J. P., Delarue M., Corringer P. J. (2009) X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111–114 [DOI] [PubMed] [Google Scholar]
- 52. Hilf R. J., Dutzler R. (2009) Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457, 115–118 [DOI] [PubMed] [Google Scholar]
- 53. Hilf R. J., Dutzler R. (2008) X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379 [DOI] [PubMed] [Google Scholar]
- 54. Gonzalez-Gutierrez G., Grosman C. (2010) Bridging the gap between structural models of nicotinic receptor superfamily ion channels and their corresponding functional states. J. Mol. Biol. 403, 693–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gonzalez-Gutierrez G., Lukk T., Agarwal V., Papke D., Nair S. K., Grosman C. (2012) Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals. Proc. Natl. Acad. Sci. U.S.A. 109, 6331–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Velisetty P., Chalamalasetti S. V., Chakrapani S. (2012) Conformational transitions underlying pore opening and desensitization in membrane-embedded GLIC. J. Biol. Chem. 287, 36864–36872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang H. L., Cheng X., Sine S. M. (2012) Intramembrane proton binding site linked to activation of bacterial pentameric ion channel. J. Biol. Chem. 287, 6482–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bocquet N., Prado de Carvalho L., Cartaud J., Neyton J., Le Poupon C., Taly A., Grutter T., Changeux J. P., Corringer P. J. (2007) A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature 445, 116–119 [DOI] [PubMed] [Google Scholar]