Abstract
Hypertension is a common disorder and the leading risk factor for cardiovascular disease and premature deaths worldwide. Genome-wide association studies (GWASs) in the European population have identified multiple chromosomal regions associated with blood pressure, and the identified loci altogether explain only a small fraction of the variance for blood pressure. The differences in environmental exposures and genetic background between Chinese and European populations might suggest potential different pathways of blood pressure regulation. To identify novel genetic variants affecting blood pressure variation, we conducted a meta-analysis of GWASs of blood pressure and hypertension in 11 816 subjects followed by replication studies including 69 146 additional individuals. We identified genome-wide significant (P < 5.0 × 10−8) associations with blood pressure, which included variants at three new loci (CACNA1D, CYP21A2, and MED13L) and a newly discovered variant near SLC4A7. We also replicated 14 previously reported loci, 8 (CASZ1, MOV10, FGF5, CYP17A1, SOX6, ATP2B1, ALDH2, and JAG1) at genome-wide significance, and 6 (FIGN, ULK4, GUCY1A3, HFE, TBX3-TBX5, and TBX3) at a suggestive level of P = 1.81 × 10−3 to 5.16 × 10−8. These findings provide new mechanistic insights into the regulation of blood pressure and potential targets for treatments.
INTRODUCTION
Hypertension is a common disorder and the leading risk factor for cardiovascular disease and premature deaths worldwide (1). It is a highly heritable trait, and around 40–60% of the interindividual variation in hypertension is attributable to genetic factors (2). Genome-wide association studies (GWASs) have identified multiple genetic loci associated with blood pressure (3–7). However, the vast majority of these loci were identified in European ancestry populations, and few such genetic studies have assessed Asian populations, especially Chinese populations. Moreover, the identified loci altogether explain only a small fraction of the risk for hypertension (3). Lifestyle factors, including sodium intake, excess alcohol drinking, elevated body mass index (BMI), and lack of exercise, are known to increase blood pressure (8). The differences in environmental exposures and genetic background between Chinese and European populations might suggest potential different pathways of blood pressure regulation. Therefore, large scale studies in Chinese are needed not only to evaluate whether the previous reported loci could be generalized to the Chinese population, but also to identify new susceptibility loci for blood pressure.
Here, we conducted a large scale GWAS of blood pressure and hypertension that included a meta-analysis of GWAS from 11 816 samples at the discovery stage and additional 69 146 samples in three independent replication studies, involving a total of 80 962 subjects from Chinese Han ancestry. We subsequently investigated whether the blood pressure variants would contribute to the traditional cardiovascular risk factors including lipid levels, plasma glucose, and BMI.
RESULTS
The discovery and replication study
The discovery stage included six independent GWASs in 11 816 individuals from Chinese populations (Table 1). The discovery meta-analysis separately evaluated associations for systolic and diastolic blood pressure (SBP and DBP) and hypertension, with 2 485 448 genotyped or imputed autosomal single-nucleotide polymorphisms (SNPs). All genotyped and imputed autosomal SNPs passed quality control filters in each of the six datasets prior to conducting the meta-analysis (Supplementary Material, Table S1). Quantile–quantile plots for SBP, DBP, and hypertension are presented in Supplementary Material, Figure S1. The genomic control inflation factor (λ) for the meta-analysis was modest (λ = 1.04), indicating that population stratification effects were negligible in our study samples. As shown in the Manhattan plots of the −log10 P-values for SBP, DBP, and hypertension in Supplementary Material, Figure S2, the meta-analysis identified two well-established loci (FGF5 and CYP17A1) at genome-wide significance (defined as P < 5.0 × 10−8).
Table 1.
Study sample characteristics
| Study | n | Male/female (% male) | Age, years (S.D.) | SBP, mmHg (S.D.) | DBP, mmHg (S.D.) | BMI, kg/m2 (S.D.) | HTN (%)a | Antihypertensive therapy (%) |
|---|---|---|---|---|---|---|---|---|
| Discovery meta-analysis (n = 11 816) | ||||||||
| InterASIA-1 | 3998 | 1870/2128 (46.8) | 52.5 (7.7) | 140.7 (24.7) | 88.1 (14.8) | 25.2 (3.8) | 61.3 | 26.7 |
| Beijing | 466 | 363/103 (77.9) | 51.1 (9.5) | 120.6 (12.8) | 77.9 (8.4) | 24.6 (3.4) | 4.5 | 0 |
| GenSalt | 1881 | 993/888 (52.8) | 38.7 (9.5) | 116.9 (14.2) | 73.7 (10.3) | 23.4 (3.2) | 9.8 | 0.4 |
| NHAPC | 2893 | 1251/1642 (43.2) | 58.6 (6.0) | 140.2 (22.7) | 80.2 (10.9) | 24.5 (3.6) | 55.0 | 28.3 |
| Nanjing | 1146 | 820/326 (71.6) | 59.0 (9.7) | 129.6 (21.1) | 81.0 (10.7) | 23.1 (3.2) | 39.5 | 18.7 |
| DFTJ | 1432 | 1118/314 (78.1) | 63.1 (8.1) | 130.0 (18.9) | 78.3 (11.0) | 24.7 (3.3) | 39.8 | 30.8 |
| Replication 1 | ||||||||
| InterASIA-2 | 12 108 | 5872/6236 (48.5) | 52.6 (8.3) | 136.4 (24.8) | 84.0 (13.5) | 25.0 (3.7) | 48.7 | 20.5 |
| Replication 2 | ||||||||
| CCHS-1 | 22 896 | 12794/10102 (55.9) | 52.7 (9.5) | 130.3 (21.6) | 80.9 (11.7) | 23.8 (3.5) | 35.5 | 14.0 |
| Replication 3 | ||||||||
| CCHS-2 | 34 142 | 13199/20943 (38.7) | 52.2 (12.1) | 133.2 (20.1) | 81.4 (11.2) | 24.3 (3.7) | 46.2 | 15.3 |
InterASIA, International Collaborative Study of Cardiovascular Disease in Asia (InterASIA in China); Beijing, Beijing Cardiovascular Risk Factors study; GenSalt, the Genetic Epidemiology Network of Salt-Sensitivity study; NHAPC, The Nutrition and Health of Aging Population in China; Nanjing, Noncommunicable Diseases Screening study in Nanjing; DFTJ, The Dongfeng-Tongji cohort study; CCHS-1, China Cardiovascular Health Study project (Jiangsu); CCHS-2, China Cardiovascular Health Study project (Shandong and Henan); n, sample size; S.D., standard deviation; SBP, systolic blood pressure; DBP, diastolic blood pressure; HTN, hypertension; BMI, body mass index.
Hypertension is defined as SBP ≥140 mmHg and/or DBP ≥90 mmHg or taking antihypertensive medication.
We then selected 15 new SNPs that were associated with blood pressure at P < 1.0 × 10−5 and 24 SNPs located in previously reported loci at P < 5.0 × 10−3 in the discovery analysis (see Materials and Methods and Supplementary Material, Fig. S3) and genotyped them in the first replication study comprising 12 108 Chinese individuals (replication 1). Twenty-two SNPs showed nominal significant association (P < 0.05) with SBP, DBP, and/or hypertension in the replication 1 and were further genotyped in an independent sample of 22 896 individuals (replication 2). Of these 22 SNPs, 14 SNPs represented the replication of previously reported associations with blood pressure, while the remaining 8 SNPs were located in novel regions for blood pressure or independent of the reported association. The detailed results for each study were provided in Supplementary Material, Table S2. We evaluated the evidence of association by combined results of the discovery, replication 1, and replication 2 studies.
Associations at loci previously identified by GWAS
Our Chinese samples replicated 14 loci previously identified in GWAS in European descent or East Asians at various strengths of associations [SOX6 was initially identified using gene-centric array (9,10), and confirmed by GWAS in African-Ancestry individuals (7)] (Table 2). Those included eight loci (CASZ1, MOV10, FGF5, CYP17A1, SOX6, ATP2B1, ALDH2, and JAG1) at the level of genome-wide significance (P < 5.0 × 10−8), four loci (ULK4, GUCY1A3, HFE, and TBX3) with suggestive significance (P < 5.0 × 10−5), and two loci (FIGN and TBX3-TBX5) at a less significant level of P < 5.0 × 10−3.
Table 2.
Blood pressure and hypertension susceptibility loci identified by GWAS in the Chinese
| Nearby gene | SNP | CHR | Position | Risk/no. of risk allele | n | RAF | SBP | DBP | HTN | Previously reported trait(s) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (S.E.) | P-value | β (S.E.) | P-value | β (S.E.) | P-value | ||||||||
| Newly identified loci | |||||||||||||
| CACNA1D | rs9810888 | 3 | 53 610 635 | G/T | 77 555 | 0.39 | 0.53 (0.10) | 5.46 × 10−8 | 0.39 (0.06) | 4.00 × 10−12 | 0.056 (0.012) | 3.60 × 10−6 | – |
| CYP21A2 | rs2021783 | 6 | 32 152 829 | C/T | 78 911 | 0.79 | 0.68 (0.12) | 3.19 × 10−9 | 0.49 (0.07) | 2.18 × 10−12 | 0.094 (0.014) | 3.53 × 10−11 | – |
| MED13L | rs11067763 | 12 | 114 682 724 | A/G | 79 651 | 0.62 | 0.81 (0.10) | 5.68 × 10−16 | 0.51 (0.06) | 2.00 × 10−18 | 0.061 (0.012) | 1.37 × 10−7 | – |
| Independent association signal in Chinese at the previous reported locus | |||||||||||||
| SLC4A7 | rs820430 | 3 | 27 523 904 | A/G | 79 318 | 0.32 | 0.76 (0.11) | 1.36 × 10−12 | 0.27 (0.06) | 7.57 × 10−6 | 0.059 (0.012) | 1.12 × 10−6 | – |
| SNPs with suggestive significance | |||||||||||||
| HLA-B | rs9266359 | 6 | 31 440 718 | C/T | 78 365 | 0.60 | 0.44 (0.10) | 7.07 × 10−6 | 0.29 (0.06) | 1.76 × 10−7 | 0.050 (0.012) | 1.33 × 10−5 | – |
| Previous reported loci in GWAS | |||||||||||||
| CASZ1 | rs880315 | 1 | 10 719 453 | C/T | 42 355 | 0.63 | 0.97 (0.16) | 5.52 × 10−10 | 0.46 (0.09) | 7.94 × 10−8 | 0.093 (0.016) | 1.99 × 10−9 | DBP |
| MOV10 | rs10745332 | 1 | 112 990 576 | A/G | 46 269 | 0.82 | 0.96 (0.18) | 2.52 × 10−9 | 0.53 (0.10) | 7.70 × 10−8 | 0.114 (0.020) | 2.70 × 10−9 | SBP, DBP |
| FIGN | rs16849225 | 2 | 164 615 066 | C/T | 45 833 | 0.59 | 0.45 (0.14) | 1.03 × 10−3 | 0.10 (0.08) | 2.07 × 10−1 | 0.040 (0.015) | 9.69 × 10−3 | SBP |
| ULK4 | rs9815354 | 3 | 41 887 655 | A/G | 45 998 | 0.19 | 0.10 (0.18) | 5.47 × 10−1 | 0.43 (0.10) | 1.34 × 10−5 | 0.007 (0.019) | 7.94 × 10−1 | DBP |
| FGF5 | rs1902859 | 4 | 81 376 727 | C/T | 45 856 | 0.41 | 1.34 (0.14) | 1.76 × 10−22 | 0.71 (0.08) | 3.75 × 10−20 | 0.130 (0.015) | 7.61 × 10−18 | SBP, DBP |
| GUCY1A3 | rs13143871 | 4 | 156 838 654 | T/C | 45 737 | 0.80 | 0.96 (0.18) | 5.16 × 10−8 | 0.49 (0.10) | 5.52 × 10−7 | 0.089 (0.019) | 1.24 × 10−6 | DBP |
| HFE | rs1799945 | 6 | 26 199 158 | G/C | 45 026 | 0.04 | 0.95 (0.36) | 4.51 × 10−3 | 0.88 (0.20) | 2.20 × 10−5 | 0.162 (0.039) | 3.98 × 10−5 | SBP, DBP, HTN |
| CYP17A1 | rs4409766 | 10 | 104 606 653 | T/C | 46 030 | 0.71 | 1.24 (0.15) | 6.08 × 10−17 | 0.59 (0.09) | 5.69 × 10−13 | 0.116 (0.017) | 7.33 × 10−13 | SBP, DBP |
| SOX6 | rs4757391 | 11 | 16 259 515 | C/T | 46 336 | 0.28 | 0.88 (0.15) | 5.20 × 10−9 | 0.49 (0.09) | 4.95 × 10−9 | 0.074 (0.017) | 3.33 × 10−6 | SBP, DBP |
| ATP2B1 | rs17249754 | 12 | 88 584 717 | G/A | 45 581 | 0.64 | 1.03 (0.15) | 3.66 × 10−12 | 0.52 (0.08) | 2.13 × 10−10 | 0.084 (0.016) | 2.08 × 10−8 | SBP, DBP, HTN |
| ALDH2 | rs11066280 | 12 | 111 302 166 | T/A | 45 860 | 0.81 | 0.96 (0.17) | 9.80 × 10−8 | 0.62 (0.10) | 3.19 × 10−10 | 0.108 (0.020) | 5.78 × 10−8 | SBP, DBP |
| TBX3-TBX5 | rs1991391 | 12 | 113 837 049 | G/A | 46 186 | 0.85 | 0.60 (0.20) | 1.81 × 10−3 | 0.21 (0.11) | 4.52 × 10−2 | 0.004 (0.021) | 9.57 × 10−1 | DBP |
| TBX3 | rs35444 | 12 | 114 036 820 | A/G | 45 917 | 0.77 | 0.83 (0.16) | 2.17 × 10−7 | 0.36 (0.09) | 6.73 × 10−5 | 0.075 (0.018) | 2.86 × 10−5 | DBP |
| JAG1 | rs1887320 | 20 | 10 913 998 | A/G | 46 123 | 0.53 | 0.78 (0.14) | 1.48 × 10−8 | 0.43 (0.08) | 2.13 × 10−8 | 0.058 (0.015) | 5.13 × 10−5 | SBP, DBP |
SNP IDs and chromosomal positions are based on NCBI Build 36 of the genome.
CHR, chromosome; RAF, risk allele frequency.
Effect size estimates (β) correspond to mmHg per risk allele for SBP and DBP, and log(odds) per risk allele for hypertension (HTN).
New blood pressure loci and Chinese-specific variants
The meta-analysis of the discovery and replication 1 and replication 2 studies found four novel regions for blood pressure. Among them, rs9810888 at the CACNA1D locus, rs2021783 at CYP21A2, and rs11067763 at MED13L achieved genome-wide significance (P < 5.0 × 10−8), while rs9266359 at the HLA-B at a suggestive significance level of P < 5.0 × 10−6. Also, we detected four potential Chinese-specific variants (P < 5.0 × 10−5) in SLC4A7, GUCY1A3, FLJ32810, and FURIN, which were not in linkage disequilibrium (LD; r2 < 0.2 in HapMap JPT + CHB) with the previously reported SNPs.
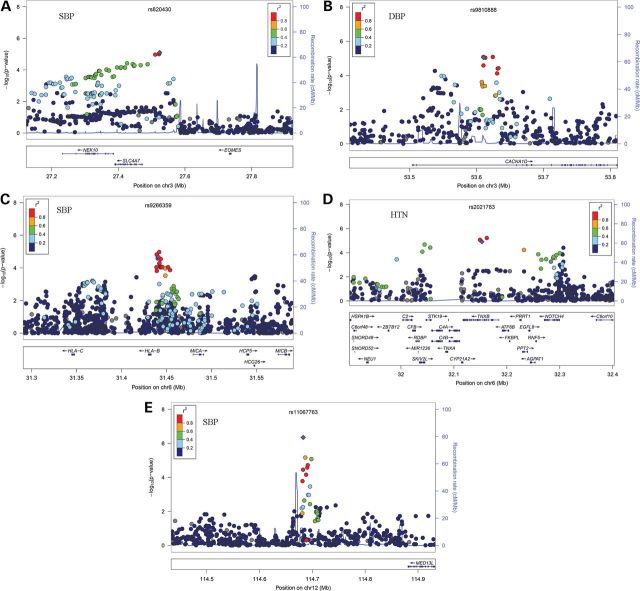
To minimize the chance of false discovery, we carried these eight novel or Chinese-specific variants forward to replication 3 study comprising 34 142 individuals. In replication 3 analyses, four SNPs (SLC4A7, CACNA1D, CYP21A2, and MED13L) showed significant associations with blood pressure after adjustment for multiple testing (P < 6.25 × 10−3 = 0.05/8), whereas rs9266359 at the HLA-B locus showed nominal significance (P < 0.05). After meta-analysis combining results of the discovery and all three replication studies, we identified three new blood pressure loci (Fig. 1). These include SNPs at 3p21.1 in CACNA1D (DBP, P = 4.00 × 10−12), 6p21.32 near CYP21A2 (SBP, P = 3.19 × 10−9; DBP, P = 2.18 × 10−12; hypertension, P = 3.53 × 10−11), and 12q24.21 near MED13L (SBP, P = 5.68 × 10−16; DBP, P = 2.00 × 10−18) (Table 2). We also detected a Chinese-specific variant in previous reported regions in European populations [rs820430 at 3p24.1 near SLC4A7 (SBP, P = 1.36 × 10−12)]. The reported lead SNP rs13082711 at SLC4A7 associated with blood pressure in Europeans had low minor allele frequency (MAF = 0.05) in the Chinese Han population. The SNP rs13082711 in Europeans and rs820430 identified in this study are not in LD (r2 = 0.06 in HapMap JPT + CHB) (Supplementary Material, Fig. S4). In addition, the SNP rs9266359 at HLA-B reached the near genome-wide significance of P = 1.76 × 10−7, and by the consistent evidences showing the same direction of association across the discovery and all replication studies. There was no evidence of between-study heterogeneity of effect-size estimates for all these new variants (all P > 0.11; I2 < 41%; Supplementary Material, Table S3).
Figure 1.
Regional association plots of blood pressure and hypertension loci. (A–E) Results are shown for rs820430 (A), rs9810888 (B), rs9266359 (C), rs2021783 (D) and rs11067763 (E). In the top panel of each, SNPs are plotted according to their chromosomal positions (NCBI build36) with their P-values (as −log10 P-values) of the discovery meta-analysis. The lead SNP is represented by a purple diamond. The r2 values of LD between the lead SNP and the other SNPs are indicated by different colors. The estimated recombination rates from 1000 Genomes June 2010 CHB + JPT samples are plotted in cyan to reflect the local LD structure. The bottom panels illustrate the locations of known genes.
Pleiotropic effects of blood pressure loci on established cardiovascular risk factors
To gain further understanding of the blood pressure susceptibility loci identified and validated in the Chinese population summarized in Table 2, we tested their associations with lipid levels, plasma glucose, and BMI in the replication samples. After Bonferroni correction for 19 independent tests, three loci showed significant associations (P < 2.6 × 10−3≈ 0.05/19) with plasma lipid traits in the expected direction (the blood pressure risk allele at CYP21A2 was associated with higher total cholesterol concentrations, at ALDH2 with higher triglyceride concentrations, and at CASZ1 with lower high-density lipoprotein cholesterol concentration; Supplementary Material, Table S4). Similarly, significant associations with BMI (P < 2.6 × 10−3) were observed in the six loci (FIGN, SLC4A7, CYP21A2, HLA-B, CYP17A1, and ALDH2). Except for FIGN and CYP17A1, the risk alleles were associated with increased BMI in a direction consistent with their associations with high blood pressure risk. Of note, the association of ALDH2 with BMI reached genome-wide significance (P = 1.21 × 10−15), which represented an unreported locus for BMI.
Cumulative impact of risk alleles on blood pressure and hypertension
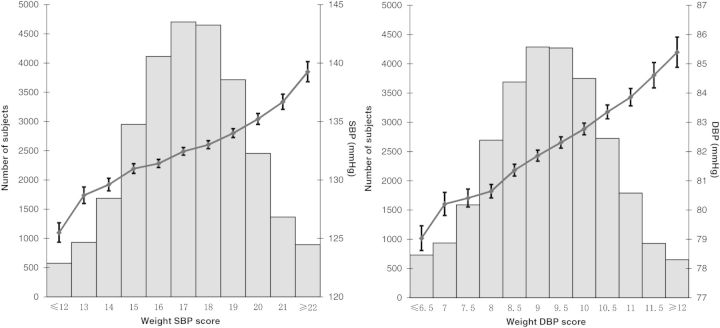
Weighted risk scores incorporating the 19 variants (Table 2) were calculated to examine the aggregate effect of risk alleles on blood pressure levels and risk of hypertension. Blood pressure levels increased linearly with an increase of weighted risk scores (Fig. 2). Regression estimates of slope (β) and its standard error (S.E.) were obtained across risk score groups for blood pressure levels. The null hypothesis of a zero slope across risk score groups was tested using two-tailed P-values [from Z = β/S.E.(β)]. The P-values for slope across risk score groups were 4.73 × 10−67 for SBP and 2.03 × 10−69 for DBP. Logistic regression models were applied to examine the relationship between weighted risk score and risk of hypertension. Individuals in the top quintile of genotype risk score had a 66% increased risk for hypertension compared with those in the bottom quintile (OR = 1.66, 95% CI = 1.54–1.79; Supplementary Material, Table S5).
Figure 2.
Cumulative effects of risk alleles on blood pressure levels. This figure shows blood pressure levels as a solid dot with whiskers extending to ±1 S.E. across the risk score categories. The gray bars represent the number of individuals in each risk score category.
Functional potential of blood pressure loci by expression quantitative trait loci analysis
To clarify the possible transcriptional mechanisms underlying the identified loci in associations with blood pressure and hypertension, we investigated the relationships of lead SNPs and proxies with expression quantitative trait loci (eQTLs) in lymphoblastoid cell lines (LCLs), adipose, liver, skin, and other tissues. Cis-eQTLs effects were found at CACNA1D (Supplementary Material, Table S6).
Pathway analysis
We applied the Gene Relationships Among Implicated Loci (GRAIL) (11) text-mining algorithm to investigate connections between 19 loci (Table 2). This analysis revealed significant (PGRAIL < 0.05) connections among blood pressure-associated genes (Supplementary Material, Fig. S5). We further investigated excessive connectivity between the 19 loci and those loci that showed a level of significance for association (P < 0.0001) in the discovery study. The less stringent threshold yielded 177 suggestive regions for blood pressure, among which we observed that 26 genes were connected to the 19 loci at PGRAIL < 0.05 (Supplementary Material, Table S7). These included genes such as FGF14 (fibroblast growth factor 14, PGRAIL = 4.8 × 10−4), HLA-DPB1 (major histocompatibility complex, class II, DP beta 1, PGRAIL = 2.0 × 10−3), and SCNN1B (natriuretic peptide A, PGRAIL = 9.7 × 10−3).
We applied MAGENTA (Meta-Analysis Gene-set Enrichment of variaNT Associations) (12) to detect enrichment of association signals in gene sets compiled from known biological pathways. The MAGENTA analysis implicated 17 biological pathways and molecular functions with a nominal P-value of <0.01 for SBP, DBP, and/or hypertension (Supplementary Material, Table S8). Most of these gene sets contain genes capturing or near a validated blood pressure SNP. For example, the KEGG MAPK signaling pathway included the newly identified CACNA1D.
DISCUSSION
This study describes the largest GWAS of blood pressure and hypertension in Chinese to date involving a total of 80 962 individuals. We identified three new loci (CACNA1D, CYP21A2, and MED13L) and a newly discovered variant near SLC4A7 at the genome-wide significance level. We also replicated 14 loci previously reported loci, 8 (CASZ1, MOV10, FGF5, CYP17A1, SOX6, ATP2B1, ALDH2, and JAG1) at genome-wide significance, and 6 (FIGN, ULK4, GUCY1A3, HFE, TBX3-TBX5, and TBX3) at a suggestive level of P = 1.81 × 10−3 to 5.16 × 10−8.
The identification of new genes for blood pressure can help further our understanding of new mechanistic insights into the regulation of blood pressure and potential targets for treatments. CACNA1D encodes a member of the alpha-1 subunit family of voltage-dependent calcium channels, several of which have effects on blood pressure regulation and serve as targets of calcium channel blockers. More recently, somatic and germline mutations in the CACNA1D gene in aldosterone-producing adenomas and primary aldosteronism were identified. These mutations resulted in channel activation at more hyperpolarized membrane potentials, implicating increased Ca2+ influx in disease pathogenesis (13). Another calcium channel encoding gene, CACNB2, has been reported to be associated with DBP (4). This emphasizes the importance of calcium channels in the regulation of blood pressure. CYP21A2 encodes a member of the cytochrome P450 superfamily required for the synthesis of steroid hormones including cortisol and aldosterone. Aldosterone regulates the amount of salt retained by the kidneys, and thus affects blood volume in the body and blood pressure. Mutations in CYP21A2 can cause congenital adrenal hyperplasia (14), a common autosomal recessive disorder caused by deficiency in the steroid 21-hydroxylase. The other two members of the family, CYP17A1 and CYP1A2, were also found associated with blood pressure (5). MED13L encodes a subunit of the mediator complex co-activating RNA polymerase II-induced transcription and is active in peroxisome proliferator-activated receptor alpha-related pathways for fatty acid metabolism that are important to hypertensive heart diseases. Missense mutations of MED13L were associated with transposition of the great arteries (15). It is also involved in Rb/E2F-mediated control of cell growth (16) and to promote proliferation of vascular smooth muscle cells(17), which participate in the progression of hypertension.
Our results showed that both shared and population-specific blood pressure susceptibility were commonly present. Of five new blood pressure variants identified in our Chinese samples, three (CACNA1D, MED13L, and SLC4A7) showed associations at various significance levels (P = 8.66 × 10−3 to 4.16 × 10−4) in the International Consortium for Blood Pressure Genome-Wide Association Studies (ICBP) (Supplementary Material, Table S9). For the remaining two SNPs, one is monomorphic in Europeans and the other has much difference in risk allele frequency between the two populations. Conversely, of the 34 loci identified in GWAS meta-analysis in individuals of European descent and East Asian, 17 (50%) showed significant associations at various significance levels in Chinese (Supplementary Material, Table S10). Of note, the effect sizes of blood pressure variants differ between the Chinese and European populations. For those loci that were not significant in our Chinese samples, six SNPs were monomorphic or had a low MAF (MAF < 0.05) in the Chinese Han population, suggesting that some of the discrepancies in association between the European and Chinese populations may be due to different LD structure. Therefore, studies conducted in Chinese populations, in addition to allowing an evaluation of the extent to which genetic markers of blood pressure identified in European populations can be generalized, also might lead to the identification of new blood pressure-associated loci.
We successfully identified new blood pressure loci by testing SNPs with P < 1.0 × 10−5 in the discovery meta-analysis. However, many blood pressure risk alleles with small-to-modest effects will still be missed at that significance threshold. We expected that some true disease loci at P < 0.0001 might share the same pathways with those detected at the genome-wide significance, and as such, could be detected through their connections with the 19 genome-wide significant loci. We found 26 such loci with a significant degree of functional connectivity at PGRAIL < 0.05 (Supplementary Material, Table S7). Besides, MAGENTA analysis identified suggestive pathways, some of which contained genes at validated blood pressure loci. These findings likely represent true associations and novel pathways, but additional studies will be necessary for definitive confirmation.
Our results should be interpreted in the context of potential limitations. First, our discovery sample size is not large enough compared with those in the previous consortia of European descent (3–5). This means that some signals especially with a low MAF and/or weaker genetic effects would not be initially detected in our study. Secondly, we adjusted for use of antihypertensive therapy by adding 10 and 5 mmHg to SBP and DBP, respectively. This approach is relatively simple and widely adopted. However, it is noteworthy that despite increased rates of blood pressure lowering treatment, few have their hypertension effectively controlled in China (18,19). We have addressed this issue by performing a sensitivity analysis and verified that our results were not substantially modified by blood pressure adjustment or no adjustment (Supplementary Material, Table S11).
In conclusion, we identified previously unreported association signals for blood pressure (CACNA1D, CYP21A2, and MED13L). Near SLC4A7, we also identified some evidence for allelic heterogeneity in Chinese compared with Europeans in relation to blood pressure associations. We replicated 14 previously reported loci in the Chinese samples. Increasing efforts in conducting the studies in multiple diverse populations will be critical for better understanding of the genetic architecture of blood pressure.
MATERIALS AND METHODS
Study design and population
A detailed description of the sample characteristics and phenotype measurements for each study are provided in Table 1 and Supplementary Material, Methods. The discovery stage was a meta-analysis consisted of 11 816 Han Chinese who underwent standardized collection of blood pressure measurements in six GWASs. These studies, named the Genetic Epidemiology Network of Cardiovascular Disease in China (GENECDC), included the International Collaborative Study of Cardiovascular Disease in Asia (InterASIA in China) (20), Beijing Cardiovascular Risk Factors study (Beijng) (21), Genetic Epidemiology Network of Salt-Sensitivity study (GenSalt) (22), the Nutrition and Health of Aging Population in China (NHAPC) (23), Noncommunicable Diseases Screening study in Nanjing (Nanjing) (24), and the Dongfeng-Tongji cohort (DFTJ) study (25). Three blood pressure measurements were measured by using standard mercury sphygmomanometer or electronic BP monitor. For individuals who were taking antihypertensive medication, blood pressure was imputed by adding 10 and 5 mmHg for SBP and DBP, respectively. Hypertension was defined by the presence of SBP ≥140 mmHg or DBP ≥90 mmHg, or self-reported of taking a medication for the treatment of hypertension. Normotensive controls were defined as individuals not taking any antihypertensives and having a SBP of <140 mmHg and a DBP of <90 mmHg.
Each study obtained approval from the institutional review boards of Fuwai Hospital, the Chinese Academy of Medical Sciences and Peking Union Medical College, and other medical institutions. All participants gave written informed consent.
Genotype imputation
Detailed descriptions of genotyping arrays and quality control filters applied to the six discovery studies are provided in Supplementary Material, Table S1. To facilitate combining results of genome-wide association scans based on the different genotyping platforms, we imputed missing genotypes based on reference haplotypes from the phased CHB + JPT HapMap data release 22 reference dataset using MACH or IMPUTE (26,27). Only imputed SNPs with high genotype information content (proper info ≥0.5 for IMPUTE and Rsq ≥0.3 for MACH) were used for the association analysis. After quality control, we obtained up to 2.5 million genotyped or imputed autosomal SNPs for subsequent association analysis.
Selection of replication SNPs and genotyping
After genome-wide association analyses for each of the six discovery studies and meta-analysis in the combined sample, SNPs representing the independent association signal were taken forward to replication 1 if they (i) showed potential association (P < 1.0 × 10−5) for SBP, DBP, or/and hypertension in the discovery meta-analysis; (ii) associated at P < 5.0 × 10−3 in the meta-analysis discovery stage in a locus previously reported to show association with genome-wide significance. If a SNP could not be genotyped, alternative tagging SNPs (with r2 of >0.8) were considered. In total, 39 SNPs were selected and genotyped using the iPLEX Sequenom MassARRAY platform (Sequenom) for replication 1. Considering that replication 1 sample has ∼ 80% power to detect a common SNP with an effect size of 1 mmHg for SBP or 0.5 mmHg for DBP using a P-value threshold of 0.05, we selected potential significant SNPs by the conservative criterion (P < 0.05) and further genotyped them in replication 2 using the iPLEX MassARRAY platform. To assess genotyping reproducibility, 380 duplicate samples were genotyped, and the concordance rate was determined to be >99.8%. We sought further evidence for association with eight SNPs in replication 3 using the TaqMan genotyping platform (ABI 7900HT Real Time PCR system, Applied Biosystems). To evaluate the quality of the genotype data between different genotyping platforms, eight SNPs in 96 random replication samples genotyped on the iPLEX Sequenom MassARRAY platform were also genotyped on the TaqMan genotyping platform, and the concordance rate between the genotypes from the two platforms was found to be 99.7%. Cluster patterns of genotyping data from the Sequenom and TaqMan analyses were examined to confirm high quality.
Statistical analysis
Within each of the six discovery studies, continuous SBP and DBP were adjusted for age, age-squared, gender, and BMI in linear regression models under an additive model, using the software as specified in Supplementary Material, Table S1. Analysis of hypertension was conducted within four studies (InterASIA, NHAPC, Nanjing, and DFTJ). Within each study, logistic regression was fitted for hypertension, adjusting for sex, age, age-squared, and BMI. We used allele dosages from imputation to account for uncertainty in imputed genotypes. A fixed-effects inverse variance-weighted meta-analysis implemented in METAL (28) was used to combine the six studies in discovery stage and to obtain results for each SNP. A quantile–quantile plot generated using R was used to evaluate the overall significance of the GWAS results and the potential impact of population stratification. The genomic inflation factor (λ) was estimated from the median of the χ2 statistic divided by 0.456. We detected the associations of SNPs in the replication populations, and additionally, we carried out meta-analysis of the discovery and replications results and considered associations genome-wide significant if they attained P < 5.0 × 10−8.
Associations of loci with established cardiovascular risk factors were examined in replication 1, 2, and 3 (if applicable) samples. High-density lipoprotein, low-density lipoprotein, total cholesterol, triglycerides (naturally logarithm transformed), fasting plasma glucose, and BMI were adjusted for age and gender in linear regression models with an additive model (1 d.f.) We combined the regression estimates from each study in a meta-analysis using inverse variance weighting.
Assessment of cumulative effect of risk variants
Blood pressure risk scores incorporating the associated SNPs were calculated for individuals in replication 1 and 2 samples. The doses of risk alleles were weighted by their effect sizes based on all discovery and replication meta-analyses and summed for each individual to calculate risk score. A risk score was rounded to 1 mmHg for SBP (groups ≤12 to ≥22) and 0.5 mmHg for DBP (groups ≤6.5 to ≥12). For SBP and DBP, we estimated the mean blood pressure level and standard error within each risk score group. For hypertension, odds ratio was calculated using a logistic regression model, with the reference group being those with weighted hypertension risk score of bottom quintile.
eQTL analysis
For each of our newly discovered loci, all proxies (r2 > 0.8) were identified using data from HapMap CHB + JPT (release 23). Cis-eQTL (defined as genes within 1 Mb) of lead SNP and their proxies were investigated in public databases including the following tissues and cell lines: liver (29), four brain regions (cerebellum, frontal cortex, temporal cortex, and pons) (30), LCL (31–35), adipose (32,35), skin (32,35), fibroblast (34), and T-cell (34). For each transcript associated with a lead blood pressure SNP or proxy, the lead cis-eQTL SNP was further identified and then LD between them was estimated using the HapMap CHB + JPT data.
Text-mining and pathway analysis
We used GRAIL (11) to evaluate whether loci associated with blood pressure were enriched for connectivity between genes representing particular pathways or molecular processes. GRAIL is a text-mining tool that identifies non-random, evidence-based links between genes using PubMed abstracts. Genes are given a significance score. PubMed abstracts published after December 2006 were omitted from the analysis to reduce confounding by results from GWAS. We performed two analyses. First, we took 19 loci (Table 2) as a seed and queried loci to investigate biological connectivity among those loci. Secondly, we investigated connectivity between the 19 loci (as seed regions) and those that were suggestively associated with SBP, DBP, and/or hypertension (P < 0.0001) (as query regions). Query regions were defined by taking all SNPs more significant than P < 0.0001, removing those at the 19 loci and pruning SNPs of r2 > 0.2 in each region using PLINK. As GRAIL tests the connectivity of regions, we also removed any duplicates where a region was represented by more than one SNP. This approach identified 177 query regions (representing 401 genes).
We tested whether the overall genome-wide association results were enriched in gene sets derived from known biological pathways using MAGENTA (12). MAGENTA calculates a corrected gene association P-value based on the most significant SNP association P-value of all SNPs in the gene region (110 kb upstream and 40 kb downstream of the gene's most extreme transcript boundaries), accounting for gene size, number of SNPs/gene, and recombination. Pathway terms from five databases within MAGENTA v2.4 [KEGG (June 2008), Panther Signaling Pathways (June 2008), PANTHER Biological Processes (January 2010), PANTHER Molecular Function (January 2010), and Ingenuity Pathway (June 2008)] were attached to each gene. For each pathway, enrichment of highly ranked gene scores above the given significance threshold (95th percentile of all gene scores) was evaluated. This statistic is then compared with 10 000 randomly permuted gene sets of identical size from the genome. Analysis was performed for the discovery meta-analysis results for SBP, DBP, and hypertension.
WEB RESOURCES
PLINK v1.07, http://pngu.mgh.harvard.edu/~purcell/plink/; R, http://www.r-project.org/; METAL, http://www.sph.umich.edu/csg/abecasis/metal/; The International HapMap Project, http://hapmap.ncbi.nlm.nih.gov/; GRAIL, http://www.broadinstitute.org/mpg/grail/grail.php; MAGENTA, http://www.broadinstitute.org/mpg/magenta/; Genevar (GENe Expression VARiation), http://www.sanger.ac.uk/resources/software/genevar/; GTEx (Genotype-Tissue Expression) eQTL Browser, http://www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi.
SUPPLEMENTARY MATERIAL
FUNDING
This study was funded by the High-Tech Research and Development Program of China (863 Plan; 2012AA02A516 and 2009AA022703) and the National Basic Research Program of China (973 Plan; 2011CB503901) from the Ministry of Science and Technology of China. InterASIA was funded by the National Science Foundation of China (30930047) and grants (2011BAI11B03) from the Ministry of Science and Technology of China. Beijing was funded by the High-Tech Research and Development Program of China (863 Plan; 2006AA02A406) and Biomedical Project from the Council of Science and Technology, Beijing (H020220030130). GenSalt was supported by research grants (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health (Bethesda, Maryland, USA). NHAPC was supported by the National High Technology Research and Development Program (863 Program; 2009AA022704), the National Basic Research Program of China (973 Program; 2012CB524900), the National Natural Science Foundation of China (30930081, 81170734, and 81021002), the Chinese Academy of Sciences (KSCX2-EW-R-10 and SIBS2008006), and Key Discipline of Shanghai Public Health-Food and Nutritional Sciences (12GWZX0702). Nanjing was funded by the China National High-Tech Research and Development Program Grant (2009AA022705), the National Natural Science Foundation of China (81273165), and the Natural Science Foundation of Jiangsu Province (BK2011776). DFTJ was supported by grants from the National Basic Research Program Grant (2011CB503806) and the Program of Introducing Talents of Discipline to Universities to T. Wu and Program for New Century Excellent Talents in University and the National Natural Science Foundation of China (81172751) to M.H. CCHS was funded by the grants (2011BAI11B03 and 2011BAI09B03) from the Ministry of Science and Technology of China.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the role of the Genetic Epidemiology Network of Cardiovascular Disease in China (GENECDC) in developing this manuscript. GENECDC members include the International Collaborative Study of Cardiovascular Disease in Asia (InterASIA in China), Beijing Cardiovascular Risk Factors study (Beijing), Genetic Epidemiology Network of Salt-Sensitivity study (GenSalt), The Nutrition and Health of Aging Population in China (NHAPC), Noncommunicable Diseases Screening study in Nanjing (Nanjing), and Dongfeng-Tongji Cohort Study (DFTJ). We also thank the ICBP Consortium for providing online blood pressure association results.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kearney P.M., Whelton M., Reynolds K., Muntner P., Whelton P.K., He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 2.Kupper N., Willemsen G., Riese H., Posthuma D., Boomsma D.I., de Geus E.J. Heritability of daytime ambulatory blood pressure in an extended twin design. Hypertension. 2005;45:80–85. doi: 10.1161/01.HYP.0000149952.84391.54. [DOI] [PubMed] [Google Scholar]
- 3.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.J., et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy D., Ehret G.B., Rice K., Verwoert G.C., Launer L.J., Dehghan A., Glazer N.L., Morrison A.C., Johnson A.D., Aspelund T., et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newton-Cheh C., Johnson T., Gateva V., Tobin M.D., Bochud M., Coin L., Najjar S.S., Zhao J.H., Heath S.C., Eyheramendy S., et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato N., Takeuchi F., Tabara Y., Kelly T.N., Go M.J., Sim X., Tay W.T., Chen C.H., Zhang Y., Yamamoto K., et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in East Asians. Nat. Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franceschini N., Fox E., Zhang Z., Edwards T.L., Nalls M.A., Sung Y.J., Tayo B.O., Sun Y.V., Gottesman O., Adeyemo A., et al. Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am. J. Hum. Genet. 2013;93:545–554. doi: 10.1016/j.ajhg.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whelton P.K., He J., Appel L.J., Cutler J.A., Havas S., Kotchen T.A., Roccella E.J., Stout R., Vallbona C., Winston M.C., et al. Primary prevention of hypertension: clinical and public health advisory from The National High Blood Pressure Education Program. JAMA. 2002;288:1882–1888. doi: 10.1001/jama.288.15.1882. [DOI] [PubMed] [Google Scholar]
- 9.Ganesh S.K., Tragante V., Guo W., Guo Y., Lanktree M.B., Smith E.N., Johnson T., Castillo B.A., Barnard J., Baumert J., et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum. Mol. Genet. 2013;22:1663–1678. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson T., Gaunt T.R., Newhouse S.J., Padmanabhan S., Tomaszewski M., Kumari M., Morris R.W., Tzoulaki I., O'Brien E.T., Poulter N.R., et al. Blood pressure loci identified with a gene-centric array. Am. J. Hum. Genet. 2011;89:688–700. doi: 10.1016/j.ajhg.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raychaudhuri S., Plenge R.M., Rossin E.J., Ng A.C., International Schizophrenia C., Purcell S.M., Sklar P., Scolnick E.M., Xavier R.J., Altshuler D., et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segre A.V., Consortium D., Investigators M., Groop L., Mootha V.K., Daly M.J., Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholl U.I., Goh G., Stolting G., de Oliveira R.C., Choi M., Overton J.D., Fonseca A.L., Korah R., Starker L.F., Kunstman J.W., et al. Somatic and germline CACNA1D calcium channel mutations in aldosterone-producing adenomas and primary aldosteronism. Nat. Genet. 2013;45:1050–1054. doi: 10.1038/ng.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H.H., Chao H.T., Ng H.T., Choo K.B. Direct molecular diagnosis of CYP21 mutations in congenital adrenal hyperplasia. J. Med. Genet. 1996;33:371–375. doi: 10.1136/jmg.33.5.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muncke N., Jung C., Rudiger H., Ulmer H., Roeth R., Hubert A., Goldmuntz E., Driscoll D., Goodship J., Schon K., et al. Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries) Circulation. 2003;108:2843–2850. doi: 10.1161/01.CIR.0000103684.77636.CD. [DOI] [PubMed] [Google Scholar]
- 16.Angus S.P., Nevins J.R. A role for mediator complex subunit MED13L in Rb/E2F-induced growth arrest. Oncogene. 2012;31:4709–4717. doi: 10.1038/onc.2011.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blaschke F., Leppanen O., Takata Y., Caglayan E., Liu J., Fishbein M.C., Kappert K., Nakayama K.I., Collins A.R., Fleck E., et al. Liver X receptor agonists suppress vascular smooth muscle cell proliferation and inhibit neointima formation in balloon-injured rat carotid arteries. Circ. Res. 2004;95:e110–e123. doi: 10.1161/01.RES.0000150368.56660.4f. [DOI] [PubMed] [Google Scholar]
- 18.Gu D., Reynolds K., Wu X., Chen J., Duan X., Muntner P., Huang G., Reynolds R.F., Su S., Whelton P.K., et al. Prevalence, awareness, treatment, and control of hypertension in china. Hypertension. 2002;40:920–927. doi: 10.1161/01.hyp.0000040263.94619.d5. [DOI] [PubMed] [Google Scholar]
- 19.Wu Y., Huxley R., Li L., Anna V., Xie G., Yao C., Woodward M., Li X., Chalmers J., Gao R., et al. Prevalence, awareness, treatment, and control of hypertension in China: data from the China National Nutrition and Health Survey 2002. Circulation. 2008;118:2679–2686. doi: 10.1161/CIRCULATIONAHA.108.788166. [DOI] [PubMed] [Google Scholar]
- 20.Gu D., Reynolds K., Wu X., Chen J., Duan X., Reynolds R.F., Whelton P.K., He J. Prevalence of the metabolic syndrome and overweight among adults in China. Lancet. 2005;365:1398–1405. doi: 10.1016/S0140-6736(05)66375-1. [DOI] [PubMed] [Google Scholar]
- 21.Hou L., Chen S., Yu H., Lu X., Chen J., Wang L., Huang J., Fan Z., Gu D. Associations of PLA2G7 gene polymorphisms with plasma lipoprotein-associated phospholipase A2 activity and coronary heart disease in a Chinese Han population: the Beijing atherosclerosis study. Hum. Genet. 2009;125:11–20. doi: 10.1007/s00439-008-0587-4. [DOI] [PubMed] [Google Scholar]
- 22.The GenSalt Collaborative Research Group. GenSalt: rationale, design, methods and baseline characteristics of study participants. J. Hum. Hypertens. 2007;21:639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye X., Yu Z., Li H., Franco O.H., Liu Y., Lin X. Distributions of C-reactive protein and its association with metabolic syndrome in middle-aged and older Chinese people. J. Am. Coll. Cardiol. 2007;49:1798–1805. doi: 10.1016/j.jacc.2007.01.065. [DOI] [PubMed] [Google Scholar]
- 24.Hu Z., Wu C., Shi Y., Guo H., Zhao X., Yin Z., Yang L., Dai J., Hu L., Tan W., et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat. Genet. 2011;43:792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 25.Wang F., Zhu J., Yao P., Li X., He M., Liu Y., Yuan J., Chen W., Zhou L., Min X., et al. Cohort profile: the Dongfeng-Tongji cohort study of retired workers. Int. J. Epidemiol. 2013;42:731–740. doi: 10.1093/ije/dys053. [DOI] [PubMed] [Google Scholar]
- 26.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 28.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genome wide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S.L., Arepalli S., Dillman A., Rafferty I.P., Troncoso J., et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grundberg E., Small K.S., Hedman A.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.P., Meduri E., Barrett A., et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stranger B.E., Montgomery S.B., Dimas A.S., Parts L., Stegle O., Ingle C.E., Sekowska M., Smith G.D., Evans D., Gutierrez-Arcelus M., et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M., et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nica A.C., Parts L., Glass D., Nisbet J., Barrett A., Sekowska M., Travers M., Potter S., Grundberg E., Small K., et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.